Protecting fungal cells from damage from solar UVB irradiation is critical for development and application of fungal insecticides but is mechanistically not understood in Beauveria bassiana, a classic insect pathogen. We unveil that two intranuclear photolyases, Phr1 and Phr2, play essential roles in repairing UVB-induced dsDNA lesions through respective decomposition of cytotoxic cyclobutane pyrimidine dimers and (6-4)-pyrimidine-pyrimidine photoproducts, hence reactivating UVB-inactivated cells effectively under visible light. Our findings shed light on the high potential of both photolyase genes for use in improving UVB resistance and application strategy of fungal insecticides.
KEYWORDS: DNA damage repair, entomopathogenic fungi, UV resistance, biological control potential, gene expression and regulation, photolyase, photoreactivation
ABSTRACT
Fungal conidia serve as active ingredients of fungal insecticides but are sensitive to solar UV irradiation, which impairs double-stranded DNA (dsDNA) by inducing the production of cytotoxic cyclobutane pyrimidine dimers (CPDs) and (6-4)-pyrimidine-pyrimidine photoproducts (6-4PPs). This study aims to elucidate how CPD photolyase (Phr1) and 6-4PP photolyase (Phr2) repair DNA damage and photoreactivate UVB-inactivated cells in Beauveria bassiana, a main source of fungal insecticides. Both Phr1 and Phr2 are proven to exclusively localize in the fungal nuclei. Despite little influence on growth, conidiation, and virulence, singular deletions of phr1 and phr2 resulted in respective reductions of 38% and 19% in conidial tolerance to UVB irradiation, a sunlight component most harmful to formulated conidia. CPDs and 6-4PPs accumulated significantly more in the cells of Δphr1 and Δphr2 mutants than in those of a wild-type strain under lethal UVB irradiation and were largely or completely repaired by Phr1 in the Δphr2 mutant and Phr2 in the Δphr1 mutant after optimal 5-h exposure to visible light. Consequently, UVB-inactivated conidia of the Δphr1 and Δphr2 mutants were much less efficiently photoreactivated than were the wild-type counterparts. In contrast, overexpression of either phr1 or phr2 in the wild-type strain resulted in marked increases in both conidial UVB resistance and photoreactivation efficiency. These findings indicate essential roles of Phr1 and Phr2 in photoprotection of B. bassiana from UVB damage and unveil exploitable values of both photolyase genes for improved UVB resistance and application strategy of fungal insecticides.
IMPORTANCE Protecting fungal cells from damage from solar UVB irradiation is critical for development and application of fungal insecticides but is mechanistically not understood in Beauveria bassiana, a classic insect pathogen. We unveil that two intranuclear photolyases, Phr1 and Phr2, play essential roles in repairing UVB-induced dsDNA lesions through respective decomposition of cytotoxic cyclobutane pyrimidine dimers and (6-4)-pyrimidine-pyrimidine photoproducts, hence reactivating UVB-inactivated cells effectively under visible light. Our findings shed light on the high potential of both photolyase genes for use in improving UVB resistance and application strategy of fungal insecticides.
INTRODUCTION
Filamentous fungal insect pathogens, such as Beauveria bassiana and Metarhizium spp., are a large resource of mycoinsecticides and mycoacaricides that have been widely integrated into arthropod pest management programs (1). Unlike chemical pesticides, fungal pesticides rely upon the active ingredients of formulated conidia or cells and hence are sensitive to solar UV irradiation and high temperature, which often occur in the seasons of pest infestation and affect the efficacy and persistency of formulated cells applied for pest control in the field (2, 3). Thus, it is a great challenge to develop fungal pesticides with enhanced stress tolerance or to optimize an application strategy that reduces or avoids stress damages of fungal cells in the field. This challenge makes it necessary to understand molecular mechanisms involved in fungal response and tolerance to environmental stresses.
UV irradiation is a common outdoor stress consisting of UVB (290 to 320 nm) and UVA (320 to 400 nm) wavelengths from sunlight, in which shorter and more detrimental UVC wavelengths (<290 nm) are removed by atmospheric ozone before solar irradiation reaches the Earth's surface (4) and the viability of formulated fungal cells applied for arthropod pest control is threatened (5, 6). Fungal cells are usually much more sensitive to UVB than to UVA (7). An exposure to UVB may result in the lesions of intracellular macromolecules, including RNA, DNA, proteins, ribosomes, and biomembranes (8, 9). Despite weaker absorbance by most biomolecules, UVA irradiation also causes cellular damage by generating reactive oxygen species (ROS), such as singlet oxygen acting on chromophores (9). Excessive UV irradiation results in the formation of covalent linkages between adjacent bases in normal DNA duplex and the generation of cyclobutane pyrimidine dimers (CPDs) and (6-4)-pyrimidine-pyrimidine photoproducts (6-4PPs) (10). These cytotoxic products may lead to growth defects, gene mutation, and even cell death (11–13).
Photoreactivation and nucleotide excision repair (NER) are two distinct mechanisms that enable decomposition of CPDs and 6-4PPs generated under UV irradiation (14). Shorter UV wavelength-induced covalent linkages in the DNA duplex can be rapidly split through a process called photorepair, which is characterized by direct transfer of electrons to cytotoxic CPDs or 6-4PPs by exposure to longer UV wavelength or visible light (15, 16). Thus, photoreactivation of impaired cells by photorepair of UV-induced DNA lesions is superior to the NER that works slowly in a complicated manner independent of light (14, 17). The photorepair process relies upon photolyases, which were first found in Escherichia coli (18). Such DNA repair enzymes exist across organisms (19), including fungi, and play important roles in UV damage repair of plants (20).
Filamentous fungi possess a cryptochrome (CRY)/photolyase family (CPF) that consists of CPD photolyases, 6-4PP photolyases, and cry-DASHs (Drosophila, Arabidopsis, Synechocystis, and Homo CRYs) (21). In Neurospora crassa, the deficiency of a CPD photolyase not involved in blue-light sensing led to no change in either UV sensitivity or survival with photoreactivation (22). Nonetheless, the CPD photolyase Phr1 of Trichoderma atroviride is evidently required for photorepair of DNA lesions in UVC-irradiated spores (23) and is mediated by noncanonical light response elements at the transcriptional level (24). The 6-4PP photolyase Cry1, but not cry-DASH, was also reported to play a significant role in photorepair of UVC-induced DNA damage in T. atroviride (25). Four CPF members exist in Ustilago maydis, a phytopathogenic fungus highly resistant to UV and ionizing irradiation, and the fungal DNA damage repair is dependent on two photolyases (Phr1 and Phr2) rather than two cry-DASHs (Cry1 and Cry2) (26). In Botrytis cinerea, the CPD photolyase BcCRY1 has been characterized as the major enzyme of light-driven DNA repair, while the cry-DASH BcCRY2 serves as a repressor of conidiation under white and black/near-UV lights but is dispensable for photoreactivation (21). These studies indicate important, but differential, roles for homologous CPD or 6-4PP photolyases in DNA damage repair and UV tolerance of filamentous fungi, but such roles are rarely attributed to cry-DASH homologs in the studied fungi. In previous studies, however, CPD and 6-4PP DNA lesions repaired by photolyases were often induced by the irradiation of undefined UV or UVC that cannot reach the Earth's surface. A methodology is needed to evaluate the efficiencies for different photolyases to repair DNA damages in fungal cells exposed to specified UVB irradiation, a main sunlight component harmful to fungal cells in nature, and to reactivate UVB-impaired fungal cells under field conditions.
Photolyase-dependent photoreactivation is of great value for formulated fungal cells applied for field control of arthropod pests. Previously, Metarhizium robertsii expressing an archaeal CPD photolyase at high levels was markedly improved in sunlight resistance, photorepair capability, and virulence, although its own CPD and 6-4PP photolyases played insufficient roles in DNA damage repair (27). Phylogenomic analysis has revealed that the B. bassiana/Cordyceps lineage could have evolutionarily gained entomopathogenicity 130 million years earlier than the Metarhizium lineage on the Earth (28–30). As a pathogen attacking the broadest spectrum of arthropod hosts known to date, B. bassiana could have evolved a more efficient photorepair system to ensure its adaptation to a wider host spectrum and diverse host habitats. This study seeks to test this hypothesis by subcellular localization and functional analysis of three CPF genes encoding CPD photolyase (Phr1), 6-4PP photolyase (Phr2), and cry-DASH (CryD) in B. bassiana. An emphasis is placed upon the roles of Phr1 and Phr2 in conidial resistance to UVB irradiation, photorepair of UVB-induced DNA lesions, and photoreactivation of UVB-inactivated conidia by means of an experimental system in which the dose of UVB irradiation is precisely controlled.
RESULTS
Bioinformatic analysis of three CPF homologs in B. bassiana.
The sequences of three CPF homologs located in the genomic database of B. bassiana (30) share a DNA photolyase domain at the N terminus and a flavin adenine dinucleotide (FAD)-binding domain at the C terminus (see Fig. S1A in the supplemental material). The two domains are located in the respective regions of residues 96 to 262 and 388 to 583 for Phr1, residues 4 to 163 and 317 to 550 for Phr2, and residues 5 to 192 and 350 to 542 for CryD. Phr1 and Phr2 consist of 587 and 623 amino acids (molecular masses, 70.15 and 66.97 kDa; isoelectric points, 7.00 and 8.68) and share an N-terminal nuclear localization signal (NLS) motif at residues 15 to 49 and 3 to 12, respectively. However, such an NLS motif is absent in the located CryD, which is composed of 720 amino acids with a molecular mass of 79.79 kDa and an isoelectric point of 7.68. The Phr1, Phr2, and CryD sequences of B. bassiana are phylogenetically closer to the counterparts of other entomopathogenic fungi, including Cordyceps militaris, Metarhizium acridum, and Metarhizium robertsii, than those of nonentomopathogenic filamentous fungi (Fig. S1B). Notably, CryD is absent in the genomes of C. militaris, M. acridum, and M. robertsii but has two homologs in U. maydis, as reported previously (26).
Transcriptional profiles and subcellular localization of three CPF homologs.
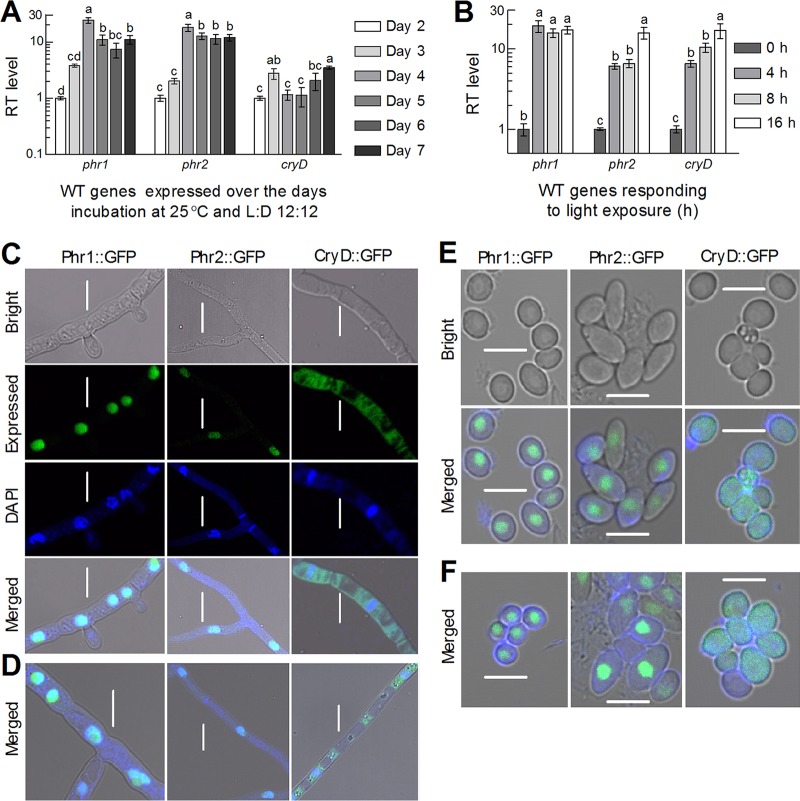
Both the phr and phr2 genes were increasingly transcribed in the wild-type strain B. bassiana ARSEF 2860 (designated wild type [WT] here) during normal incubation on Sabouraud dextrose agar plus yeast extract (SDAY) at 25°C in a light/dark (L:D) cycle of 12:12 h (12:12). As illustrated in Fig. 1A, the transcript levels of the two genes increased by 23- and 13-fold on day 4 with respect to the standard on day 2, and they maintained relatively high levels in the following 3 days. The transcript level of cryD increased by 1.8-fold on day 3 and 2.5-fold on day 7 and fluctuated around the standard in the rest days. Moreover, a 4-h exposure to light after a 3-day continuous dark incubation resulted in 18-fold upregulation of phr1 in the WT in comparison to no light exposure, whereas a 16-h light exposure was required for transcriptional peak of phr2 (15-fold) or cryD (16-fold) (Fig. 1B). Apparently, transcriptional response of phr1 to light cue was much more prompt than similar responses of phr2 and cryD in B. bassiana. The transcript level of phr2 increasing with the time of light exposure is consistent with the transcript accumulation of the 6-4PP photolyase gene cry1 in the Trichoderma reesei cells exposed to light (31).
FIG 1.
Transcriptional profiles and subcellular localization of three CPF homologs in B. bassiana. (A and B) Relative transcript (RT) levels of three CPF genes in the WT cultures grown at 25°C for 2 to 7 days at L:D 12:12 with respect to the standard at the end of day 2 and exposed to white light for 4 to 16 h with respect to the standard not exposed to light after 3-day dark incubation, respectively. Different lowercase letters marked on the bars of each group denote significant differences (Tukey's HSD test, P < 0.05). Error bars denote the SD from the results from three cDNA samples (replicates) assessed in qPCR experiments. (C to F) LSCM images (scale bars = 5 μm) for the signals of Phr1::GFP, Phr2::GFP, and CryD::GFP fusion proteins expressed in the hyphal cells and conidia of transgenic strains, respectively. The hyphal cells were collected from the 2-day-old cultures of a conidial suspension in SDB shaken at 25°C under continuously illuminated or full-dark conditions and stained with the nucleus-specific dye DAPI (shown in blue), followed by visualization through LSCM. The conidia were collected from the SDAY cultures near the respective ends of light and dark exposures during conidiation at L:D 12:12 and stained with DAPI. Note that the green signal of GFP-tagged Phr1 and Phr2 fusion proteins merges well with the stained color in the nuclei but is absent in the cytoplasm, in which the GFP-tagged CryD fusion proteins are present, irrespective of the hyphal cells and conidia exposed to light (C and E) or dark (D and F).
A possible nuclear localization of either Phr1 or Phr2 suggested by the N-terminal NLS motif was confirmed by subcellular localization experiments. As revealed by laser scanning confocal microscopy (LSCM), the green signals of green fluorescent protein (GFP)-tagged Phr1 and Phr2 fusion proteins in the hyphal cells stained by a nucleus-specific dye merged very well with the stained color (shown in blue) in the nuclei but were not present in the cytoplasm, irrespective of the hyphal cells taken from the cultures growing in Sabouraud dextrose broth (SDB) under illuminated (Fig. 1C) or full-dark (Fig. 1D) conditions. In contrast, the green signal of the NLS-free CryD fusion protein tagged by GFP appeared consistently in the cytoplasm of the stained cells and seemed to weakly overlap the stained color in the nuclei. The subcellular localization of each fusion protein in conidia was the same as observed in the hyphal cells, regardless of the conidia taken from the SDAY cultures near the end of exposure to light (Fig. 1E) or dark (Fig. 1F) during conidiation at L:D 12:12. These observations clarified the matter that either Phr1 or Phr2 was exclusively localized in the nuclei, whereas CryD was localized mainly in the cytoplasm, implicating more involvements of Phr1 and Phr2 than of CryD in the nuclear events of B. bassiana, including DNA damage repair.
Roles of three CPF homologs in UVB resistance and antioxidant response.
Compared to the WT and complementary strains (control strains), the Δphr1, Δphr2, and ΔcryD mutants displayed little changes in colony growth rates on rich SDAY, minimal Czapek-Dox agar (CDA), and modified CDAs with different carbon or nitrogen sources (Fig. S2A). The mutants also did not show significant changes in conidial yields estimated from the 8-day-old SDAY cultures at the optimal regime of 25°C and L:D 12:12; however, the median time for 50% conidial germination (GT50) at 25°C was shortened by 1.5 h in the Δphr1 mutant but prolonged by 1.1 h in the Δphr2 and ΔcryD mutants (Fig. S2B). In standardized bioassays, the estimates of median lethal time (LT50) for the mutants against Galleria mellonella larvae were also similar to those of their control strains regardless of whether a conidial suspension was topically applied for normal cuticle infection or injected into hemocoel for cuticle-bypassing infection (Fig. S2C). These results indicate no substantial role of each CPF gene in the fungal growth, conidiation, and virulence.
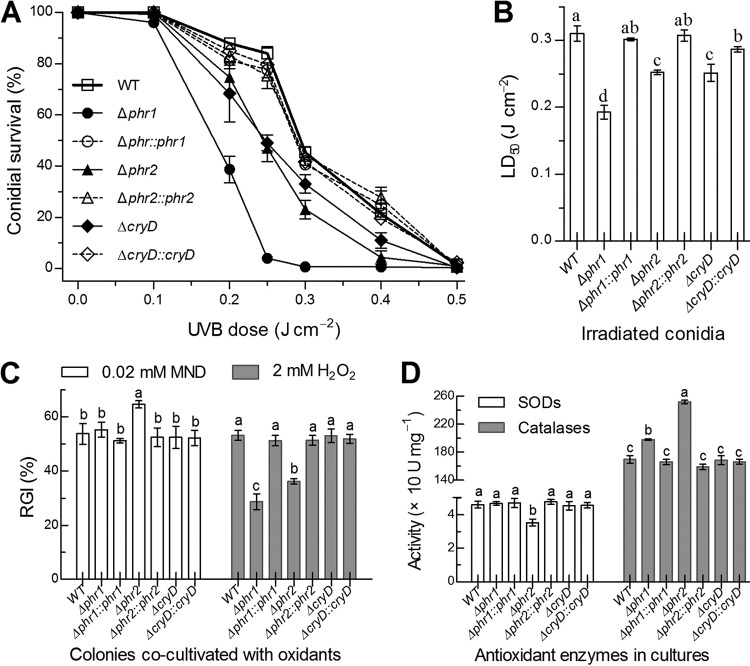
Despite little influence on the above-mentioned traits, deletion of phr1, phr2, or cryD resulted in more rapid decline of conidial survival at the irradiated UVB doses of 0.1 to 0.5 J cm−2 than in the conidial survival trends of the control strains (Fig. 2A). Consequently, the median lethal dose (LD50) indicative of UVB resistance decreased by 38% in the Δphr1 mutant and by ∼19% in both the Δphr2 and ΔcryD mutants versus the WT, and the decrease was well restored by targeted gene complementation (Fig. 2B). Since cellular UVB resistance is often correlated with antioxidant responses (32, 33), we assayed the response of each strain to the oxidant menadione (superoxide anion-generating compound) or H2O2 during colony growth on CDA. The relative growth inhibition of each strain by each oxidant demonstrated an increased sensitivity of only the Δphr1 mutant to menadione but an unexpectedly enhanced tolerance of both the Δphr1 and Δphr2 mutants to H2O2 (Fig. 2C). Intriguingly, these antioxidant responses correlated quite well with reduced superoxide dismutase (SOD) activity in the Δphr1 mutant and increased catalase activity in both the Δphr1 and Δphr2 mutants (Fig. 2D). In contrast, the ΔcryD mutant showed no significant change in either response to each oxidant or antioxidant enzyme activity. These results implicated a linkage of Phr1 with the activity of SODs as scavengers of superoxide anions (32, 34) and of both Phr1 and Phr2 with the activity of catalases that decompose H2O2 in B. bassiana (35).
FIG 2.
Roles of Phr1, Phr2, and CryD in UVB resistance and antioxidant response of B. bassiana. (A and B) Conidial survival trends of targeted gene deletion mutants and control strains over the doses of UVB irradiation and LD50 estimates for their UVB resistance, respectively. (C) Relative growth inhibition (RGI) of fungal colonies by 0.02 M menadione (MND) or 2 mM H2O2 after 8 days of cocultivation at 25°C on CDA plates, on which 1-μl aliquots of a 106 conidia ml−1 suspension were spotted for initiation of colony growth. (D) Total activities of SODs and catalases in the protein extracts isolated from 3-day-old SDAY cultures. Different lowercase letters marked on the bars of each group denote significant differences (Tukey’s HSD test, P < 0.05). Error bars denote the standard error (SE) (A) or SD (B to D) from the results from three replicates.
Roles of three CPF homologs in photoreactivation of UVB-inactivated conidia.
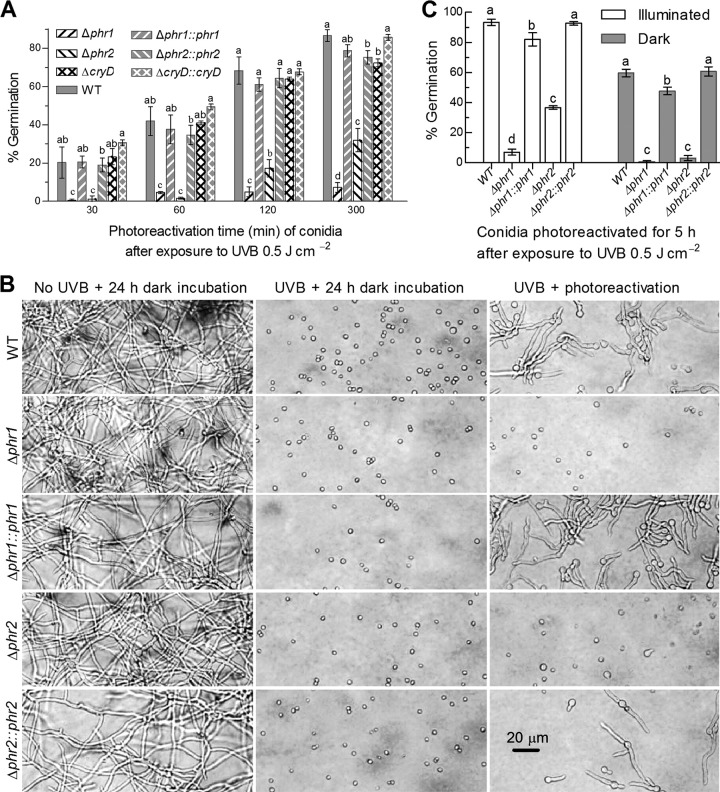
In the experiments for optimal time of photoreactivation right after exposure to the lethal UVB dose of 0.5 J cm−2, conidia from the 8-day-old SDAY cultures of control strains grown at the optimal regime germinated at rates increasing with the time of exposure to white (visible) light for photoreactivation, which was followed by dark incubation for a total period of 24 h, including the time of light exposure at 25°C. The irradiated conidia of the control strains showed mean (± standard deviation [SD]) germination rates of 20% (± 0.9%), 38% (± 3.7%), 65% (± 3.7%), and 80% (± 5.8%) at the end of dark incubation after 0.5-, 1-, 2- and 5-h light exposures (Fig. 3A and B), respectively; however, the germination rates were extremely low (∼1%), irrespective of 10-min photoreactivation provided or not (data not shown). In contrast, germination was hardly observed for the irradiated conidia of the Δphr1 and Δphr2 mutants unless light exposure was prolonged for at least 1 h. Increasing the light exposure to 2 and 5 h resulted in the germination rates of 5% and 7% in the Δphr1 mutant and of 17% and 32% in the Δphr2 mutant, respectively. Apparently, the fungal capability of reactivating UVB-inactivated conidia under visible light was much more impaired in the absence of phr1 than of phr2. The UVB-irradiated conidia of the ΔcryD mutant were reactivated at rates similar or close to those of the counterparts of the control strains irrespective of shorter or longer exposure to light. The results indicated a more prominent role of Phr1 than of Phr2 but little role of CryD in photoreactivation of UVB-inactivated conidia. Previously, cry-DASH homologs in bacteria, fungi, plants, and animals were shown to repair CPD lesions of single-stranded DNAs in vitro (36–39). Since fungal DNA is double-stranded, the cryD mutants showing little change in photoreactivation efficiency were not included in the experiments relating to double-stranded DNA (dsDNA) lesions in vivo.
FIG 3.
Factors affecting photoreactivation of B. bassiana conidia inactivated at the lethal UVB dose of 0.5 J cm−2. (A) Germination percentages of UVB-inactivated conidia reactivated by time course exposure to white light. All conidia were collected from the 8-day-old cultures grown at 25°C and L:D 12:12, irradiated at the UVB dose, reactivated for 30 to 300 min under white light, and then incubated for 19 to 23.5 h at 25°C in darkness. (B) Microscopic images for the germination status of irradiated conidia at the end of a 19-h dark incubation after a 5-h incubation under white light (photoreactivation) or in darkness (NER) at 25°C. The conidia not irradiated at the UVB dose (left column) were incubated as controls for 24 h in darkness. (C) Germination percentages of irradiated conidia after 5 h of photoreactivation and 19 h of incubation in darkness at 25°C. Conidia were collected from the 8-day-old cultures grown under continuously illuminated or full-dark conditions. Different lowercase letters marked on the bars of each group denote significant differences (Tukey’s HSD test, P < 0.05). Error bars denote the SD from the results from three replicates of each strain.
Moreover, conidia from the 8-day-old SDAY cultures grown under continuous illumination or dark conditions were irradiated at the lethal UVB dose and assayed for their viability after optimal 5-h photoreactivation. After irradiation, conidia from the illuminated cultures of each strain tended to be more readily photoreactivated than those from the dark cultures (Fig. 3C). The irradiated conidia of all control strains were photoreactivated by 82% to 93% and 48% to 60% for the sources of illuminated and dark cultures, respectively. In contrast, the photoreactivated conidia of the Δphr1 and Δphr2 mutants decreased to 7% and 37% for the illuminated source and to only 0.7% and 3% for the dark source, respectively. These data indicated conspicuous impacts of conidiation conditions on the photoreactivation efficiency of UVB-inactivated conidia and a greater role of Phr1 than of Phr2 in photoreactivation, irrespective of the conidia being produced under light or dark conditions.
Photorepair activities of Phr1 and Phr2 against CPDs and 6-4PPs.
The lethal UVB dose of 0.5 J cm−2 was used to irradiate the hyphal cells suspended in a 0.8-mm layer of 0.01% yeast extract in petri dishes for induction of CPD and 6-4PP DNA lesions, followed by enzyme-linked immunosorbent assays (ELISA) with anti-CPD and anti-6-4PP antibodies for quantification of accumulated CPDs and 6-4PPs. As a consequence of ELISA, the amounts of CPDs and 6-4PPs in the impaired DNA of the irradiated cells increased by 60% and 10% in the Δphr1 mutant and by 43% and 25% in the Δphr2 mutant versus the WT (Fig. 4A), respectively. After a 5-h exposure to white light for photoreactivation, the amount of CPDs in the impaired DNA of the Δphr1 mutant was similar to that prior to photoreactivation, contrasting with a marked CPD decrease (27%) in the Δphr2 mutant. Inversely, the amount of 6-4PPs was restored to the WT level in the Δphr1 mutant but showed little change in the Δphr2 mutant after optimal photoreactivation. In contrast, the 5-h NER treatment in darkness resulted in no significant CPD or 6-4PP change in either the Δphr1 or Δphr2 mutant in comparison to the control strains. These data indicated that Phr1 and Phr2 played main roles in decomposing CPDs and 6-4PPs, respectively, and also collaborate in CPD photorepair, since the UVB-induced CPDs were not fully repaired by Phr1 in the Δphr2 mutant.
FIG 4.
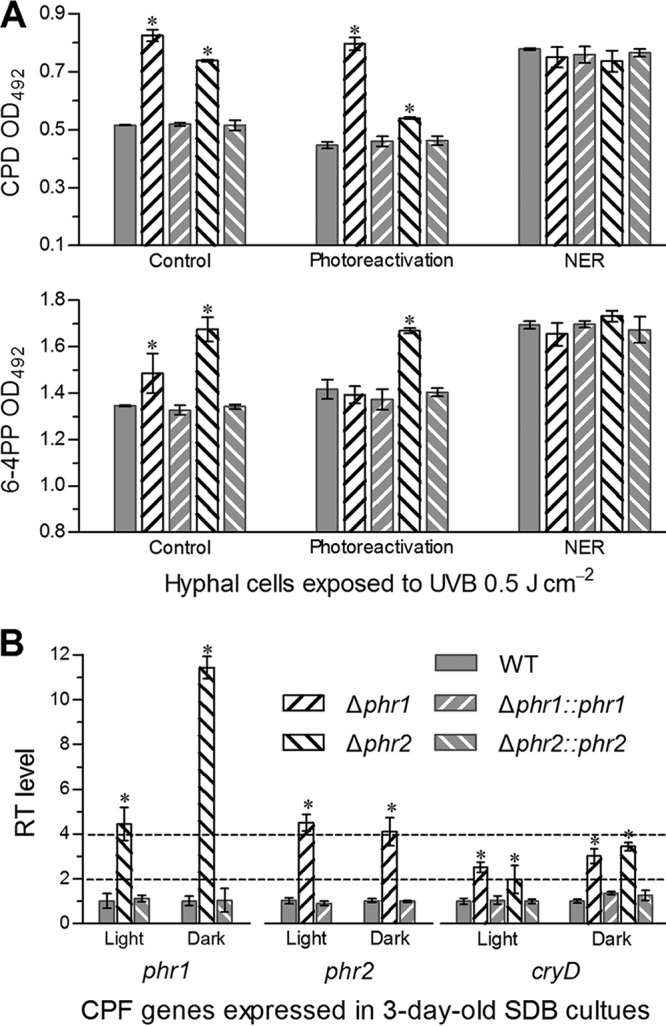
Activities of Phr1 and Phr2 in photorepair of B. bassiana DNA lesions induced by UVB irradiation. (A) ELISA readings for the amounts of UVB-induced CPDs and 6-4PPs probed by anti-CPD and 6-4PP antibodies in the impaired DNAs of two Δphr mutants and control strains. Hyphal cells from 3-day-old SDB cultures were suspended in a 0.8-mm layer of 0.01% yeast extract, irradiated at the UVB dose of 0.5 J cm−2, and incubated for 5 h under white light (photoreactivation) or in darkness (NER), followed by DNA extraction. The DNA samples extracted from the irradiated cells not exposed to white light or incubated in darkness were used as controls. (B) Relative transcript (RT) levels of partner CPF genes in the 3-day-old SDB cultures of phr mutants with respect to the WT standard. The asterisked bars in each bar group differ significantly from those that are unmarked (Tukey’s HSD test, P < 0.05). Error bars denote the SD from three DNA (A) or cDNA (B) samples of each strain.
Next, each of three CPF genes in each Δphr mutant was transcriptionally profiled through real-time quantitative PCR (qPCR). As illustrated in Fig. 4B, transcript levels of phr2 and cryD in the Δphr1 mutant with respect to the WT standard were elevated by 3.5- and 1.5-fold under white light and 3- and 2-fold in full darkness, respectively. The phr2 deletion resulted in transcriptional upregulation of phr1 by 3.4- and 10.4-fold and of cryD by 0.98- and 2.4-fold under the two culture conditions, respectively. The upregulated transcripts were well restored to WT levels by targeted phr1 or phr2 complementation, implicating a complementary or compensatory role of two other CPF partner genes for the absence of phr1 or phr1.
Responses of Phr1 and Phr2 to light spectra during photoreactivation.
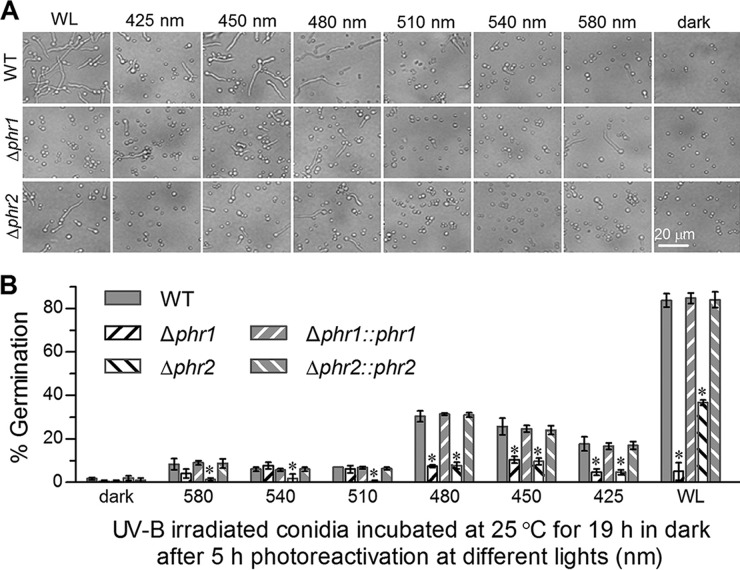
The impacts of light spectra (425 to 580 nm) on the 2-h photoreactivation efficiencies of Phr1 and Phr2 toward UVB-inactivated conidia were evaluated using white light as a positive control and full darkness as a negative control. For all control strains, the conidia irradiated at the lethal UVB dose were best reactivated by white light (∼84%), followed by the blue light at 480 nm (∼31%), 450 nm (∼25%), or 425 nm (∼17%) and green lights of 510 to 580 nm (6% to 9%), in order, but they were not significantly reactivated in full darkness (Fig. 5). Intriguingly, the irradiated Δphr1 mutant conidia were better reactivated by the blue light at 450 nm (10.3%) than by white light (5%) and other tested spectra (4% to 7.6%). In contrast, the irradiated Δphr2 mutant conidia were reactivated to a maximum of 37% under white light and to the same levels as occurred in the Δphr1 mutant under the blue lights but at lower levels under the green lights. These data implied that Phr1 could absorb visible light more efficiently than Phr2 for photoreactivation despite an equal efficiency of both in blue-light absorbance. The marked difference in the efficiency of white-light reactivation between the Δphr1 and Δphr2 mutants indicated a much more important role of Phr1 than of Phr2 in reactivating the UVB-inactivated conidia under visible light.
FIG 5.
Impact of light spectra on photoreactivation of UVB-inactivated conidia by Phr1 and Phr2 in B. bassiana. Microscopic images (A) and germination percentages (B) were taken from the conidia irradiated at the lethal UVB dose of 0.5 J cm−2, reactivated for 5 h under the lights of different spectra (425 to 580 nm), white light (positive control), and full-dark conditions (negative control), and incubated for 19 h in darkness at 25°C. The asterisked bars in each bar group differ significantly from those unmarked (Tukey's HSD test, P < 0.05). Error bars denote the SD from the results from three replicates of each strain.
Overexpression of phr1 or phr2 enhances conidial UVB resistance and photoreactivation efficiency.
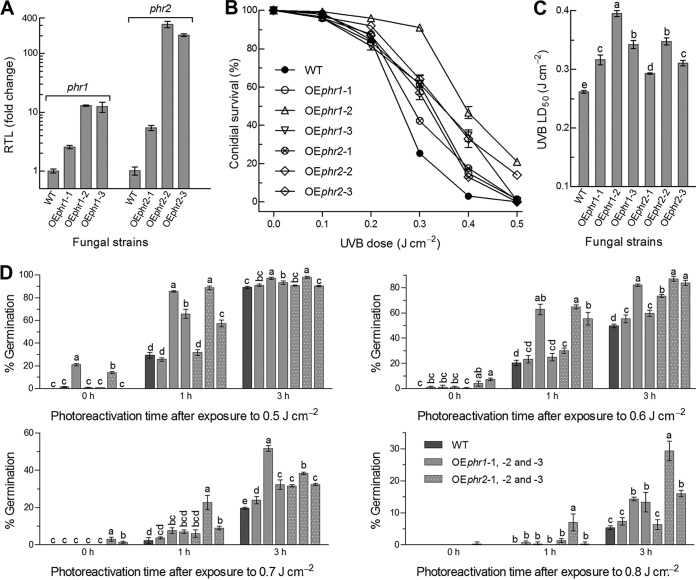
To explore exploitable values of phr1 and phr2 against UVB irradiation, three strains overexpressing phr1 (OEphr1) or phr2 (OEphr2) in the WT were evaluated for their UVB resistance and photoreactivation efficiency in parallel with WT. As shown in Fig. 6A, phr1 was transcriptionally upregulated by 1.5-, 12.0-, and 11.3-fold in the three OEphr1 strains, while phr2 was upregulated by 4.4-, 310-, and 204-fold in the three OEphr2 strains, respectively. Consequently, conidial UVB resistance was significantly enhanced by 21% to 51% in the OEphr1 strains and 12% to 33% in the OEphr1 strains in comparison to the WT (Fig. 6B and C). The most UVB-resistant strain was OEphr1-2 or OEphr2-2 among strain OEphr1 or OEphr2, respectively. However, the increased UVB resistance correlated weakly with the upregulated levels of phr1 or phr2. In photoreactivation assays, the conidia of OEphr1-2 and OEphr2-2 irradiated at the high UVB doses of 0.5 to 0.8 J cm−2 were more rapidly reactivated than those of other OE strains after a 1- or 3-h exposure to white light (Fig. 6D). For OEphr1-2 and OEphr2-2, the conidia irradiated at 0.5 J cm−2 were reactivated by 87% and 89% after a 1-h light exposure and 97% and 98% after a 3-h light exposure, respectively. Their 3-h photoreactivation efficiencies decreased to 82% and 87% at the UVB dose of 0.6 J cm−2, 52% and 38% at 0.7 J cm−2, and 14% and 29% at 0.8 J cm−2, respectively. These data highlight that either phr1 or phr2 can be exploited for enhancing UVB resistance and photoreactivation efficiency by overexpression of each in B. bassiana.
FIG 6.
Conidial UVB resistance and photoreactivation efficiency enhanced by upregulated expression of phr1 or phr2 in B. bassiana. (A) Relative transcript levels (RTL) of phr1 and phr2 in three OEphr1 and OEphr2 strains versus WT. (B and C) Conidial survival trends of WT and OEphr strains over the doses of UVB irradiation and LD50 estimates for their UVB resistance, respectively. (D) Germination percentages of the conidia exposed to white light for a 1- or 3-h photoreactivation and then incubated for 23 or 21 h in full darkness at 25°C after UVB irradiation at the doses of 0.5, 0.6, 0.7, and 0.8 J cm−2, respectively. The irradiated conidia not reactivated under white light were incubated for 24 h in darkness and used as controls. Different lowercase letters marked on the bars of each denote significant differences (Tukey’s HSD, P < 0.05). Error bars denote the SD from the results from three replicates.
DISCUSSION
Both Phr1 and Phr2 were proven to exclusively localize in the nuclei of hyphal cells and conidia regardless of being cultivated under illuminated or dark conditions, contrasting with a main localization of CryD in the cytoplasm of B. bassiana. The nuclear localization of both photolyases in B. bassiana is distinct from subcellular localization of some homologs in other eukaryotes, such as mitochondrial localization of the photolyase mCRY1 in mouse cells (40), and chloroplast, mitochondrial, or nuclear localization of CPD photolyase in rice cells (41) and of 6-4PP photolyase in tobacco cells (42). The nucleus-specific localization of Phr1 and Phr2 implicates that both of them function in the nuclei of B. bassiana, and their main functions are confirmed to resist UVB irradiation by photorepair of UVB-induced DNA lesions, as discussed below.
The Δphr1 mutant suffered a larger decrease in conidial UVB resistance than did the Δphr2 mutant despite little changes in their growth rate, conidiation capacity, and virulence. The decreased UVB resistance of the Δphr1 and Δphr2 mutants was hardly attributable to alterations in antioxidant responses and total SOD or catalase activities of both mutants. More severe DNA damages could have occurred in the UVB-inactivated conidia of the Δphr1 mutant than of the Δphr2 mutant. This speculation was verified by the levels of CPDs and 6-4PPs quantified from the UVB-impaired DNAs of both mutants and their photorepair activities against the two cytotoxic products. In B. bassiana, the main roles of Phr1 and Phr2 in the decomposition of CPDs and 6-4PPs are well shown by the high DNA repair activity of Phr1 in the Δphr2 mutant and Phr2 in the Δphr1 mutant, respectively, and are partially consistent with the reported roles of their homologs in B. cinerea (21), M. robertsii (27), T. atroviride (23–25), and U. maydis (26). In the present study, intriguingly, both 6-4PP and CPD levels were significantly higher in the UVB-irradiated cells of the Δphr1 mutant and Δphr2 mutant than of the control strains, respectively. Moreover, the DNA CPD lesions in the irradiated Δphr2 mutant cells were insufficiently repaired in the presence of phr1. Such phenomena suggest an insufficient role of either Phr1 or Phr2 alone in preventing cells from the generation of CPDs or 6-4PPs under UVB irradiation and hence a possible collaboration of both photolyases in DNA damage repair of B. bassiana. This possibility is revealed at transcriptional level by upregulated expression of phr2 in the Δphr1 mutant and of phr1 in the Δphr2 mutant under either light or dark conditions. The compensatory role of one photolyase gene for the absence of another implicates that either photolyase is involved in assisting the decomposition of a cytotoxic product targeted mainly by another partner. Despite a significant contribution to conidial UVB resistance, the cytoplasmic CryD was proven to have no role in photoreactivation of UVB-inactivated conidia in B. bassiana, like cry-DASH homologs characterized previously in B. cinerea (21), T. atroviride (25), and U. maydis (26). This implicates an unlikely involvement of CryD in photorepair of UVB-induced in vivo CPD and 6-4PP dsDNA lesions due to its localization in cytoplasm.
Furthermore, Phr1 and Phr2 are highly efficient in reactivation of UVB-inactivated conidia since the irradiated conidia of all control strains germinated more than 80% after optimal photoreactivation. Three factors were found to be influential on the efficiency of photorepair. Although the photocycle of DNA repair by a photolyase was reported to fall into subnanosecond timescale (43), we found a dependence of efficiency on the time of exposure to visible light and a requirement of 5 h of light exposure for sufficient reactivation of the UVB-inactivated conidia. This suggests a need of optimal photoreactivation for full absorbance of light by Phr1 and Phr2 in B. bassiana. Light quality is another influential factor. Visible light resulted in the best photoreactivation among the tested lights, likely due to the DNA damage repair that is achieved by electron transfer to CPDs or 6-4PPs upon the absorbance of visible light by photolyase (10). Also, expression levels of Phr1 and Phr2 under visible light could be different from those expressed under other lights tested, affecting the levels of light absorbance and photoreactivation by either photolyase. In our study, the Δphr1 mutant lost much more ability to photoreactivate the UVB-inactivated conidia than did the Δphr2 mutant and showed only 5% germination in contrast with 37% for the Δphr2 mutant after photoreactivation. Compared to an 84% photoreactivation efficiency in the presence of both Phr1 and Phr2, Phr1 in the Δphr2 mutant reactivated a greater proportion of UVB-inactivated conidia than did Phr2 in the Δphr1 mutant under visible light in B. bassiana. Importantly, both conidial UVB resistance and photoreactivation efficiency were greatly enhanced in OEphr1-2 and OEphr2-2, in which phr1 and phr2 were upregulated by 12- and 310-fold in the WT strain, respectively. Compared to sharply unregulated phr2, much less upregulated phr1 resulted in higher UVB resistance and equal or greater photoreactivation efficiency at all tested UVB doses except 0.8 J cm−2. Apparently, Phr1 plays more prominent role than Phr2 in normal photorepair of DNA lesions in the irradiated conidia. The difference in photoreactivation efficiencies between the two photolyases could be partially due to more prompt response of phr1 than of phr2 to light at transcriptional level. In addition, the presence of light during conidiation was also recognized as an influential factor. The conidia from continuously illuminated cultures of either Δphr mutant or the control strain were more readily reactivated under visible light than those from the cultures grown at L:D 12:12 and far more readily than those from the full-dark cultures.
Altogether, Phr1 and Phr2 are required for photorepair of DNA lesions in UVB-impaired cells, and Phr1 plays more important role than Phr2 in photoprotection of B. bassiana from UVB damage. For practical purposes, we developed the methodology to evaluate the roles of two photolyases in the repair of UVB-induced DNA lesions and the reactivation of UVB-impaired conidia under visible light. Previously, the completely exposed surface of a lawn at the site of 30°17′57″N, 12°5′7″E was shown to receive a total UVB dose of 2.439 J cm−2 from solar irradiation in a typically sunny day of summer (5:00 a.m. to 7:00 pm), and the majority of the UVB irradiation occurred from 9:00 a.m. to 3:00 p.m. (6). Without doubt, the solar UVB irradiation is far weaker under a crop canopy than on the exposed lawn surface. As unveiled by phenotypic analyses of both Δphr mutants and OEphr strains, the roles of Phr1 and Phr2 in resisting UVB irradiation and photoreactivating UVB-inactivated conidia are prominent in B. bassiana in comparison to the insufficient roles of their homologs in M. robertsii (27). Therefore, this study has validated the hypothesis that B. bassiana has evolved a more efficient photorepair system than has the Metarhizium lineage for adaptation to a wider host spectrum and more complicated host habitats. Our findings have shed light upon the merits of phr1 and phr2 for improved UVB resistance and application strategy of the fungal insecticides based on B. bassiana.
MATERIALS AND METHODS
Bioinformatic analysis of CPF homologs in B. bassiana.
Three CPF homologs were located in the genomic database of B. bassiana (30) under the NCBI RefSeq accession no. NZ_ADAH00000000 by Blast analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) based on the queries of all CPF sequences of Aspergillus fumigatus and U. maydis in the NCBI protein databases. The sequences of located Phr1, Phr2, and CryD (GenBank accession numbers EJP69699, EJP70165, and EJP68422, respectively) were aligned for structural comparison by means of the SMART program (http://smart.embl-heidelberg.de). The phylogenetic relationships of each with the corresponding homologs of some other entomopathogenic or nonentomopathogenic fungi were analyzed using a neighbor-joining method in the MEGA7 software (http://www.megasoftware.net).
Subcellular localization of CPF homologs in B. bassiana.
The open reading frames (ORFs) of phr1, phr2, and cryD and the promoter region of the homogenous gene tef encoding translation elongation factor 1 alpha (GenBank accession no. EJP68386) were amplified from the WT cDNA and genomic DNA with paired primers (Table S1), respectively. The vector pAN52-C-bar in which the C cassette 5′-PmeI-SpeI-EcoRV-EcoRI-BamHI-3′ follows the Aspergillus nidulans promoter PtrpC (34) was digested with BglII/NcoI for a replacement of PtrpC by Ptef. The resultant cassette was digested with EcoRI/BamHI and ligated to GFP (GenBank accession no. U55763). The resultant vector pAN52-C-GFP-bar was digested with EcoRV. Each amplified ORF was then inserted into the N terminus of GFP in the linearized vector using a one-step cloning kit (Vazyme, Nanjing, China), forming pAN52-x::GFP-bar (x = phr1, phr2, or cryD). The final plasmid was transformed into WT through blastospore transformation (44). Putative transformants were screened by the bar resistance to phosphinothricin (200 μg ml−1) and examined under a fluorescence microscope. For each transformation, a strain showing maximal green signal was grown on SDAY (4% glucose, 1% peptone, and 1.5% agar plus 1% yeast extract) for full conidiation. The conidia were suspended in SDB (i.e., agar-free SDAY) and incubated at 25°C for 2 days on a shaking (150 rpm) bed under continuously illuminated and full-dark conditions, respectively. Hyphal cells harvested from each culture were stained with the nucleus-specific dye DAPI (4′,6′-diamidine-2′-phenylindole dihydrochloride) and visualized for subcellular localization of each GFP-tagged fusion protein through LSCM.
Construction of phr1, phr2, and cryD mutants.
Our previous backbone plasmids and protocols (32, 45) were used to delete each of the CPF-coding genes from the WT strain by homogeneous recombination of its 5′ and 3′ coding/flanking fragments separated by the bar marker (Fig. S3A) and rescue it in the corresponding deletion mutant by ectopic integration of a cassette consisting of its full-length coding/flanking sequence and sur marker. Briefly, the 5′ and 3′ fragments of each target gene were amplified from the genomic DNA of WT with paired primers (Table S1) and inserted into p0380-bar at appropriate enzyme sites using the one-step cloning kit (Vazyme), forming p0380-5′x-bar-3′x (x = phr1, phr2, or cryD) for targeted gene deletion. The full-length sequence of each target gene with flanking regions was amplified from the WT DNA and inserted into p0380-sur-gateway to exchange for the gateway fragment under the action of Gateway BP Clonase II enzyme mix (Invitrogen, Shanghai, China), yielding p0380-sur-x for targeted gene complementation. The resultant plasmids were propagated in Escherichia coli TOP10 or E. coli DH5a (Invitrogen) and transformed into the WT strain or the corresponding deletion mutant via Agrobacterium-mediated transformation. Putative mutants were screened by the bar resistance to phosphinothricin (200 μg ml−1) or the sur resistance to chlorimuron ethyl (15 μg ml−1) in a selective medium and identified through PCR and Southern blot analyses with paired primers and designed probes (Table S1). All genomic DNAs used for Southern blotting of phr1, phr2, and cryD were digested with EcoRV, XhoI, and HindIII, respectively. Positive Δx mutants (Fig. S3B to D) and control strains (WT and Δx::x mutants) were used in the following experiments of three replicates.
Phenotypic experiments.
For each fungal strain, 1-μl aliquots of a 106 conidia ml−1 suspension were centrally spotted on the plates of SDAY, CDA (3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, and 0.001% FeSO4 plus 1.5% agar), and CDAs amended with different carbon (3% of glucose, trehalose, glycerol, or sodium acetate) or nitrogen (0.3% of NH4Cl, NaNO2, or NH4NO3) sources. The spotted plates were incubated for 8 days at the optimal regime of 25°C and L:D 12:12, followed by estimation of the diameter of each colony as an index of growth rate, with two measurements taken perpendicular to each other across its center. The same spotting method was used to initiate colony growth at the same regime on CDA alone (control) or supplemented with a sensitive concentration of H2O2 (2 mM) or menadione (0.02 mM) for oxidative stress. After an 8-day incubation, the diameter of each colony was estimated as mentioned above. Relative growth inhibition of each strain by each oxidant was computed as (Sc – St)/Sc × 100, where Sc and St denote the areas of colonies grown in the control and each treatment, respectively. In addition, an SOD activity assay kit (Sigma-Aldrich, St. Louis, MO, USA) and catalase activity assay kit (Jiancheng Biotech, Nanjing, China) were used to quantify the total activities of SODs and catalases, respectively, in the protein extracts isolated from 3-day-old SDAY cultures, as described previously (46). One unit of enzymatic activity was defined as the SOD amount required for inhibition of 50% pyrogallol autoxidation rate or 1 mM H2O2 consumed per min. The total SOD or catalase activity was expressed as units per milligram of protein extract.
To assess conidiation capacity, 100-μl aliquots of a 107 conidia ml−1 suspension were evenly spread on SDAY plates (9-cm diameter), followed by 8-day incubation at 25°C and L:D 12:12 for full conidiation. Three plugs were then taken from each plate culture using a cork borer (5-mm diameter). The conidia on each plug were released into 1 ml of 0.02% Tween 80 via ultrasonic vibration. The conidial concentration in the suspension was assessed using a hemocytometer and converted to the number of conidia per unit area of plate culture.
Conidia harvested from the SDAY cultures of each strain were assessed for their viability and resistance to UVB irradiation. Conidial viability was quantified as GT50 (in hours) for 50% germination on the plates of a germination medium (GM; 2% sucrose, 0.5% peptone, and 1.5% agar) at optimal temperature of 25°C (46). Conidial UVB resistance was quantified according to our previous protocols (6, 7). Briefly, 80-μl aliquots of a 107 conidia ml−1 suspension were evenly smeared onto GM plates (7-cm diameter), followed by ∼10 min of air-drying. The uncovered plates were placed in the sample tray of a Bio-Sun++ UV irradiation chamber (Vilber Lourmat, Marne-la-Vallée, France) and irradiated with a weighted wavelength of 312 nm at the UVB doses of 0.1 to 0.6 J cm−2 or not irradiated (control). The UVB irradiation of the weighted wavelength was emitted from two 30-W fluorescent tubes (T-20.M) above the sample tray and automatically adjusted four times per second for an error control of no more than 1 μJ cm−2 at a fixed dose (according to the manufacturer’s guide). The irradiated plates were immediately covered with lids and incubated for 24 h at 25°C. The germination percentage on each plate was assessed with the counts of germinated and nongerminated conidia in three fields of microscopic view (×100 magnification). The LD50 (J cm−2) indicative of UVB resistance was estimated from the fitted survival trends of conidia over the applied doses (6).
The conidial virulence of each strain was assayed on G. mellonella larvae (∼300 mg each) through two infection modes. Briefly, cohorts of ∼35 larvae were immersed for ∼10 s in 40-ml aliquots of a 107 conidia ml−1 suspension for normal cuticle infection. Alternatively, 5 μl of a 105 conidia ml−1 suspension was injected into the hemocoel of each larva in each cohort (i.e., ∼500 conidia injected per larva). All the treated cohorts were separately maintained in plastic containers for 10 days at 25°C and monitored daily for survival/mortality records. The resultant time-mortality trends were subjected to probit analysis, yielding estimates of LT50 as an index of fungal virulence.
Assays for photorepair activities of Phr1 and Phr2 against CPDs and 6-4PPs.
Hyphal cells were collected from the 3-day-old cultures of Δphr1 mutant, Δphr2 mutant, and control strains initiated with 106 conidia ml−1 SDB, rinsed repeatedly with distilled water, and resuspended in 0.01% yeast extract. Aliquots of a 3-ml cell suspension were poured into 7-cm-diameter petri dishes, forming a suspension layer of 0.8 mm in depth. The dishes were irradiated at the high UVB dose of 0.5 J cm−2 in the UV chamber for generation of CPDs and 6-4PPs in the DNA of each strain. The irradiated dishes were immediately covered with lids and incubated at 25°C for 5 h under white light (photoreactivation) or in full darkness (NER treatment), followed by the collection of hyphal cells through centrifugation. The white light was controlled in a 280-liter illuminated growth chamber (Laifu Tech Co., Ningbo, China), which is installed vertically with eight 26-W 95-cm-long white fluorescent tubes in each side and presents a white-light intensity of 12,000 lx (see the manufacturer's guide). The dishes for NER treatment were wrapped with two layers of light-proof foil and incubated in the same growth chamber. Subsequently, total DNA of each strain was extracted from the hyphal cells of either treatment using the Biospin fungus genomic DNA extraction kit (Bioer Technology Co. Ltd., Hangzhou, China). The DNA immediately isolated from the UVB-irradiated cells without photoreactivation or NER treatment was used as a control for each strain. ELISAs with anti-CPD and anti-6-4PP antibodies were performed to quantify photorepair activities against the UVB-induced CPDs and 6-4PP in three DNA samples diluted to 0.4 and 4 μg ml−1, respectively, following the user’s guides of the CPD and 6-4PP ELISA kits (Cosmo Bio Co. Ltd., Japan). The absorbance of each DNA dilution at 492 nm was used to represent the amount of CPDs or 6-4PPs as an index of photorepair activity.
Transcriptional profiling of three CPF genes.
To profile the transcript levels of three CPF genes during normal incubation or in response to the time of light exposure, the WT strain was grown by spreading 100-μl aliquots of a 107 conidia ml−1 suspension on cellophane-overlaid SDAY plates and incubated at 25°C for 2 to 7 days in the fixed L:D 12:12 cycle or 3 days in full darkness. The dark cultures were then exposed to white light for 4, 8, and 16 h or not exposed (control). Total RNAs were extracted daily from the SDAY cultures during a 7-day incubation or from the 3-day-old dark cultures exposed to light for different hours under the action of an RNAiso Plus reagent (TaKaRa, Dalian, China) and reversely transcribed into cDNAs with a PrimeScript RT reagent kit (TaKaRa) at 37°C. To reveal the impact of each phr deletion on the expression of other CPF genes, total RNAs were extracted from the 3-day-old SDAY cultures of Δphr1 mutant, Δphr2 mutant, and control strains grown at the regime of 25°C and L:D 12:12 and transcribed into cDNAs, as mentioned above. The qPCR analysis with paired primers (Table S1) was performed under the action of SYBR Premix Ex Taq (TaKaRa) to assess transcript levels of phr1, phr2, and cryD in three cDNA samples (with a content standardized by dilution) derived from the WT cultures of each treatment (time length of incubation or light exposure) or from the 3-day-old cultures of each strain using the β-actin gene as an internal standard. The 2−ΔΔCT method (47) was used to compute the relative transcript level of each CPF gene in the WT strain during the 7-day period of cultivation with respect to the standard on day 2 or over the time of light exposure with respect to the standard not exposed to light after continuous dark incubation and in each Δphr mutant or complementary strain with respect to the WT standard.
Assays for photoreactivation of UVB-impaired conidia.
Conidia were collected from the 8-day-old SDAY cultures of deletion mutants and control strains grown at the optimal regime of 25°C and L:D 12:12, spread on the GM plates, and irradiated at the high UVB dose of 0.5 J cm−2 in the aforementioned UV chamber. To optimize the time of light exposure, the irradiated plates were immediately covered with lids and incubated at 25°C for reactivation of 0, 10, 30, 60, 120, and 300 min under white light in the illuminated growth chamber, as mentioned above. After light exposure, all of the plates were transferred to a fully dark growth chamber for incubation of 19 to 24 h at 25°C. To assess conidial responses to different lights during photoreactivation, the irradiated dishes were covered with the lids, to which transmission spectrum-specific filters (425, 450, 480, 510, 540, and 580 nm [fluctuating ∼15 nm around each central transmission spectrum]; Jinhezhong Co., Beijing, China) were attached, and the lower parts (bottoms) of the dishes were tightly wrapped with light-proof foil to prevent lateral light from transmission but to not affect aeration, as described previously (48). The dishes attached with the filters or not attached for the white-light control and dark control (dishes completely wrapped with foil) were photoreactivated at 25°C for the optimal time of 5 h in the illuminated chamber, followed by a 19-h incubation in full darkness. At the end of the dark incubation, the germination percentage of irradiated conidia on each plate was estimated using microscopic counts, as mentioned above. To evaluate the impact of conidiation conditions on photoreactivation, conidia from the 8-day-old SDAY cultures of the tested strains grown at 25°C under continuous illumination and dark conditions were also irradiated at the same UVB dose, followed by estimation of their germination percentages after a 5-h photoreactivation and 19-h dark incubation.
Validation for enhancement of UVB resistance and photoreactivation efficiency by phr1 or phr2 overexpression.
The strong promoter Ptef was ligated into the XmaI site of p0380-bar digested with XmaI, forming the backbone plasmid p0380-Ptef-bar for targeted gene overexpression. Subsequently, the ORFs of phr1 and phr2 amplified from the WT cDNA were inserted into the XmaI/BamHI sites of the backbone plasmid digested by XmaI and BamHI, respectively. The resultant plasmids were propagated in E. coli DH5a (Invitrogen) and transformed into the WT strain via Agrobacterium-mediated transformation. Putative OEphr colonies were screened by bar resistance to phosphinothricin (200 μg ml−1) and sequentially identified through PCR and qPCR with paired primers (Table S1). Three OEphr1 or OEphr2 strains were selected based on upregulated expression levels of phr1 or phr2 and evaluated in parallel with WT for conidial UVB resistance and photoreactivation efficiency, as described in the experiments of the Δphr mutant and control strains. In photoreactivation assays, particularly, the conidia of each strain were irradiated at the UVB doses of 0.5, 0.6, 0.7, and 0.8 J cm−2, followed by 1 or 3 h of reactivation under white light and 23 or 21 h of dark incubation for germination at 25°C. The irradiated conidia not exposed to light were used as a control.
Statistical analysis.
All data from the experiments of three replicates were subjected to one-factor (strain) analysis of variance, followed by Tukey’s honestly significant difference (HSD) test for the difference of means between the deletion mutants and control strains.
Supplementary Material
ACKNOWLEDGMENTS
She-Long Zhang (Core Facilities, College of Life Sciences, ZJU) is acknowledged for technical assistance with LSCM.
Funding of this work was provided by the Ministry of Science and Technology of the People's Republic of China (grant 2017YFD0201202), the National Natural Science Foundation of China (grants 31772218 and 31801795), and the Fundamental Research Funds for the Central Universities of China (grant 2018FZA6003).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02459-18.
REFERENCES
- 1.de Faria MR, Wraight SP. 2007. Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol Control 43:237–256. doi: 10.1016/j.biocontrol.2007.08.001. [DOI] [Google Scholar]
- 2.Rangel DEN, Braga GUL, Fernandes ÉKK, Keyser CA, Hallsworth JE, Roberts DW. 2015. Stress tolerance and virulence of insect-pathogenic fungi are determined by environmental conditions during conidial formation. Curr Genet 61:383–404. doi: 10.1007/s00294-015-0477-y. [DOI] [PubMed] [Google Scholar]
- 3.Zhang LB, Feng MG. 2018. Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl Microbiol Biotechnol 102:4995–5004. doi: 10.1007/s00253-018-9033-2. [DOI] [PubMed] [Google Scholar]
- 4.Madronich S. 1993. UV radiation in the natural and perturbed atmosphere, p 17−69. In Tevini M. (ed), UV-B radiation and ozone depletion. Lewis, Boca Raton, FL. [Google Scholar]
- 5.Alves RT, Bateman RP, Prior C, Leather SR. 1998. Effects of simulated solar radiation on conidial germination of Metarhizium anisopliae in different formulations. Crop Prot 17:675–679. doi: 10.1016/S0261-2194(98)00074-X. [DOI] [Google Scholar]
- 6.Huang BF, Feng MG. 2009. Comparative tolerances of various Beauveria bassiana isolates to UV-B irradiation with a description of a modeling method to assess lethal dose. Mycopathologia 168:145–152. doi: 10.1007/s11046-009-9207-7. [DOI] [PubMed] [Google Scholar]
- 7.Yao SL, Ying SH, Feng MG, Hatting JL. 2010. In vitro and in vivo responses of fungal biocontrol agents to the gradient doses of UV-B and UV-A irradiation. Biocontrol 55:413–422. doi: 10.1007/s10526-009-9265-2. [DOI] [Google Scholar]
- 8.Engelberg D, Klein C, Martinetto H, Struhl K, Karin M. 1994. The UV response involving the Ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell 77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths HR, Mistry P, Herbert KE, Lunec J. 1998. Molecular and cellular effects of ultraviolet light-induced genotoxicity. Crit Rev Clin Lab Sci 35:189–237. doi: 10.1080/10408369891234192. [DOI] [PubMed] [Google Scholar]
- 10.Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 11.Yasui A, Eker APM, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. 1994. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J 13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancar A. 1996. No “end of history” for photolyases. Science 272:48–49. doi: 10.1126/science.272.5258.48. [DOI] [PubMed] [Google Scholar]
- 13.de Laat WL, Jaspers NGJ, Hoeijmakers JHJ. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev 13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 14.Suter B, Wellinger RE, Thoma F. 2000. DNA repair in a yeast origin of replication: contributions of photolyase and nucleotide excision repair. Nucleic Acids Res 28:2060–2068. doi: 10.1093/nar/28.10.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker AP, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JHJ, van der Horst GTJ. 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol 15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 17.Smerdon MJ, Thoma F. 1990. Site-specific DNA-repair at the nucleosome level in a yeast minichromosome. Cell 61:675–684. doi: 10.1016/0092-8674(90)90479-X. [DOI] [PubMed] [Google Scholar]
- 18.Sancar A, Rupert CS. 1978. Cloning of the phr gene and amplification of photolyase in Escherichia coli. Gene 4:295–308. doi: 10.1016/0378-1119(78)90047-1. [DOI] [PubMed] [Google Scholar]
- 19.Goosen N, Moolenaar GF. 2008. Repair of UV damage in bacteria. DNA Repair (Amst) 7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Fortunato AE, Annunziata R, Jaubert M, Bouly JP, Falciatore A. 2015. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J Plant Physiol 172:42–54. doi: 10.1016/j.jplph.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Cohrs KC, Schumacher J. 2017. The two cryptochrome/photolyase family proteins fulfill distinct roles in DNA photorepair and regulation of conidiation in the gray mold fungus Botrytis cinerea. Appl Environ Microbiol 83:e00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimura M, Ito Y, Ishii C, Yajima H, Linden H, Harashima T, Yasui A, Inoue H. 1999. Characterization of a Neurospora crassa photolyase-deficient mutant generated by repeat induced point mutation of the phr gene. Fungal Genet Biol 28:12–20. doi: 10.1006/fgbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- 23.Berrocal-Tito GM, Esquivel-Naranjo EU, Horwitz BA, Herrera-Estrella A. 2007. Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction. Eukaryot Cell 6:1682–1692. doi: 10.1128/EC.00208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cervantes-Badillo MG, Muñoz-Centeno T, Uresti-Rivera EE, Argüello-Astorga GR, Casas-Flores S. 2013. The Trichoderma atroviride photolyase-encoding gene is transcriptionally regulated by non-canonical light response elements. FEBS J 280:3697–3708. doi: 10.1111/febs.12362. [DOI] [PubMed] [Google Scholar]
- 25.García-Esquivel M, Esquivel-Naranjo EU, Hernandez-Onate MA, Ibarra-Laclette E, Herrera-Estrella A. 2016. The Trichoderma atroviride cryptochrome/photolyase genes regulate the expression of blr1-independent genes both in red and blue light. Fungal Biol 120:500–512. doi: 10.1016/j.funbio.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Brych A, Mascarenhas J, Jaeger E, Charkiewicz E, Pokorny R, Bolker M, Doehlemann G, Batschauer A. 2016. White collar 1-induced photolyase expression contributes to UV-tolerance of Ustilago maydis. Microbiologyopen 5:224–243. doi: 10.1002/mbo3.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang WG, St. Leger RJ. 2012. Enhanced UV resistance and improved killing of malaria mosquitoes by photolyase transgenic entomopathogenic fungi. PLoS One 7:e43069. doi: 10.1371/journal.pone.0043069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Q, Jin K, Ying SH, Zhang YJ, Xiao GH, Shang YF, Duan ZB, Hu X, Xie XQ, Zhou G, Peng GX, Luo ZB, Huang W, Wang B, Fang WG, Wang SB, Zhong Y, Ma LJ, St. Leger RJ, Zhao GP, Pei Y, Feng MG, Xia YX, Wang CS. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet 7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng P, Xia YL, Xiao GH, Xiong CH, Zhang SW, Zheng HJ, Zheng HJ, Huang Y, Zhou Y, Wang SY, Zhao GP, Liu XZ, St. Leger RJ, Wang CS. 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol 12:R116. doi: 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao GH, Ying SH, Zheng P, Wang ZL, Zhang SW, Xie XQ, Shang YF, Zheng HJ, Zhou Y, St. Leger RJ, Zhao GP, Wang CS, Feng MG. 2012. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep 2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzmán-Moreno J, Flores-Martínez A, Brieba LG, Herrera-Estrella A. 2014. The Trichoderma reesei Cry1 protein is a member of the cryptochrome/photolyase family with 6-4 photoproduct repair activity. PLoS One 9:e100625. doi: 10.1371/journal.pone.0100625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie XQ, Li F, Ying SH, Feng MG. 2012. Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PLoS One 7:e30298. doi: 10.1371/journal.pone.0030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie XQ, Guan Y, Ying SH, Feng MG. 2013. Differentiated functions of Ras1 and Ras2 proteins in regulating the germination, growth, conidiation, multi-stress tolerance and virulence of Beauveria bassiana. Environ Microbiol 15:447–462. doi: 10.1111/j.1462-2920.2012.02871.x. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Shi HQ, Ying SH, Feng MG. 2015. Distinct contributions of one Fe- and two Cu/Zn-cofactored superoxide dismutases to antioxidation, UV tolerance and virulence of Beauveria bassiana. Fungal Genet Biol 81:160–171. doi: 10.1016/j.fgb.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang ZL, Zhang LB, Ying SH, Feng MG. 2013. Catalases play differentiated roles in the adaptation of a fungal entomopathogen to environmental stresses. Environ Microbiol 15:409–418. doi: 10.1111/j.1462-2920.2012.02848.x. [DOI] [PubMed] [Google Scholar]
- 36.Selby CP, Sancar A. 2006. A cryptochrome photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci U S A 103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO. 2008. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci U S A 105:21023–21027. doi: 10.1073/pnas.0805830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castrillo M, Bernhardt A, Avalos J, Batschauer A, Pokorny R. 2015. Biochemical caracterization of the DASH-type cryptochrome CryD from Fusarium fujikuroi. Photochem Photobiol 91:1356–1367. doi: 10.1111/php.12501. [DOI] [PubMed] [Google Scholar]
- 39.Tagua VG, Pausch M, Eckel M, Gutierrez G, Miralles-Duran A, Sanz C, Eslava AP, Pokorny R, Corrochano LM, Batschauer A. 2015. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc Natl Acad Sci U S A 112:15130–15135. doi: 10.1073/pnas.1514637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi K, Kanno S, Smit B, van der Horst GT, Takao M, Yasui A. 1998. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res 26:5086–5092. doi: 10.1093/nar/26.22.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi M, Teranishi M, Ishida H, Kawasaki J, Takeuchi A, Yamaya T, Watanabe M, Makino A, Hidema J. 2011. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA. Plant J 66:433–442. doi: 10.1111/j.1365-313X.2011.04500.x. [DOI] [PubMed] [Google Scholar]
- 42.Banaś AK, Hermanowicz P, Sztatelman O, Labuz J, Aggarwal C, Zglobicki P, Jagiello FD, Strzalka W. 2018. 6,4-PP photolyase encoded by AtUVR3 is localized in nuclei, chloroplasts and mitochondria and its expression is down-regulated by light in a photosynthesis-dependent manner. Plant Cell Physiol 59:44–57. doi: 10.1093/pcp/pcx159. [DOI] [PubMed] [Google Scholar]
- 43.Kao YT, Saxena C, Wang L, Sancar A, Zhong D. 2005. Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci U S A 102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ying SH, Feng MG. 2006. Novel blastospore-based transformation system for integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl Microbiol Biotechnol 72:206–210. doi: 10.1007/s00253-006-0447-x. [DOI] [PubMed] [Google Scholar]
- 45.Zhou G, Wang J, Qiu L, Feng MG. 2012. A Group III histidine kinase (mhk1) upstream of high-osmolarity glycerol pathway regulates sporulation, multi-stress tolerance and virulence of Metarhizium robertsii, a fungal entomopathogen. Environ Microbiol 14:817–829. doi: 10.1111/j.1462-2920.2011.02643.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhang LB, Tang L, Ying SH, Feng MG. 2016. Regulative roles of glutathione reductase and four glutaredoxins in glutathione redox, antioxidant activity and iron homeostasis of Beauveria bassiana. Appl Microbiol Biotechnol 100:5907–5917. doi: 10.1007/s00253-016-7420-0. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Tong SM, Zhang AX, Guo CT, Ying SH, Feng MG. 2018. Daylight length-dependent translocation of VIVID photoreceptor in cells and its essential role in conidiation and virulence of Beauveria bassiana. Environ Microbiol 20:169–185. doi: 10.1111/1462-2920.13951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.