Abstract
Previous research has linked high mobility group box 3 (HMGB3) overexpression to the malignant progression and poor prognosis of non-small cell lung cancer (NSCLC). The present study investigated the role of HMGB3 in cell survival and colony formation of NSCLC cells. Stable knockdown of HMGB3 in A549 cells was achieved by lentiviral-based shRNA interference and verified by detection of the transcriptional and translational level of HMGB3 with reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. The influence of HMGB3 knockdown on A549 cell viability and apoptotic rate was evaluated by Cell Counting Kit-8 assay and flow cytometry following annexin V staining, respectively. The proliferative capacity of A549 cells with or without HMGB3 knockdown was compared by measuring their colony forming efficiency. The results of the current study revealed that HMGB3 knockdown significantly reduced cell viability and colony forming efficiency while promoting apoptosis in A549 cells, indicating that HMGB3 may be pivotal for the survival and colony formation of A549 cells, serving a notable role in NSCLC progression.
Keywords: high mobility group box 3, non-small cell lung cancer, A549, cell viability, apoptosis, colony forming efficiency
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide, with 80–90% of all lung cancer patients diagnosed with non-small cell lung cancer (NSCLC) (1–3). Great progress has been made in understanding the carcinogenesis of NSCLC; however, owing to the heterogeneity and genetic instability of NSCLC cells, therapeutic methods that are being used in the clinic, including chemotherapy, radiation therapy or targeted therapy such as epithelial growth factor receptor (EGFR)-tyrosine kinase inhibitors, often exhibit short-lived treatment response (4). Novel therapeutic targets or methods are therefore required.
A previous study revealed that high mobility group box 3 (HMGB3) overexpression is an independent risk factor for NSCLC progression, lymph node metastasis and poor survival rates of patients (5), indicating the prognostic value and the oncogenic role of HMGB3 overexpression in NSCLC development. HMGB3 is a less studied member in the evolutionarily conserved HMGB protein family, alongside HMGB1 and HMGB2 (6,7). Initially reported to be a modulator involved in tissue development (8–10) and hematopoietic stem cell differentiation (11–13), HMGB3 has also been linked to cancer development by certain studies (14–19); however, its influence on in the cell biological behaviors of NSCLC cells remains unrevealed.
The present study investigated the role of HMGB3 on the cell viability and colony forming ability of NSCLC cells by constructing an A549 cell line that was stably knocked down for HMGB3 expression using a lentiviral short hairpin RNA (shRNA) interference technique. The data generated by the present study clearly demonstrated that HMGB3 is necessary for A549 cell survival and colony formation, indicating its role in NSCLC progression and its potential as a novel therapeutic target.
Materials and methods
Cell culture
The human NSCLC A549 cell line was a gift from Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). The human NSCLC H1299 cell line was purchased from Chinese Academy of Sciences Cell Bank (Shanghai, China). A549 and H1299 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, and 100 U/ml of penicillin and streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), in a humidified incubator at 37°C with 5% CO2. Cells in the logarithmic growth phase were subject to each experiment at a density of 0.5–1×106/ml.
HMGB3 knockdown in NSCLC cells by shRNA transfection
Lentiviral-based shRNA vectors (containing puromycin and enhanced green fluorescent protein) were constructed by Genecopoeia, Inc. (Rockville, MD, USA). Specifically, three shRNAs with different target sequence in the HMGB3 coding DNA sequence and one non-targeting shRNA as negative control were involved in this experiment (referred to as shHMGB3-1, −2 and −3, and shNC, respectively). The stable knockdown of HMGB3 in NSCLC cells (A549 and H1299 cells) was performed by shRNA transfection using an EndoFectin transfection (Genecopoeia, Inc.) following manufacturer's protocol. Transfection with HMGB3 (250 µg/ml) was conducted for 24 h by EndoFectin transfection. Subsequent experiments were performed after transfection 36 h. Additionally, positive transfection was determined by detecting the green fluorescence under fluorescent microscope. The efficiency of HMGB3 knockdown in each cell line transfected with different shRNAs was evaluated by measuring the mRNA and protein expression level of HMGB3 by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting, respectively.
Cell viability, apoptosis and colony formation assay
The viability (at 1, 12 and 24 h) of NSCLC cells with or without HMGB3 knockdown was compared using Cell Counting Kit-8 (Beyotime Institute of Biotechnology, Shanghai, China), following the manufacturer's protocol.
Apoptosis of NSCLC cells with or without HMGB3 knockdown was compared by flow cytometry using a Attune NxT flow cytometer (Thermo Fisher Scientific, Inc.) and FlowJo software (version 7.6; FlowJo LLC, Ashland, OR, USA) following staining with the Annexin V-EGFP Apoptosis Detection kit (Beyotime Institute of Biotechnology) following the manufacturer's protocol. NSCLC cells with or without HMGB3 knockdown were cultured to logarithmic phase, followed by 0.25% trypsin treatment and pipetting to make single cell suspension in complete culture medium.
Approximately 200 cells in each experimental group were seeded in 10 ml RPMI-1640 in a cell culture dish and maintained in a humidified incubator at 37°C with 5% CO2 for 2–3 weeks until macroscopic colonies formed. Following removal of the culture medium, cells in each culture dish were fixed with 4% paraformaldehyde solution (room temperature) for 15 min followed by incubation with 1% crystal violet staining solution (20 min at room temperature). The number of colonies of >50 cells were counted under low-magnification light microscope (×100), and the colony forming efficiency of each experimental group was calculated with the following formula: Clonal formation rate=number of clones formed/number of inoculated cells ×100%.
RT-qPCR
Total RNA was isolated by TRIzol®, chloroform and isopropanol (Takara Bio, Inc., Otsu, Japan) extraction. Human HMGB3 and GAPDH mRNA expression in cells of each experimental group was evaluated by RT-qPCR using a custom-made RT-qPCR kit (cat. no. QP001) purchased from Genecopoeia, Inc., following the manufacturer's instructions. Briefly, poly-A RNAs in the cell lysate in each experimental group were first reversely transcribed to cDNAs, and HMGB3 and GAPDH cDNAs were subject to amplification by qPCR monitored by SYBRGreen fluorescent dye. GAPDH was used as reference gene for HMGB3 quantification. HMGB3 expression levels were normalized to levels of GAPDH using the 2−ΔΔCq method (20). Primers for HMGB3 and GAPDH cDNA were as follows: HMGB3 forward, 5′-GACCAGCTAAGGGAGGCAA-3′ and reverse 5′-ACAGGAAGAATCCAGACGGT-3′; and GAPDH forward, 5′-CCTGCACCACCAACTGCTTA-3′ and reverse, 5′-GGCCATCCACAGTCTTCTGAG-3′. Thermocycling conditions for amplifying HMGB3 and GAPDH cDNAs were as follows: Initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation 95°C for 10 sec, 55°C for 20 sec and 72°C for 15 sec.
Western blot analysis
Anti-HMG4 (cat. no. ab201569), anti-β-actin (cat. no. ab6276) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG (cat. no. ab6728) were purchased from Abcam (Cambridge, UK). Antibodies against Bax (cat. no. D2E11), Bcl-2 (cat. no. D55G8), cleaved caspase-9 (cat. no. D2D4), cleaved caspase-8 (cat. no. 11G10) and anti-biotin (D5A7) rabbit mAb (HRP conjugate; cat. no. 5571S) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibodies were applied in western blotting analysis following manufacturer's instructions with minor modifications. Cells were homogenized in lysis buffer [10 mM Tris base, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1% Triton X-100; 0.5% NP-40; and protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Inc.)]. Protein concentrations were determined using a BCA assay kit (Thermo Fisher Scientific, Inc.) with bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.) as a standard. Briefly, following separation of proteins (20 µl/cell group) in cell lysates by SDS-PAGE (15%) and transferred onto nitrocellulose membranes. Following blocking at 4°C overnight with non-fat milk and preparation with PBS, the membranes were incubated for 1 h at room temperature with specific primary antibodies diluted in bovine serum albumin-dissolved Tris-buffered saline diluent with final dilution concentration of 1:1,000; secondary antibodies were diluted in the same diluent at 1:10,000 ratio to probe the specific primary antibodies by incubation for 30 min at room temperature and colorized using an enhanced chemiluminescent substrate (Thermo Fisher Scientific, Inc.). Quantified grayscale analysis of western blotting result was performed using ImageJ software (version 1.48, National Institutes of Health, Bethesda, MD, USA); β-actin was used as loading control for each analysis.
Statistical analysis
According to the recommended protocol by Szklarczyk et al (21), the proteins interacting with HMGB3 were investigated using the STRING online database (search term, HMGB3; https://string-db.org/cgi/input.pl; version 10.5), with the confidence score set at 0.4. HMGB3-associated protein analysis was conducted through Gene ontology (GO) enrichment analysis (http://www.geneontology.org/). Statistical analysis was performed using SPSS 17 software (SPSS, Inc., Chicago, IL, USA). Quantitative data were presented as mean ± standard deviation and analyzed by analysis of one-way analysis o variance. Comparison between experimental groups and control group in each graph was performed with Dunnett's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
HMGB3 knockdown in NSCLC cells using shRNA
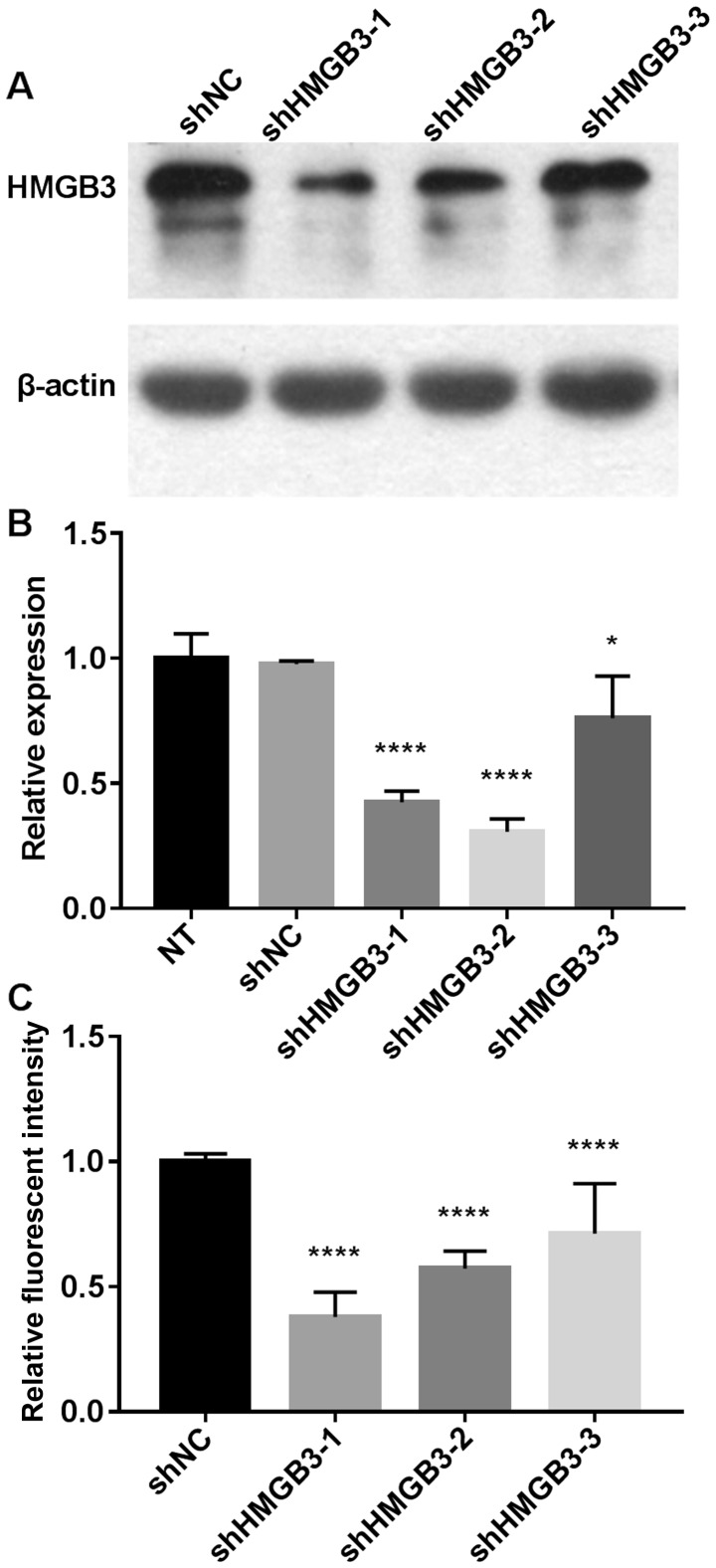
To investigate the role of HMGB3 on the tumor biology of human NSCLC A549 and H1299 cell lines with stable knockdown of HMGB3 were constructed using different shRNAs (Fig. 1). The data revealed that A549 cells transfected with shHMGB3-1 and shHMGB3-2 exhibited a >50% decrease in HMGB3 expression at the transcriptional and translational level (P<0.0001), compared with non-transfected control cells (NT) or cells transfected with non-targeting shRNA control (shNC). A549 cells transfected with shHMGB3-3 exhibited a moderate decrease in HMGB3 expression, whereas those transfected with shHMGB3-1 exhibited the lowest HMGB3 protein expression. It was therefore decided to use only the A549 cell line with HMGB3 knockdown achieved using shHMGB3-1 (referred to as shHMGB3 hereafter) for further experimentation. A similar degree of HMGB3 knockdown using shHMGB3-1 was also achieved in H1299 NSCLC cells (data not shown).
Figure 1.
HMGB3 knockdown in A549 cells by shRNA transfection. (A) Western blotting result demonstrating that transfection with shHMGB3-1 or shHMGB3-2 significantly decreased the protein expression level of HMGB3 in A549 cells. (B) Reverse transcription-quantitative polymerase chain reaction result showing that transfection with shHMGB3-1 or shHMGB3-2 significantly decreased HMGB3 mRNA expression level in A549 cells. The mRNA expression level in each transfection group was normalized to that in NT cells. (C) Fluorescent intensity analysis of the western blotting result. The fluorescent intensity of the HMGB3 band in each shHMGB3 transfection group was normalized to that in control cells transfected with non-targeting shRNA (shNC). Data in panel (B) and (C) represents three independent repeats. *P<0.05; ****P<0.0001. shRNA, short hairpin RNA; HMGB3, high mobility group box 3; NT, non-transfected control; shHMGB3, shRNA targeting HMGB3.
Decrease in cell viability and increase in apoptosis of NSCLC cells following HMGB3 knockdown
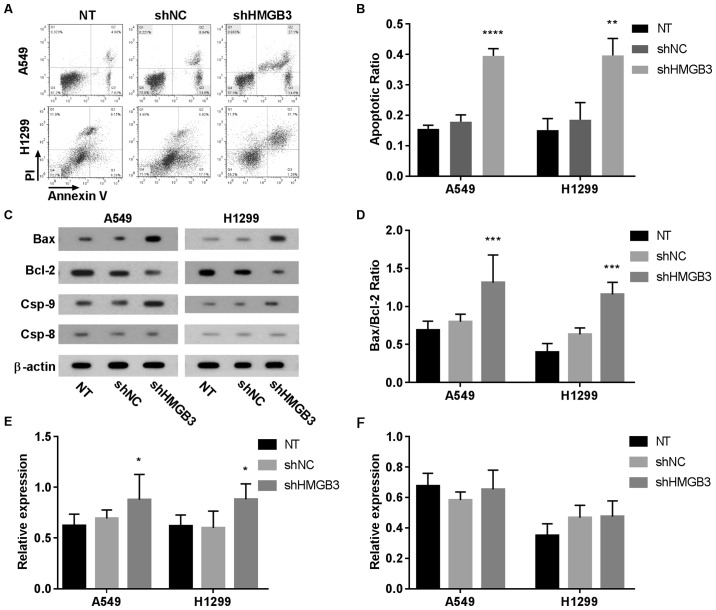
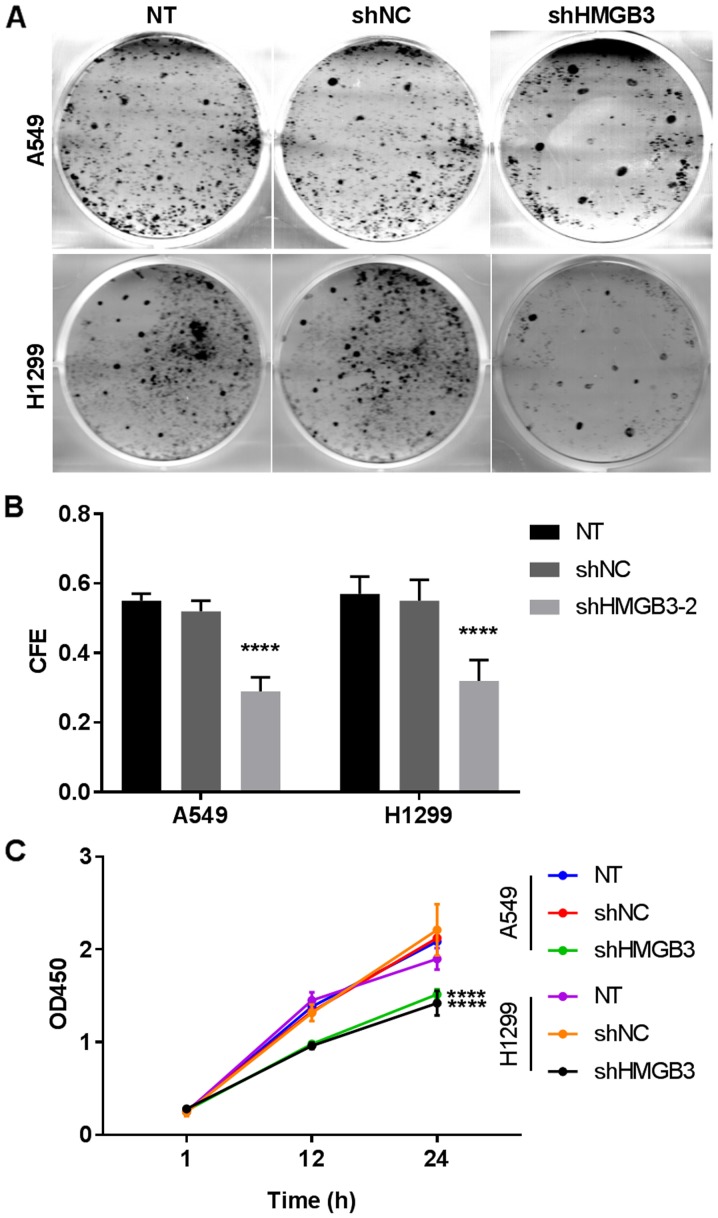
The role of HMGB3 in the self-maintenance of NSCLC cells was assessed by comparing the viability and apoptosis of A549 or H1299 cells with or without HMGB3 knockdown. The flow cytometry data revealed a >2-fold increase in cell apoptosis following HMGB3 knockdown in A549 (P<0.0001) or H1299 (P<0.01) cells (Fig. 2A and B. These results indicated that HMGB3 may be vital for the in vitro survival of A549 or H1299 cells. To investigate the apoptotic inducing mechanism of HMGB3 knockdown in A549 or H1299 cells, the expression levels or activation status of certain apoptosis-associated proteins, namely Bax, Bcl-2, caspase-9 and caspase-8, under different HMGB3 expression conditions by western blotting (Fig. 2C-F). These data clearly indicated that HMGB3 knockdown significantly increased the Bax/Bcl-2 ratio by nearly 2-fold in A549 or H1299 cells (P<0.001), which has been suggested as a key step during the initiation of apoptosis; HMGB3 knockdown also increased caspase-9 activation in A549 and H1299 cells by ~40% (P<0.05) compared with NT, as evidenced by the increased caspase-9 cleavage, but not that of caspase-8. Considering that caspase-9 and caspase-8 belong to the intrinsic and extrinsic apoptosis pathway, respectively, these data indicated that HMGB3 knockdown primarily activates the intrinsic apoptosis pathway in NSCLC cells in vitro. To investigate the influence of HMGB3 on the colony formation capacity of NSCLC cells, the colony-forming efficiency of A549 or H1299 cells with or without HMGB3 knockdown was assessed (Fig. 3A and B). The data revealed that the colony forming efficiency of A549 or H1299 cells decreased by >40% after HMGB3 knockdown (P<0.0001), indicating that it serves a notable role in the colony formation of NSCLC cells. The results of the CCK-8 assay exhibited a decrease in cell viability of almost 50% (P<0.0001) in the two HMGB3-knockdown cell lines, compared with the NT (Fig. 3C). Cell viability of A549 or H1299 was decreased following HMGB3 knockdown.
Figure 2.
Increasing apoptosis in A549 or H1299 NSCLC cells following HMGB3 knockdown. (A) Representative flow cytometry data showing an increased population of apoptotic cells in A549 or H1299 cells following HMGB3 knockdown. Annexin-V positively stained cells were considered apoptotic cells. (B) Percentage of apoptotic cells was increased in A549 OR H1299 cells following HMGB3 knockdown. (C) Western blotting detecting the expression level of apoptosis-associated proteins in A549 or H1299 cells with different treatment. (D) Bax over Bcl-2 ratio was increased by HMGB3 knockdown in A549 or H1299 cells. (E and F), increased (E) caspase-9 activation and (F) unchanged caspase-8 activation in A549 or H1299 cells following HMGB3 knockdown. Western blotting data analysis was performed by comparing the gray scale of protein bands revealed by western blotting following normalization to β-actin. Data in panel (B) and (D-F) each represents at least 3 independent repeats. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. shRNA, short hairpin RNA; HMGB3, high mobility group box 3; NT, non-transfected control; shHMGB3, shRNA targeting HMGB3; shNC, negative control shRNA.
Figure 3.
Colony forming ability and cell viability of non-small cell lung cancer cells were decreased following HMGB3 knockdown. (A) Typical result of colony forming assay showing a decreased colony forming ability of A549 or H1299 cells following HMGB3 knockdown. (B) The CFE of A549 cells was decreased following HMGB3 knockdown. (C) Cell viability of A549 or H1299 was decreased following HMGB3 knockdown. Data in (B and C) are from three independent repeats. Statistical significance test was performed on data obtained on the last time point. ****P<0.0001. shRNA, short hairpin RNA; HMGB3, high mobility group box 3; NT, non-transfected control; shHMGB3, shRNA targeting HMGB3; shNC, negative control shRNA; OD, optical density; CFE, colony formation efficiency.
HMGB3 facilitates NSCLC survival in vitro possibly by interacting with FACT complex protein SSRP1 and SUPT16H
To investigate the possible mechanism underlying the supporting effect of HMGB3 on NSCLC survival, the proteins that have been suggested to interact with HMGB3 were investigated using the STRING online database. There were 20 proteins retrieved with a confidence score >0.4 who have either been experimentally confirmed or presumed by text mining to interact with HMGB3 (Fig. 4A). GO enrichment analysis revealed that the majority of these proteins were involved in DNA replication, repair and regulation of mRNA translation, although a small portion of these proteins belonged to the Toll-like receptor signaling pathways (Fig. 4B; Table I). Notably, among these proteins FACT complex subunit SPT16 (SUPT16H) and structure specific recognition protein 1 (SSRP1) exhibited the highest confidence score. These two proteins are the two subunits of facilitates chromatin transcription (FACT) complex, which has been confirmed to be frequently upregulated in different types of malignant cancer. These findings indicate that HMGB3 may facilitate NSCLC survival by interacting with FACT complex proteins SUPT16H and SSRP1 or other proteins involved in the regulation of DNA metabolism.
Figure 4.
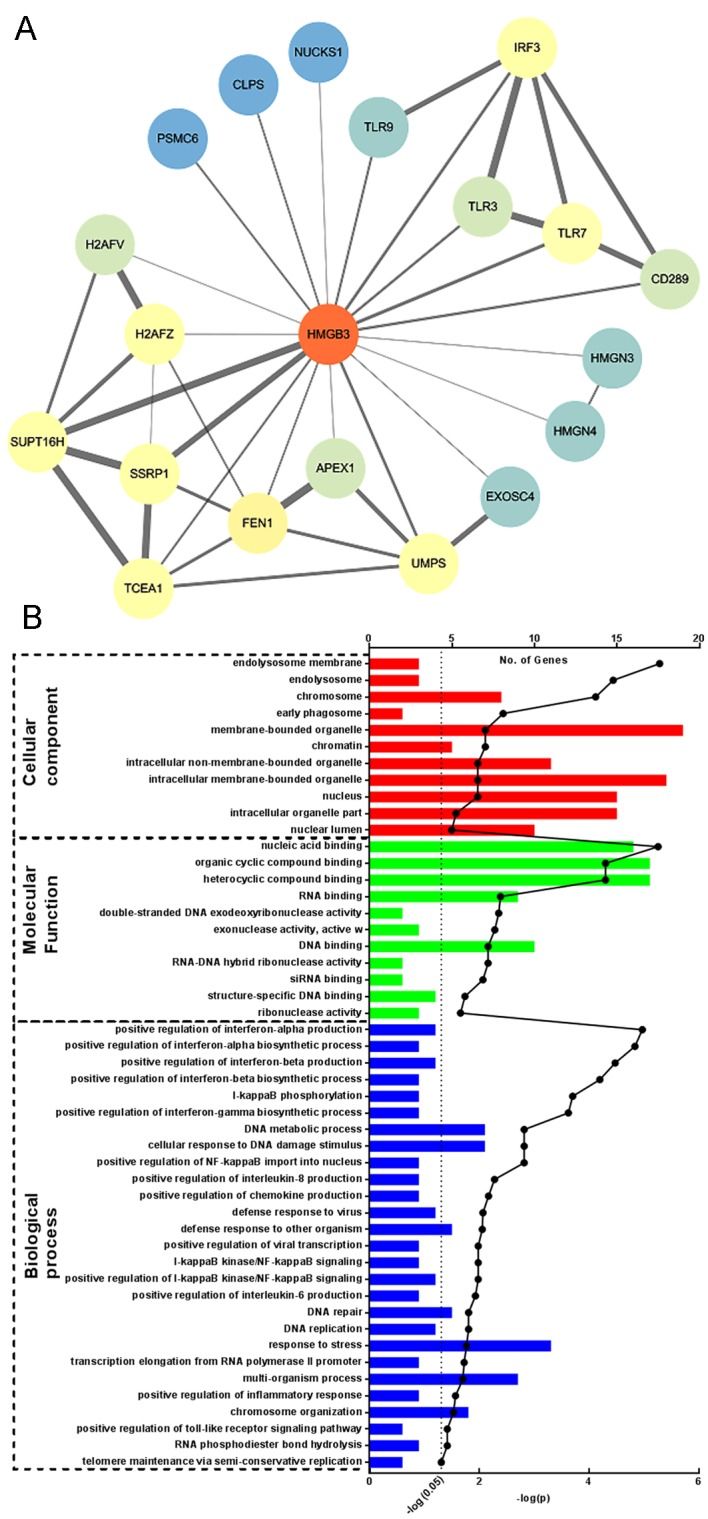
Investigation on proteins potentially interacting with HMGB3. (A) Proteins that may interact with HMGB3 were obtained using STRING online database with the confidence score set at 0.4; each node represent a protein, and each edge between two nodes represents the protein-protein interaction. The width of each edge represents the confidence score of that protein-protein interaction. (B) GO annotation of predicted HMGB3 interacting proteins; number of genes enriched to each GO term was plotted to the upper x-axis in box-plot fashion, and the -log2 (p) of each annotated term was plotted to the lower x-axis in dotted-linear graph fashion. HMGB3, high mobility group box 3; GO, Gene Ontology.
Table I.
Gene Ontology enrichment analysis of predicted HMGB3 interacting proteins.
| Category | Description | Enriched genes | FDR P-value |
|---|---|---|---|
| Cellular component | Endolysosome membrane | TLR3, TLR9, TLR7 | 5.23×10−6 |
| Endolysosome | TLR3, TLR9, TLR7 | 3.65×10−5 | |
| Chromosome | SUPT16H, SSRP1, H2AFZ, H2AFV, EXOSC4, HMGN4 | 7.63×10−5 | |
| HMGN3, HMGB3 | |||
| Early phagosome | TLR9, TLR7 | 0.00371 | |
| Membrane-bounded organelle | SUPT16H, APEX1, UMPS, CLPS, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, TLR9, NUCKS1, HMGB3, TLR7, PSMC6, TCEA1 | 0.00785 | |
| Chromatin | H2AFZ, H2AFV, EXOSC4, HMGN4, HMGN3 | 0.00785 | |
| Intracellular non-membrane-bounded organelle | SUPT16H, APEX1, SSRP1, H2AFZ, FEN1, H2AFV, EXOSC4, HMGN4, HMGN3, HMGB3, TCEA1 | 0.0108 | |
| Intracellular membrane-bounded organelle | SUPT16H, APEX1, UMPS, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, TLR9, NUCKS1, HMGB3, TLR7, PSMC6, TCEA1 | 0.0108 | |
| Nucleus | SUPT16H, APEX1, UMPS, SSRP1, H2AFZ, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, NUCKS1, HMGB3, PSMC6, TCEA1 | 0.0108 | |
| Intracellular organelle part | SUPT16H, APEX1, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, TLR9, TLR7, PSMC6, TCEA1 | 0.0268 | |
| Nuclear lumen | SUPT16H, APEX1, SSRP1, H2AFZ, FEN1, IRF3, EXOSC4, HMGN3, PSMC6, TCEA1 | 0.0321 | |
| Molecular | Nucleic acid binding function | SUPT16H, APEX1, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, TLR9, NUCKS1, HMGB3, TLR7, TCEA1, | 5.57×10−6 |
| Organic cyclic compound binding | SUPT16H, APEX1, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3, EXOSC4, HMGN4, HMGN3, TLR9, NUCKS1, HMGB3, TLR7, PSMC6, TCEA1 | 5.04×10−5 | |
| Heterocyclic compound binding | SUPT16H, APEX1, SSRP1, H2AFZ, TLR3, FEN1, H2AFV, IRF3 EXOSC4, HMGN4, HMGN3, TLR9, NUCKS1, HMGB3, TLR7, PSMC6, TCEA1 | 5.04×10−5 | |
| RNA binding | SUPT16H, APEX1, SSRP1, TLR3, EXOSC4, TLR9, NUCKS1 | 0.00416 | |
| HMGB3, TLR7 | |||
| Double-stranded DNA exodeoxyribonuclease activity | APEX1, FEN1 | 0.00448 | |
| Exonuclease activity, active with either ribo-or deoxyribonucleic acids and producing 5′-phosphomonoesters | APEX1, FEN1, EXOSC4 | 0.00529 | |
| DNA binding | APEX1, SSRP1, H2AFZ, FEN1, H2AFV, IRF3 | 0.00699 | |
| HMGN4, HMGN3, HMGB3, TCEA1 | |||
| RNA-DNA hybrid ribonuclease activity | APEX1, FEN1 | 0.00699 | |
| siRNA binding | TLR9, TLR7 | 0.00869 | |
| Structure-specific DNA binding | APEX1, H2AFZ, FEN1, HMGB3 | 0.0185 | |
| Ribonuclease activity | APEX1, FEN1, EXOSC4 | 0.0223 | |
| Biological process | Positive regulation of interferon-alpha production | TLR3, IRF3, TLR9, TLR7 | 1.08×10−5 |
| Positive regulation of interferon-α biosynthetic process | TLR3, TLR9, TLR7 | 1.47×10−5 | |
| Positive regulation of interferon-β production | TLR3, IRF3, TLR9, TLR7 | 3.38×10−5 | |
| Positive regulation of interferon-β biosynthetic process | TLR3, TLR9, TLR7 | 6.41×10−5 | |
| I-κB phosphorylation | TLR3, TLR9, TLR7 | 2.01×10−4 | |
| Positive regulation of interferon-γ biosynthetic process | TLR3, TLR9, TLR7 | 2.41×10−4 | |
| DNA metabolic process | SUPT16H, APEX1, SSRP1, FEN1, EXOSC4, HMGB3, TCEA1 | 0.00154 | |
| Cellular response to DNA damage stimulus | SUPT16H, APEX1, SSRP1, FEN1, IRF3, PSMC6, TCEA1 | 0.00154 | |
| Positive regulation of NF-κB import into nucleus | TLR3, TLR9, TLR7 | 0.00154 | |
| Positive regulation of interleukin-8 production | TLR3, TLR9, TLR7 | 0.00535 | |
| Positive regulation of chemokine production | TLR3, TLR9, TLR7 | 0.00686 | |
| Defense response to virus | TLR3, IRF3, EXOSC4, TLR7 | 0.00869 | |
| Defense response to other organism | TLR3, IRF3, EXOSC4, TLR9, TLR7 | 0.00895 | |
| Positive regulation of viral transcription | SUPT16H, SSRP1, TCEA1 | 0.0106 | |
| I-κB kinase/NF-κB signaling | TLR3, TLR9, TLR7 | 0.0106 | |
| Positive regulation of I-κB kinase/NF-κB signaling | TLR3, IRF3, TLR9, TLR7 | 0.0106 | |
| Positive regulation of interleukin-6 production | TLR3, TLR9, TLR7 | 0.0119 | |
| DNA repair | SUPT16H, APEX1, SSRP1, FEN1, TCEA1 | 0.0159 | |
| DNA replication | SUPT16H, APEX1, SSRP1, FEN1, | 0.0159 | |
| Response to stress | SUPT16H, APEX1, SSRP1, TLR3, FEN1, IRF3, EXOSC4, TLR9, TLR7, PSMC6, TCEA1 | 0.0173 | |
| Transcription elongation from RNA polymerase II promoter | SUPT16H, SSRP1, TCEA1 | 0.0191 | |
| Multi-organism process | SUPT16H, UMPS, SSRP1, TLR3, EXOSC4, TLR9, TLR7, PSMC6, TCEA1 | 0.0202 | |
| Positive regulation of inflammatory response | TLR3, TLR9, TLR7 | 0.0275 | |
| Chromosome organization | SUPT16H, APEX1, SSRP1, FEN1, HMGN3, HMGB3 | 0.0298 | |
| Positive regulation of Toll-like receptor signaling pathway | TLR3, TLR9 | 0.0387 | |
| RNA phosphodiester bond hydrolysis | APEX1, FEN1, EXOSC4 | 0.0388 | |
| Telomere maintenance via semi-conservative replication | APEX1, FEN1 | 0.05 |
HMGB3, high mobility group box 3.
Discussion
Several members of the HMGB family have been found to promote self-renewal and colony formation in stem cells as well as in cancer cells (22–28), indicating the presence of an association between these two cell types; however, the detailed molecular mechanisms underlying their cancer-promoting activity remain obscure. HMGB3 belongs to the HMGB family, members of which are proposed to regulate gene transcription by affecting DNA structure and influencing transcription complex formation or by altering gene availability (29). On the basis of previous results, which found that HMGB3 overexpression was closely associated with to the development of NSCLC, this protein was selected for further investigation.
The present study constructed lentiviral plasmids encoding three different shRNAs with distinct target sequences picked from the coding sequence of human HMGB3 gene. Verified by RT-qPCR and western blotting, shRNA3-1 and shRNA3-2 exhibited superior suppression of HMGB3 expression than shHMGB3-3, so these two shRNAs were used to construct stable HMGB3-knockdown A549 cell lines. The effect of HMGB3 knockdown on the cell viability and apoptosis was then assessed in A549 and H1299 NSCLC cells. The CCK-8 assay result revealed that HMGB3 knockdown could significantly decrease A549 or H1299 cell viability, whereas annexin V staining followed by flow cytometry analysis demonstrated that there was an increase in the apoptosis rate of A549 or H1299 cells following HMGB3 knockdown, indicating that HMGB3 is involved in the in vitro survival of NSCLC cells. Western blot analysis further indicated that HMGB3 knockdown induces apoptosis in A549 and H1299 cells, likely by activating the intrinsic apoptosis pathway. A clonogenic assay is a widely used technique for evaluating the influence of different experimental conditions, including compound treatment, UV-irradiation or gene manipulation, on the reproductive viability (or colony-forming ability) of adherent cells (30); therefore, this technique was used to assess the influence of HMGB3 knockdown on the colony formation of A549 or H1299 cells. The data from this assay revealed that HMGB3 knockdown significantly decreased the colony-forming efficiency of these two NSCLC cells. Collectively, these data indicated that HMGB3 has a pivotal role in NSCLC cell survival and colony formation.
Oncogenesis is the result of uncontrolled cell proliferation led by multiple gene deregulation, including the abnormal activation of oncogenes and deactivation of tumor-suppressor genes that are involved in cancer initiation, local infiltration and metastasis (31). HMG proteins are the second most abundant chromatin proteins, serving notable roles in regulating chromatin structure and gene expression (32). HMG proteins can be sub-grouped into HMGA, HMGB and HMGN super-families; among which the HMGB family proteins contain two HMG-box DNA binding motifs that mediate specific non-sequential DNA binding and has been identified in different DNA-binding proteins, including transcriptions factors (33). Besides, the HMG-box motif has also been found to involve in protein-protein interactions. HMGB3 is a member of the HMGB super-family (33). Genecards records show that HMGB3 expression is extremely low in lung, liver, brain and urinary tissues compared with that in other tissues and is upregulated in the liver cancer HuH-7 and HepG2 cell lines, as well as in the NSCLC A549 cell line and in 293 cells (https://www.genecards.org/). Initial study on HMGB3 revealed that it participated in regulating the balance between self-renewal and differentiation in hematopoietic stem cells (11). The oncogenic potential and prognostic value of HMGB3 in different cancer types has also been appreciated (34,35).
The mechanisms of HMGB3 overexpression and promoting cancer development remain unclear. A study conducted by Zhang et al (35) proposed that the WNT/β-catenin pathway, a known oncogenic signaling pathway, could be activated by HMGB3 overexpression. The results of bioinformatics analysis performed in the present study by retrieving and analyzing HMGB3-interacting proteins using the STRING online database indicated that HMGB3 may interact with the FACT complex proteins SUPT16H and SSRP1. Previous studies have suggested that the FACT complex can have an oncogenic role in multiple types of malignant cancer. For instance, Garcia et al (36) suggested that expression of FACT complex proteins was associated with cancer cells' poor differentiation and stemness. Dinant et al (37) reported that SUPT16H facilitates the repair of DNA damage induced by UV-irradiation and promote mRNA restart by increasing chromatin dynamics. Dermawan et al (38) further demonstrated that the pharmaceutical inhibition of FACT complex enhanced the anticancer effect of an EGFR/human epidermal growth factor receptor 2 inhibitor on glioblastoma cells. On the basis of these results, we hypothesize that HMGB3 facilitates NSCLC cell survival and development, presumably by interacting with and increasing the function of FACT complex proteins SUPT16H and SSRP1. However, this hypothesis requires further investigation in future.
Collectively, the results of the present study provided evidence of the molecular function of HMGB3 in the initiation and development of NSCLC, contributing to research into the molecular mechanism of its cellular features and the development of an early diagnostic method or targeted therapeutic agents.
Acknowledgements
Not applicable.
Funding
Hebei medical science research project 2015 Key scientific and technological research plan (grant no. 20150210).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NS wrote the manuscript. BW and GF participated in cell culture and cell transfection. LD and SY participated in cell viability, apoptosis and colony formation assay. WJ and YL participated in RT-qPCR assay and western blot. NS participated in statistical analysis and revised the manuscript. All authors read and approved the final the manuscript.
Ethics approval and consent to publish
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Meza R, Meernik C, Jeon J, Cote ML. Lung cancer incidence trends by gender, race and histology in the United States, 1973–2010. PLoS One. 2015;10:e0121323. doi: 10.1371/journal.pone.0121323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Riaz SP, Lüchtenborg M, Coupland VH, Spicer J, Peake MD, Møller H. Trends in incidence of small cell lung cancer and all lung cancer. Lung Cancer. 2012;75:280–284. doi: 10.1016/j.lungcan.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song N, Liu B, Wu JL, Zhang RF, Duan L, He WS, Zhang CM. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34:2599–2603. doi: 10.1007/s13277-013-0807-y. [DOI] [PubMed] [Google Scholar]
- 6.Barreiro-Alonso A, Lamas-Maceiras M, Rodriguez-Belmonte E, Vizoso-Vázquez Á, Quindós M, Cerdán ME. High mobility group B proteins, their partners, and other redox sensors in ovarian and prostate cancer. Oxid Med Cell Longev. 2016;2016:5845061. doi: 10.1155/2016/5845061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst) 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Oka T, Sasakawa T, Komori N, Miyamoto K, Suzuki I, Sassa T, Natori Y. Developmental changes in the expression of HMG 2a protein. FEBS Lett. 1993;316:20–22. doi: 10.1016/0014-5793(93)81728-I. [DOI] [PubMed] [Google Scholar]
- 9.Oka T, Endo Y, Ito M, Miyamoto K, Sasakawa T, Suzuki I, Natori Y. Molecular cloning of chick liver HMG 2a cDNA and developmental expression of HMG 2a mRNA. Biochim Biophys Acta. 1992;1130:224–226. doi: 10.1016/0167-4781(92)90534-7. [DOI] [PubMed] [Google Scholar]
- 10.Gordon JS, Rosenfeld BI, Kaufman R, Williams DL. Evidence for a quantitative tissue-specific distribution of high mobility group chromosomal proteins. Biochemistry. 1980;19:4395–4402. doi: 10.1021/bi00560a003. [DOI] [PubMed] [Google Scholar]
- 11.Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci USA. 2006;103:13783–13788. doi: 10.1073/pnas.0604006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood. 2005;105:627–634. doi: 10.1182/blood-2004-07-2551. [DOI] [PubMed] [Google Scholar]
- 13.Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, Cline AP, Bodine DM. Hmgb3: An HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102:1298–1306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- 14.Cai X, Ding H, Liu Y, Pan G, Li Q, Yang Z, Liu W. Expression of HMGB2 indicates worse survival of patients and is required for the maintenance of Warburg effect in pancreatic can. Acta Biochim Biophys Sin (Shanghai) 2017;49:119–127. doi: 10.1093/abbs/gmw124. [DOI] [PubMed] [Google Scholar]
- 15.Yu JW, Mai W, Cui YL, Kong LY. Genes and pathways identified in thyroid carcinoma based on bioinformatics analysis. Neoplasma. 2016;63:559–568. doi: 10.4149/neo_2016_409. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Wang Y, Gao Y, Zhang Y, Chen M, Xu M, Hu L, Jing Y, Jing F, Li C, et al. Knockdown of high mobility group-box 3 (HMGB3) expression inhibits proliferation, reduces migration, and affects chemosensitivity in gastric cancer cells. Med Sci Monit. 2016;22:3951–3960. doi: 10.12659/MSM.900880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Cai Y, Zhao H, Xu Z, Sun Q, Luo M, Gu L, Meng M, Han X, Sun H. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol. 2015;36:4785–4792. doi: 10.1007/s13277-015-3130-y. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang Y, Luo X, Liu C, Xie J, Cai C. Increased expression of HMGB3: A novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:345–352. [PMC free article] [PubMed] [Google Scholar]
- 19.Elgamal OA, Park JK, Gusev Y, Azevedo-Pouly AC, Jiang J, Roopra A, Schmittgen TD. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One. 2013;8:e76402. doi: 10.1371/journal.pone.0076402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45((D1)):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xian L, Huso T, Belton A, Huso D, Resar LM. High mobility group A1 chromatin remodeling protein expands the intestinal stem cell compartment and Paneth cell niche through Wnt/β-catenin signaling and Sox9. Am Assoc Cancer Res. 2016;76:16–20. [Google Scholar]
- 23.Wu T, Zhang W, Yang G, Li H, Chen Q, Song R, Zhao L. HMGB1 overexpression as a prognostic factor for survival in cancer: A meta-analysis and systematic review. Oncotarget. 2016;7:50417–50427. doi: 10.18632/oncotarget.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumter TF, Xian L, Huso T, Koo M, Chang YT, Almasri TN, Chia L, Inglis C, Reid D, Resar LM. The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr Mol Med. 2016;16:353–393. doi: 10.2174/1566524016666160316152147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resar L, Xian L, Huso T, Belton A, Cope L, Huso D. High mobility group A1 chromatin remodeling protein regulates self-renewal, niche formation and regenerative function in adult stem cells through wnt/β-catenin signaling. Am Soc Hematol. 2016;128:2647. [Google Scholar]
- 26.Kalomoiris S, Cicchetto AC, Lakatos K, Nolta JA, Fierro FA. Fibroblast growth factor 2 regulates high mobility group A2 expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2016;117:2128–2137. doi: 10.1002/jcb.25519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallante P, Sepe R, Puca F, Fusco A. High mobility group a proteins as tumor markers. Front Med (Lausanne) 2015;2:15. doi: 10.3389/fmed.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa K, Pham LD, Arai K, Lo EH. High-mobility group box 1: An amplifier of stem and progenitor cell activity after stroke. Acta Neurochir Suppl. 2013;118:31–38. doi: 10.1007/978-3-7091-1434-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stros M. HMGB proteins: Interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Rafehi H, Orlowski C, Georgiadis GT, Ververis K, El-Osta A, Karagiannis TC. Clonogenic assay: Adherent cells. J Vis Exp. 2011;pii:2573. doi: 10.3791/2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postnikov YV, Bustin M. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim Biophys Acta. 2016;1859:462–467. doi: 10.1016/j.bbagrm.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stros M, Launholt D, Grasser KD. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Nishikawa R, Kato M, Okato A, Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T, Seki N. Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell aggressiveness and is involved in prostate cancer pathogenesis. J Hum Genet. 2018;63:195–205. doi: 10.1038/s10038-017-0371-1. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L, Hu Y, Zhang F, Li X, Zhang W, Li X. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/β-catenin pathway. PLoS One. 2017;12:e0179741. doi: 10.1371/journal.pone.0179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia H, Miecznikowski JC, Safina A, Commane M, Ruusulehto A, Kilpinen S, Leach RW, Attwood K, Li Y, Degan S, et al. Facilitates chromatin transcription complex is an ‘accelerator’ of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013;4:159–173. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Dermawan JK, Hitomi M, Silver DJ, Wu Q, Sandlesh P, Sloan AE, Purmal AA, Gurova KV, Rich JN, Lathia JD, et al. Pharmacological targeting of the histone chaperone complex FACT preferentially eliminates glioblastoma stem cells and prolongs survival in preclinical models. Cancer Re. 2016;76:2432–2442. doi: 10.1158/0008-5472.CAN-15-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.