Abstract
Chemoresistance is a major therapeutic obstacle in the treatment of human pancreatic ductal adenocarcinoma (PDAC). As an oxidative stress responsive transcription factor, nuclear factor erythroid 2-related factor 2 (Nrf2) regulates the expression of cytoprotective genes. Nrf2 not only plays a critical role in chemoprevention, but also contributes to chemoresistance. In this study, we found that digoxin markedly reversed drug resistance of gemcitabine by inhibiting Nrf2 signaling in SW1990/Gem and Panc-1/Gem cells. Further research revealed that digoxin regulated Nrf2 at transcriptional level. In in vivo study, we found that digoxin and gemcitabine in combination inhibited tumor growth more substantially when compared with gemcitabine treatment alone in SW1990/Gem-shControl cells-derived xenografts. In the meantime, SW1990/Gem-shNrf2 cells-derived xenografts responded to gemcitabine and combination treatment similarly, suggesting that digoxin sensitized gemcitabine-resistant human pancreatic cancer to gemcitabine, which was Nrf2 dependent. These results demonstrated that digoxin might be used as a promising adjuvant sensitizer to reverse chemoresistance of gemcitabine-resistant pancreatic cancer to gemcitabine via inhibiting Nrf2 signaling.
Abbreviations: CHX, cycloheximide; SW1990/Gem, gemcitabine-resistant SW1990; Panc-1/Gem, gemcitabine-resistant Panc-1; PDAC, pancreatic ductal adenocarcinoma; Nrf2, nuclear factor erythroid 2-related factor 2; Keap1, Kelch-like ECH-associated protein 1; AREs, antioxidant-response elements; GCL, glutamate cysteine ligase; NQO, NADP(H), quinone oxidoreductase; HO-1, heme oxygenase-1; PI3K, phosphatidylinositol-3 kinase; MAPKs, Mitogen-activated protein kinases; mRNA, messenger RNA; shRNA, short hairpin RNA; RI, resistant index; RF, reversal fold; qRT-PCR, quantitative reverse transcription-PCR
Keywords: Digoxin, Pancreatic cancer cells, Gemcitabine, Chemoresistance, Nrf2
Highlights
-
•
Digoxin could reverse drug resistance of gemcitabine in gemcitabine-resistant pancreatic cancer cells.
-
•
Digoxin significantly inhibited Nrf2 signaling in gemcitabine-resistant pancreatic cancer cells.
-
•
Digoxin-mediated reversing drug resistance of gemcitabine in gemcitabine-resistant pancreatic cancer cells was Nrf2 dependent.
1. Introduction
PDAC is one of the most fatal human malignant cancers, because it is often diagnosed at middle or late stage. It is currently the fourth leading cause of cancer death worldwide with a less than 5% 5-year survival rate [1], [2]. Although some effective treatment measures are used, PDAC death rate is still on the rise. The development of chemoresistance is a major reason leading to chemotherapy failure in pancreatic cancer. Gemcitabine, a deoxycytidine analog that inhibits DNA replication and thereby arrests tumor growth, is widely used single-agent chemotherapy for pancreatic cancer, but high rate of chemoresistance reduce the effectiveness of its clinical treatment [3]. Therefore, it is necessary to find potential adjuvants to reverse the gemcitabine resistance in gemcitabine-resistant pancreatic cancer.
Nrf2, a basic leucine zipper transcription factor, participates in protecting cells from electrophilic or oxidative stresses through regulating cellular redox homeostasis [4], [5]. Nrf2 regulates the expression of its downstream genes such as glutamate cysteine ligase (GCL), NADP(H): quinone oxidoreductase (NQO), heme oxygenase-1 (HO-1) and several ATP-dependent drug efflux pumps through binding to antioxidant-response elements (AREs) [6], [7], [8], [9]. Kelch-like ECH-associated protein 1 (Keap1), a substrate adaptor protein, connects Nrf2 and Cul3-dependent E3 ubiquitin ligase to form complex, suppresses Nrf2 activity under basal condition [10]. When the intracellular stable environment is changed, electrophiles and oxidants inhibit the Keap1-mediated proteasomal degradation, causing the translocation of Nrf2 to the nucleus. Then Nrf2 binds to AREs and enhances transcription of its target genes.
Recently, some studies suggested that overactivation of Nrf2 signaling was one of the reasons for the drug resistance during chemotherapy [11], [12]. Frequent mutations of Keap1 in human cancers such as breast and lung cancer result in the upregulation of Nrf2 signaling [13]. We previously reported that Nrf2 and its downstream genes were highly expressed in MCF-7/DOX cells, and using Nrf2 siRNA to knockdown Nrf2 could reverse chemoresistance [14]. Similarly, tamoxifen and imatinib-resistant cancer cells also exhibited overactivation of Nrf2 signaling [15], [16]. Moreover, Hong et al. found that drug resistance was increased or decreased in pancreatic cancer cells with overexpression or knockdown of Nrf2, respectively [17]. Therefore, Nrf2 may be expected to become a pharmacological target to reverse chemoresistance in drug-resistant cancers with overactivation of Nrf2 signaling. It is necessary to find adjuvants that have inhibitory effect of Nrf2 activity and such adjuvants combined with chemotherapy drugs might be useful to reverse chemoresistance.
Cardiac glycosides, a class of glycosides with strong cardiac functions, are mainly used in the treatment of chronic cardiac insufficiency and heart failure through inhibiting plasma membrane Na+/K+-ATPase. Among them, digoxin is mainly used to treat heart failure [18], [19] and several studies have reported that digoxin exerted anti-tumor activities by inhibition of proliferation, induction of apoptosis, supporting its potential use for cancer therapy [20], [21]. Choi et al. found that digoxin was able to inhibit activity of the Nrf2-ARE luciferase reporter gene in A549-ARE cells [22], suggesting that digoxin may be a potent Nrf2 inhibitor.
Here, we demonstrated that digoxin, a potent Nrf2 inhibitor, reversed drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. Mechanistically, digoxin inhibited the activity of Nrf2 through suppressing phosphatidylinositol-3 kinase (PI3K)/Akt signaling pathway. Thus, digoxin might be a promising agent to reverse gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells via inhibiting Nrf2 signaling.
2. Materials and methods
2.1. Materials
Gemcitabine (purity > 98%), cycloheximide (purity > 93%) and MTT (purity > 98%) were purchased from Sigma-Aldrich (St. Louis, USA). LY294002 (purity > 98%) was purchased from Beyotime Institute of Biotechnology (Shanghai, China). Digoxin (purity > 97%), etoposide (purity > 99%), paclitaxel (purity > 99%), cisplatin (purity > 99%), 5-Fluorouracil (5-FU, purity > 99%), cytarabine (ara-C, purity > 99%), doxorubicin (purity > 99%) and MG132 (purity > 97%) were purchased from Selleck Chemicals (Houston, USA). Actinomycin D (purity > 95%) was purchased from KeyGen (Nanjing, China). Anti-NQO1, anti-HO-1 and anti-GCLC antibodies were obtained from Santa Cruz Biotechnology (Texas, USA). Anti-Keap1, anti-Nrf2, anti-p-Akt, anti-Akt, anti-p-P38 and anti-P38, anti-p-ERK1/2, anti-ERK1/2, anti-p-JNK and anti-JNK antibodies were purchased from Cell Signaling Technology (Danvers, USA). Anti-ABCC1 and anti-ABCC5 antibodies were purchased from ABclonal (Wuhan, China). Anti-β-actin and anti-ubiquitin antibodies were obtained from Bioworld (Minnesota, USA). Anti-lamin A antibody was obtained from Sigma-Aldrich (St. Louis, USA).
2.2. Cell culture
Panc-1 cells were obtained from Cell Bank of the Chinese Academic of Sciences (Shanghai, China). SW1990 and gemcitabine-resistent SW1990 (SW1990/Gem) cells were kindly provided by Prof. Feng Qian (Tsinghua University, Beijing, China). Gemcitabine-resistent Panc-1 (Panc-1/Gem) cells were established by our laboratory.
Briefly, Panc-1 cells were initially exposed to 5 μM of gemcitabine and then concentrations of gemcitabine were gradually increased (5, 10, 20, 40, 80, 160 and 320 μM) every 2 weeks until cells became resistant to 320 μM of gemcitabine. SW1990, Panc-1, SW1990/Gem and Panc-1/Gem cells were incubated in DMEM medium (Gibco, Grand Island, NY) containing 10% heat-inactivated FBS (Gibco, Grand Island, NY), 100 U/mL of penicillin, and 100 mg/mL of streptomycin under a humidified 5% CO2 environment at 37 °C.
2.3. MTT assay
Cells were plated into 96-well plates at approximately 5000 cells/well, and then treated with different doses of drugs for 24 h. Cell viability was detected by MTT assay according to manufacturer's instruction. Cell viability = Atreated/Acontrol × 100%. SPSS statistical software was used to calculate the value of IC50. Resistant index (RI) = IC50 of gemcitabine-resistant pancreatic cancer cells/IC50 of pancreatic cancer cells. Reversal fold (RF) = IC50 of gemcitabine in the absence of digoxin/IC50 of gemcitabine in the presence of digoxin. All assays were performed in triplicate.
2.4. Flow cytometric
Annexin V/PI staining kit (KeyGen Biotechology, Nanjing, China) was used to detect cell apoptosis following the manufacturer's instruction. Apoptotic cells were measured by using FACS Calibur flow cytometry (Accuri® C6, BD Biosciences, Franklin Lakes, NJ, USA) and the data was analyzed using Cell Quest software (BD Biosciences).
2.5. Colony formation assay
About 500 cells were plated into six-well plates for 24 h. Subsequently, cells were treated with 80 nM of digoxin, 40 µM of gemcitabine or in combination for 24 h. Drug-free growth medium was added and cells were further incubated for 14 days before fixation with 4% formaldehyde and staining with 0.5% crystal violet. The number of colonies were then counted microscopically.
2.6. Western blot
Total and nuclear protein samples were prepared. Protein samples were detected by western blot using a standard protocol. Anti-actin and anti-lamin A antibodies were used as loading control antibodies for total protein samples and nuclear protein samples, respectively. The relative levels of Nrf2, Keap1, NQO1, HO-1, GCLC, ABCC1 and ABCC5 were analysis by using Image J software.
2.7. Immunofluorescence
Cells were incubated with Nrf2 primary antibody in PBS containing 1% BSA (1:200) for 1 h at 37 °C and then treated with Alexa Fluors 488 goat anti-rabbit antibody (KeyGen, Nanjing, China). Nuclei were visualized with DAPI (Santa Cruz, Texas, USA). Images were acquired by inverted fluorescence microscope (Nikon, Japan).
2.8. CHX-chase analysis
The half-time of Nrf2 in SW1990/Gem and Panc-1/Gem cells was detected by CHX-chase analysis. SW1990/Gem and Panc-1/Gem cells were pre-treated with or without 80 nM of digoxin for 6 h. Subsequently, cells were incubated with 25 μM of cycloheximide. Total cell extracts were prepared at indicated time point after following treatment with cycloheximide, and then cell extracts were detected by western blot. Image J software was used to quantify the intensity of the bands.
2.9. Ubiquitination assay
To detect ubiquitin-conjugated endogenous Keap1, cells were treated with or without 80 nM of digoxin for 6 h. The cell lysate was incubated with Keap1 antibody overnight at 4 °C, and then incubated with protein A/G-agarose beads (Santa Cruz, Texas, USA) for another 4 h at 4 °C. Immunoprecipitated proteins were analyzed by western blot with ubiquitin antibody. The band intensities of ubiquitinated Keap1 was quantified using Image J software.
2.10. Quantitative real-time PCR
RNA samples were reverse transcribed to cDNA, and then quantitative PCR reaction was performed using AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) in the LightCycler1 96 Real-Time PCR System (Roche, Basel, Swiss). The primer sequences used in this study were shown in Table 1.
Table 1.
The sequences of primers used in the study.
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| Nrf2 | CAGCTTTTGGCGCAGACATT | GACTGGGCTCTCGATGTGAC |
| Keap1 | ATCGATGGCCACATCTATG | GATCCTTCGTGTCAGCATTG |
| HO-1 | CTTTCAGAAGGGCCAGGTGA | GTAGACAGGGGCGAAGACTG |
| NQO1 | GGTTTGGAGTCCCTGCCATT | TTGCAGAGAGTACATGGAGCC |
| PRDX1 | CCCACGGAGATCATTGTT | CGAGATGCCTTCATCAGCCT |
| PRDX2 | GAAGCTGTCGGACTACAAAGG | TCGGTGGGGCACACAAAAG |
| PRDX3 | GCCGTTGTCAATGGAGAGTT | CAACAGCGTCAGAGTCACCT |
| PRDX4 | AGAGGAGTGCCACTTCTACG | GGAAATCTTCGCTTTGCTTAGGT |
| SRXN1 | AGTTTTAGGGTACAGTTTGGCTAGGTATC | AGTGGTACTTGTGCTAGGCAT ATTAGTAA |
| SOD2 | TGGGGTTGGCTTGGTTTCAA | GGAATAAGGCCTGTTGTTCCTTG |
| CAT | CGGAGATTCAACACTGCCAATG | TTCTTGACCGCTTTCTTCTGGA |
| GCLC | GGACAAGAATACACCATCTCCA | ATACTGCAGGCTTGGAATGTC |
| GCLM | GGGAACCTGCTGAACTGG | CTGGGTTGATTTGGGAACTC |
| GSR | AGGAGCTGGAGAACGCTGGC | CAATGGCCCAGAGCAGGCA |
| GSTA2 | GGCTGCAGCTGGAGTAGAGT | AAGGCAGGGAAGTAGCGATT |
| GSTA4 | GGCAGCAAGGCCCAAGCTCCACT | GGCCTAAAGATGTTGTAGACGG |
| GSTM2 | ACA ACCTGTGCGGGGAATC | AGCTTCAGCATTTCAGGGAGTG |
| GSTM3 | GACTTTCCTAATCTGCCCTACCTC | TTCTTCTTCAGTCTCACCACACAT |
| AKR1B1 | TATTCACTGGCCGACTGGCTTTA | GAACCACATTGCCCGACTCA |
| AKR1B10 | GCAGGACGTGAGACTTCTACC | ATCCTGCATCAATGGCCACC |
| AKR1C1 | TAGCCTGTGAGGGAGGAAGAA | TTGCCAATTTGGTGGCCTCT |
| AKR1C3 | GGATTTGGCACCTATGCACCTC | CTATATGGCGGAACCCAGCTTCTA |
| ALDH1A1 | ACTCCCAAGCACGCTTAGTGCTC | TCGTCATGTCTTAGCCAGCT |
| ALDH3A1 | ACTGGGCGTGGTCCTCGTCATTGG | GTGAGGATGGTGGGGGCTATGTAG |
| ABCC1 | CATTGGCGAGCCTGGTAG | TCGTAGGAGTGTCCGTGGAT |
| ABCC2 | CTTGGGCTTCCTATGGCTCC | ATCGAACAGCAGGGACTGTG |
| ABCC3 | CAGAGAAGGTGCAGGTGACA | CTAAAGCAGCATAGACGCCC |
| ABCC5 | GTTCAGGAGAACTCGACCGTTGG | TTTGGAAGTAGTCCGGATGGGCTT |
| IDH1 | TGCAAAAATATCCCCCGGCT | TACATCCCCATGGCAACACC |
| ME1 | CTGCCTGTCATTCTGGATGT | ACCTCTTACTCTTCTCTGCC |
| PGD | ATTCTCAAGTTCCAAGACACCG | GTGGTAAAACAGGGCATGGGA |
| GAPDH | CTGACTTCAACAGCGACACC | TGCTGTAGCCAAATTCGTTGT |
2.11. Measurement of PI3K activity
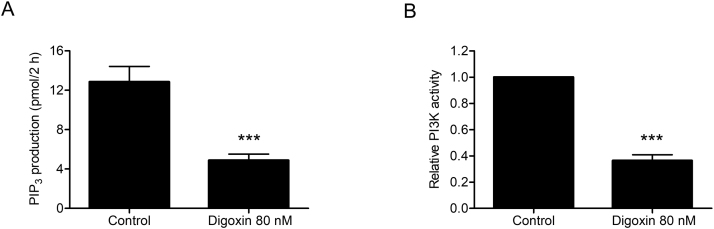
PI3K activity was detected by a PI3K activity ELISA kit (Echelon Biosciences Inc., Salt Lake City, UT, USA) according to the manufacturer's instructions. Briefly, PI3K protein (Sigma-Aldrich, St. Louis, USA) was reacted with PI(4,5)P2 as a substrate in a reaction buffer without and with 80 nM of digoxin at 30 ℃ for 2 h. After termination of the reaction, a primary PI(3,4,5)P3 detector was added to the reaction mixture and incubated for 60 min at room temperature. The mixture was transferred to a PI3K ELISA plate, and 60 min later a second detector was added and further incubated at room temperature for 30 min. A tetramethylbenzidine solution was added to the final reaction mixture, and absorbance were measured at 450 nm using a microplate reader. PI(3,4,5)P3 production (pmol/2 h) was regarded as an index of PI3K activity.
2.12. Transfection of Nrf2 shRNA and Nrf2 plasmid
For transfection of shRNA, lentiviral particles encoding Nrf2 or non-target shRNA (Sigma-Aldrich, St. Louis, USA) were diluted in OptiMEM (Gibco, Grand Island, NY) containing 6 μg/mL polybrene, and then were added to SW1990/Gem and Panc-1/Gem cells. After 3 days, 5 μg/mL of puromycin were used to select transfected cells. Cells transfected with the shRNA lentiviral particles were seeded into six-well plates and western blot analysis were used to detect the protein level of Nrf2. Nrf2 plasmid was obtained from Prof. Siwang Yu from Peking University (Beijing, China) and transfected into cells according to the manufacturer's instruction of ExFect transfection reagent (Vazyme, Nanjing, China).
2.13. In vivo study
Female BALB/c nude mice (18 ± 2 g, 6 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). All animals were maintained under standard environment on a 12 h light/dark cycle and allowed access adlibitum to water and diet. The Animal Ethics Committee of China Pharmaceutical University approved all protocols for animal. Control non-specific shRNA (shControl)-, or Nrf2-targeting shRNA (shNrf2)-transfected SW1990/Gem cells (1 × 106 cells/200 μl) were injected into subdermal space of mice on the right flanks. When the tumors volume reached 80–100 mm3, mice were randomly allocated into four groups, and then treated with vehicle, digoxin (0.1 mg/kg, daily, i.g.), gemcitabine (50 mg/kg, Once every four days, i.p.) and in combination for 24 days. Tumor volume and body weight were recorded every four days. Tumor volume = (a × b × b)/2 (a, the largest diameter; b, the smallest diameter). Tumors were removed from mice, and western blot was used to detect tissue extracts. Tumor tissues were fixed with formalin and embedded with paraffin.
2.14. Immunohistochemistry and TUNEL assay
Immunohistochemical stains against Nrf2, NQO1, HO-1, GCLC, ABCC5 and Ki-67 were performed using immunohistochemistry kit (KeyGen, Nanjing, China) according to the manufacturer's instructions. In situ cell apoptosis were detected by using TUNEL apoptosis detection kit (Beyotime Institute of Biotechnology, Shanghai, China) according to the manufacturer's instructions. Inverted fluorescence microscope (Nikon, Japan) was used to photograph all sections.
2.15. Statistical analysis
All results were expressed as mean ± SD. Statistical analysis was performed with the t-test for two groups or one-way ANOVA for multiple groups. P < 0.05 was considered to be a statistically significant difference.
3. Results
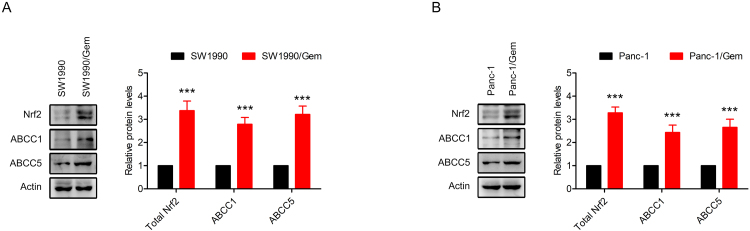
3.1. Determination of drug resistance and the Nrf2 signaling pathway was upregulated in SW1990/Gem and Panc-1/Gem cells
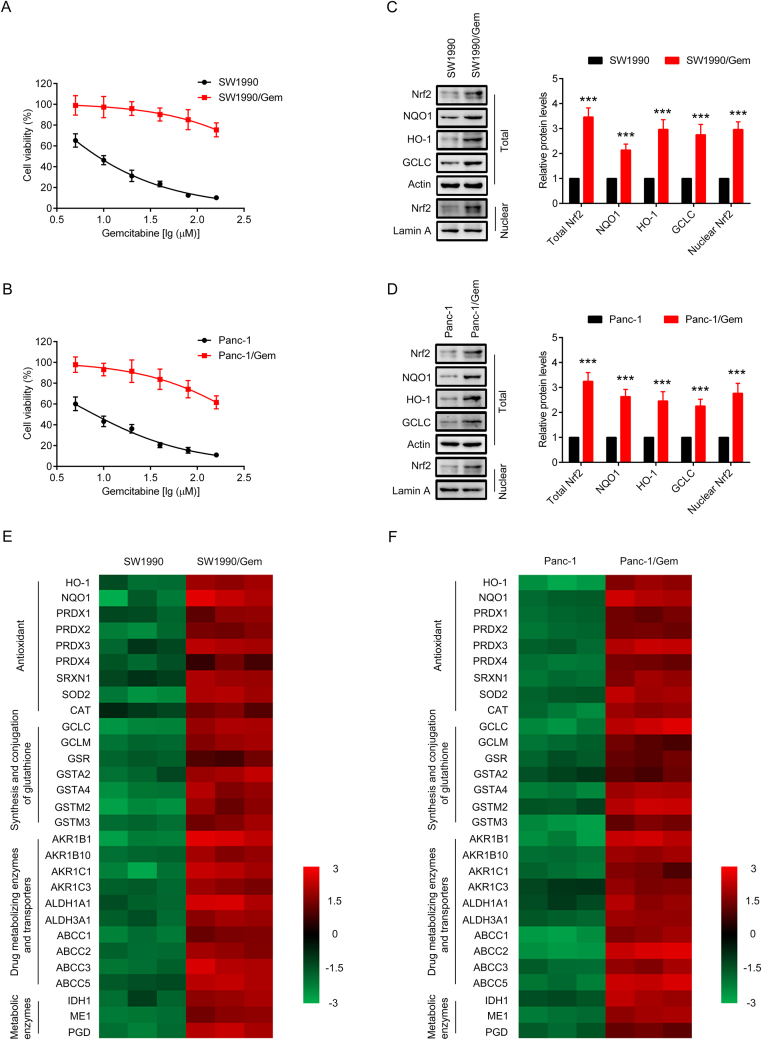
Resistance index is used to evaluate the drug resistance of drug-resistant cancer cells to chemotherapy drugs. In order to verify SW1990/Gem and Panc-1/Gem cells were gemcitabine-resistant, MTT assay was performed, and resistance index was calculated. As shown in Fig. 1A-B and Table 2, the IC50 values of gemcitabine in SW1990/Gem and Panc-1/Gem cells increased dramatically compared with SW1990 and Panc-1 cells. RI in SW1990/Gem and Panc-1/Gem cells were 57.04 and 37.75 respectively, indicating that the SW1990/Gem and Panc-1/Gem cells were gemcitabine-resistant. Moreover, we found that Nrf2, NQO1, HO-1, GCLC, ABCC1 and ABCC5 protein levels were markedly increased in SW1990/Gem and Panc-1/Gem cells (Fig. 1C–D and Supplementary Fig. S1A–B). In addition, Nrf2 target genes encoding antioxidant enzymes, enzymes of glutathione synthesis and conjugation, drug metabolizing enzymes and transporters, metabolic enzymes were also highly expressed in SW1990/Gem and Panc-1/Gem cells (Fig. 1E–F). We also investigated whether SW1990/Gem and Panc-1/Gem cells were resistant to other anticancer agents including etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin. As shown in Table 2, the IC50 values of etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin in SW1990/Gem and Panc-1/Gem cells markedly increased compared with their parental cells. The RI of etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin were 5.32, 8.41, 3.19, 5.56, 11.67 and 6.4 in SW1990/Gem cells, and the RI of etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin were 4.28, 6.58, 2.42, 4.49, 9.65 and 5.06 in Panc-1/Gem cells, respectively. These results demonstrated that gemcitabine-resistant pancreatic cancer cell lines also exhibited chemoresistance to other anticancer agents.
Fig. 1.
Nrf2 signaling was upregulated in SW1990/Gem and Panc-1/Gem cells. (A–B) The cytotoxicity of gemcitabine to SW1990, SW1990/Gem, Panc-1 and Panc-1/Gem cells. (C–D) Western blot was used to detect the protein level of Nrf2, NQO1, HO-1, and GCLC in SW1990, SW1990/Gem, Panc-1 and Panc-1/Gem cells. (E–F) The expression levels of Nrf2 target genes in SW1990, SW1990/Gem, Panc-1 and Panc-1/Gem cells. The colors of the heatmap reflect log2-expression levels of Nrf2 target genes in SW1990, SW1990/Gem, Panc-1 and Panc-1/Gem cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. SW1990 or Panc-1 cells.
Table 2.
Determination of IC50 of different anticancer drugs.
| Group | SW1990 | SW1990/Gem | RI | Panc-1 | Panc-1/Gem | RI |
|---|---|---|---|---|---|---|
| Gemcitabine (μM) | 9.33 ± 0.9 | 532.15 ± 26.13 | 57.04 | 7.85 ± 0.56 | 296.34 ± 22.17 | 37.75 |
| Etoposide (μM) | 117.46 ± 14.02 | 624.52 ± 44.89 | 5.32 | 102.7 ± 9.35 | 439.24 ± 37.58 | 4.28 |
| Paclitaxel (nM) | 35.43 ± 4.34 | 298.15 ± 37.68 | 8.41 | 32.67 ± 2.97 | 214.82 ± 16.45 | 6.58 |
| Cisplatin (μM) | 58.74 ± 4.82 | 187.23 ± 21.09 | 3.19 | 65.38 ± 7.74 | 158.39 ± 13.29 | 2.42 |
| 5-FU (μM) | 33.05 ± 2.85 | 183.68 ± 8.4 | 5.56 | 39.14 ± 1.99 | 175.74 ± 11.55 | 4.49 |
| Ara-C (μM) | 1.38 ± 0.23 | 16.11 ± 1.43 | 11.67 | 1.21 ± 0.09 | 11.68 ± 0.7 | 9.65 |
| Doxorubicin (μM) | 4.35 ± 0.59 | 27.85 ± 1.89 | 6.4 | 4.66 ± 0.29 | 23.59 ± 1.64 | 5.06 |
IC50 values of different anticancer drugs were detected by using MTT assay. Data were expressed as mean ± SD of three independent experiments.
3.2. Digoxin enhanced the sensitivity of SW1990/Gem and Panc-1/Gem cells to gemcitabine
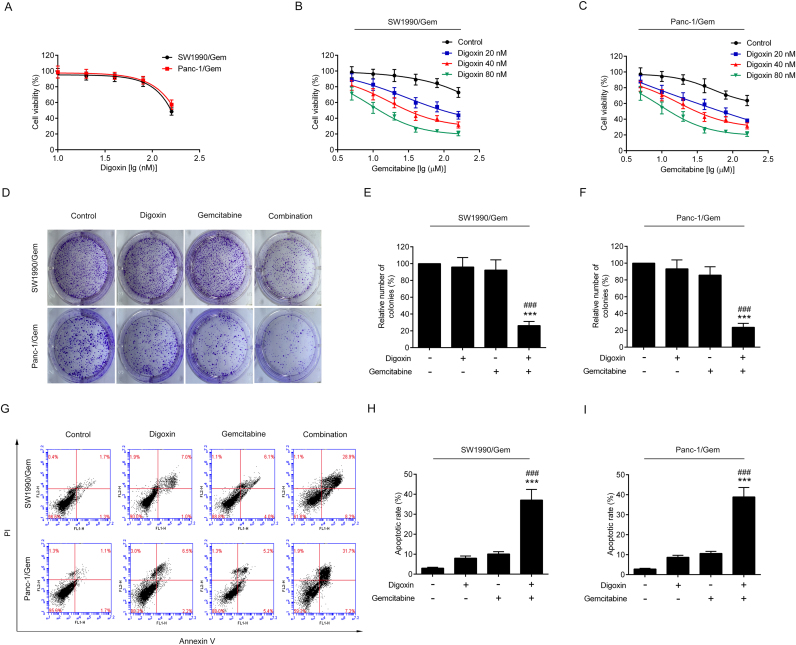
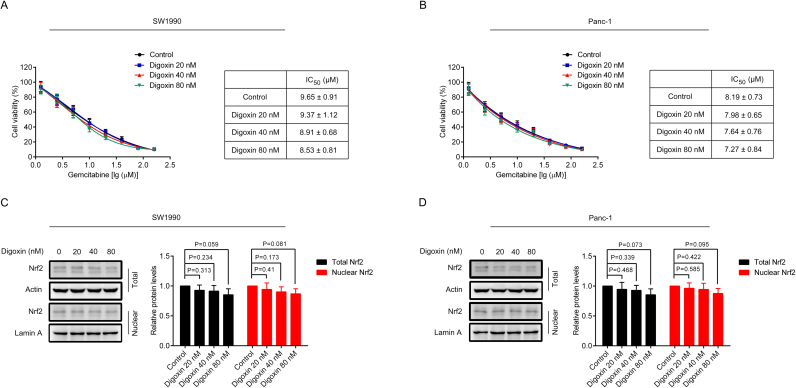
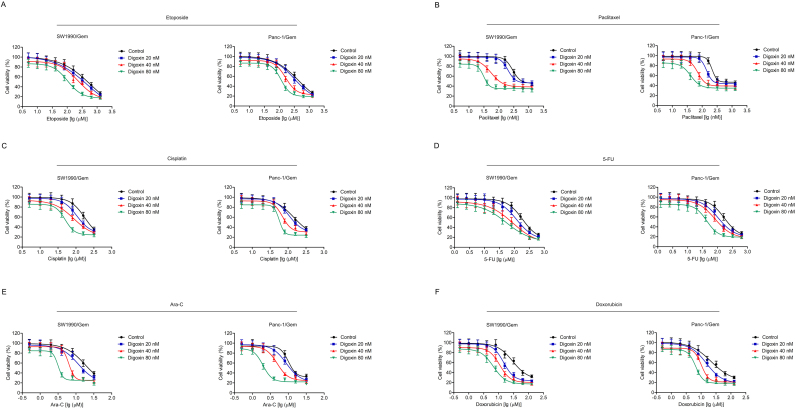
MTT assay was used to test the toxicity of digoxin in SW1990/Gem and Panc-1/Gem cells. As shown in Fig. 2A, 20 and 40 nM of digoxin had no significant cytotoxicity, and 80 nM of digoxin exhibited only slight cytotoxicity. To eliminate the effect of digoxin on SW1990/Gem and Panc-1/Gem cells, nontoxic doses (20, 40 and 80 nM) of digoxin were used in the following experiments. Digoxin at the doses of 20, 40 and 80 nM enhanced the cytotoxicity of gemcitabine in SW1990/Gem and Panc-1/Gem cells (Fig. 2B–C). Digoxin increased the sensitivity of SW1990/Gem cells to gemcitabine by 5.27-fold, 13.97-fold and 35.83-fold at the doses of 20, 40 and 80 nM, while it increased the sensitivity of Panc-1/Gem cells to gemcitabine by 3.91-fold, 7.17-fold and 20.43-fold at the doses of 20, 40 and 80 nM, respectively (Table 3). Meanwhile, our results demonstrated that gemcitabine in combination with digoxin dramatically inhibited cell colony formation and increased the number of cell undergoing apoptosis when compared with gemcitabine alone (Fig. 2D–I). Interestingly, digoxin at the doses of 20, 40 and 80 nM could not significantly enhance the cytotoxicity of gemcitabine in SW1990 and Panc-1 cells (Supplementary Fig. S2A–B). These results indicated that digoxin could enhance the chemosensitivity of gemcitabine in SW1990/Gem and Panc-1/Gem cells, but not in their parental cells. In addition, we evaluated the effect of digoxin on the sensitivity of SW1990/Gem and Panc-1/Gem cells to other anticancer agents including etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin and found that digoxin could significantly enhance the sensitivity of SW1990/Gem and Panc-1/Gem cells to these anticancer agents (Table 3 and Supplementary Fig. S3).
Fig. 2.
Digoxin enhanced the sensitivity of SW1990/Gem and Panc-1/Gem cells to gemcitabine. (A) The cytotoxicity of digoxin to SW1990/Gem and Panc-1/Gem cells. (B–C) Reverse effects of digoxin on drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. (D–F) Colony formation assay. (G–I) Flow cytometric analysis of apoptosis. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group, ###P < 0.001 vs. gemcitabine group.
Table 3.
Effects of digoxin on the sensitivity of SW1990/Gem and Panc-1/Gem cells to different anticancer drugs.
| Group | SW1990/Gem |
Panc-1/Gem |
|||
|---|---|---|---|---|---|
| IC50 | RF | IC50 | RF | ||
| Gemcitabine (μM) | Control | 519.59 ± 31.14 | 1 | 283.47 ± 21.7 | 1 |
| Digoxin 20 nM | 98.55 ± 6.12 | 5.27 | 72.47 ± 6 | 3.91 | |
| Digoxin 40 nM | 37.18 ± 4.75 | 13.97 | 39.54 ± 4.61 | 7.17 | |
| Digoxin 80 nM | 14.5 ± 2.03 | 35.83 | 13.87 ± 2.16 | 20.43 | |
| Etoposide (μM) | Control | 633.19 ± 40.77 | 1 | 445.82 ± 30.85 | 1 |
| Digoxin 20 nM | 445.77 ± 22.04 | 1.42 | 327.28 ± 20.82 | 1.36 | |
| Digoxin 40 nM | 232.98 ± 18.38 | 2.72 | 172.46 ± 10.57 | 2.59 | |
| Digoxin 80 nM | 110.95 ± 13.63 | 5.71 | 119.34 ± 12.85 | 3.74 | |
| Paclitaxel (nM) | Control | 293.07 ± 19.72 | 1 | 216.53 ± 16.27 | 1 |
| Digoxin 20 nM | 209.57 ± 17.65 | 1.4 | 144.27 ± 11.57 | 1.5 | |
| Digoxin 40 nM | 61.04 ± 4.99 | 4.8 | 67.82 ± 4.05 | 3.19 | |
| Digoxin 80 nM | 35.77 ± 4.31 | 8.19 | 37.01 ± 4.99 | 5.85 | |
| Cisplatin (μM) | Control | 186.47 ± 11.41 | 1 | 155.53 ± 11.78 | 1 |
| Digoxin 20 nM | 112.87 ± 8.52 | 1.65 | 107.17 ± 7.35 | 1.45 | |
| Digoxin 40 nM | 86.24 ± 6.39 | 2.16 | 68.15 ± 6.24 | 2.28 | |
| Digoxin 80 nM | 51.73 ± 6.28 | 3.6 | 58.44 ± 6.06 | 2.66 | |
| 5-FU (μM) | Control | 185.38 ± 14.67 | 1 | 168.39 ± 10.8 | 1 |
| Digoxin 20 nM | 118.23 ± 10.08 | 1.57 | 110.88 ± 7.6 | 1.52 | |
| Digoxin 40 nM | 86.55 ± 7.17 | 2.14 | 73.46 ± 7.73 | 2.29 | |
| Digoxin 80 nM | 55.25 ± 7.06 | 3.36 | 38.89 ± 4.72 | 4.33 | |
| Ara-C (μM) | Control | 16.88 ± 1.15 | 1 | 10.93 ± 0.96 | 1 |
| Digoxin 20 nM | 11.4 ± 1.07 | 1.48 | 8.5 ± 0.74 | 1.29 | |
| Digoxin 40 nM | 6.65 ± 0.5 | 2.54 | 4.66 ± 0.3 | 2.35 | |
| Digoxin 80 nM | 2.99 ± 0.32 | 5.65 | 1.61 ± 0.33 | 6.79 | |
| Doxorubicin (μM) | Control | 29.24 ± 2.74 | 1 | 24.4 ± 1.84 | 1 |
| Digoxin 20 nM | 14.03 ± 1.54 | 2.08 | 15.02 ± 1.68 | 1.62 | |
| Digoxin 40 nM | 11.06 ± 1.19 | 2.64 | 10.82 ± 1.16 | 2.26 | |
| Digoxin 80 nM | 6.74 ± 0.47 | 4.34 | 7.52 ± 0.92 | 3.24 | |
Effects of digoxin on the sensitivity of SW1990/Gem and Panc-1/Gem cells to different anticancer drugs were detected by MTT assay. Data were expressed as mean ± SD of three independent experiments.
3.3. Digoxin inhibited the Nrf2 signaling in SW1990/Gem and Panc-1/Gem cells
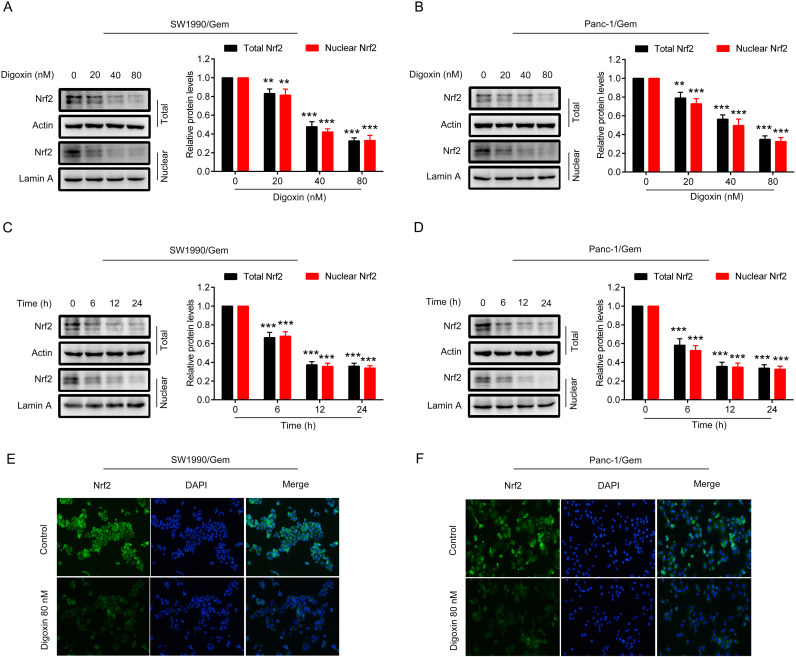
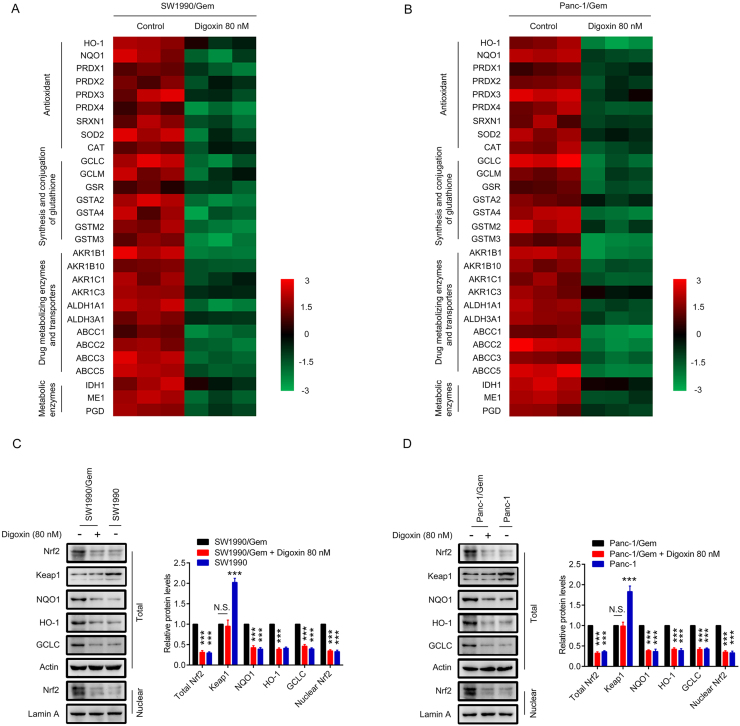
As upregulation of Nrf2 protein levels were observed in SW1990/Gem and Panc-1/Gem cells, we evaluated the inhibitory effect of digoxin on the protein levels of Nrf2. We found that total and nuclear Nrf2 protein levels were markedly decreased in SW1990/Gem and Panc-1/Gem cells treated with 20, 40, and 80 nM of digoxin for 24 h (Fig. 3A–B). Subsequently, we observed the effect of 80 nM digoxin on Nrf2 protein level in SW1990/Gem and Panc-1/Gem cells at different time points. We found that the protein level of Nrf2 began to reduce from 6 h after digoxin treatment (Fig. 3C–D). Endogenous Nrf2 immunostaining further proved the inhibitory effects of digoxin on the protein expression of Nrf2 in SW1990/Gem and Panc-1/Gem cells (Fig. 3E–F). Interestingly, digoxin at the doses of 20, 40 and 80 nM could not significantly inhibit total and nuclear Nrf2 protein levels in SW1990 and Panc-1 cells (Supplementary Fig. S2C–D). Next, we investigated whether digoxin could inhibit the expressions of Nrf2 target genes in SW1990/Gem and Panc-1/Gem cells. We found that Nrf2 target genes encoding antioxidant enzymes, enzymes of glutathione synthesis and conjugation, drug metabolizing enzymes and transporters, metabolic enzymes were markedly decreased in SW1990/Gem and Panc-1/Gem cells treated with 80 nM of digoxin (Fig. 4A–B). Meanwhile, 80 nM of digoxin significantly suppressed protein expressions of NQO1, HO-1, GCLC and ABCC5 (Fig. 4C–D and Supplementary Fig. S4A–B). These results indicated that digoxin could inhibit Nrf2 signaling in SW1990/Gem and Panc-1/Gem cells.
Fig. 3.
Digoxin inhibited the Nrf2 signaling in SW1990/Gem and Panc-1/Gem cells. (A–B) Effects of digoxin on the protein level of Nrf2 in SW1990/Gem and Panc-1/Gem cells. (C–D) Effects of digoxin on the protein level of Nrf2 in SW1990/Gem and Panc-1/Gem cells at different time points. (E–F) Immunostaining of endogenous Nrf2 in SW1990/Gem and Panc-1/Gem cells. All images were shown at ×200. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as **P < 0.01, ***P < 0.001 vs. control group. N.S., no significant.
Fig. 4.
Digoxin inhibited the expressions of Nrf2 target genes in SW1990/Gem and Panc-1/Gem cells. (A–B) Effects of digoxin on the expressions of Nrf2 target genes in SW1990/Gem and Panc-1/Gem cells. The colors of the heatmap reflect log2-expression levels of Nrf2 target genes in SW1990/Gem and Panc-1/Gem cells. (C–D) Effects of digoxin on protein levels of NQO1, HO-1 and GCLC in SW1990/Gem and Panc-1/Gem cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as **P < 0.01, ***P < 0.001 vs. control group. N.S., no significant.
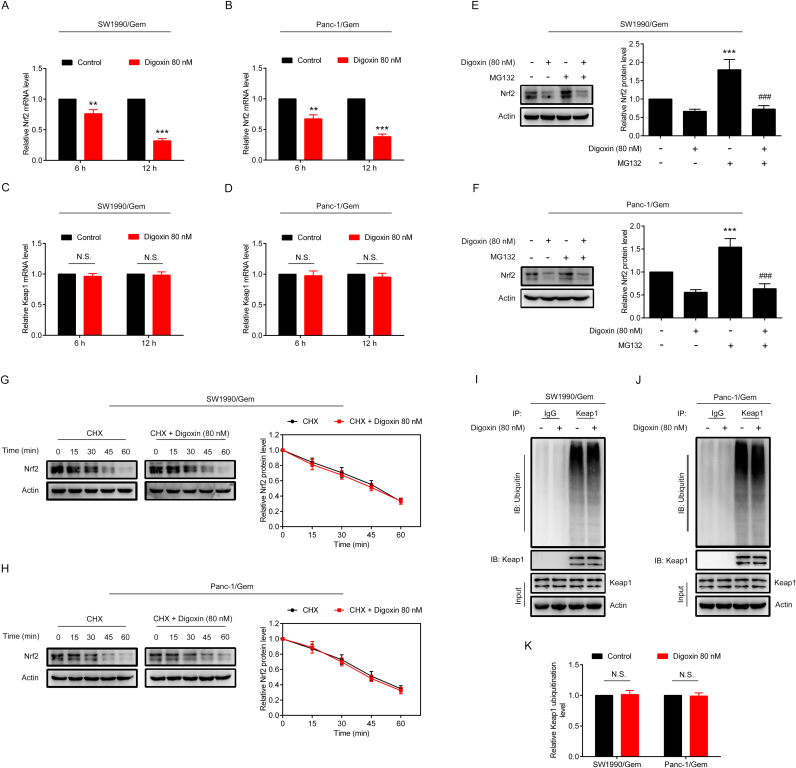
3.4. Digoxin inhibited Nrf2 by decreasing Nrf2 mRNA at transcriptional level in SW1990/Gem and Panc-1/Gem cells
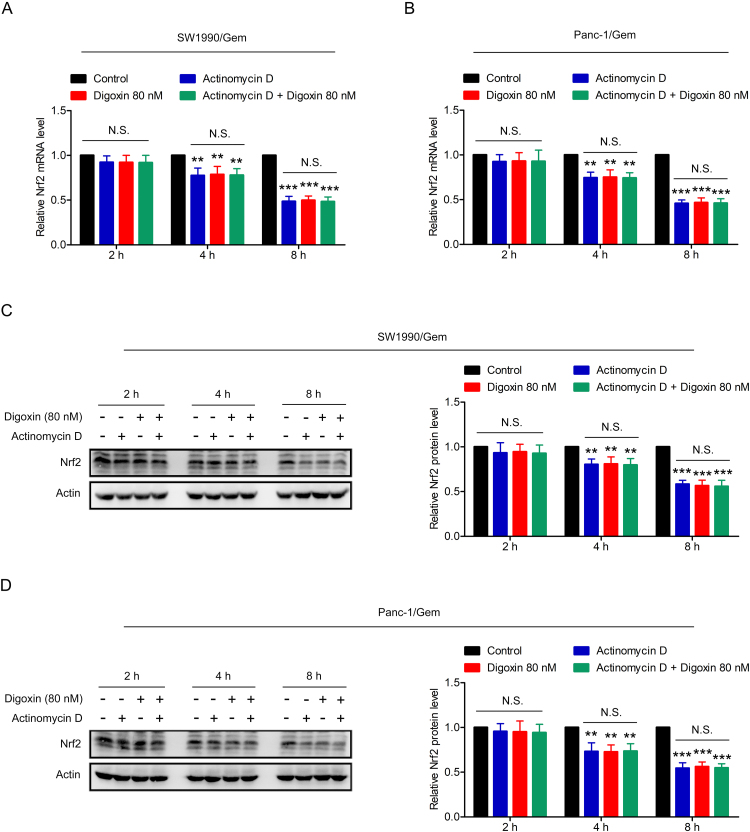
To test whether digoxin decreased the protein level of Nrf2 via a transcriptional mechanism, Nrf2 and Keap1 mRNA levels were measured by qRT-PCR. The results showed that the Keap1 mRNA level had no change, while the Nrf2 mRNA level was markedly decreased in SW1990/Gem and Panc-1/Gem cells treated with digoxin (Fig. 5A–D). To determine whether the reduction of Nrf2 mRNA was due to decreased Nrf2 mRNA stability, digoxin was incubated with or without actinomycin D (a transcription inhibitor) in SW1990/Gem and Panc-1/Gem cells for 2 h, 4 h and 8 h. As shown in Supplementary Fig. S5A–B, no significant difference of Nrf2 mRNA level was found between actinomycin D alone and combination group, indicating that digoxin did not affect stability of Nrf2 mRNA. Moreover, Nrf2 protein level in combination group had no significant change compared with actinomycin D alone group (Supplementary Fig. S5C–D), indicating that digoxin did not affect translation and degradation of Nrf2 protein. We also found that MG132 treatment alone increased the Nrf2 protein level, while digoxin and MG132 cotreatment significantly decreased it (Fig. 5E–F). Moreover, CHX assay and western blot assay were used to test the effect of digoxin on the half-life of Nrf2 in SW1990/Gem and Panc-1/Gem cells, and it was found that digoxin did not change the half-life of Nrf2 (Fig. 5G–H). In addition, ubiquitination analysis revealed that ubiquitination of Keap1 did not change in the presence or absence of digoxin (Fig. 5I–K). These results demonstrated that digoxin inhibited the Nrf2 signaling through decreasing Nrf2 mRNA at transcriptional level without affecting stability of Nrf2 mRNA and translation and degradation of Nrf2 protein in SW1990/Gem and Panc-1/Gem cells.
Fig. 5.
Digoxin inhibited Nrf2 by decreasing Nrf2 mRNA at transcriptional level in SW1990/Gem and Panc-1/Gem cells. (A–D) Quantitative real-time PCR analysis. (E–F) SW1990/Gem and Panc-1/Gem cells were treated with 80 nM of digoxin, 20 µM of MG132, or a combination of digoxin and MG132 for 6 h, the protein level of Nrf2 were detected by Western blot. (G–H) CHX-chase analysis. (I–K) Ubiquitination assay. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as **P < 0.01, ***P < 0.001 vs. control group, ###P < 0.001 vs. MG132 group. N.S., no significant.
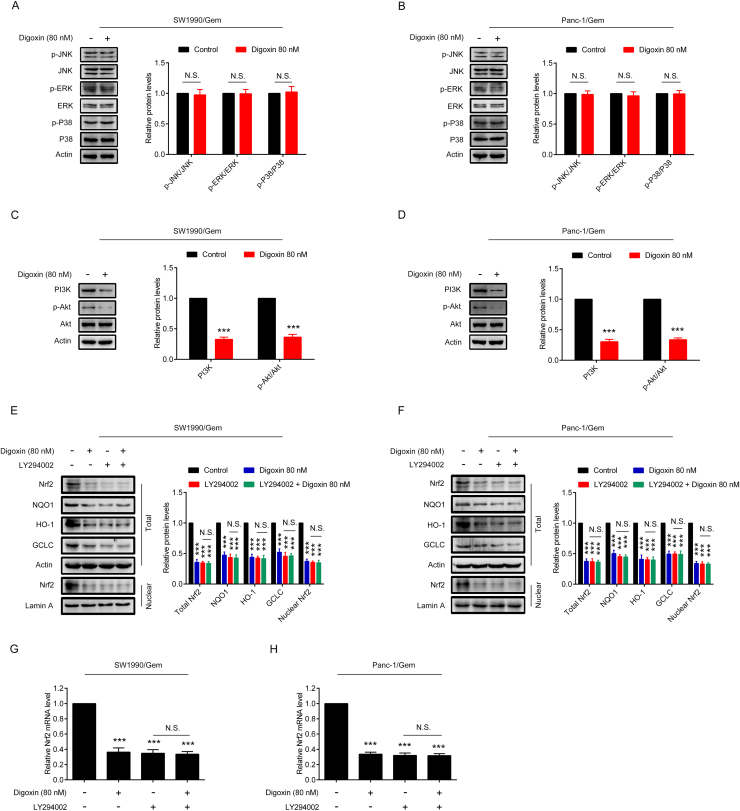
3.5. Digoxin decreased Nrf2 at transcriptional level through inhibiting PI3K/Akt pathway in SW1990/Gem and Panc-1/Gem cells
To determine the mechanism of digoxin in decreasing Nrf2 mRNA level, effects of digoxin on the MAPKs and PI3K/Akt signaling pathways were studied. We found that digoxin attenuated PI3K and p-Akt protein levels, without affecting p-P38, p-ERK1/2 and p-JNK protein levels in SW1990/Gem and Panc-1/Gem cells (Fig. 6A–D). In addition, we found that digoxin could significantly inhibit PI3K kinase activity (Supplementary Fig. S6A–B). LY294002, a PI3K inhibitor, inhibited the protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 (Fig. 6E–F and Supplementary Fig. S4C–D). Importantly, digoxin did not markedly change LY294002-induced reduction of these proteins (Fig. 6E–F and Supplementary Fig. S4C–D). Furthermore, LY294002 could reduce the Nrf2 mRNA level, and digoxin was unable to decrease the Nrf2 mRNA level further (Fig. 6G–H). These results suggested that digoxin inhibited Nrf2 signaling through suppressing PI3K/Akt signaling pathway in SW1990/Gem and Panc-1/Gem cells.
Fig. 6.
Digoxin decreased Nrf2 at transcriptional level through inhibiting PI3K/Akt pathway in SW1990/Gem and Panc-1/Gem cells. (A–B) Effects of digoxin on protein levels of p-JNK, JNK, p-ERK1/2, ERK1/2, p-P38 and P38. (C–D) Effects of digoxin on protein levels of PI3K, p-Akt, Akt. (E–F) SW1990/Gem and Panc-1/Gem cells were treated with 80 nM of digoxin, 20 µM of LY294002, or a combination of digoxin and LY294002 for 24 h, the protein levels of Nrf2, NQO1, HO-1, and GCLC were detected by Western blot. (G–H) SW1990/Gem and Panc-1/Gem cells were treated with 80 nM of digoxin, 20 µM of LY294002, or a combination of digoxin and LY294002 for 24 h, Nrf2 mRNA levels were detected by qRT-PCR. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group. N.S., no significant.
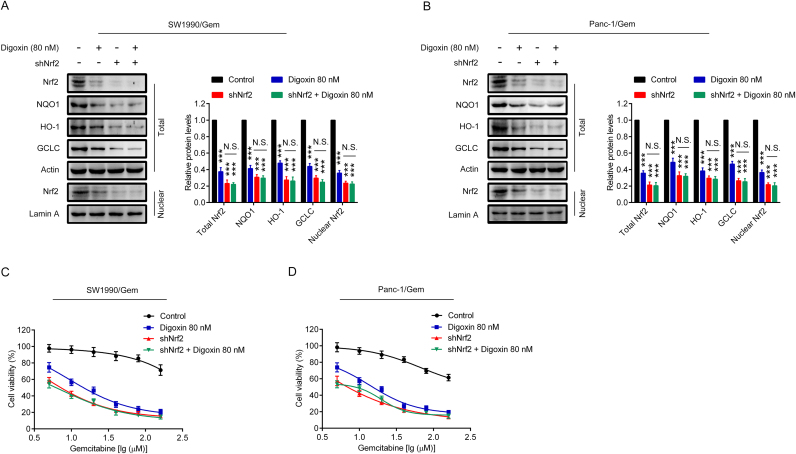
3.6. Digoxin increased the sensitivity of SW1990/Gem and Panc-1/Gem cells to gemcitabine by inhibiting Nrf2 signaling
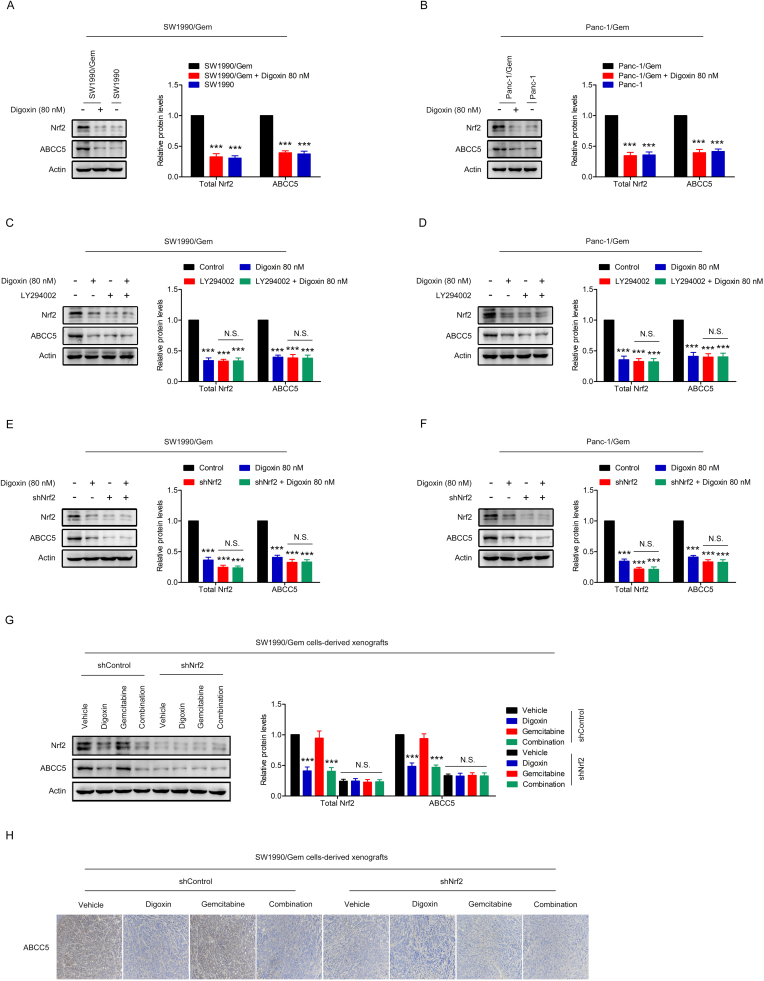
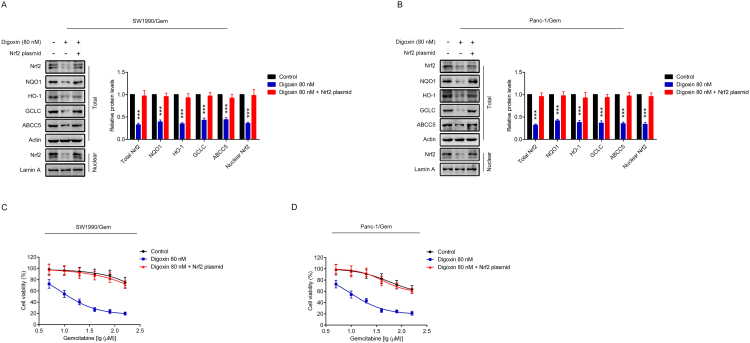
To determine if Nrf2 signaling was involved in digoxin-mediated reversing of gemcitabine resistance in SW1990/Gem and Panc-1/Gem cells, cells were transfected with Nrf2 shRNA (SW1990/Gem-shNrf2 and Panc-1/Gem-shNrf2). The protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 were markedly reduced in SW1990/Gem-shNrf2 and Panc-1/Gem-shNrf2 cells (Fig. 7A–B and Supplementary Fig. S4E–F). Moreover, we found that the protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 were no markedly changed in SW1990/Gem-shNrf2 and Panc-1/Gem-shNrf2 cells treated with digoxin (Fig. 7A–B and Supplementary Fig. S4E–F). In addition, we tested the sensitivity of SW1990/Gem-shNrf2 and Panc-1/Gem-shNrf2 cells to gemcitabine. Our results showed that SW1990/Gem and Panc-1/Gem cells with Nrf2 knockdown were more sensitive to gemcitabine than SW1990/Gem and Panc-1/Gem cells, respectively (Fig. 7C–D). However, the effects of digoxin were diminished in SW1990/Gem-shNrf2 and Panc-1/Gem-shNrf2 cells (Fig. 7C–D). In addition, Nrf2 overexpression plasmid were used to restore Nrf2 protein level in digoxin-treated gemcitabine-resistant pancreatic cancer cells, and found that gemcitabine resistance was restored (Supplementary Fig. S7). These data demonstrated that digoxin increased the sensitivity of SW1990/Gem and Panc-1/Gem cells to gemcitabine through inhibiting Nrf2 signaling.
Fig. 7.
Digoxin increased the sensitivity of SW1990/Gem and Panc-1/Gem cells to gemcitabine by inhibiting Nrf2 signaling. (A–B) Effects of Nrf2 knockdown on protein levels of Nrf2, NQO1, HO-1, and GCLC in SW1990/Gem and Panc-1/Gem cells. (C–D) Effects of Nrf2 knockdown on reversing drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group. N.S., no significant.
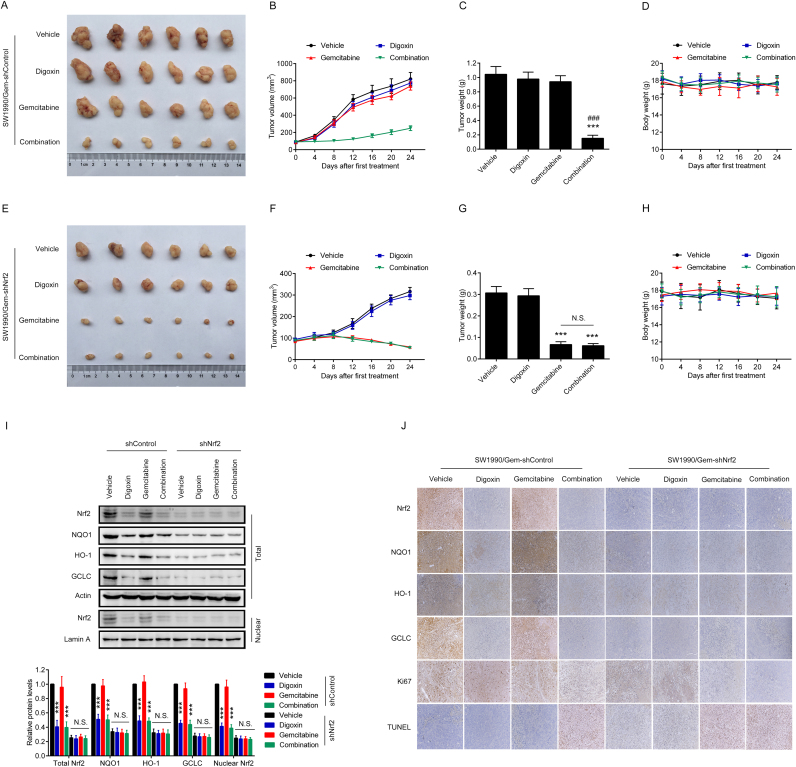
3.7. Digoxin sensitized SW1990/Gem cells-derived xenografts to gemcitabine treatment by inhibiting Nrf2 signaling
To confirm whether digoxin-mediated reversing of gemcitabine resistance in SW1990/Gem cells-derived xenografts was Nrf2 dependent, SW1990/Gem-shControl cells and SW1990/Gem-shNrf2 cells were injected into the subdermal space of nude mice on the right flanks. As shown in Fig. 8A–C, SW1990/Gem-shControl cells-derived xenografts in the combination group had smaller tumor volume and lighter tumor weight than that in the gemcitabine group. In contrast, SW1990/Gem-shNrf2 cells-derived xenografts responded to gemcitabine and combination treatment similarly (Fig. 8E–G). Interesting, the body weight of nude mice was no significant changed in this experiment (Fig. 8D, H). Moreover, compared with SW1990/Gem-shControl cells-derived xenografts, SW1990/Gem-shNrf2 cells-derived xenografts had lower protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 (Fig. 8I–J and Supplementary Fig. S4G–H). In addition, digoxin could markedly decrease the protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 in SW1990/Gem-shControl cells-derived xenografts, but no significantly change of these protein levels were observed in SW1990/Gem-shNrf2 cells-derived xenografts (Fig. 8I–J and Supplementary Fig. S4G–H). Furthermore, compared with gemcitabine group, the combined treatment of gemcitabine with digoxin markedly increased cell apoptosis only in SW1990/Gem-shControl cells-derived xenografts, but not in SW1990/Gem-shNrf2 cells-derived xenografts which had a high rate of cell apoptosis with gemcitabine treatment (Fig. 8J). Interestingly, gemcitabine and digoxin in combination significantly inhibited cell proliferation compared with gemcitabine treatment alone in SW1990/Gem-shControl cells-derived xenografts (Fig. 8J). These results showed that digoxin could sensitize gemcitabine-resistant pancreatic cancer cell xenografts to gemcitabine through inhibiting Nrf2 signaling.
Fig. 8.
Digoxin sensitized SW1990/Gem cells-derived xenografts to gemcitabine treatment by inhibiting Nrf2 signaling. (A–D) Digoxin sensitized SW1990/Gem-shControl cells-derived xenografts to gemcitabine treatment. (E–H) Digoxin could not sensitize SW1990/Gem-shNrf2 cells-derived xenografts to gemcitabine treatment. (I) Effects of digoxin on the protein levels of Nrf2, NQO1, HO-1, and GCLC in tumor tissues. (J) Tumor tissues were subjected to IHC-Nrf2, IHC-NQO1, IHC-HO-1, IHC-GCLC, IHC-Ki67 and TUNEL staining. All images were shown at ×200. Data were expressed as mean ± SD, n = 6. Significant differences were indicated as ***P < 0.001 vs. vehicle group, ###P < 0.001 vs. gemcitabine group. N.S., no significant.
4. Discussion
Chemoresistance is one of the major difficulties during cancer chemotherapy. Mechanisms such as inducing activity of efflux transporter proteins, facilitating detoxification by metabolizing enzymes, enhancing DNA repairment and altering oncogenes were involved in the process of chemoresistance [23], [24], [25], [26]. Previous studies have suggested Nrf2 was a novel therapeutic target to reverse drug resistance [27], [28]. Nrf2 could regulate the expression of its target genes, such as NQO1, HO-1, GCLC and ABCC5, et al. It was reported that the overexpression of NQO1 was associated with the late clinical stage, lymph node metastasis and poor prognosis in breast and cervical cancers [29], [30]. HO-1 overexpression could improve proliferation, angiogenesis and metastasis of melanoma cells and decrease survival of tumor-bearing mice [31]. GCLC, a glutamate cysteine ligase subunit, has been reported to increase cisplatin resistance in NSCLC xenografts [32]. ABCC5 was reported to mediate the ATP-dependent transport of several anticancer agents and antiviral nucleosides [33] and confer resistance to gemcitabine [34]. These findings suggested that chemoresistance is associated, at least in part, with the activation of Nrf2 signaling. Here, we found SW1990/Gem and Panc-1/Gem cells had remarkable higher expression of Nrf2 and its target genes compared to their parent cells. Furthermore, Nrf2 shRNA could partially reverse drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. We therefore suggested that the activation of Nrf2 signaling in gemcitabine-resistant pancreatic cancer cells was one of the reasons that result in gemcitabine resistance. Therefore, combination of Nrf2 small molecule inhibitors and gemcitabine may effectively reverse gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells.
Previous studies have identified some small-molecule inhibitors of Nrf2. Ren et al. found that brusatol improved the sensitivity of various of cancer cells and A549 xenografts to cisplatin and other anticancer drugs through inhibiting the protein level of Nrf2 [35]. Luteolin, a potent Nrf2 small-molecule inhibitor, enhanced sensitivity of human lung cancer cells to chemotherapeutic drugs through reducing cellular GSH level [36]. Epigallocatechin 3-gallate was reported to decrease the activity of Nrf2 and HO-1 expression and promote apoptosis in A549 cells [37]. We have previously reported that wogonin was able to reverse drug resistance of doxorubicin in MCF-7/DOX cells by inhibiting the activity of Nrf2 and decreasing the expression of NQO1 and HO-1 [14]. Except for natural products, Singh et al. found a potent Nrf2 selective small-molecule inhibitor, ML385, through performing a cell-based high throughput screen from Molecular Libraries Small Molecule Repository. They also found ML385 was able to treat alone or combine with carboplatin for the treatment of non-small cell lung cancer harboring Keap1 mutations [38]. In the present study, we found that digoxin could significantly inhibit the activity of Nrf2 in nanomolar concentrations. However, luteolin, epigallocatechin 3-gallate, wogonin and ML385 exhibited the inhibition of Nrf2 activity in micromolar concentrations, indicating that digoxin had better bioactivity on inhibiting Nrf2 activity when compared with these Nrf2 inhibitors. Although brusatol could inhibit the activity of Nrf2 in nanomolar concentrations, the rapid and transient reduction of Nrf2 was reversible and the effective concentration of brusatol was reported to lead to weight loss in nude mice, which may limit its clinical use. However, clinical trials of digoxin in the treatment of cancer have been carried out. Lin et al. found that digoxin could inhibit human prostate cancer growth and disease progression and no patients had significant digoxin toxicity [39], while Frankel et al. found that digoxin plus trametinib could control melanoma in patients and no toxicities attributable to digoxin were observed [40].
Here, we evaluated the effects of digoxin in combination with gemcitabine on reversing the drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. We found that digoxin could reverse drug resistance of gemcitabine in SW1990/Gem and Panc-1/Gem cells. Recently, digoxin was reported to inhibit HIF-1α protein level in different types of cancer cells including C4-2 prostate cancer cell, MDA-MB-231 breast cancer cell, Hep3B hepatoma cell and A549 lung cancer cell [41], [42], [43], [44]. Moreover, studies have shown that HIF-1α was involved in the process of chemoresistance, and inhibition of HIF-1α will increase the sensitivity of cancer cell to chemotherapeutic drugs [45], [46], [47]. In addition, Lee et al. found that digoxin had a novel function as a phosphatase 2A (PP2A) inhibitor, and enhanced the radiosensitivity of radioresistant NSCLC cells through inhibiting PP2A [48]. These previous studies indicated that digoxin-mediated reversing of gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells might be associated with inhibition of HIF-1α and PP2A. However, we found that SW1990/Gem and Panc-1/Gem cells with Nrf2 knockdown exhibited high sensitivity to gemcitabine and digoxin was unable to further improve sensitivity of these cells to gemcitabine, indicating that digoxin-mediated reversing of gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells was Nrf2 dependent.
Previous study suggested that Nrf2 activity was regulated by continuously 26S proteasome-mediated Nrf2 protein degradation in a Keap1-dependent manner [49], indicating that Nrf2 protein degradation might be involved in digoxin-mediated downregulation of Nrf2 signaling. Moreover, digoxin might inhibit Nrf2 protein level by decreasing Nrf2 protein translation. However, our study demonstrated that digoxin was unable to increase Nrf2 protein degradation and inhibit Nrf2 protein translation, suggesting the existence of other regulatory mechanisms. It was reported that the activity of Nrf2 was regulated by several upstream protein kinases signaling pathways, such as PI3K/Akt and MAPKs [50], [51], [52], [53]. In this study, we confirmed that digoxin reduced the protein and mRNA levels of Nrf2 by inhibiting PI3K/Akt signaling pathway. Numerous studies have suggested that a variety of transcription factors could inhibit Nrf2 activity through competing with Nrf2 for AREs binding or physical association with Nrf2. The immediate early proteins and Small MAF proteins have been reported to compete with Nrf2 for AREs binding [54], [55]. In addition, several other transcription factors, including peroxisome proliferator-activated receptor [56], estrogenreceptor α [57], activating transcription factor 3 [58], short-form estrogen-related receptor β [59] and retinoic acid receptor α [60] have been suggested to form complexes with Nrf2 to inhibit Nrf2 activity. Nuclear factor-κB inhibited Nrf2 signaling through depriving CBP from Nrf2 and promoting recruitment of HDAC3 to MafK [61]. Whether these transcription factors were involved in digoxin-mediated regulation of Nrf2 signaling pathway remained to be investigated.
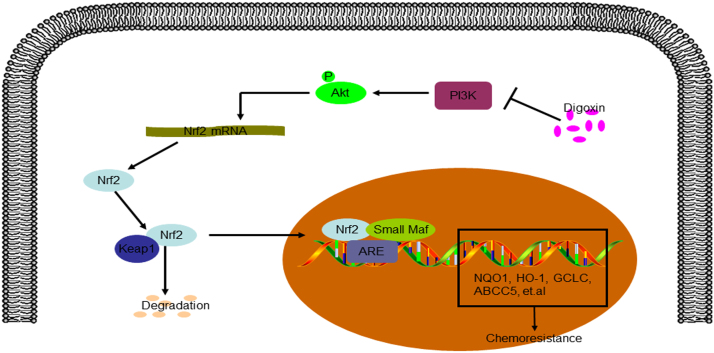
Our results suggested that digoxin could significantly reverse gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells. In addition, digoxin inhibited the activity of Nrf2 through suppressing PI3K/Akt signaling pathway in gemcitabine-resistant pancreatic cancer cells (Fig. 9). Further studies were essential to exploit digoxin as a chemotherapeutic adjuvant for chemoresistance pancreatic cancer.
Fig. 9.
The pathway involved in gemcitabine-resistant pancreatic cancer cells with digoxin treatment.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81372268, 81672816 and 81872337), the Program for Jiangsu Province Innovative Research (KYLX16_1120), the Natural Science Foundation for Distinguished Young Scholars of Jiangsu Province (No. BK20130026), the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (No. ZJ11173).
Acknowledgments
Conflict of interest
There is no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101131.
Appendix A. Supplementary material
Supplementary Fig. S1.
Protein expressions of ABCC1 and ABCC5 in SW1990, SW1990/Gem, Panc-1 and Panc-1/Gem cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. SW1990 or Panc-1 cells.
Supplementary Fig. S2.
Digoxin could not enhance the cytotoxicity of gemcitabine and inhibit the Nrf2 signaling in SW1990 and Panc-1 cells. (A–B) Effects of digoxin on chemosensitivity of SW1990 and Panc-1 cells to gemcitabine. (C–D) Effects of digoxin on the protein levels of Nrf2 in SW1990 and Panc-1 cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments.
Supplementary Fig. S3.
Digoxin enhanced the sensitivity of SW1990/Gem and Panc-1/Gem cells to etoposide, paclitaxel, cisplatin, 5-FU, ara-C and doxorubicin. Data were expressed as mean ± SD, and the results were representative of three independent experiments.
Supplementary Fig. S4.
Digoxin inhibited the protein expression of ABCC5 in SW1990/Gem and Panc-1/Gem cells and in SW1990/Gem cells-derived xenografts. (A–B) Effect of digoxin on protein levels of ABCC5 in SW1990/Gem and Panc-1/Gem cells. (C–D) SW1990/Gem and Panc-1/Gem cells were treated with 80 nM of digoxin, 20 µM of LY294002, or a combination of digoxin and LY294002 for 24 h, the protein levels of Nrf2 and ABCC5 were detected by Western blot. (E-F) Effects of Nrf2 knockdown on protein levels of Nrf2 and ABCC5 in SW1990/Gem and Panc-1/Gem cells. (G) Effects of digoxin on the protein levels of Nrf2 and ABCC5 in tumor tissues. (H) Tumor tissues were subjected to IHC-ABCC5 staining. All images were shown at ×200. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group or vehicle group. N.S., no significant.
Supplementary Fig. S5.
Digoxin inhibited Nrf2 not through affecting stability of Nrf2 mRNA and translation and degradation of Nrf2 protein in SW1990/Gem and Panc-1/Gem cells. (A–B) 80 nM of digoxin was incubated with or without actinomycin D (5 μg/mL) in SW1990/Gem and Panc-1/Gem cells for 2 h, 4 h and 8 h. Nrf2 mRNA levels at different time points (2 h, 4 h and 8 h) were detected using qRT-PCR. (C–D) 80 nM of digoxin was incubated with or without actinomycin D (5 μg/mL) in SW1990/Gem and Panc-1/Gem cells for 2 h, 4 h and 8 h. Nrf2 protein levels at different time points (2 h, 4 h and 8 h) were detected using Western blot. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as **P < 0.01, ***P < 0.001 vs. control group. N.S., no significant.
Supplementary Fig. S6.
Effect of digoxin on PI3K kinase activity. PI3K was reacted with PI(4,5)P2 in the presence and absence of digoxin for 2 h and PI(3,4,5)P3 production was measured with a competitive ELISA. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group.
Supplementary Fig. S7.
(A–B) Effects of Nrf2 overexpression plasmid on protein levels of Nrf2, NQO1, HO-1, GCLC and ABCC5 in digoxin-treated SW1990/Gem and Panc-1/Gem cells. (C–D) Nrf2 overexpression plasmid could restore drug resistance of gemcitabine in digoxin-treated SW1990/Gem and Panc-1/Gem cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. Significant differences were indicated as ***P < 0.001 vs. control group.
References
- 1.Stathis A., Moore M.J. Advanced pancreatic carcinoma: current treatment and future challenges. Nat. Rev. Clin. Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.de Sousa C.L., Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 4.Moon E.J., Giaccia A. Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radic. Biol. Med. 2015;79:292–299. doi: 10.1016/j.freeradbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motohashi H., Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Jaiswal A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi A., Suzuki H., Itoh K., Yamamoto M., Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem. Biophys. Res. Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 8.Vollrath V., Wielandt A.M., Iruretagoyena M., Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem. J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahaffey C.M., Zhang H., Rinna A., Holland W., Mack P.C., Forman H.J. Multidrug-resistant protein-3 gene regulation by the transcription factor Nrf2 in human bronchial epithelial and non-small-cell lung carcinoma. Free Radic. Biol. Med. 2009;46:1650–1657. doi: 10.1016/j.freeradbiomed.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh K., Mimura J., Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid. Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 11.Wang X.J., Sun Z., Villeneuve N.F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G.T., Wong P.K., Zhang D.D. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X.F., Yao J., Gao S.G., Wang X.S., Peng X.Q., Yang Y.T., Feng X.S. Nrf2 overexpression predicts prognosis and 5-FU resistance in gastric cancer. Asian Pac. J. Cancer Prev. 2013;14:5231–5235. doi: 10.7314/apjcp.2013.14.9.5231. [DOI] [PubMed] [Google Scholar]
- 13.Hayes J.D., McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Y., Zhang F., Sun Z., Zhou W., Li Z.Y., You Q.D., Guo Q.L., Hu R. Drug resistance associates with activation of Nrf2 in MCF-7/DOX cells, and wogonin reverses it by down-regulating Nrf2-mediated cellular defense response. Mol. Carcinog. 2013;52:824–834. doi: 10.1002/mc.21921. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.K., Yang J.W., Kim M.R., Roh S.H., Kim H.G., Lee K.Y., Jeong H.G., Kang K.W. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic. Biol. Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Tarumoto T., Nagai T., Ohmine K., Miyoshi T., Nakamura M., Kondo T., Mitsugi K., Nakano S., Muroi K., Komatsu N., Ozawa K. Ascorbic acid restores sensitivity to imatinib via suppression of Nrf2-dependent gene expression in the imatinib-resistant cell line. Exp. Hematol. 2004;32:375–381. doi: 10.1016/j.exphem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Hong Y.B., Kang H.J., Kwon S.Y., Kim H.J., Kwon K.Y., Cho C.H., Lee J.M., Kallakury B.V., Bae I. Nuclear factor (erythroid-derived 2)-like 2 regulates drug resistance in pancreatic cancer cells. Pancreas. 2010;39:463–472. doi: 10.1097/MPA.0b013e3181c31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madelaire C., Schou M., Nelveg-Kristensen K.E., Schmiegelow M., Torp-Pedersen C., Gustafsson F., Kober L., Gislason G. Use of digoxin and risk of death or readmission for heart failure and sinus rhythm: a nationwide propensity score matched study. Int. J. Cardiol. 2016;221:944–950. doi: 10.1016/j.ijcard.2016.07.111. [DOI] [PubMed] [Google Scholar]
- 19.Al-Khateeb M., Qureshi W.T., Odeh R., Ahmed A.M., Sakr S., Elshawi R., Bdeir M.B., Al-Mallah M.H. The impact of digoxin on mortality in patients with chronic systolic heart failure: a propensity-matched cohort study. Int. J. Cardiol. 2017;228:214–218. doi: 10.1016/j.ijcard.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Winnicka K., Bielawski K., Bielawska A., Surazynski A. Antiproliferative activity of derivatives of ouabain, digoxin and proscillaridin A in human MCF-7 and MDA-MB-231 breast cancer cells. Biol. Pharm. Bull. 2008;31:1131–1140. doi: 10.1248/bpb.31.1131. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y.T., Yan J.Y., Han X.C., Niu F.L., Zhang J.H., Hu W.N. Anti-proliferative effect of digoxin on breast cancer cells via inducing apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2017;21:5837–5842. doi: 10.26355/eurrev_201712_14032. [DOI] [PubMed] [Google Scholar]
- 22.Choi E.J., Jung B.J., Lee S.H., Yoo H.S., Shin E.A., Ko H.J., Chang S., Kim S.Y., Jeon S.M. A clinical drug library screen identifies clobetasol propionate as an NRF2 inhibitor with potential therapeutic efficacy in KEAP1 mutant lung cancer. Oncogene. 2017;36:5285–5295. doi: 10.1038/onc.2017.153. [DOI] [PubMed] [Google Scholar]
- 23.Szakacs G., Paterson J.K., Ludwig J.A., Booth-Genthe C., Gottesman M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 24.Tew K.D. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–4320. [PubMed] [Google Scholar]
- 25.Wosikowski K., Schuurhuis D., Kops G.J., Saceda M., Bates S.E. Altered gene expression in drug-resistant human breast cancer cells. Clin. Cancer Res. 1997;3:2405–2414. [PubMed] [Google Scholar]
- 26.Rosell R., Taron M., Ariza A., Barnadas A., Mate J.L., Reguart N., Margel M., Felip E., Mendez P., Garcia-Campelo R. Molecular predictors of response to chemotherapy in lung cancer. Semin. Oncol. 2004;31:20–27. doi: 10.1053/j.seminoncol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 27.No J.H., Kim Y.B., Song Y.S. Targeting nrf2 signaling to combat chemoresistance. J. Cancer Prev. 2014;19:111–117. doi: 10.15430/JCP.2014.19.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao A.M., Ke Z.P., Wang J.N., Yang J.Y., Chen S.Y., Chen H. Apigenin sensitizes doxorubicin-resistant hepatocellular carcinoma BEL-7402/ADM cells to doxorubicin via inhibiting PI3K/Akt/Nrf2 pathway. Carcinogenesis. 2013;34:1806–1814. doi: 10.1093/carcin/bgt108. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Zhang Y., Wu Q., Cui X., Lin Z., Liu S., Chen L. Clinical implications of high NQO1 expression in breast cancers. J. Exp. Clin. Cancer Res. 2014;33:14. doi: 10.1186/1756-9966-33-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y., Kong J., Yan G., Ren X., Jin D., Jin T., Lin L., Lin Z. NQO1 overexpression is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. Bmc Cancer. 2014;14:414. doi: 10.1186/1471-2407-14-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Was H., Cichon T., Smolarczyk R., Rudnicka D., Stopa M., Chevalier C., Leger J.J., Lackowska B., Grochot A., Bojkowska K., Ratajska A., Kieda C., Szala S., Dulak J., Jozkowicz A. Overexpression of heme oxygenase-1 in murine melanoma: increased proliferation and viability of tumor cells, decreased survival of mice. Am. J. Pathol. 2006;169:2181–2198. doi: 10.2353/ajpath.2006.051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimori S., Abe Y., Nishi M., Hamamoto A., Inoue Y., Ohnishi Y., Nishime C., Matsumoto H., Yamazaki H., Kijima H., Ueyama Y., Inoue H., Nakamura M. The subunits of glutamate cysteine ligase enhance cisplatin resistance in human non-small cell lung cancer xenografts in vivo. Int. J. Oncol. 2004;25:413–418. [PubMed] [Google Scholar]
- 33.Wijnholds J., Mol C.A., van Deemter L., de Haas M., Scheffer G.L., Baas F., Beijnen J.H., Scheper R.J., Hatse S., De Clercq E., Balzarini J., Borst P. Multidrug-resistance protein 5 is a multispecific organic anion transporter able to transport nucleotide analogs. Proc. Natl. Acad. Sci. USA. 2000;97:7476–7481. doi: 10.1073/pnas.120159197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oguri T., Achiwa H., Sato S., Bessho Y., Takano Y., Miyazaki M., Muramatsu H., Maeda H., Niimi T., Ueda R. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol. Cancer Ther. 2006;5:1800–1806. doi: 10.1158/1535-7163.MCT-06-0025. [DOI] [PubMed] [Google Scholar]
- 35.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang X., Wang H., Fan L., Wu X., Xin A., Ren H., Wang X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011;50:1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Kweon M.H., Adhami V.M., Lee J.S., Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J. Biol. Chem. 2006;281:33761–33772. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 38.Singh A., Venkannagari S., Oh K.H., Zhang Y.Q., Rohde J.M., Liu L., Nimmagadda S., Sudini K., Brimacombe K.R., Gajghate S., Ma J., Wang A., Xu X., Shahane S.A., Xia M., Woo J., Mensah G.A., Wang Z., Ferrer M., Gabrielson E., Li Z., Rastinejad F., Shen M., Boxer M.B., Biswal S. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem. Biol. 2016;11:3214–3225. doi: 10.1021/acschembio.6b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J., Zhan T., Duffy D., Hoffman-Censits J., Kilpatrick D., Trabulsi E.J., Lallas C.D., Chervoneva I., Limentani K., Kennedy B., Kessler S., Gomella L., Antonarakis E.S., Carducci M.A., Force T., Kelly W.K. A pilot phase II study of digoxin in patients with recurrent prostate cancer as evident by a rising PSA. Am. J. Cancer Ther. Pharmacol. 2014;2:21–32. [PMC free article] [PubMed] [Google Scholar]
- 40.Frankel A.E., Eskiocak U., Gill J.G., Yuan S., Ramesh V., Froehlich T.W., Ahn C., Morrison S.J. Digoxin plus trametinib therapy achieves disease control in BRAF wild-type metastatic melanoma patients. Neoplasia. 2017;19:255–260. doi: 10.1016/j.neo.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gayed B.A., O'Malley K.J., Pilch J., Wang Z. Digoxin inhibits blood vessel density and HIF-1a expression in castration-resistant C4-2 xenograft prostate tumors. Clin. Transl. Sci. 2012;5:39–42. doi: 10.1111/j.1752-8062.2011.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong C.C., Zhang H., Gilkes D.M., Chen J., Wei H., Chaturvedi P., Hubbi M.E., Semenza G.L. Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. 2012;90:803–815. doi: 10.1007/s00109-011-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Qian D.Z., Tan Y.S., Lee K., Gao P., Ren Y.R., Rey S., Hammers H., Chang D., Pili R., Dang C.V., Liu J.O., Semenza G.L. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc. Natl. Acad. Sci. USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei D., Peng J.J., Gao H., Li H., Li D., Tan Y., Zhang T. Digoxin downregulates NDRG1 and VEGF through the inhibition of HIF-1alpha under hypoxic conditions in human lung adenocarcinoma A549 cells. Int. J. Mol. Sci. 2013;14:7273–7285. doi: 10.3390/ijms14047273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowa T., Menju T., Chen-Yoshikawa T.F., Takahashi K., Nishikawa S., Nakanishi T., Shikuma K., Motoyama H., Hijiya K., Aoyama A., Sato T., Sonobe M., Harada H., Date H. Hypoxia-inducible factor 1 promotes chemoresistance of lung cancer by inducing carbonic anhydrase IX expression. Cancer Med. 2017;6:288–297. doi: 10.1002/cam4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y., Guan Z., Liang L., Cheng Y., Zhou J., Li J., Xu Y. HIF-1alpha/MDR1 pathway confers chemoresistance to cisplatin in bladder cancer. Oncol. Rep. 2016;35:1549–1556. doi: 10.3892/or.2015.4536. [DOI] [PubMed] [Google Scholar]
- 47.Chen J., Ding Z., Peng Y., Pan F., Li J., Zou L., Zhang Y., Liang H. HIF-1alpha inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PLoS One. 2014;9:e98882. doi: 10.1371/journal.pone.0098882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J.Y., Kim M.S., Lee M.S., Ju J.E., Chung N., Jeong Y.K. Digoxin enhances radiation response in radioresistant A549 cells by reducing protein phosphatase 2A. Biosci. Rep. 2017;37 doi: 10.1042/BSR20171257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMahon M., Itoh K., Yamamoto M., Hayes J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y.J., Jeong H.Y., Kim Y.B., Lee Y.J., Won S.Y., Shim J.H., Cho M.K., Nam H.S., Lee S.H. Reactive oxygen species and PI3K/Akt signaling play key roles in the induction of Nrf2-driven heme oxygenase-1 expression in sulforaphane-treated human mesothelioma MSTO-211H cells. Food Chem. Toxicol. 2012;50:116–123. doi: 10.1016/j.fct.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 51.Lee D.S., Jeong G.S. Butein provides neuroprotective and anti-neuroinflammatory effects through Nrf2/ARE-dependent haem oxygenase 1 expression by activating the PI3K/Akt pathway. Br. J. Pharmacol. 2016;173:2894–2909. doi: 10.1111/bph.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J., Zhang L., Zhang Y., Luo M., Wu Q., Yu L., Chu H. Transcriptional upregulation centra of HO-1 by EGB via the MAPKs/Nrf2 pathway in mouse C2C12 myoblasts. Toxicol. Vitr. 2015;29:380–388. doi: 10.1016/j.tiv.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 53.Ma B., Wang J., Tong J., Zhou G., Chen Y., He J., Wang Y. Protective effects of Chaenomeles thibetica extract against carbon tetrachloride-induced damage via the MAPK/Nrf2 pathway. Food Funct. 2016;7:1492–1500. doi: 10.1039/c5fo01430a. [DOI] [PubMed] [Google Scholar]
- 54.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen T., Huang H.C., Pickett C.B. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 56.Ikeda Y., Sugawara A., Taniyama Y., Uruno A., Igarashi K., Arima S., Ito S., Takeuchi K. Suppression of rat thromboxane synthase gene transcription by peroxisome proliferator-activated receptor gamma in macrophages via an interaction with NRF2. J. Biol. Chem. 2000;275:33142–33150. doi: 10.1074/jbc.M002319200. [DOI] [PubMed] [Google Scholar]
- 57.Ansell P.J., Lo S.C., Newton L.G., Espinosa-Nicholas C., Zhang D.D., Liu J.H., Hannink M., Lubahn D.B. Repression of cancer protective genes by 17beta-estradiol: ligand-dependent interaction between human Nrf2 and estrogen receptor alpha. Mol. Cell. Endocrinol. 2005;243:27–34. doi: 10.1016/j.mce.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Brown S.L., Sekhar K.R., Rachakonda G., Sasi S., Freeman M.L. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res. 2008;68:364–368. doi: 10.1158/0008-5472.CAN-07-2170. [DOI] [PubMed] [Google Scholar]
- 59.Zhou W., Lo S.C., Liu J.H., Hannink M., Lubahn D.B. ERRbeta: a potent inhibitor of Nrf2 transcriptional activity. Mol. Cell. Endocrinol. 2007;278:52–62. doi: 10.1016/j.mce.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 60.Wang X.J., Hayes J.D., Henderson C.J., Wolf C.R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc. Natl. Acad. Sci. USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu G.H., Qu J., Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim. Biophys. Acta. 1783;2008:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]