This study aimed at identifying the microbial mechanisms of enteric methane mitigation when linseed, nitrate, and saponins were fed to nonlactating cows alone or in a combination. Hydrogen is a limiting factor in rumen methanogenesis. We hypothesized that linseed and saponins would affect hydrogen producers and nitrate would affect hydrogen consumption, leading to reduced methane production in the rumen. Contrary to what was predicted, both linseed and nitrate had a deleterious effect on hydrogen producers; linseed also redirected hydrogen consumption toward propionate production, whereas nitrate stimulated the growth of nitrate-reducing and, hence, hydrogen-consuming bacterial taxa. This novel knowledge of microbial mechanisms involved in rumen methanogenesis provides insights for the development and optimization of methane mitigation strategies.
KEYWORDS: linseed, methane, microbiota, nitrate, rumen, saponin
ABSTRACT
Dietary supplementation with linseed, saponins, and nitrate is a promising methane mitigation strategy in ruminant production. Here, we aimed to assess the effects of these additives on the rumen microbiota in order to understand underlying microbial mechanisms of methane abatement. Two 2-by-2 factorial design studies were conducted simultaneously, which also allowed us to make a broad-based assessment of microbial responses. Eight nonlactating cows were fed diets supplemented with linseed or saponin in order to decrease hydrogen production and nitrate to affect hydrogen consumption; also, combinations of linseed plus nitrate or saponin plus nitrate were used to explore the interaction between dietary treatments. Previous work assessed effects on methane and fermentation patterns. Rumen microbes were studied by sequencing 18S and 16S rRNA genes and ITS1 amplicons. Methanogen activity was monitored by following changes in mcrA transcript abundance. Nitrate fed alone or in combination in both studies dramatically affected the composition and structure of rumen microbiota, although impacts were more evident in one of the studies. Linseed moderately modified only bacterial community structure. Indicator operational taxonomic unit (OTU) analysis revealed that both linseed and nitrate reduced the relative abundance of hydrogen-producing Ruminococcaceae. Linseed increased the proportion of bacteria known to reduce succinate to propionate, whereas nitrate supplementation increased nitrate-reducing bacteria and decreased the metabolic activity of rumen methanogens. Saponins had no effect on the microbiota. Inconsistency found between the two studies with nitrate supplementation could be explained by changes in microbial ecosystem functioning rather than changes in microbial community structure.
IMPORTANCE This study aimed at identifying the microbial mechanisms of enteric methane mitigation when linseed, nitrate, and saponins were fed to nonlactating cows alone or in a combination. Hydrogen is a limiting factor in rumen methanogenesis. We hypothesized that linseed and saponins would affect hydrogen producers and nitrate would affect hydrogen consumption, leading to reduced methane production in the rumen. Contrary to what was predicted, both linseed and nitrate had a deleterious effect on hydrogen producers; linseed also redirected hydrogen consumption toward propionate production, whereas nitrate stimulated the growth of nitrate-reducing and, hence, hydrogen-consuming bacterial taxa. This novel knowledge of microbial mechanisms involved in rumen methanogenesis provides insights for the development and optimization of methane mitigation strategies.
INTRODUCTION
Methane emissions associated with ruminant livestock production are an important contributor to global greenhouse gas emissions (1). Rumen methanogenesis is a naturally occurring process that involves methanogenic archaea consuming hydrogen to reduce carbon dioxide. Hydrogen and carbon dioxide production occurs during feed fermentation by bacteria, protozoa, and fungi; hydrogen availability is a limiting factor for methane production. In addition, there is a significant linear relationship between protozoan concentration in the rumen and methane emissions (2). Among the measures that have been undertaken to reduce methane production by ruminants, diet composition and inclusion of feed additives have received the most attention (3). Among them, nitrate added to ruminants’ diets consistently and persistently lowers methane emissions (4). Linseed oil, which is rich in linoleic acid, has proven to be one of the most efficient lipid sources used in methane mitigation strategies (4). Saponins are natural phytogenic feed additives used to improve animal feeding and production characteristics (5). Theoretically, these three additives lead to decreased methane production via different modes of action. Nitrate is an alternative electron acceptor, as its reduction competes with methane production for hydrogen (6). Additionally, nitrate or its reduced forms might be toxic to rumen methanogens and protozoa (7), but this effect was not systematically reported (8, 9). Lipids from linseed (and fats in general) added to diets replace a proportion of dietary carbohydrates and, as rumen microbes do not ferment them, less hydrogen is produced. Protozoal numbers have been reported to decrease with supplementary linseed oil (8, 10), although this effect was not always observed (11). Saponins can reduce methanogenesis by a toxic effect on rumen protozoa (5), but in vivo results indicate otherwise, as rumen microbes can deglycosylate and, thus, inactivate saponins (12).
Based on available information, we hypothesized that linseed oil and saponins would mainly affect hydrogen production (by a toxic effect on protozoa or by providing alternative substrates for rumen fermentation) and nitrate would mainly modulate hydrogen consumption pathways (by providing an alternative hydrogen sink). We performed amplicon-type sequencing analysis of rumen contents, sampled during two previous studies (8, 13); the first one reported the effect of linseed, nitrate, and linseed plus nitrate supplementation on enteric methane production; tea saponin replaced linseed in the second one. The primary aim of the current study was to search for changes in rumen microbiota structure and methanogenic activity that could explain observed reductions in methane emissions.
Minor but significant changes induced by treatment can be masked by spurious between-group differences unrelated to the treatment but rather to the host animal, the diet, or sample management. Moreover, it is not unusual to find reports on nitrate and fatty acid supplementation where methane decreased in a similar way, but effects on rumen microbiota were contrasting (14–17). On the other hand, it was recently shown that combination of microbial data from multiple sets of hosts with supposed similar microbiota should increase specificity and allow identification of causal microbes (18). Therefore, we took advantage of the data available from two independent studies, analyzed it separately but by following the same procedures, and made an integrated interpretation. Our secondary objective was to try to find clues to explain inconsistency in results from published studies.
(This article was submitted to an online preprint archive [19].)
RESULTS
Eight nonlactating dairy cows were randomly allocated to two 2-by-2 factorial designs. In study 1, dietary treatments consisted of control (CTL) diet, supplemented alternatively with linseed oil (LIN), nitrate (NIT), and linseed plus nitrate (LIN+NIT); in study 2, tea saponin (TEA) replaced linseed oil. In order to achieve adequate statistical power, the statistical model for both studies included cow as random effect, and fixed effects were experimental period and the following: (i) in study 1, linseed (CTL and NIT versus LIN and LIN+NIT), nitrate (CTL and LIN versus NIT and LIN+NIT), and their interaction, termed linseed×nitrate (or lin×nit), and (ii) in study 2, saponin (CTL and NIT versus TEA and TEA+NIT), nitrate (CTL and TEA versus NIT and TEA+NIT), and their interaction, saponin×nitrate (sap×nit). Throughout the text, linseed, nitrate, and saponin will refer to diet contrasts detailed above.
In study 1, compared to CTL, dietary treatments LIN, NIT, and LIN+NIT decreased methane production (g/day) by 22%, 29%, and 33%, respectively, and methane yield (g/kg of dry matter intake [DMI]) by 25%, 29%, and 32% (8). In study 2, NIT and TEA+NIT decreased methane production by 42% and 34% and methane yield by 36% and 29%, respectively, compared to CTL (13). TEA alone had no effect on methane production or on volatile fatty acid (VFA) profiles.
In both studies, Bacteroidales and Clostridiales were the dominant bacterial orders and accounted for more than 88% of the classified reads, regardless of the dietary treatment (see Fig. S1 in the supplemental material). Sequences affiliated with the Methanobrevibacter genus accounted for 80% of all archaeal sequences in both studies, followed by Methanosphaera, unclassified methanogens, and three Methanomassiliicoccaceae genera (Fig. S1). In both studies, Piromyces represented more than 60% of rumen fungi, followed by Orpinomyces and Caecomyces. Dietary treatments did not affect fungal community composition or its structure (Fig. S2), and we are not going to discuss it further.
Linseed moderately affected bacterial community composition with no effect on rumen methanogens and protozoa.
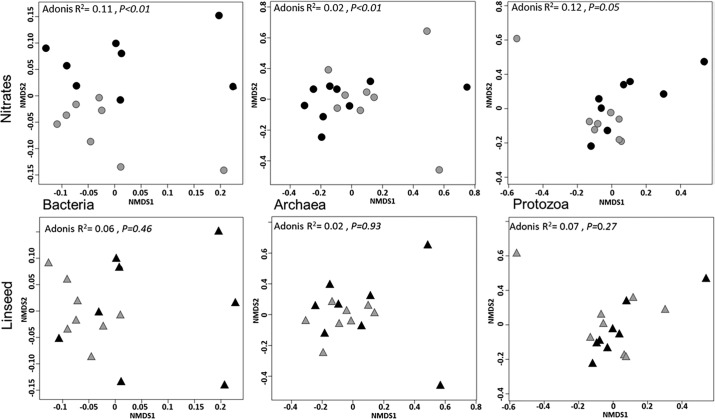
Nonmetric multidimensional scaling (NMDS) did not reveal any distinct clustering of bacterial communities (Fig. 1), and total bacterial numbers were similar (Table 1) in cows receiving or not receiving linseed-supplemented diets. Accordingly, CowPI predictions showed no changes in metabolic profiles (Table S1). However, the richness index was reduced by the linseed treatment (Table S2), and linseed increased (P < 0.05) relative abundance of Selenomonadales, Synergistales, Elusimicrobiales, and Micrococcales (Table 2). Moreover, indicator species analysis showed that Ruminococcaceae-related operational taxonomic units (OTUs) characterized the bacterial community of cows not receiving linseed supplementation (Fig. 2 and Table S3).
FIG 1.
Structure and composition of bacterial, archaeal, and protozoal communities in study 1, related to nitrate or linseed treatments (black symbols) and respective controls (gray symbols), were examined by multivariate analysis. NMDS plots derived from Bray-Curtis dissimilarities between cows are shown. Each symbol is representative of a single cow. Samples are plotted along the first two-component axes. Microbial composition was compared using Adonis.
TABLE 1.
Abundance of total bacteria and abundance and activity of methanogenic archaea in the rumen of nonlactating cows fed methane-reducing additives
| Parameter | Abundance and activity |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study 1a |
Effect |
Study 2b |
Effect |
|||||||||||||
| CTL | LIN | NIT | LIN+NIT | SEM | Linseed | Nitrate | lin×nit | CTL | TEA | NIT | TEA+NIT | SEM | Saponin | Nitrate | sap×nit | |
| Total bacterial concn (log10 rrs copies/ng DNA) | 5.09 | 4.94 | 4.95 | 4.98 | 0.051 | 0.50 | 0.54 | 0.33 | 5.04 | 4.96 | 5.02 | 5.15 | 0.057 | 0.64 | 0.12 | 0.05 |
| Methanogen concn (log10 mcrA copies/ng DNA) | 2.95 | 2.68 | 2.72 | 2.76 | 0.051 | 0.36 | 0.15 | <0.1 | 2.97 | 2.84 | 2.97 | 3.07 | 0.081 | 0.84 | 0.25 | 0.24 |
| Methanogen activityc (2−ΔCT × 106) | 23.91 | 21.54 | 10.49 | 8.19 | 3.384 | 0.51 | <0.01 | 0.99 | 18.67 | 16.08 | 7.40 | 8.28 | 4.463 | 0.76 | <0.01 | 0.53 |
In study 1, cows were fed a control (CTL) diet and CTL diet supplemented with linseed (LIN), nitrate (NIT), and linseed plus nitrate (LIN+NIT). Tested effects were linseed (CTL and NIT versus LIN and LIN+NIT) and nitrate (CTL and LIN versus NIT and LIN+NIT) and their interaction, lin×nit.
In study 2, cows were fed a control (CTL) diet and control diet supplemented with tea saponin (TEA), nitrate (NIT), and tea saponin plus nitrate (TEA+NIT). Tested effects were saponin (CTL and NIT versus TEA and TEA+NIT) and nitrate (CTL and TEA versus NIT and TEA+NIT) and their interaction, sap×nit.
Methanogen activity is measured as mcrA expression levels.
TABLE 2.
Bacterial orders significantly affected by at least one dietary treatment in the rumen of nonlactating cows fed methane-reducing additivesc
|
Bacterial order |
Relative abundance (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study 1a |
Effect |
Study 2b |
Effect |
|||||||||||||
| CTL | LIN | NIT | LIN+NIT | SEM | Linseed | Nitrate | lin×nit | CTL | TEA | NIT | TEA+NIT | SEM | Saponin | Nitrate | sap×nit | |
| Bacteroidales | 46.5 | 44.0 | 46.5 | 44.0 | 0.010 | 0.88 | 0.54 | 0.35 | 39.98 | 37.55 | 45.92 | 42.18 | 0.012 | 0.12 | 0.01 | 0.76 |
| Selenomonadales | 1.54 | 2.27 | 1.77 | 2.76 | 0.002 | 0.03 | 0.42 | 0.82 | 2.14 | 1.95 | 2.11 | 2.66 | 0.001 | 0.40 | 0.14 | 0.11 |
| Coriobacteriales | 0.29 | 0.20 | 0.39 | 0.36 | 0.000 | 0.16 | 0.01 | 0.39 | 0.22 | 0.19 | 0.32 | 0.37 | 0.000 | 0.73 | 0.00 | 0.18 |
| Gastranaerophilales | 0.18 | 0.17 | 0.07 | 0.07 | 0.000 | 0.99 | 0.09 | 0.83 | 0.26 | 0.21 | 0.11 | 0.10 | 0.000 | 0.34 | 0.00 | 0.57 |
| Unclassified | 0.14 | 0.20 | 0.14 | 0.05 | 0.000 | 0.30 | 0.02 | 0.03 | 0.17 | 0.18 | 0.14 | 0.11 | 0.000 | 0.59 | 0.10 | 0.47 |
| Synergistales | 0.05 | 0.06 | 0.03 | 0.06 | 0.000 | 0.04 | 0.26 | 0.26 | 0.05 | 0.03 | 0.05 | 0.03 | 0.000 | 0.07 | 0.96 | 0.60 |
| Elusimicrobiales | 0.02 | 0.10 | 0.06 | 0.08 | 0.000 | 0.04 | 0.56 | 0.22 | 0.03 | 0.04 | 0.07 | 0.04 | 0.000 | 0.71 | 0.87 | 0.46 |
| Burkholderiales | 0.01 | 0.01 | 0.06 | 0.06 | 0.000 | 0.82 | 0.00 | 0.62 | 0.01 | 0.01 | 0.11 | 0.04 | 0.000 | 0.29 | 0.01 | 0.44 |
| Unclassified Deltaproteobacteria (×10−3) | 7.08 | 1.42 | 2.03 | 2.66 | 0.000 | 0.09 | 0.26 | 0.15 | 4.36 | 0.31 | 2.97 | 1.00 | 0.000 | 0.03 | 0.93 | 0.43 |
| Victivallales (×10−3) | 6.44 | 4.65 | 1.09 | 6.17 | 0.000 | 0.28 | 0.27 | 0.04 | 1.38 | 3.44 | 1.54 | 4.18 | 0.000 | 0.12 | 0.97 | 0.92 |
| Xanthomonadales (×10−3) | 4.68 | 2.37 | 3.64 | 8.64 | 0.000 | 0.98 | 0.45 | 0.48 | 7.74 | 1.28 | 2.77 | 9.45 | 0.000 | 0.99 | 0.37 | 0.00 |
| Micrococcales (×10−3) | 4.26 | 13.5 | 5.16 | 10.2 | 0.000 | 0.01 | 0.59 | 0.65 | 8.22 | 6.51 | 15.9 | 9.85 | 0.000 | 0.19 | 0.13 | 0.62 |
| Opitutae vadin HA64 (×10−3) | 0.36 | 7.24 | 1.53 | 3.94 | 0.000 | 0.06 | 0.78 | 0.31 | 0.68 | 1.16 | 3.14 | - | 0.000 | 0.40 | 0.97 | 0.14 |
In study 1, cows were fed a control (CTL) diet and CTL diet supplemented with linseed (LIN), nitrate (NIT) and linseed plus nitrate (LIN+NIT). Tested effects were linseed (CTL and NIT versus LIN and LIN+NIT) and nitrate (CTL and LIN versus NIT and LIN+NIT) and their interaction, lin×nit.
In study 2, cows were fed a control (CTL) diet and control diet supplemented with tea saponin (TEA), nitrate (NIT) and tea saponin plus nitrate (TEA+NIT). Tested effects were saponin (CTL and NIT versus TEA and TEA+NIT) and nitrate (CTL and TEA versus NIT and TEA+NIT) and their interaction, sap×nit.
Values are the means from four observations, and analysis was performed on square root-transformed taxonomic tables using the aov function in R.
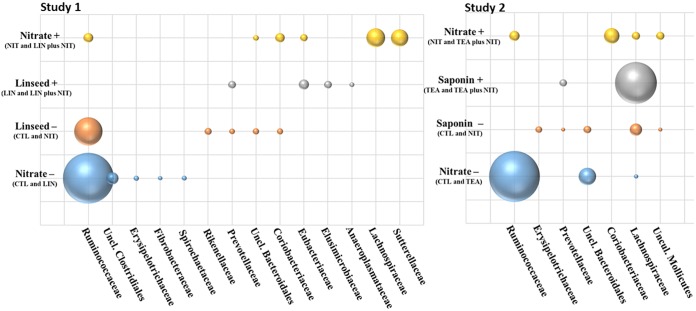
FIG 2.
Bubble charts showing indicator OTU distribution by dietary treatment in the rumen of nonlactating cows fed methane-reducing additives. Bubble size reflects the count number in the rarefied data set.
Regarding methanogen concentration, mcrA copy numbers per nanogram of extracted DNA were not affected by linseed supplementation (Table 1), and neither was overall community structure (Fig. 1 and Table 3).
TABLE 3.
Archaeal species detected in the rumen of nonlactating cows fed methane-reducing additivesc
| Archaeal species | Relative abundance (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study 1a |
Effect |
Study 2b |
Effect |
|||||||||||||
| CTL | LIN | NIT | LIN+NIT | SEM | Linseed | Nitrate | lin×nit | CTL | TEA | NIT | TEA+NIT | SEM | Saponin | Nitrate | sap×nit | |
| Methanobacterium alkaliphilum | 0.04 | 0.71 | 0.03 | 0.01 | 0.002 | 0.166 | 0.126 | 0.138 | 0.01 | 0.56 | 0.05 | 0.00 | 0.001 | 0.184 | 0.173 | 0.120 |
| Methanobrevibacter acididurans | 0.01 | 0.00 | 0.01 | 0.01 | 0.000 | 0.966 | 0.506 | 0.649 | 0.00 | 0.00 | 0.00 | 0.01 | 0.000 | 0.448 | 0.876 | 0.623 |
| Methanobrevibacter boviskoreani clade | 2.72 | 1.44 | 2.72 | 1.86 | 0.018 | 0.502 | 0.891 | 0.887 | 3.89 | 0.50 | 0.40 | 2.46 | 0.018 | 0.776 | 0.740 | 0.234 |
| Methanobrevibacter gottschalkii clade | 38.08 | 45.63 | 21.10 | 19.28 | 0.051 | 0.584 | <0.001 | 0.371 | 39.39 | 38.46 | 29.32 | 27.01 | 0.051 | 0.862 | 0.203 | 0.824 |
| Methanobrevibacter oralis | 0.47 | 0.34 | 0.32 | 0.25 | 0.001 | 0.158 | 0.115 | 0.641 | 0.27 | 0.46 | 0.30 | 0.27 | 0.001 | 0.271 | 0.250 | 0.137 |
| Methanobrevibacter ruminantium clade | 36.28 | 33.06 | 54.18 | 62.10 | 0.043 | 0.742 | <0.01 | 0.365 | 33.80 | 37.75 | 47.87 | 47.66 | 0.043 | 0.766 | 0.107 | 0.735 |
| Methanobrevibacter sp. strain RT | 0.02 | 0.03 | 0.01 | 0.00 | 0.000 | 0.999 | 0.059 | 0.102 | 0.04 | 0.02 | 0.00 | 0.01 | 0.000 | 0.673 | 0.078 | 0.168 |
| Other Methanobrevibacter | 2.54 | 1.64 | 1.70 | 1.13 | 0.002 | 0.109 | 0.136 | 0.712 | 1.56 | 2.05 | 1.52 | 1.50 | 0.002 | 0.331 | 0.220 | 0.296 |
| Methanosphaera cuniculi | 0.22 | 0.04 | 0.25 | 0.23 | 0.001 | 0.575 | 0.561 | 0.668 | 0.05 | 0.09 | 0.08 | 0.10 | 0.001 | 0.641 | 0.760 | 0.774 |
| Methanosphaera sp. strain A4 | 0.08 | 0.03 | 0.05 | 0.04 | 0.000 | 0.193 | 0.746 | 0.456 | 0.05 | 0.03 | 0.02 | 0.03 | 0.000 | 0.912 | 0.089 | 0.066 |
| Methanosphaera sp. strain ISO3-F5 | 7.97 | 6.41 | 8.60 | 8.36 | 0.011 | 0.691 | 0.581 | 0.777 | 9.43 | 8.59 | 6.26 | 11.61 | 0.011 | 0.143 | 0.937 | 0.053 |
| Other Methanosphaera | 0.83 | 0.35 | 0.88 | 0.34 | 0.001 | 0.081 | 0.934 | 0.913 | 0.45 | 0.57 | 0.25 | 0.50 | 0.001 | 0.211 | 0.375 | 0.639 |
| Other Methanobacteriaceae | 0.07 | 0.05 | 0.08 | 0.05 | 0.000 | 0.255 | 0.976 | 0.806 | 0.04 | 0.05 | 0.04 | 0.06 | 0.000 | 0.418 | 0.841 | 0.565 |
| Other Methanococcales | 0.01 | 0.00 | 0.00 | 0.00 | 0.000 | 0.337 | 0.337 | 0.337 | 0.00 | 0.01 | 0.00 | 0.00 | 0.000 | 0.337 | 0.337 | 0.337 |
| Methanomicrobium mobile | 0.24 | 0.08 | 0.01 | 0.00 | 0.003 | 0.320 | 0.081 | 0.376 | 0.51 | 0.46 | 0.62 | 0.04 | 0.003 | 0.399 | 0.679 | 0.477 |
| Other | 0.09 | 0.07 | 0.00 | 0.01 | 0.001 | 0.918 | 0.156 | 0.849 | 0.00 | 0.23 | 0.00 | 0.01 | 0.001 | 0.257 | 0.306 | 0.311 |
| Group 10 species | 0.60 | 0.00 | 0.16 | 0.00 | 0.003 | 0.166 | 0.405 | 0.405 | 0.99 | 0.84 | 0.61 | 0.56 | 0.005 | 0.820 | 0.435 | 0.908 |
| Candidatus “Methanomethylophilus alvus” | 0.02 | 0.01 | 0.02 | 0.01 | 0.000 | 0.360 | 0.642 | 0.689 | 0.00 | 0.02 | 0.01 | 0.01 | 0.000 | 0.243 | 0.845 | 0.110 |
| Group 12 species | 1.99 | 2.21 | 2.50 | 2.05 | 0.006 | 0.902 | 0.878 | 0.745 | 1.51 | 2.41 | 3.27 | 2.36 | 0.006 | 0.999 | 0.244 | 0.218 |
| Group 8 species | 0.10 | 0.09 | 0.03 | 0.00 | 0.000 | 0.797 | 0.234 | 0.876 | 0.02 | 0.00 | 0.00 | 0.00 | 0.000 | 0.214 | 0.156 | 0.297 |
| Group 9 species | 0.03 | 0.02 | 0.09 | 0.16 | 0.001 | 0.677 | 0.244 | 0.648 | 0.04 | 0.21 | 0.03 | 0.02 | 0.001 | 0.052 | 0.025 | 0.039 |
| Other Methanomassiliicoccaceae | 2.96 | 3.12 | 3.87 | 1.58 | 0.000 | 0.842 | 0.687 | 0.226 | 2.74 | 1.51 | 3.44 | 1.67 | 0.000 | 0.987 | 0.859 | 0.883 |
| Other | 4.59 | 4.66 | 3.37 | 2.52 | 0.001 | 0.918 | 0.156 | 0.849 | 5.18 | 5.15 | 5.92 | 4.11 | 0.001 | 0.257 | 0.306 | 0.311 |
In study 1, cows were fed a control (CTL) diet or CTL diet supplemented with linseed (LIN), nitrate (NIT), or linseed plus nitrate (LIN+NIT). Tested effects were linseed (CTL and NIT versus LIN and LIN+NIT) and nitrate (CTL and LIN versus NIT and LIN+NIT) and their interaction, lin×nit.
In study 2, cows were fed a control (CTL) diet or CTL diet supplemented with tea saponin (TEA), nitrate (NIT), or tea saponin plus nitrate (TEA+NIT). Tested effects were saponin (CTL and NIT versus TEA and TEA+NIT) and nitrate (CTL and TEA versus NIT and TEA+NIT) and their interaction, sap×nit.
Values are the means from four observations, and analysis was performed on square root-transformed taxonomic tables using the aov function in R.
Feeding linseed did not modify protozoan community structure and composition compared to the respective control treatment (Fig. 1 and Table 4). There were 3 indicator OTUs identified, 2 associated with CTL diet and 1 with LIN diet, but they all represented less than 0.01% of the rarefied data set (13,809 reads per individual).
TABLE 4.
Protozoa genera detected in the rumen of nonlactating cows fed methane-reducing additivesc
| Protozoa genus | Relative abundance (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study 1a |
Effect |
Study 2b |
Effect |
|||||||||||||
| CTL | LIN | NIT | LIN+NIT | SEM | Linseed | Nitrate | lin×nit | CTL | TEA | NIT | TEA+NIT | SEM | Saponin | Nitrate | sap×nit | |
| Entodinium | 59.84 | 49.66 | 46.52 | 33.94 | 0.036 | 0.67 | 0.05 | 0.10 | 46.21 | 51.05 | 47.55 | 50.55 | 0.040 | 0.68 | 0.90 | 0.88 |
| Polyplastron | 11.57 | 8.50 | 17.51 | 10.43 | 0.021 | 0.75 | 0.26 | 0.27 | 7.94 | 9.73 | 10.39 | 7.19 | 0.016 | 0.62 | 0.92 | 0.63 |
| Unclassified Trichostomatia | 9.93 | 11.59 | 15.26 | 16.70 | 0.023 | 0.97 | 0.36 | 0.75 | 22.28 | 9.34 | 14.59 | 14.16 | 0.032 | 0.23 | 0.72 | 0.47 |
| Isotricha | 1.52 | 3.92 | 6.00 | 4.34 | 0.009 | 0.15 | 0.06 | 0.68 | 5.43 | 5.27 | 6.72 | 6.43 | 0.008 | 0.56 | 0.65 | 0.76 |
| Dasytricha | 1.31 | 0.80 | 1.86 | 4.01 | 0.006 | 0.65 | 0.10 | 0.76 | 0.90 | 1.92 | 1.67 | 1.77 | 0.004 | 0.19 | 0.34 | 0.13 |
| Unclassified | 0.17 | 0.17 | 0.06 | 0.19 | 0.000 | 0.08 | 0.15 | 0.06 | 0.14 | 0.18 | 0.17 | 0.14 | 0.000 | 0.85 | 0.82 | 0.57 |
| Ophryoscolex | 0.16 | 0.00 | 0.46 | 1.33 | 0.002 | 0.06 | 0.00 | 0.67 | 3.26 | 3.08 | 0.41 | 1.47 | 0.010 | 0.64 | 0.52 | 0.78 |
| Unclassified Ciliophora | 0.04 | 0.09 | 0.04 | 0.06 | 0.000 | 0.59 | 0.50 | 0.27 | 0.03 | 0.07 | 0.06 | 0.05 | 0.000 | 0.09 | 0.81 | 0.09 |
| Unclassified SARd | 0.04 | 0.02 | 0.01 | 0.02 | 0.000 | 0.09 | 0.38 | 0.66 | 0.04 | 0.04 | 0.01 | 0.02 | 0.000 | 0.15 | 0.00 | 0.57 |
| Trichostomatia | 0.04 | 0.28 | 0.02 | 0.02 | 0.000 | 0.04 | 0.01 | 0.06 | 0.10 | 0.08 | 0.24 | 0.03 | 0.000 | 0.41 | 0.94 | 0.34 |
| Pseudoplatyophyra | 0.00 | 0.05 | 0.02 | 0.00 | 0.000 | 0.00 | 0.18 | 0.12 | 0.03 | 0.00 | 0.00 | 0.000 | 0.21 | 0.50 | 0.17 | |
In study 1, cows were fed a control (CTL) diet and CTL diet supplemented with linseed (LIN), nitrate (NIT) and linseed plus nitrate (LIN+NIT). Tested effects were linseed (CTL and NIT versus LIN and LIN+NIT) and nitrate (CTL and LIN versus NIT and LIN+NIT) and their interaction, lin×nit.
In study 2, cows were fed a control (CTL) diet and control diet supplemented with tea saponin (TEA), nitrate (NIT) and tea saponin plus nitrate (TEA+NIT). Tested effects were saponin (CTL and NIT versus TEA and TEA+NIT) and nitrate (CTL and TEA versus NIT and TEA+NIT) and their interaction, sap×nit.
Values are the means from four observations, and analysis was performed on square root-transformed taxonomic tables using the aov function in R.
SAR, Stramenopiles-Alveolata-Rhizaria cluster.
Tea saponins had only minor effects on rumen microbial population.
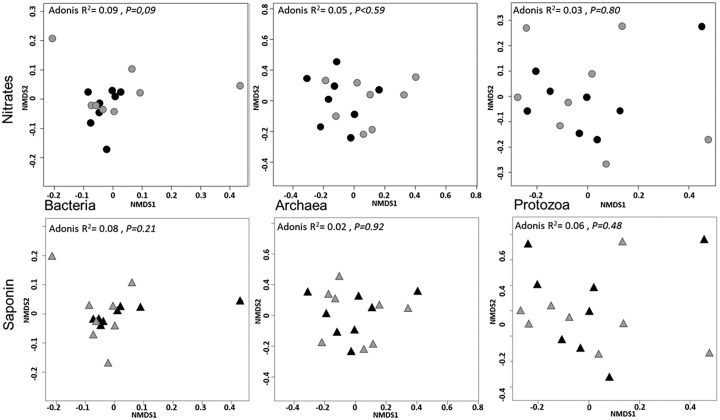
Adding tea saponin to diets only affected the low-abundance order of unclassified Deltaproteobacteria (Table 2). No changes in diversity indices were noticed (Table S2). NMDS (Fig. 3) and permutational multivariate analysis of variance (PERMANOVA) analysis did not reveal significant changes in bacterial community, although Lachnospiraceae were highly abundant in cows supplemented with saponin (Fig. 2). Similarly, concentration and taxonomic composition of the archaeal community were not influenced by tea saponin (Fig. 3 and Table 3), and neither was the protozoan community structure (Fig. 3 and Table 4).
FIG 3.
Structure and composition of bacterial, archaeal, and protozoal communities in study 2, related to nitrate or saponin treatments (black symbols) and respective controls (gray symbols), were examined by multivariate analysis. NDMS plots derived from Bray-Curtis dissimilarities between cows are shown. Each symbol is representative of a single cow. Samples are plotted along the first two-component axes. Microbial composition was compared using Adonis.
Nitrate remodels bacterial and archaeal communities.
In order to have an integrated discussion on the effects of nitrate on microbes from both studies, we needed to compare like to like. This is why we compared microbial communities of cows fed CTL diet in each study. Bacterial communities of these cows were similar (Adonis R2 of 0.16 and P value of 0.26). A small numerical difference was noted in the Bacteroidales/Clostridiales ratios, which were 1.02 and 0.81 in study 1 and study 2, respectively. Similar to bacteria, methanogenic communities in animals fed CTL diets were similar between studies (Adonis R2 of 0.028 and P value of 0.942). Regarding protozoa, some differences were revealed by NMDS and PERMANOVA analyses between the two control groups. NMDS graphs (Fig. S3) showed only a small overlap between the protozoan population fed CTL in each study, which was confirmed by an Adonis test (P < 0.1). Also, Entodinium-related sequences accounted for 60% of total classified sequences in study 1, whereas they represented 46% of sequences in study 2 (Table 4). Although this difference was not statistically significant, it was accompanied by significantly higher numbers of Trichostomatia- and Isotricha-related sequences in study 1 than study 2 (Table 4).
Feeding nitrate, in both studies, increased Coriobacteriales and Burkholderiales relative abundance and decreased (study 2), or tended to decrease (study 1), abundance of Gastranaerophilales (Table 2). In addition, in study 2, nitrate supplementation increased the relative abundance of Bacteroidales (Table 2). Diversity indices were not influenced by dietary treatment (Table S2) in any study. NMDS analysis (Fig. 1) revealed that while in study 1 nitrate supplementation was the major driver of phylogenetic dissimilarity among bacterial communities (Adonis R2 of 0.11, P value of <0.01), in study 2 nitrate only moderately affected community structure (Adonis R2 of 0.09, P value of 0.09). Indicator species analysis revealed that 10 OTUs in study 1 and 21 in study 2 were differentially abundant between cows fed and not fed nitrate (P value of <0.05 and indicator value of >0.7; Table S3 and Table S4). Lachnospiraceae and Sutterellaceae characterized nitrate-supplemented diets (Fig. 2) in study 1, and Coriobacteriaceae and the uncultured Mollicutes family were identified as indicator OTUs for nitrate-supplemented diets in study 2. More interestingly, in both studies Ruminococcaceae-related OTUs characterized the bacterial community of control cows (Fig. 2).
CowPI predictive analysis suggested that nitrogen metabolism was increased in both studies when nitrate was fed to cows (Table S1). Nitrate supplementation induced numerous changes in metabolic profiles. Regarding carbohydrate metabolism, nitrate supplementation would affect most of the described pathways, but observed changes were different in each study. Predictions regarding lipid metabolism were more consistent between studies and suggested that dietary supplementation with nitrates would decrease biosynthesis of fatty acids.
In both studies, feeding nitrate had no effect on methanogen concentration in the rumen (mcrA copy numbers) but reduced methanogen activity (mcrA expression levels) (Table 1). When cows were fed nitrate, Shannon and Simpson diversity indices decreased or tended to decrease (Table S2), although the overall taxonomic composition was not affected (Table 3). NMDS and PERMANOVA analyses showed that feeding nitrates deeply modified archaeal community structure in study 1 but had no effect on community structure in study 2 (Fig. 1 and 3).
In study 1, Entodinium relative abundance tended to decrease and Isotricha tended to increase in animals receiving nitrate-supplemented diets (Table 4). Diversity indices remained similar between diets and contrasts (Table S2). However, there was some evidence (Adonis R2 of 0.12, P value of 0.05) that nitrate modulated the rumen protozoan population in cows (Fig. 1). In contrast, in study 2, nitrate had no effect on protozoan community in the rumen of nonlactating dairy cows (Fig. 3).
Correlation patterns of microbial population.
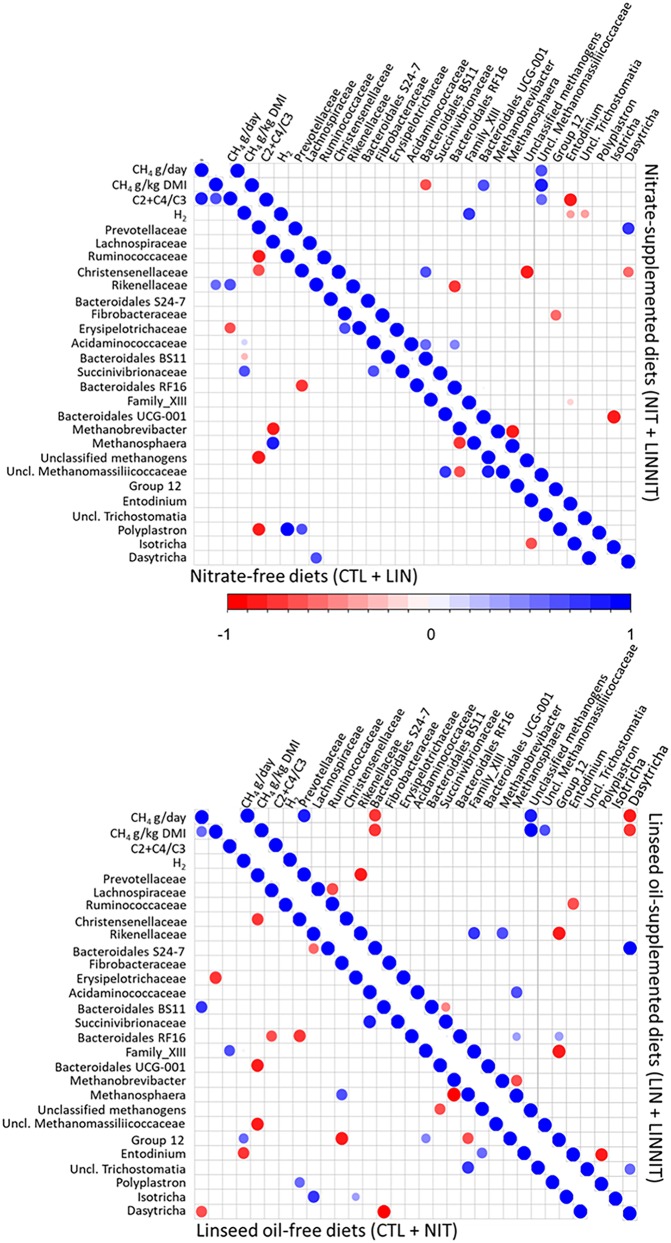
We analyzed the correlation between bacterial families and genera of methanogens and protozoa (Fig. 4 and 5). Values for methane production (g/day), yield (g/kg DMI), hydrogen production (only for study 1), and (acetate + butyrate)/propionate ratio from the data sets of Guyader et al. (8, 13) were also included in the analysis. Only significant correlations are discussed.
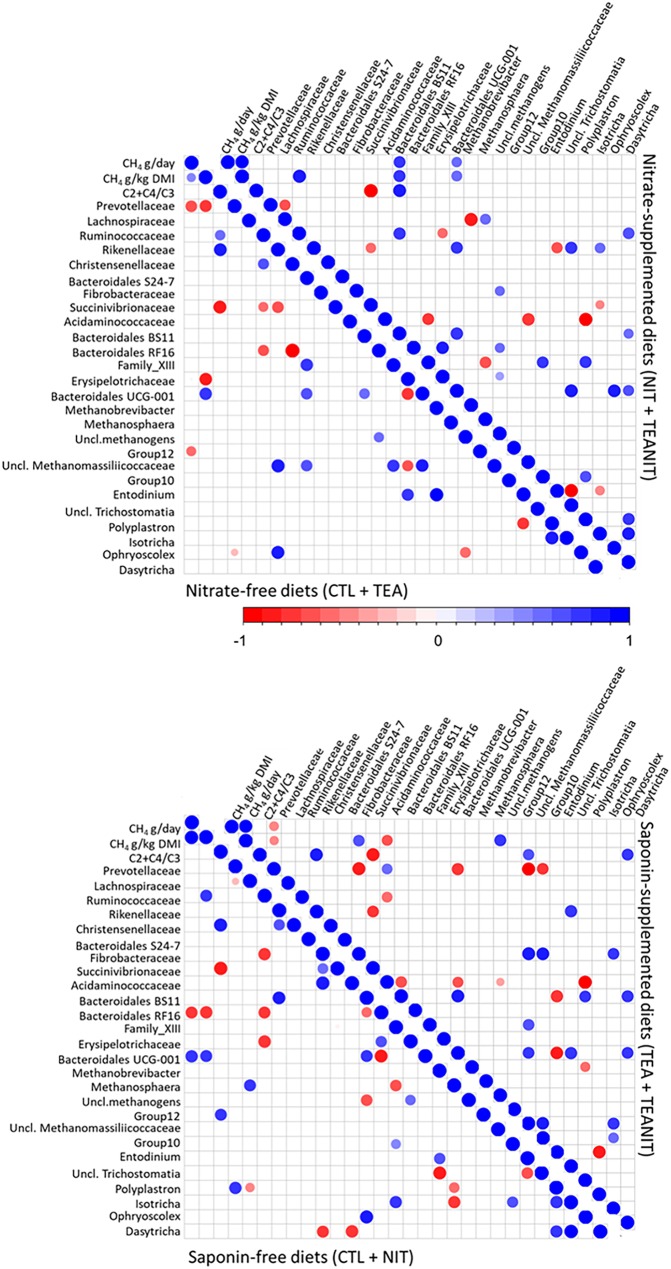
FIG 4.
Spearman’s rank correlation matrix of the dominant ruminal bacterial families, genera of archaea and protozoa, and fermentation parameters in study 1. Illustrated correlation patterns are for nitrate and linseed supplementations. Listed microbial populations were detected in at least 50% of the rumen samples analyzed and represent at least 1% of the bacterial, archaeal, protozoal, or fungal communities. Strong correlations are indicated by large circles, whereas weak correlations are indicated by small circles. The colors of the scale bar denote the nature of the correlation, with 1 indicating perfect positive correlation (dark blue) and −1 indicating perfect negative correlation (dark red) between two microbial populations.
FIG 5.
Spearman’s rank correlation matrix of the dominant ruminal bacterial families, genera of archaea and protozoa, and fermentation patterns in study 2. Illustrated correlation patterns are for nitrate and tea saponin supplementations. Listed microbial populations were detected in at least 50% of the rumen samples analyzed and represent at least 1% of the bacterial, archaeal, protozoal, or fungal communities. Strong correlations are indicated by large circles, whereas weak correlations are indicated by small circles. The colors of the scale bar denote the nature of the correlation, with 1 indicating perfect positive correlation (dark blue) and −1 indicating perfect negative correlation (dark red) between two microbial populations.
In study 1 (Fig. 4), methane production (g/day) and yield (g/kg DMI) were positively correlated (R2 = 0.83 and R2 = 0.69, respectively) with the (acetate + butyrate)/propionate ratio when cows were not fed nitrate; in these animals, methane yield correlated positively with Rikenellaceae (R2 = 0.56). In the absence of nitrate, Methanobrevibacter negatively correlated with unclassified Methanomassiliicoccaceae (R2 = −0.64), and Ruminococcaceae correlated positively with members of the protozoal Polyplastron genus (R2 = 0.97). When diets were supplemented with nitrate, methane production and yield as well as (acetate + butyrate)/propionate ratio were strongly correlated with a group of unclassified Methanomassiliicoccaceae (R2 = 0.69, R2 = 0.85, and R2 = 0.59, respectively). In addition, when diets were nitrate supplemented, a positive correlation was established between Prevotellaceae and Dasytricha (R2 = 0.70 and R2 = 0.73). There was a strong negative correlation between Methanobrevibacter and Methanosphaera independently of nitrate supplementation (R2 of −0.76 in cows not fed nitrate and R2 of −0.83 in cows fed nitrate).
Methane production and yield when linseed was fed to cows correlated negatively with the Bacteroidales S 24.7 group (R2 = −0.76) and Dasytricha (R2 = −0.83) populations and positively with an unclassified archaeal taxon (R2 = 0.83) and unclassified Methanomassiliicoccaceae (R2 = 0.61). Independently of linseed supplementation, a negative correlation between Methanobrevibacter and Methanosphaera was observed (R2 = −0.64 and R2 = −0.95).
In study 2 (Fig. 5), when diets were not supplemented with nitrate, methane production correlated negatively with Prevotellaceae (R2 = −0.69) and methanogen group 12 (R2 = −0.50). When diet was supplemented with tea saponin, Prevotellaceae correlated negatively with methane production (R2 = −0.47) and yield (R2 = −0.43) as well as with Fibrobacteraceae (R2 = −0.88), Bacteroidales (R2 = −0.76), and two families of Methanomassiliicoccaceae (R2 of −0.90 for unclassified Methanomassiliicoccaceae and R2 of −0.70 for group 10).
DISCUSSION
Guyader et al. (8) showed that combining dietary strategies acting theoretically on hydrogen production (lipids) and consumption (nitrate) can have an additive effect on methane reduction. In a second study, they confirmed the antimethanogenic potential of nitrate supplementation but observed no effect of tea saponin on methane production (13). These studies were conducted simultaneously; cows were selected at random from the same experimental herd and were randomly allocated to a study. Given the consistency of results for methane production and fermentation patterns reported in the two articles of Guyader et al. (8, 13), we decided to analyze the rumen microbiota from both studies at the same time (from DNA extraction up to statistical tests). Although linseed and nitrate have a medium to high potential methane-mitigating effect (the effect of saponins being less reproductive) (20), microbial data are scarce and inconsistent between studies. This could be explained by different methodologies for rumen sample collection, conservation, and nucleic acid extraction, as well as on how data were obtained and analyzed (21, 22). Thus, second, we compiled the microbial data in order to get insight into the mode of action of nitrate on the rumen microbial ecosystem.
To this aim, we first checked that the microbiota of the two groups of cows was comparable; hence, we performed a detailed analysis of microbial community structure and composition in rumen contents sampled during the period when CTL diet was fed to each animal. No major differences in bacterial communities were observed, except a nonsignificant shift in the Bacteroidales/Clostridiales ratio, which is known to vary widely across individual animals (23). However, we observed numerical differences in the relative abundance of Entodinium (60% in study 1 versus 46% in study 2), which is consistent with enumeration results reported previously (5.71 and 5.38 log10 cells/ml in study 1 and study 2, respectively [8, 13]), showing more abundant ciliate populations in cows from study 1.
In study 1, dietary supplementation with linseed increased the relative abundance of Selenomonadales. This is in accordance with our previous work exploring the effects of linseed plus nitrate on rumen microbiota (24) in bulls, where we reported increased numbers in sequences affiliated with three Selenomonas genera and one unclassified Selenomonadales genus. As these microbes are potential nitrate reducers (25), we hypothesized that their growth was supported by the higher nitrate availability, but the present study suggests that it is a linseed effect. Oleic acid (representing, on average, 20% of linseed oil fatty acids) stimulated the growth of Selenomonas ruminantium in pure cultures (26). However, for in vivo studies, results are contrasting: Selenomonas was among the genera explaining differences in bacterial community structure between lambs fed a linseed diet and those fed a control diet (27), but there was no change in Selenomonas abundance when cows were fed sunflower oil (30% oleic acid) (23). Members of the Selenomonadales order are also known to reduce succinate to propionate, which is in agreement with a higher molar proportion of propionate in the rumen of cows fed linseed (8). Linseed supplementation also increased abundance of uncultured Bacteroidetes, and the Bacteroidales S27-7 family was negatively correlated with methane production and yield. On the other hand, linseed diets were characterized by decreased abundance of Ruminococcaceae, which is in agreement with previous findings that fatty acids are toxic to these cellulolytic microbes (23, 26, 28). We observed no effect on rumen protozoan numbers (8) and diversity, although Dasytricha correlated negatively with methane emissions and positively with the Bacteroidales S27-7 family. Linseed oil supplementation also had no effect on the abundance or diversity of the rumen methanogenic community. In accordance with previous results (9, 24), the antimethanogenic potential of linseed oil fatty acids was not related to archaeal numbers in the rumen but rather to a lower metabolic activity of these microbes, which could be explained by lower availability of hydrogen.
Adding tea saponins to the diet had no effect on microbial numbers or on diversity. This is consistent with the lack of changes in methane production or VFA profiles reported by Guyader et al. (13). The efficacy of saponins in suppressing methane production varies considerably depending on the chemical structure, source, dose, and diet (29). Saponins have been reported to inhibit rumen protozoa (5) and, thus, limit hydrogen production in the rumen. However, in our previous work (13) and the study of Ramírez-Restrepo et al. (30), adding tea saponins to ruminants’ diets had the opposite effect on protozoan numbers. Saponins break down the membrane of protozoa by interacting with their sterols. However, rumen microbes can degrade the sugar moiety of saponins, rendering them inactive. To improve the antiprotozoal effect of saponins, changing their chemical structure and, thus, protecting them from microbial degradation, was recently proposed (12).
Nitrate supplementation induced changes in the relative abundance of CowPI-generated functional profiles of bacteria, although metabolic pathways were affected in a dissimilar way between studies. This was expected, as predictions are based on 16S rRNA gene data and multivariate analysis of OTU tables also show differences between studies. In study 1, multivariate analysis revealed that nitrate supplementation altered bacterial and archaeal communities. However, in study 2, NMDS and PERMANOVA results were less conclusive, although reductions of methane emissions and changes in fermentation parameters were comparable between experiments. Nevertheless, both studies pinpointed a limited number of taxa associated with decreased methane emissions in nitrate-fed cows. Nitrate supplementation increased the abundance of Coriobacteriales and Burkholderiales orders, which contain taxa with known nitrate-reducing activity (31–33). This coincides with predicted higher nitrogen metabolism functions and is in accordance with the numerically higher nitrite concentrations measured by Guyader et al. (8, 13) in nitrate-fed cows. Also, cows not fed nitrate presented an enhanced cellulolytic community, which is in accordance with our previous results showing a toxic effect on Ruminococcaceae in animals fed linseed plus nitrate diets (24). Ruminococcus flavefaciens and Ruminococcus albus populations decreased in the rumen of goats when nitrate was added to the diet (25). An in vitro study (34) showed that the growth of these two cellulolytic bacteria was inhibited by nitrite at a level of 3 mmol/liter, but measured nitrite levels in our studies rarely exceeded 0.08 mmol/liter (8, 13). Lower concentrations could still be toxic, as another study showed that the specific growth rate of R. flavefaciens, but not R. albus, was decreased by less than 0.03 mmol/liter of nitrate (35). Marais et al. (35) also argued that nitrite inhibits electron transport systems (R. flavefaciens), so bacteria not possessing an electron transport system (R. albus) are less affected. R. flavefaciens and R. albus are the only cultured Ruminococcus species able to degrade cellulose (36), making them an important part of a functional rumen ecosystem. In vitro, R. albus produces acetate, hydrogen, and carbon dioxide, and its metabolic activity is stimulated by the presence of methanogens (37). Thus, reducing Ruminococcaceae numbers by nitrate supplementation would decrease the amount of hydrogen produced, which could indirectly reduce methane production. This conclusion is also supported by the decreased expression levels of the methanogenic mcrA gene, which has been shown to correlate with methane emissions (24, 38, 39). However, Ruminococcaceae are an important group of bacteria inhabiting the rumen and are able to degrade plant cell wall polysaccharides into metabolizable energy. This implies that inhibition of the rumen fibrolytic community decreases fiber degradation. In the present studies, nitrate supplementation did not affect total tract digestibility (8, 13), but linseed tended to reduce fiber digestibility (8).
We also observed a strong positive correlation between unclassified Methanomassiliicoccaceae and methane production when cows were fed nitrate-supplemented diets. Veneman et al. (9) also reported an increase in the abundance of Methanomassiliicoccaceae-related methanogens in the rumen of nitrate-fed animals. Methanomassiliicoccaceae are obligate hydrogen-dependent methylotrophic methanogens (40), whereas most of the other rumen methanogens perform methanogenesis from hydrogen and carbon dioxide. They are part of a unique methanogen order with a characteristic set of genes involved in the methanogenesis pathway (40). It is likely that their particular physiology confers on them a competitive advantage when the activity of other methanogens is affected in a nitrate/nitrite-enriched environment.
We conducted this study to understand how the rumen microbial ecosystem responds to dietary methane mitigation by linseed, saponin, and nitrate supplementation alone or in combination. We hypothesized that adding linseed or saponins to the diet reduces hydrogen production by a toxic effect on rumen protozoa and by replacing dietary carbohydrates with nonfermentable fatty acids; additionally, we were expecting that nitrate supplementation would redirect hydrogen consumption toward nitrate reduction rather than methanogenesis. Changes in the rumen microbial ecosystem were monitored using archaeon-, bacterium-, eukaryote-, and fungus-specific primers targeting either 16S or 18S rRNA genes and ITS1. Our sequencing strategy allowed us to accurately draw the parallel between changes in methane emissions and microbiota structure. Our study showed that linseed oil decreases methane emissions by reducing the number of hydrogen producers (cellulolytic Ruminococcaceae) and by stimulating propionate producers (Selenomonas), thereby diverting hydrogen from methanogenesis. Nitrate supplementation favored the development of nitrate-reducing bacteria (Coriobacteriales and Burkholderiales) and had a negative effect on cellulolytic Ruminococcaceae; as a consequence, nitrate supplementation also significantly affected methanogen community structure and activity. In contrast, we did not show any shifts in rumen microbiota structure and activity due to dietary supplementation with tea saponins.
In a secondary aim of our work, we capitalized on data available from two independent studies, expecting to draw relevant conclusions. It is common that studies exploring microbial mechanisms of the same methane abatement strategy come to dissimilar conclusions. Authors generally argue that these differences are due to differences in diet, animal species, physiologic stage, and different sample processing or bioinformatics pipelines. In the present work, we minimized the impact of study design on data interpretation, despite some inconsistent results being observed for nitrate-supplemented diets from study 1 and study 2. Nitrate reduced methanogen activity and stimulated nitrate-reducing bacterial populations in both studies. Similarly, Ruminococcaceae-related OTUs characterized nitrate-free diets in both studies. In contrast, multivariate analysis showed that nitrate altered bacterial and archaeal communities in study 1, whereas only a moderate effect on bacteria was observed in study 2. In both experiments, each experimental period lasted 5 weeks. It is possible that microbiota shifted as a result of imposed dietary treatments and did not completely migrate back to the initial state. In a massive rumen contents exchange study, Weimer et al. (41) found that cows almost completely reconstructed their microbiota in 3 weeks, with a complete return to its original host-specific state in 9 weeks. However, pH and VFA profiles returned to the original values much more quickly, within 1 day. We could argue that changes induced by nitrate supplementation were at the level of microbe function rather than species composition. This is supported by the fact that reductions in methane emissions and shifts in VFA profiles were comparable between studies. A metatranscriptomic approach will be more fruitful to further explore microbial mechanisms of methane mitigation using linseed and/or nitrate.
MATERIALS AND METHODS
The experiments were conducted at the animal facilities of INRA Herbipôle Unit (Saint-Genès Champanelle, France). All procedures involving animals were conducted in accordance with the French Ministry of Agriculture guidelines for animal research and all applicable European guidelines and regulations on animal experimentation. The experiments were approved by the Auvergne Regional Ethics Committee for Animal Experimentation, approval number CE50-12.
Animals, experimental design, and feeding management.
Animals and experimental design were described by Guyader et al. (8, 13). Briefly, eight nonlactating Holstein cows were separated into two groups conducted in parallel according to a two-by-two factorial design. Within each study, four cows were randomly assigned to four dietary treatments during 5-week experimental periods. In study 1, diets were on a dry matter (DM) basis: control diet (CTL; 50% natural grassland hay and 50% concentrate), control diet with 4% linseed oil (LIN; 2.6% added fat), control diet with 3% calcium nitrate (NIT; 2.3% nitrate), and control diet with 4% linseed oil plus 3% calcium nitrate (LIN+NIT; 2.6% added fat plus 2.3% nitrate) (8). In study 2, diets were on a DM basis: control diet (CTL; 50% natural grassland hay and 50% concentrate), control diet with 0.77% tea saponin (TEA; 0.5% saponin), control diet with 3% calcium nitrate (NIT; 2.3% nitrate), and control diet with 0.77% tea saponin plus 3% calcium nitrate (TEA+NIT; 0.5% saponin plus 2.3% nitrate) (13). The chemical compositions of the diets CTL and NIT were similar between the two studies. Methane emissions and fermentation parameters are those described in companion papers of Guyader et al. (8, 13).
Rumen content sampling for microbial analysis.
At the end of each experimental period, whole rumen content samples (200 g) were taken, through cannula, from multiple sites within the rumen. Sampling was done 3 h after the morning feeding, when methane emission differences between diets measured in the same animal were maximal (42). A part of each sample (∼30 g) was mixed with 30 ml ice-cold phosphate-buffered saline (PBS), pH 6.8, and homogenized using a Polytron grinding mill (Kinematica GmbH, Steinhofhalde, Switzerland) for three cycles of 1 min with intervals of 1 min on ice. Approximately 0.5 g was transferred to a 2.5-ml Eppendorf tube and mixed with 1 ml of RNAlater stabilization solution (Applied Biosystems, Austin, TX, USA). Tubes were immediately stored at −80°C until further processing.
Total nucleic acid extraction and cDNA synthesis.
Total nucleic acids (DNA and RNA) were coextracted from all samples by bead beating and phenol-chloroform extraction, followed by saline-alcohol precipitation (43). The yield and purity of extracted DNA and RNA were assessed using a NanoDrop lite spectrophotometer (Thermo Fisher Scientific, Wilmington, DE); RNA integrity was estimated with an Agilent RNA 6000 Nano kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. Following extraction and quality assessment, RNA was reverse transcribed using a reverse transcriptase kit with random primers (Promega, Madison, WI), according to the manufacturer’s instructions, on a T-100 thermocycler (Bio-Rad, Hercules, CA).
Quantification and gene expression of microbial communities.
Samples from each cow from the two sampling days of each experimental period were pooled by mixing an equal quantity of DNA or equal volumes of cDNA. Quantification of gene targets was performed on microbial DNA and cDNA by quantitative PCR (qPCR) using a StepOnePlus apparatus (Applied Biosystems, Villebon sur Yvette, France). Reactions were run in triplicate in 96-well plates, using 15.5 μl of 1× TaKaRa SYBR Ex Taq premix (Lonza, France), 0.25 μmol each forward and reverse primer, and 20 ng of DNA or 2 μl of cDNA in a final volume of 20 μl. Primer description, average amplification efficiency, slope, and R2 values from qPCR are described in Table S4 in the supplemental material as required by MIQE guidelines for PCR (44). Negative controls without templates were run in each assay to assess overall specificity.
Abundances of total bacteria (based on 16S rRNA gene copies) and methanogens (based on mcrA DNA copies) were assessed using absolute quantification as previously described (39). The level of expression of the functional mcrA gene (based on mcrA cDNA copies) was assessed using the 2−ΔCT method (45) (CT is threshold cycle) with 16S rRNA gene copies as an internal reference: 2−ΔCT = 2−(CT mcrA × CT rrs).
Technical triplicates were averaged while checking overlaying of amplification plots at the CT value. Absolute quantification of total bacteria and methanogenic archaea was expressed as log10 16S rRNA gene and mcrA copies/ng extracted DNA, respectively.
Sequencing strategy and data analysis.
Approximately 3 μg of extracted DNA was sent to the Roy J. Carver Biotechnology Center (Urbana, IL, USA) for fluidigm amplification and sequencing of bacterial and archaeal 16S rRNA genes, eukaryotic 18S ribosomal DNA (rDNA) for protozoa, and internal transcribed region 1 (ITS1) for fungi (Table S5). The libraries were sequenced on a 250-paired-end MiSeq run and generated 8,249,698 raw reads for bacterial 16S rRNA genes, 1,778,521 for archaeal 16S rRNA genes, 836,803 for ITS1, and 2,245,531 for eukaryotic 18S rDNA (Table S6). Data were analyzed on an in-house Galaxy-based graphic user interface for QIIME (46), PIPITS (47), and IM Tornado (48) (Table S5). All pipelines included a quality control step, removing sequences with Phred scores of <33 and trimming based on expected amplicon lengths, as well as merging paired reads, chimera search, and removal and OTU picking (Table S6). Merging paired-end archaeal 16S rRNA gene reads was performed by mothur’s (49) make.contigs command before input in the QIIME pipeline. Taxonomic classification for Bacteria and Protozoa was based on the SILVA v123 database (50), for Archaea on RIM-DB (51), and for fungi on the UNITE database (52). CowPI (53), the rumen microbiome-focused version of PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (54), was used to predict dietary treatment-induced changes in bacterial metabolic profiles.
Statistical analysis.
Results from qPCR quantification, relative abundance (after square root transformation) of microbes at different taxonomic levels, diversity indices, and CowPI functional gene relative abundances were analyzed by analysis of variance (ANOVA) in R (version 3.4.0). The statistical model included the random effect of cow (n = 4); fixed effect of period (n = 4); contrasts for nitrate (CTL and LIN versus NIT and LIN+NIT in study 1, CTL and TEA versus NIT and TEA+NIT in study 2), linseed (CTL and NIT versus LIN and LIN+NIT in study 1), and tea saponin (CTL and NIT versus TEA and TEA+NIT in study 2); and the interaction linseed×nitrate or saponin×nitrate. Significance was considered at a P value of ≤0.05. Trends were discussed at 0.05 < P ≤ 0.1. Least-square means are reported throughout.
OTUs with fewer than 3 sequences were withdrawn from further analysis. OTU tables were imported in R and rarefied to minimize the variations created by different sample depths of subsampling. Further analysis was performed using the vegan R package (55). Alpha diversity values for all microbial communities were obtained using various diversity indices (Shannon and Simpson diversity indices, richness, and evenness) and analyzed by ANOVA for the effect of contrasts and the interactions described above. NMDS was used to ordinate microbial libraries (4 cows and 4 experimental periods per study and per microbial group). We used the betadisper function to check the homogeneity of group dispersions before performing a PERMANOVA analysis via the Adonis function of vegan. The multipatt function from R package indicspecies (56) was used to find indicator OTUs using a 5% significance level for selecting indicators in cows fed linseed, tea saponin, and nitrate. The species-site group association parameter was IndVal.g.
Correlation analyses between microbial populations and some fermentation parameters (methane, hydrogen, and VFA ratio) were performed in R. Only microbial groups that represented more than 1% (average of all samples) of the total community within each of the three microbial groups (bacteria, archaea, or protozoa) and that were detected in at least 50% of rumen samples were included in the analysis. Spearman’s rank correlations and P values were calculated by the above-described contrasts and plotted using the packages hmisc (57) and corrplot (58).
Accession number(s).
Raw sequence data are available in the Sequence Read Archive (SRA) under BioProject ID PRJNA415383.
Supplementary Material
ACKNOWLEDGMENTS
J.G. was the recipient of an INRA-Région Auvergne Ph.D. scholarship. C.S. acknowledges receipt of a postdoctoral fellowship from Fundación Alfonso Martín Escudero (Madrid, Spain). We also thank NEOVIA for financial support.
We thank the INRA personnel of the Experimental Unit Herbipôle (L. Mouly, D. Roux, S. Rudel, and V. Tate) for taking care of the animals and of the UMR Herbivores (L. Genestoux and D. Graviou) for their help in sampling and performing laboratory analysis. We are grateful to the INRA MIGALE bioinformatics platform (http://migale.jouy.inra.fr) for providing computational resources. We thank David Marsh for checking and amending our English. We are grateful to Richard Dewhurst for constructive criticism of the manuscript.
Authors made the following contributions: conception or design of the work, M.P., J.G., M.S., and D.M.; data collection, M.P., J.G., M.S., C.S., and A.R.S.; data analysis and interpretation, M.P., J.G., M.S., C.S., A.R.S., C.G., C.M., D.M., and A.B.; drafting the article, M.P.; critical revision of the article, J.G., M.S., C.S., A.R.S., C.G., C.M., D.M., and A.B.; and final approval of the version to be published, M.P., J.G., M.S., C.S., A.R.S., C.G., C.M., D.M., and A.B. We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02657-18.
REFERENCES
- 1.Gerber PJ, Steinfeld H, Henderson B, Mottet A, Opio C, Dijkman J, Falcucci A, Tempio G. 2013. Tackling climate change through livestock—a global assessment of emissions and mitigation opportunities. Food and Agriculture Organization of the United Nations (FAO), Rome, Italy. [Google Scholar]
- 2.Guyader J, Eugène M, Nozière P, Morgavi DP, Doreau M, Martin C. 2014. Influence of rumen protozoa on methane emission in ruminants: a meta-analysis approach. Animal 8:1816–1825. doi: 10.1017/S1751731114001852. [DOI] [PubMed] [Google Scholar]
- 3.Veneman JB, Saetnan ER, Clare AJ, Newbold CJ. 2016. MitiGate; an online meta-analysis database for quantification of mitigation strategies for enteric methane emissions. Sci Total Environ 572:1166–1174. doi: 10.1016/j.scitotenv.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 4.Doreau M, Bamière L, Pellerin S, Lherm M, Benoit M. 2014. Mitigation of enteric methane for French cattle: potential extent and cost of selected actions. Anim Prod Sci 54:1417–1422. doi: 10.1071/AN14207. [DOI] [Google Scholar]
- 5.Patra AK, Saxena J. 2009. The effect and mode of action of saponins on the microbial populations and fermentation in the rumen and ruminant production. Nutr Res Rev 22:204–219. doi: 10.1017/S0954422409990163. [DOI] [PubMed] [Google Scholar]
- 6.Allison M, Reddy C. 1984. Adaptations of gastrointestinal bacteria in response to changes in dietary oxalate and nitrate, p 248–256. 3rd International Symposium on Microbial Ecology. International Society for Microbial Ecology, Washington, DC. [Google Scholar]
- 7.Lee C, Beauchemin KA. 2014. A review of feeding supplementary nitrate to ruminant animals: nitrate toxicity, methane emissions, and production performance. Can J Anim Sci 94:557–570. doi: 10.4141/cjas-2014-069. [DOI] [Google Scholar]
- 8.Guyader J, Eugène M, Meunier B, Doreau M, Morgavi DP, Silberberg M, Rochette Y, Gerard C, Loncke C, Martin C. 2015. Additive methane-mitigating effect between linseed oil and nitrate fed to cattle. J Anim Sci 93:3564–3577. doi: 10.2527/jas.2014-8196. [DOI] [PubMed] [Google Scholar]
- 9.Veneman JB, Muetzel S, Hart KJ, Faulkner CL, Moorby JM, Perdok HB, Newbold CJ. 2015. Does dietary mitigation of enteric methane production affect rumen function and animal productivity in dairy cows? PLoS One 10:e0140282. doi: 10.1371/journal.pone.0140282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin C, Ferlay A, Mosoni P, Rochette Y, Chilliard Y, Doreau M. 2016. Increasing linseed supply in dairy cow diets based on hay or corn silage: Effect on enteric methane emission, rumen microbial fermentation, and digestion. J Dairy Sci 99:3445–3456. doi: 10.3168/jds.2015-10110. [DOI] [PubMed] [Google Scholar]
- 11.Benchaar C, Romero-Pérez GA, Chouinard PY, Hassanat F, Eugene M, Petit HV, Côrtes C. 2012. Supplementation of increasing amounts of linseed oil to dairy cows fed total mixed rations: effects on digestion, ruminal fermentation characteristics, protozoal populations, and milk fatty acid composition. J Dairy Sci 95:4578–4590. doi: 10.3168/jds.2012-5455. [DOI] [PubMed] [Google Scholar]
- 12.Ramos-Morales E, de la Fuente G, Nash RJ, Braganca R, Duval S, Bouillon ME, Lahmann M, Newbold CJ. 2017. Improving the antiprotozoal effect of saponins in the rumen by combination with glycosidase inhibiting iminosugars or by modification of their chemical structure. PLoS One 12:e0184517. doi: 10.1371/journal.pone.0184517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guyader J, Eugene M, Doreau M, Morgavi DP, Gerard C, Loncke C, Martin C. 2015. Nitrate but not tea saponin feed additives decreased enteric methane emissions in nonlactating cows. J Anim Sci 93:5367–5377. doi: 10.2527/jas.2015-9367. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Yu Z, Meng Q. 2012. Effects of nitrate on methane production, fermentation, and microbial populations in in vitro ruminal cultures. Bioresour Technol 103:173–179. doi: 10.1016/j.biortech.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Latham EA, Anderson RC, Pinchak WE, Nisbet DJ. 2016. Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front Microbiol 7:228. doi: 10.3389/fmicb.2016.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enjalbert F, Combes S, Zened A, Meynadier A. 2017. Rumen microbiota and dietary fat: a mutual shaping. J Appl Microbiol 123:782–797. doi: 10.1111/jam.13501:782-797. [DOI] [PubMed] [Google Scholar]
- 17.Martin C, Morgavi DP, Doreau M. 2010. Methane mitigation in ruminants: from microbe to the farm scale. Animal 4:351–365. doi: 10.1017/S1751731109990620. [DOI] [PubMed] [Google Scholar]
- 18.Surana NK, Kasper DL. 2017. Moving beyond microbiome-wide associations to causal microbe identification. Nature 552:244. doi: 10.1038/nature25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popova M, Guyader J, Silberberg M, Seradj AR, Saro C, Bernard A, Gérard C, Martin C, Morgavi DP. 2018. Changes in the rumen microbiota of cows in response to dietary supplementation with nitrate, linseed, and saponin alone or in combination. bioRxiv 10.1101/383067. [DOI] [PMC free article] [PubMed]
- 20.Hristov AN, Oh J, Firkins JL, Dijkstra J, Kebreab E, Waghorn G, Makkar HPS, Adesogan AT, Yang W, Lee C, Gerber PJ, Henderson B, Tricarico JM. 2013. Mitigation of methane and nitrous oxide emissions from animal operations. I. A review of enteric methane mitigation options. Animal 91:5045–5069. doi: 10.1017/S1751731113000876. [DOI] [PubMed] [Google Scholar]
- 21.Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, Schwager E, Crabtree J, Ma S, The Microbiome Quality Control Project Consortium, Abnet CC, Knight R, White O, Huttenhower C. 2017. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol 35:3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh Y-C, Needham D, Sieradzki E, Fuhrman J. 2017. Taxon disappearance from microbiome analysis indicates need for mock communities as a standard in every sequencing run. bioRxiv doi: 10.1101/206219. [DOI] [PMC free article] [PubMed]
- 23.Zened A, Combes S, Cauquil L, Mariette J, Klopp C, Bouchez O, Troegeler-Meynadier A, Enjalbert F. 2013. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol 83:504–514. doi: 10.1111/1574-6941.12011. [DOI] [PubMed] [Google Scholar]
- 24.Popova M, McGovern E, McCabe MS, Martin C, Doreau M, Arbre M, Meale SJ, Morgavi DP, Waters SM. 2017. The structural and functional capacity of ruminal and cecal microbiota in growing cattle was unaffected by dietary supplementation of linseed oil and nitrate. Front Microbiol 8:937. doi: 10.3389/fmicb.2017.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asanuma N, Yokoyama S, Hino T. 2015. Effects of nitrate addition to a diet on fermentation and microbial populations in the rumen of goats, with special reference to Selenomonas ruminantium having the ability to reduce nitrate and nitrite. Anim Sci J 86:378–384. doi: 10.1111/asj.12307. [DOI] [PubMed] [Google Scholar]
- 26.Maczulak AE, Dehority BA, Palmquist DL. 1981. Effects of long-chain fatty acids on growth of rumen bacteria. Appl Environ Microbiol 42:856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons T, Boland T, Storey S, Doyle E. 2017. Linseed oil supplementation of lambs’ diet in early life leads to persistent changes in rumen microbiome structure. Front Microbiol 8:1656. doi: 10.3389/fmicb.2017.01656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maia MRG, Chaudhary LC, Figueres L, Wallace RJ. 2007. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek 91:303–314. doi: 10.1007/s10482-006-9118-2. [DOI] [PubMed] [Google Scholar]
- 29.Patra A, Park T, Kim M, Yu Z. 2017. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J Anim Sci Biotechnol 8:13. doi: 10.1186/s40104-017-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramírez-Restrepo CA, Barry TN, Marriner A, López-Villalobos N, McWilliam EL, Lassey KR, Clark H. 2010. Effects of grazing willow fodder blocks upon methane production and blood composition in young sheep. Anim Feed Sci Technol 55:33–43. doi: 10.1016/j.anifeedsci.2009.10.003. [DOI] [Google Scholar]
- 31.Anderson RC, Rasmussen MA, Jensen NS, Allison MJ. 2000. Denitrobacterium detoxificans gen. nov., sp. nov., a ruminal bacterium that respires on nitrocompounds. Int J Syst Evol Microbiol 50:633–638. doi: 10.1099/00207713-50-2-633. [DOI] [PubMed] [Google Scholar]
- 32.Pukall R, Lapidus A, Nolan M, Copeland A, Glavina Del Rio T, Lucas S, Chen F, Tice H, Cheng J-F, Chertkov O, Bruce D, Goodwin L, Kuske C, Brettin T, Detter JC, Han C, Pitluck S, Pati A, Mavrommatis K, Ivanova N, Ovchinnikova G, Chen A, Palaniappan K, Schneider S, Rohde M, Chain P, D'haeseleer P, Göker M, Bristow J, Eisen JA, Markowitz V, Kyrpides NC, Klenk H-P, Hugenholtz P. 2009. Complete genome sequence of Slackia heliotrinireducens type strain (RHS 1T). Stand Genomic Sci 1:234–241. doi: 10.4056/sigs.37633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashida N, Ishii S, Hayano S, Tago K, Tsuji T, Yoshimura Y, Otsuka S, Senoo K. 2010. Isolation of functional single cells from environments using a micromanipulator: application to study denitrifying bacteria. Appl Microbiol Biotechnol 85:1211–1217. doi: 10.1007/s00253-009-2330-z. [DOI] [PubMed] [Google Scholar]
- 34.Iwamoto M, Asanuma N, Hino T. 2002. Ability of Selenomonas ruminantium, Veillonella parvula, and Wolinella succinogenes to reduce nitrate and nitrite with special reference to the suppression of ruminal methanogenesis. Anaerobe 8:209–215. doi: 10.1006/anae.2002.0428. [DOI] [Google Scholar]
- 35.Marais JP, Therion JJ, Mackie RI, Kistner A, Dennison C. 1988. Effect of nitrate and its reduction products on the growth and activity of the rumen microbial population. Br J Nutr 59:301–313. [DOI] [PubMed] [Google Scholar]
- 36.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 37.Wolin MJ, Miller TL, Stewart CS. 1997. Microbe-microbe interactions, p 467–491. In Hobson PN, Stewart CS (ed), The rumen microbial ecosystem. Hall, London, United Kingdom. doi: 10.1007/978-94-009-1453-7_11. [DOI] [Google Scholar]
- 38.Shi WB, Moon CD, Leahy SC, Kang DW, Froula J, Kittelmann S, Fan C, Deutsch S, Gagic D, Seedorf H, Kelly WJ, Atua R, Sang C, Soni P, Li D, Pinares-Patino CS, McEwan JC, Janssen PH, Chen F, Visel A, Wang Z, Attwood GT, Rubin EM. 2014. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popova M, Morgavi DP, Martin C. 2013. Methanogens and methanogenesis in the rumens and ceca of lambs fed two different high-grain-content diets. Appl Environ Microbiol 79:1777–1786. doi: 10.1128/AEM.03115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borrel G, O’Toole PW, Harris HMB, Peyret P, Brugère J-F, Gribaldo S. 2013. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol 5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weimer PJ, Stevenson DM, Mantovani HC, Man SLC. 2010. Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents. J Dairy Sci 93:5902–5912. doi: 10.3168/jds.2010-3500. [DOI] [PubMed] [Google Scholar]
- 42.Guyader J, Eugène M, Doreau M, Rochette Y, Morgavi DP, Martin C. 2014. Association of nitrate and linseed oil effectively reduces methane emission in ruminants, p 87 Proceedings of the Australian Society of Animal Production. ISNH/ISRP, Canberra, Australia. doi: 10.1017/S1751731114001852. [DOI] [Google Scholar]
- 43.Popova M, Martin C, Morgavi DP. 2010. Improved protocol for high-quality co-extraction of DNA and RNA from rumen digesta. Folia Microbiol (Praha) 55:368–372. doi: 10.1007/s12223-010-0060-3. [DOI] [PubMed] [Google Scholar]
- 44.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS, Griffiths RI, Schonrogge K. 2015. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol 6:973–980. doi: 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, White B, Nelson H, Kocher J-P, Chia N. 2014. IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries. PLoS One 9:e114804. doi: 10.1371/journal.pone.0114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seedorf H, Kittelmann S, Henderson G, Janssen PH. 2014. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J 2:e494. doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson TJ, Huws SA, Edwards JE, Kingston-Smith AH, Siu-Ting K, Hughes M, Rubino F, Friedersdorff M, Creevey CJ. 2018. CowPI: a rumen microbiome focussed version of the PICRUSt functional inference software. Front Microbiol 9:1095. doi: 10.3389/fmicb.2018.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oskansen J, Blanchet F, Kindt R. 2016. vegan: community ecology package. R package version 2.0-7. https://cran.r-project.org/web/packages/vegan/index.html, accessed March 2018.
- 56.De Caceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. [DOI] [PubMed] [Google Scholar]
- 57.Harrell FE Jr, Dupont C, et al. 2017. Hmisc: Harrell miscellaneous. R package version 4.0-3. http://cran.r-project.org/web/packages/Hmisc, accessed March 2018.
- 58.Taiyun W, Viliam S. 2016. corrplot: visualization of a correlation matrix. https://cran.r-project.org/web/packages/corrplot/index.html, accessed March 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.