Abstract
Anaplastic thyroid cancer (ATC), with a mean survival time of 6 months, was reported in 2012 to account for 1–2% of all thyroid tumor cases in the US. Understanding the molecular mechanisms underlying carcinogenesis and progression in ATC would contribute to determining novel therapeutic targets. The aberrant expression of microRNA-544 (miR-544) has been demonstrated in various cancer types. However, its expression and biological function in human ATC remain largely unknown. Therefore, the present study investigated the expression, function and molecular mechanism of miR-544 in ATC. Results of reverse transcription-quantitative polymerase chain reaction demonstrated that the expression levels of miR-544 in 40 pairs of surgical specimens and human ATC cell lines were significantly decreased, compared with the normal thyroid tissues and cell line. Functional assays indicated that ectopic expression of miR-544 significantly decreased the viability, proliferation and metastasis of SW1736 cells, whereas miR-544 inhibitor significantly enhanced the viability, proliferation and metastasis of 8305C cells. Furthermore, the present study confirmed that the oncogene Yin Yang-1 (YY1) was a direct target of miR-544. It was further demonstrated that YY1 overexpression rescued the inhibitory effect of progression induced by miR-544 in ATC cells. Finally, in vivo study indicated that miR-544 suppressed the tumorigenicity of ATC cells. In conclusion, the present study demonstrated that miR-544 may function as a tumor suppressor in ATC and serve as a future therapeutic target for patients with ATC.
Keywords: microRNA-544, anaplastic thyroid cancer, Yin Yang-1, migration, invasion
Introduction
Thyroid cancer incidence is rapidly increasing in USA, with the estimated annual diagnoses and mortalities in 2017 being 56,870 and 2010, respectively (1). Thyroid cancer is generally classified as papillary, follicular and anaplastic carcinomas. Amongst these classifications, anaplastic thyroid cancer (ATC) was reported in 2012 to account for 1–2% of all thyroid tumor cases in the US (2). It is characterized by aggressive and local invasion, and frequent distant metastasis. Currently, available therapies for ATC include chemotherapy, radiotherapy and surgery (2). However, ATC remains one of the most fatal cancer types, with a mean survival time of 6 months (2). Therefore, it is crucial to identify the molecular etiology and molecular mechanisms underlying the progression and metastasis in ATC, and thus improve the therapeutic strategies and prognosis.
Previously, microRNA (miRNA), a series of small, highly-conserved, non-coding RNA molecules 18–25 nucleotides in length, are known to activate or inhibit the progression of various cancer types and have been proposed as novel targets for anticancer therapies (3). Of all miRNAs, >50% are known to be involved in human tumorigenesis by directly targeting oncogenes or tumor suppressor genes (3). For instance, miR-125b inhibits the tumor growth by directly targeting phosphatidylinositol-4.5-biphosphate 3-kinase catalytic subunit δ in ATC (4). Additionally, miR-4295 serves as an oncogene and promotes cell proliferation and invasion in ATC via cyclin dependent kinase inhibitor 1A (5). Previously, a number of studies have reported that miR-544 was significantly downregulated or upregulated in a number of human cancer types (6–10). However, miR-544 may serve as an oncogene or tumor suppressor depending on the type of tissue and the context in which it is expressed (6–10). In ATC, the expression pattern, biological roles and potential molecular mechanism of miR-544 remain largely unknown.
In the present study, the expression level of miR-544 was firstly detected in ATC tissues and cell lines. Secondly, the biological function of miR-544 in ATC proliferation, migration and invasion in vitro and in vivo was investigated. Finally, Yin Yang-1 (YY1) was identified as a direct target of miR-544. The results revealed that targeting the miR-544/YY1 axis may represent a promising therapeutic strategy for ATC treatment.
Materials and methods
Cell culture and tissue collections
The ATC cell lines (SW1736, KAT-18 and 8305C) and immortal thyroid cell line Nthy-ori3-1 were purchased from the American Type Culture Collection (Manassas, VA, USA). The cell lines were authenticated using short-tandem repeat profiling, which was performed by BMR Genomics (Padova, Italy). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (both from HyClone; GE Healthcare Life Sciences, Logan, UT, USA). Cells were incubated in a humidified atmosphere containing 5% CO2 and humidified sphere of 95% humidity at 37°C. Human ATC specimens and their adjacent normal thyroid tissues (40 pairs) were collected from 15 males and 25 female patients (mean, 62 years; range, 34–72 years) who underwent surgery between January 2016 and July 2017, according to an approved human protocol at the Yantai Laiyang Central Hospital (Yantai, China) and were used to detect the expression of miR-544 and mRNA expression of YY1. The present study was approved by the Ethics Committee of Yantai Laiyang Central Hospital. Written informed consent was obtai ned from every patient.
Cell transfection
The miR-544 mimic (sense, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAACTTT-3′) and the negative control (miR-NC; sense, 5′-ACUACUGAGUGACAGUAGA-3′) were purchased from Ambion (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The inhibitor control (anti-miR-NC; 5′-CAGUACUUUUGUGUAGUACAA-3′) was purchased from Ambion (Thermo Fisher Scientific, Inc.). The miR-544 inhibitor was obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The following sequences were used: 5′-CUUGUUAAAAAGCAGAUUCU-3′. The small RNAs, including small interfering (si)-YY1 and sicontrol, were obtained from Santa Cruz (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The small RNA sequences are as follows: siYY1-5′-GACGACUACAUUGAACAATT-3; negative control RNA-5′-UUCUCCGAACGUGUCACGUTT-3. YY1 overexpression plasmid was achieved using pcDNA3.1/YY1 transfection. A total of 10 nmol of YY1-pcDNA3.1 was transfected into the cells. Phblv-u6-puro vectors was purchased from Han Heng Biotechnology Co., Ltd. (Shanghai, China). For cell transfection, SW1736 and 8305C cells (2×105) were seeded in six-well plates and cultured until 60% confluency was reached. Transfection was performed with Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The transfection mixture was replaced in a medium containing 10% FBS following 6–8 h and after 48 h the transfection efficiency was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot.
Cell Counting Kit-8 (CCK-8)
A CCK-8 assay (Beyotime Institute of Biotechnology, Haimen, China) was used to detect the cell viability. Briefly, SW1736 and 8305C cells were seeded into 96-well plates (1×103/well) in DMEM containing 10% FBS for 0, 24, 48, 72 h at 37°C. At the indicated time 10 µl CCK-8 was added to each well. After incubation for 3 h at room temperature, the absorbance of each well was measured using Multiskan MK3.
Colony formation assay
For the colony formation assay, 4×102 SW1736 and 8305C cells were seeded in 6-well plates separately. After 10 days, the cells were washed with PBS three times, fixed with 4% paraformaldehyde for 15 min and stained with 0.5% crystal violet at room temperature (Beyotime Institute of Biotechnology). The clone number (cell population >50) was counted using a CKX41 light microscope (magnification, ×10).
Dual-luciferase assay
The fragment of the YY1 3′-untranslated region (UTR) containing the miR-544 predicted binding sequences (predicted by targetscan 7.1) or the mutant sequences was synthesized by Shanghai GenePhama Co., Ltd., and then cloned into a psiCHECK-2 vector (Promega Corporation, Madison, WI, USA). For convenience, the psiCHECK-2-YY1-3 tUTR-wild-type (Wt) and the psiCHECK-2-YY1-3 tUTR-wild-type (Mut) were designated as Wt and Mut, respectively. SW1736 cells were co-transfected with the aforementioned luciferase reporter vector with miR-544 mimic or miR-NC. 8305C cells were co-transfected with the aforementioned luciferase reporter vector with miR-544 inhibitor or anti-miR-NC. Cell transfection were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cells were cultured 37°C for 24 h and luciferase activities were analyzed using the Dual-Luciferase Reporter Assay system. Results were normalized to Renilla luciferase activity.
Wound-healing assay
The wound-healing assay was performed to assess cell migration. In brief, SW1736 and 8305C cells seeded in six-well plates (8×105 cells/well) and cultured until they reached 90% confluency at 37°C. The cell monolayer was subsequently scraped with a sterile 200 µl micropipette tip to create separate wounds, and the wells were washed three times with PBS to remove cell debris. Finally, the cells were cultured at 37°C and observed at 0 and 24 h using a CKX41 light microscope (magnification, ×10).
Transwell and Matrigel assays
Migration and invasion abilities were analyzed using Transwell chambers (Corning Incorporated, Corning, NY, USA). For the invasion assays, the Transwell inserts were coated with 50 µl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). For the assays, 5×104 SW1736 and 8305C cells were resuspended in 0.1 ml serum-free DMEM and added to the Transwell inserts. DMEM with 10% FBS was added to the lower wells. After incubation for 8 h (migration) or 12 h (invasion), the cells on the upper surface of the membrane were removed, and the cells on the lower surface were fixed with 10% methanol at room temperature for 15 min and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) at room temperature for 5 min and then counted under a light microscope (10 × magnification).
Total mRNA extraction RT-qPCR
The SW1736 and 8305C cells were dissolved in TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) to extract the total mRNA. Following spectrophotometric quantification, cDNA was synthesized using an iScript cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. Subsequently, qPCR was performed using SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. For detecting the expression of YY1, β-actin acted as internal control. For detecting the expression of miR-544, U6 served as internal control. The thermocycling conditions were as follows: 95°C for 10 min followed by 50 cycles of 95°C for 10 sec, 55°C for 10 sec, 72°C for 5 sec; 99°C for 1 sec; 59°C for 15 sec; 95°C for 1 sec; followed by cooling to 40°C. The relative expression level of miR-544 and YY1 was calculated as the inverse log of ∆∆Cq and normalized to the reference (11). The primers used for amplification were: YY1, forward, 5′-CCCCGGCAAGTGTGAGTGAA-3′, and reverse, 5′-ACAAGCCAAGGTCCTGCTGG-3′; β-actin, forward, 5′-GATCATTGCTCCTCCTGAGC-3′, and reverse, 5′-ACTCCTGCTTGCTGATCCAC-3′; miR-544, forward, 5′-GCCCGATTCTGCATTTTTAGC-3′, and reverse 5′-CGGGCTAAGACGTAAAAACG-3′; and U6, forward, 5′-TGCGGGTGCTCGCTTCGCAGC-3′, and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′.
RNA immunoprecipitation
RNA immunoprecipitation assays were performed using an Imprint RNA Immunoprecipitation kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol with the protein argonaute-2 (AGO2; dilution, 1:100; cat. no. 2897; Cell Signaling Technology, Inc., Danvers, MA, USA) or IgG antibodies (dilution, 1:100; cat. no. A6066; Sigma-Aldrich; Merck KGaA) for 6 h at 4°C. The expression of miR-544 and the YY1 mRNA levels in the immunoprecipitates were analyzed with RT-qPCR analysis, according to the aforementioned protocol.
Western blot analysis
Cells were lysed using radioimmunoprecipitation assay buffer (Beijing Solarbio Bioscience and Technology Co., Ltd., Beijing, China) and protein was quantified using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Identical amounts of proteins (20 µg) from the lysates of the SW1736 and 8305C cells were subjected to electrophoresis through 10% SDS-PAGE and were transferred onto polyvinylidene difluoride membranes (Invitrogen; Thermo Fisher Scientific, Inc.). The membranes were blocked with 5% skimmed milk for 30 min at 4°C and incubated with primary antibodies for YY1 (dilution, 1:200; cat. no. ab12132; Abcam, Cambridge, MA, USA), E-cadherin (dilution, 1:500; cat. no. sc-8426; Santa Cruz Biotechnology, Inc.), N-cadherin (dilution, 1:500; cat. no. sc-8424; Santa Cruz Biotechnology, Inc.) or β-actin (dilution, 1:500; cat. no. AF0003; Beyotime Institute of Biotechnology) overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody (dilution, 1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) at room temperature for 2 h. Signals were detected with Enhanced Chemiluminescent Plus (Beyotime Institute of Biotechnology), according to the manufacturer's protocols. The relative protein levels were calculated based on β-actin as the loading control.
Xenografted tumor model
A total of 24 female, 4-week old BALB/c nude mice (weight, range; 20–25 g) purchased from Jilin University (Changchun, China) were inoculated subcutaneously with 1×106 SW1736 cells infected with lentiviral-miR-544 mimics or lentiviral-miR-NC in the dorsal flank and maintained in a specific-pathogen free environment with constant humidity (45–50%) and constant temperature (25–27°C) under a 12/12 h light/dark cycle with free access to food and water. Measurements were obtained from the tumors every 5 days, including the greatest tumor length and width measurements with calipers, and then the tumor volumes were analyzed. Tumor volume was calculated using the equation: Volume=(length × width2)/2. After 30 days, the animals were sacrificed by cervical dislocation, and tumors were excised and weighed. The short hairpin (sh) YY1 and shControl transfected into the SW1736 cells was carried out as previously reported. To determine the function of YY1 in the growth of tumors in vivo, stable lentiviral siYY1- and siControl-SW1736 cells (1×106) were subcutaneously injected into the dorsal flank of athymic nude mice (n=6/group), studied as aforementioned statement. All animal studies were approved by the Animal Care and Welfare Committee of Yantai Laiyang Central Hospital.
Bioinformatics analysis
The prediction of the YY1 3′-UTR as a miR-544 binding target was determined using TargetScan 7.1 software (www.targetscan.org) (12). TargetScan is online software, that predicts biological targets of miRNA by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA.
Statistical analysis
Data are expressed as mean ± standard error. Statistical analysis was conducted using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). Statistical significance was determined through unpaired Student's t-tests or one-way analysis of variance with post-hoc Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-544 is downregulated in ATC tissues and cell lines
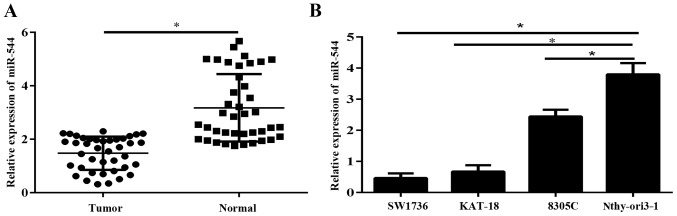
Expression of miR-544 was determined in 40 ATC samples and adjacent normal thyroid tissues via RT-qPCR. The results demonstrated that miR-544 was significantly downregulated in ATC tissues, compared with normal thyroid tissues (P<0.05; Fig. 1A). The expression level of miR-544 was significantly reduced in ATC cell lines, compared with the immortal thyroid cell line Nthy-ori3-1 (P<0.05; Fig. 1B).
Figure 1.
miR-544 is downregulated in ATC tissues and cell lines. (A) The expression level of miR-544 in ATC tissues and normal thyroid tissues was detected via RT-qPCR. (B) The expression level of miR-544 was analyzed in ATC cell lines (SW1736, KAT-18 and 8305C) and normal thyroid cell line Nthy-ori3-1 via RT-qPCR. *P<0.05. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; ATC, anaplastic thyroid cancer.
miR-544 inhibits the viability and proliferation of ATC cells in vitro
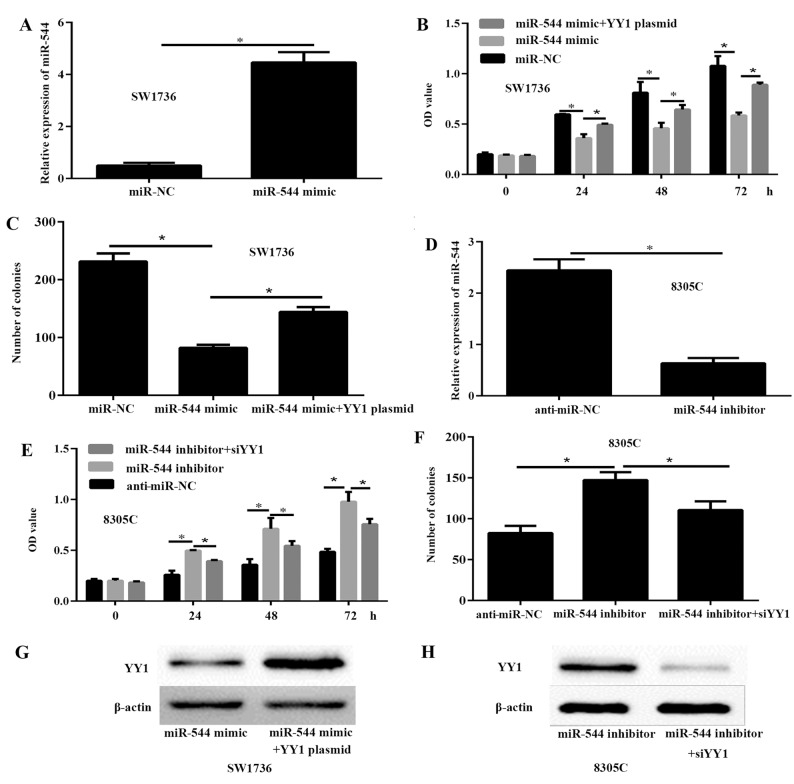
To investigate the potential functions of miR-544 in ATC, SW1736 cells were transfected with 50 nM miR-544 mimic or miR-NC to significantly increase the expression of miR-544 (Fig. 2A). The results of the CCK-8 (Fig. 2B) and colony formation assays (Fig. 2C) demonstrated that the viability and proliferation of the SW1736 cells transfected with miR-544 mimic were significantly decreased, compared with SW1736 cells transfected with miR-NC. To further investigate the effects of miR-544 on ATC progression, 50 nM miR-544 inhibitor was transfected into the 8305C cells. The RT-qPCR results demonstrated that the expression of miR-544 was significantly decreased in the 8305C cells compared with the anti-miR-NC cells (P<0.05; Fig. 2D). The CCK-8 (Fig. 2E) and colony formation assays (Fig. 2F) indicated that miR-544 knockdown significantly enhanced the viability and proliferation of 8305C cells, compared with the anti-miR-NC cells (P<0.05). These data demonstrated that miR-544 inhibited the viability and proliferation of ATC cells. To further demonstrate the association of miR-544 and YY1 in ATC, miR-544 mimic or miR-544 mimic and YY1 plasmid were transfected into SW1736 cells, and miR-544 inhibitor or miR-544 inhibitor and siYY1 into 8305C cells. The results of the western blot analysis indicated that YY1 was significantly upregulated in SW1736 cells transfected with miR-544 mimic and YY1 plasmid compared to miR-544 mimic group (Fig. 2G). In addition, the expression of YY1 was decreased in 8305C cells transfected with miR-544 inhibitor and siYY1 compared to miR-544 inhibitor (Fig. 2H).
Figure 2.
miR-544 inhibits anaplastic thyroid cancer cells growth via the expression of YY1. SW1736 cells were transfected with 50 nM miR-NC, miR-544 mimic or miR-544 mimic and YY1 plasmid, while 8305C cells were transfected with 50 nM anti-miR-NC, miR-544 inhibitor or miR-544 inhibitor and si-YY1 for 48 h prior to analysis. The expression level of miR-544 was evaluated via reverse transcription-quantitative polymerase chain reaction in (A) SW1736 and (D) 8305C cells. The cell viability was detected with a Cell Counting Kit-8 assay in (B) SW1736 and (E) 8305C cells. The proliferation was detected with a colony formation assay in (C) SW1736 and (F) 8305C cells. The protein expression of YY1 was detected by western blot analysis in (G) SW1736 and (H) 8305C cells. *P<0.05. YY1, Yin Yang-1; miR, microRNA; miR-NC, miR-negative control; anti-miR-NC, inhibitor control; OD, optical density.
miR-544 inhibits the migration and invasion of ATC cells
To investigate the effect of miR-544 on the metastatic ability of ATC cells, wound-healing, migration and invasion assays were performed in SW1736 and 8305C cells. The wound-healing assays demonstrated that the miR-544-overexpressing cells had significantly decreased migration, compared with the SW1736 cells transfected with miR-NC (P<0.05; Fig. 3A). By contrast, the miR-544-silenced 8305C cells had significantly enhanced migration capability, compared with the 8305C cells transfected with anti-miR-NC (P<0.05; Fig. 3B). Consistent with the wound-healing assay, the Transwell and Matrigel assays also demonstrated that miR-544 restoration significantly decreased the migration and invasion capabilities of SW1736 cells (Fig. 3C and D), whereas miR-544 inhibition significantly enhanced the migration and invasion capabilities of 8305C cells (P<0.05; Fig. 3E and F).
Figure 3.
miR-544 inhibits metastasis and epithelial-mesenchymal transition of anaplastic thyroid cancer cells via the expression of YY1. SW1736 cells were transfected with miR-NC, miR-544 mimic or miR-544 mimic and YY1 plasmid, while 8305C cells were transfected with anti-miR-NC, miR-544 inhibitor or miR-544 inhibitor and si-YY1 for 48 h prior to analysis. (A) wound-healing assay was used to analyze the migration capability of SW1736 cells. (B) Wound-healing assay was used to analyze the migration capability of 8305C cells. Transwell migration and Matrigel invasion assays were used to detect the migration and invasion capability, respectively. (C) Images (captured at ×100 magnification) of migration and invasion of SW1736 cells in the lower chamber (scale bars, 50 µm). (D) Quantification of migration and invasion of SW1736 cells in the lower chamber (scale bars, 50 µm). (E) ages (captured at ×100 magnification) of migration and invasion of 8305C cells in the lower chamber (scale bars, 50 µm). (F) Quantification of migration and invasion of 8305C cells in the lower chamber (scale bars, 50 µm). (G) The protein expression of E-cadherin and N-cadherin in SW1736 cells was detected with a western blot analysis assay (H) The protein expression of E-cadherin and N-cadherin in 8305C cells was detected with a western blot analysis assay. *P<0.05. YY1, Yin Yang-1; miR, microRNA; miR-NC, miR-negative control; anti-miR-NC, inhibitor control.
Epithelial-mesenchymal transition (EMT) is a potential mechanism for tumor cell metastasis (13). To investigate whether miR-544 affects EMT, the expression of the epithelial marker E-cadherin and mesenchymal marker N-cadherin were measured in SW1736 and 8305C cells by western blot analysis. The results demonstrated that miR-544 mimic notably enhanced the expression of E-cadherin and downregulated that of N-cadherin in SW1736 cells (Fig. 3G), whereas miR-544 inhibitor notably decreased the expression of E-cadherin and upregulated that of N-cadherin in 8305C cells (Fig. 3H).
YY1 is a direct target of miR-544
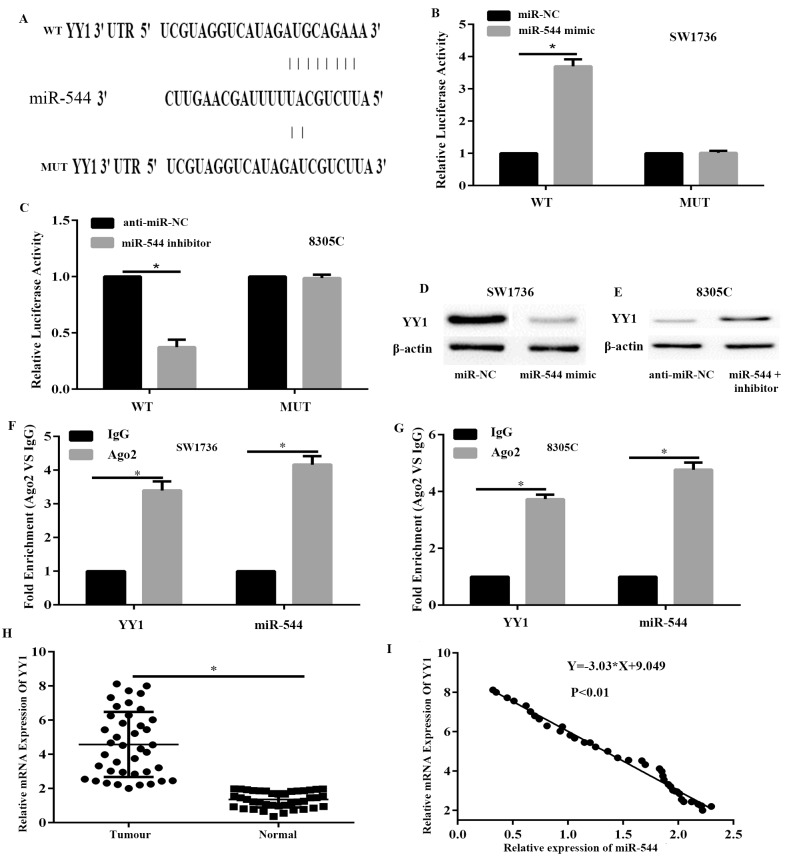
To determine the underlying molecular mechanisms by which miR-544 suppresses ATC cancer growth, a search was conducted to determine candidate targets of miR-544 that may serve a role in ATC progression with and TargetScan 7.1. Among the candidates, YY1, which is a pro-oncogene gene that is frequently upregulated in various cancer types, was predicted to be a miR-544 target and selected for further experimental verification. The predicted interaction between miR-544 and the target site in the YY1 3′-UTR is shown in Fig. 4A. Dual-luciferase reporter assays were conducted to investigate whether miR-544 targets YY1 by binding to its 3′-UTR. SW1736 cells were co-transfected with the Wt or Mut reporter vector and miR-544 mimic or miR-NC. The results demonstrated that luciferase activities were significantly decreased in the SW1736 cells transfected with the Wt reporter vector and miR-544 mimic but not in the cells with the mutant reporter vector compared with the transfected cells with the miR-NC (P<0.05; Fig. 4B). Additionally, inhibition of miR-544 significantly increased the luciferase activity of the plasmid transfected with the WT 3′-UTR of YY1 in 8305C cells compared with the transfected cells with the anti-miR-NC (P<0.05; Fig. 4C). Furthermore, the western blot assay demonstrated that miR-544 overexpression decreased the protein expression of YY1 in SW1736 cells compared with the transfected cells with the miR-NC (Fig. 4D), whereas the opposite is true in 8305C cells transfected with inhibitor compared with the transfected cells with the anti-miR-NC (Fig. 4E). To further confirm the interaction between miR-544 and YY1 3′-UTR, RNA immunoprecipitation assays were performed. In the RNA extracted from the precipitated AGO2 protein, it was possible to detect the miR-544 and YY1 3′UTR with notable enrichment, compared with IgG in SW1736 and 8305C cells (Fig. 4F and G), indicating that miR-544 and the YY1 3′-UTR existed in a RNA-induced silencing complex. These data demonstrate that miR-544 directly targets YY1 by binding to its 3′-UTR region in ATC cells.
Figure 4.
YY1 is a direct target of miR-544. (A) Sequence alignment of human miR-544 with 3′-UTR of YY1, as predicted by TargetScan. A luciferase assay in (B) SW1736 and (C) 8305C cells co-transfected with indicated miRNA mimic or miRNA inhibitor and luciferase reporter plasmids. Expression levels of YY1 were detected in (D) SW1736 and (E) 8305C cells with a western blot analysis assay. The association between miR-544/YY1 and AGO2 was analyzed. SW1736 and 8305C cellular lysates were used for RNA immunoprecipitation with an AGO2 antibody. RT-qPCR was used to detect the expression of miR-544 and YY1 in SW1736 cells (F). qPCR was used to detect the expression of miR-544 and YY1 in 8305C cells (G). miR-544 expression data was normalized to U6 small RNA expression. All data of YY1 expression were normalized to β-actin mRNA expression levels. (H) The mRNA expression levels of YY1 were significantly increased in ATC tissues, compared with normal tissues, as assessed with RT-qPCR. (I) Correlation between miR-544 expression levels and YY1 mRNA levels in ATC tissues was analyzed using Spearman's correlation analysis. *P<0.05. YY1, Yin Yang-1; miR, microRNA; miR-NC, miR-negative control; anti-miR-NC, inhibitor control; Mut, mutated; Wt, wild-type; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; ATC, anaplastic thyroid cancer.
Subsequently, the mRNA expression levels of YY1 were examined in ATC and adjacent normal tissues via RT-qPCR. The results demonstrated that the expression level of YY1 was significantly upregulated in ATC tissues, compared with adjacent normal tissues (P<0.05; Fig. 4H). Additionally, Spearman's correlation analysis indicated that the expression levels of miR-544 were significantly inversely correlated with YY1 mRNA in ATC tissues (P<0.05; Fig. 4I). Overall, these data demonstrate that miR-544 directly targets YY1 by binding to its 3′-UTR region in ATC cells.
YY1 silencing inhibits ATC cell growth in vitro and in vivo
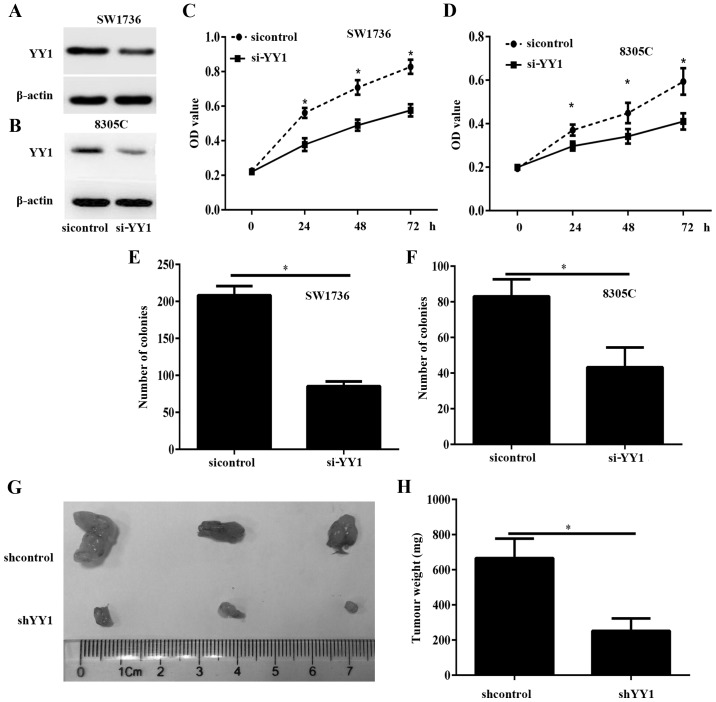
To investigate the potential functions of YY1 in ATC, SW1736 and 8305C cells with were transfected with si-YY1 or sicontrol to decrease the expression of YY1 (Fig. 5A and B). The CCK-8 assay results demonstrated that the viability of SW1736 and 8305C cells transfected with si-YY1 was significantly decreased, compared with the sicontrol cells (P<0.05; Fig. 5C and D). The colony formation assay indicated that the colony numbers in the SW1736 and 8305C cells transfected with si-YY1 were significantly decreased, compared with those in the sicontrol cells (P<0.05; Fig. 5E and F). To confirm the aforementioned data in vivo, tumor xenograft mouse models were produced. Nude mice were subcutaneously implanted with SW1736 cells stably transfected with lentiviral shYY1 and shControl. After 30 days, all mice were sacrificed and the tumor xenograft was obtained (Fig. 5G). The tumor weight significantly decreased in the shYY1 group, compared with the shControl group (P<0.05; Fig. 5H).
Figure 5.
YY1 silencing inhibits anaplastic thyroid cancer cell growth. (A and B) The western blot analysis assay demonstrated the protein expression of YY1 in SW1736 and 8305C cells. (C and D) The cell viability was detected with a Cell Counting Kit-8 assay. (E and F) The proliferation was detected with a colony formation assay. (G and H) YY1 silencing suppressed tumor growth in nude mice. (G) Representative images of tumors from the implanted mice. (H) The tumor weight was measured. *P<0.05. YY1, Yin Yang-1; si-YY1, small interfering YY1; shYY1, short hairpin YY1.
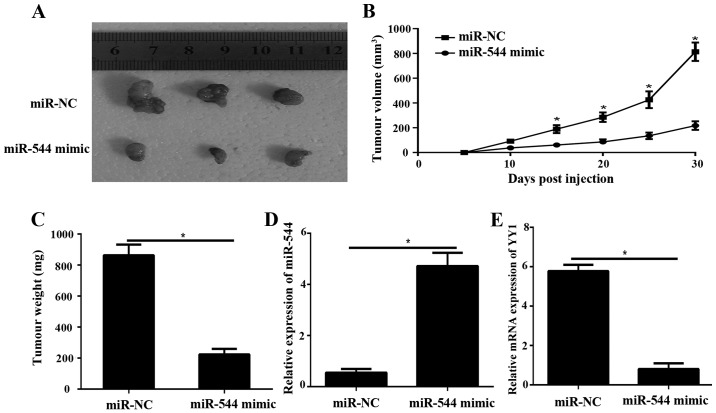
miR-544 suppresses tumor growth in nude mice by downregulating YY1
To confirm the aforementioned data in vivo, tumor xenograft mouse models were produced. Nude mice were subcutaneously implanted with SW1736 cells transfected with lentiviral miR-544 and miR-NC. After 30 days, all mice were sacrificed, and the tumor xenograft was obtained (Fig. 6A). The tumor volume miR-544 in mimic group was dramatically smaller compared with the miR-NC group (P<0.05; Fig. 6B). The tumor weight (Fig. 6C) were significantly decreased in the miR-544 mimic group, compared with the miR-NC group (P<0.05; Fig. 6C). The results of RT-qPCR demonstrated that the expression level of miR-544 in the miR-544 mimic tumor group was significantly decreased, compared with the miR-NC group (P<0.05; Fig. 6C and D). The mRNA level of YY1 was significantly decreased in the tumor-transfected miR-544 mimic compared to miR-NC group (P<0.05; Fig. 6E).
Figure 6.
miR-544 suppresses tumor growth in nude mice by downregulating YY1. (A) Representative images of tumors from the implanted mice. The (B) tumor volume and (C) tumor weight were measured. (D) RT-qPCR analysis of miR-544 expression levels in the miR-544 mimics tumor xenograft mice and miR-NC tumor xenograft mice. (E) The mRNA expression of YY1 was detected by RT-qPCR in the miR-544 mimics tumor xenograft mice and miR-NC tumor xenograft mice. *P<0.05. miR-NC, microRNA-negative control; YY1, Yin Yang-1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-544 promoted ATC progression via YY1
Finally, whether miR-544 promotes ATC progression via YY1 was investigated. The protein expression level of YY1 was detected by western blot assay (Fig. 2G and H). The results of the CCK-8 (P<0.05; Fig. 2B), colony formation assay (Fig. 2C; P<0.05), and Transwell migration and Matrigel invasion assays (P<0.05; Fig. 3C and D) demonstrated that YY1 overexpression significantly reversed the inhibitory effects of miR-544 on the viability, proliferation and metastasis of SW1736 cells compared with the miR-544 mimic group (P<0.05), whereas YY1 silencing partially reversed the promotion effects of the miR-544 inhibitor of the viability (P<0.05; Fig. 2E), proliferation (P<0.05; Fig. 2F), and metastasis (P<0.05; Fig. 3E and F) of 8305C cells compared with the miR-544 inhibitor group (P<0.05). Furthermore, western blot analysis indicated that YY1 partially reverses the inhibitory effect of EMT induced by miR-544 mimic in SW1736 cells (Fig. 3G); however, YY1 silencing can partially reverse the inhibitory effect of EMT induced by miR-544 inhibitor in 8305C cells (Fig. 3H). These data demonstrate that miR-544 inhibits tumor progression via YY1 in ATC.
Discussion
Previous studies demonstrated that the abnormal expression of miR-544 is associated with glioma, osteosarcoma, cervical cancer, breast cancer and gastric cancer (6–10). However, the expression and function of miR-544 in ATC tissues remains unclear. In the present study, it was indicated that miR-544 is downregulated in ATC and that it may act as a novel tumor suppressor in ATC. Furthermore, upregulation of miR-544 significantly suppressed tumor growth, proliferation and metastasis, while the miR-544 inhibitor enhanced the tumor progression. Additionally, it was demonstrated that miR-544 acted as a tumor suppresser via directly targeting YY1.
YY1 is a conserved multifunctional protein belonging to the GLI-Kruppel family, and it binds to the promoter of other genes primarily through the C-terminal zinc finger region (14,15). As a transcriptional regulator, YY1 is involved in multiple biological functions of cells, including embryonic formation, cell proliferation, apoptosis, DNA repair and differentiation via upregulating or downregulating different genes (16). It has been reported that YY1 is overexpressed and serves as oncogene in numerous tumor types, including breast, ovary, colon, prostate, gastric and laryngeal cancer (17–21). However, YY1 may serve an inhibitory role in pancreatic cancer (22). A recent study demonstrated that the expression of YY1 was significantly upregulated in differentiated thyroid and anaplastic cancer (23). However, the function of YY1 in ATC has not been well illustrated. In the present study, consistent with previous study, it was also determined that YY1 has a high expression level in ATC tissues. Additionally, YY1 silencing suppressed cell viability and proliferation, and significantly decreased the migration and invasion, of ATC cells. As a key transcription factor, YY1 regulates its target gene transcription in a complex manner. YY1 can activate the transcription of oncogenes and tumor suppressors in different cancer tissues (14–23). However, the mechanism regulating YY1 has not been well illustrated. In the present study, YY1 was identified as a direct target of miR-544 by dual-luciferase reporter assays. Furthermore, Transwell migration and invasion assay indicated that the most important effect exerted by miR-544 on ATC cells invasion and migration, which was partially reversed when co-transfected with the YY1 plasmid. These results demonstrated that YY1 was a functional target gene of miR-544 in ATC. It is notable that each miRNA can regulate dozens of genes and multiple miRNAs may regulate the identical gene, which affect the activities of whole pathways. Therefore, the possibility that other target genes may also be involved in the suppressive effects of miR-544 cannot be excluded.
In conclusion, it was determined that miR-544 was significantly downregulated in ATC. Furthermore, it was demonstrated, for the first time, that the role of the miR-544/YY1 axis is to regulate ATC proliferation and metastasis. This novel miR-544/YY1 association provides a new insight into the mechanisms underlying ATC development, and indicates that targeting the miR-544/YY1 axis may represent a promising therapeutic strategy for ATC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
FW and BS contributed to study design, statistical analysis, data interpretation, manuscript preparation and the literature search. ZL performed the experiments and data collection. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Yantai Laiyang Central Hospital (Yantai, China). Written informed consent was obtained from every patient. All animal studies were approved by the Animal Care and Welfare Committee of Yantai Laiyang Central Hospital.
Patient consent for publications
All patients provided written informed consent prior to participation in the present study and consent for the publication of the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.O'Neill JP, Shaha AR. Anaplastic thyroid cancer. Oral Oncol. 2013;49:702–706. doi: 10.1016/j.oraloncology.2013.03.440. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Bu Q, You F, Pan G, Yuan Q, Cui T, Hao L, Zhang J. MiR-125b inhibits anaplastic thyroid cancer cell migration and invasion by targeting PIK3CD. Biomed Pharmacother. 2017;88:443–448. doi: 10.1016/j.biopha.2016.11.090. [DOI] [PubMed] [Google Scholar]
- 5.Shao M, Geng Y, Lu P, Xi Y, Wei S, Wang L, Fan Q, Ma W. miR-4295 promotes cell proliferation and invasion in anaplastic thyroid carcinoma via CDKN1A. Biochem Biophys Res Commun. 2015;464:1309–1313. doi: 10.1016/j.bbrc.2015.07.128. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z, Wang S, Zhu J, Yang Q, Dong H, Huang J. MicroRNA-544 down-regulates both Bcl6 and Stat3 to inhibit tumor growth of human triple negative breast cancer. Biol Chem. 2016;397:1087–1095. doi: 10.1515/hsz-2016-0104. [DOI] [PubMed] [Google Scholar]
- 7.Jin S, Dai Y, Li C, Fang X, Han H, Wang D. MicroRNA-544 inhibits glioma proliferation, invasion and migration but induces cell apoptosis by targeting PARK7. Am J Transl Res. 2016;8:1826–1837. [PMC free article] [PubMed] [Google Scholar]
- 8.Mao L, Zhang Y, Deng X, Mo W, Yu Y, Lu H. Transcription factor KLF4 regulates microRNA-544 that targets YWHAZ in cervical cancer. Am J Cancer Res. 2015;5:1939–1953. [PMC free article] [PubMed] [Google Scholar]
- 9.Ma R, Zhang G, Wang H, Lv H, Fang F, Kang X. Downregulation of miR-544 in tissue, but not in serum, is a novel biomarker of malignant transformation in glioma. Oncol Lett. 2012;4:1321–1324. doi: 10.3892/ol.2012.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhi Q, Guo X, Guo L, Zhang R, Jiang J, Ji J, Zhang J, Zhang J, Chen X, Cai Q, et al. Oncogenic miR-544 is an important molecular target in gastric cancer. Anticancer Agents Med Chem. 2013;13:270–275. doi: 10.2174/1871520611313020013. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;(4) doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 15.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 16.Bonavida B, Kaufhold S. Prognostic significance of YY1 protein expression and mRNA levels by bioinformatics analysis in human cancers: A therapeutic target. Pharmacol Ther. 2015;150:149–168. doi: 10.1016/j.pharmthera.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L, Bonavida B. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol. 2005;27:131–141. [PubMed] [Google Scholar]
- 18.Kang W, Tong JH, Chan AW, Zhao J, Dong Y, Wang S, Yang W, Sin FM, Ng SS, Yu J, et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J Transl Med. 2014;12:80. doi: 10.1186/1479-5876-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinnappan D, Xiao D, Ratnasari A, Andry C, King TC, Weber HC. Transcription factor YY1 expression in human gastrointestinal cancer cells. Int J Oncol. 2009;34:1417–1423. [PubMed] [Google Scholar]
- 21.Qu SY, Sun YY, Li YH, Xu ZM, Fu WN. YY1 directly suppresses MYCT1 leading to laryngeal tumorigenesis and progress. Cancer Med. 2017;6:1389–1398. doi: 10.1002/cam4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D, Zhang J, Wu Y, Shi G, Yuan H, Lu Z, Zhu Q, Wu P, Lu C, Guo F, et al. YY1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the CDKN3/MdM2/P53/P21 signaling pathway. Int J Cancer. 2018;142:1392–1404. doi: 10.1002/ijc.31173. [DOI] [PubMed] [Google Scholar]
- 23.Arribas J, Castellví J, Marcos R, Zafón C, Velázquez A. Expression of YY1 in differentiated thyroid cancer. Endocr Pathol. 2015;26:111–118. doi: 10.1007/s12022-015-9359-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.