Abstract
Background
Fleas (Siphonaptera) are the most clinically important ectoparasites of dogs and cats worldwide. Rising levels of pet ownership, climate change and globalisation are increasing the importance of a detailed understanding of the endemicity and prevalence of flea-borne pathogens. This requires continued surveillance to detect change. This study reports a large-scale survey of pathogens in fleas collected from client-owned cats and dogs in the UK.
Methods
Recruited veterinary practices were asked to follow a standardised flea inspection protocol on a randomised selection of cats and dogs brought into the practice in April and June 2018. A total of 326 practices participated and 812 cats and 662 dogs were examined. Fleas were collected, identified to species and pooled flea samples from each host were analysed for the presence of pathogens using PCR and sequence analysis.
Results
Overall, 28.1% of cats and 14.4% of dogs were flea infested. More than 90% of the fleas on both cats and dogs were cat fleas, Ctenocephalides felis felis. Fleas of the same species from each infested host were pooled. DNA was amplified from 470 of the pooled flea samples using conventional PCR, 66 of which (14% ± 95% CI 3.14%) were positive for at least one pathogen. Fifty-three (11.3% ± 95% CI 2.85%) of the pooled flea DNA samples were positive for Bartonella spp., 35 were from cats and 4 from dogs, the remainder had no host record. Seventeen of the Bartonella spp. samples were found to be Bartonella henselae, 27 were Bartonella clarridgeiae (of two different strains), 4 samples were Bartonella alsatica and one was Bartonella grahamii; 4 samples could not be identified. Fourteen (3% ± 95% CI 1.53%) of the flea DNA samples were found to be positive for Dipylidium caninum, 10 of the D. caninum-infected samples were collected from cats and one from a dog, the other 3 positive flea samples had no host species record. Only 3 flea samples were positive for Mycoplasma haemofelis or Mycoplasma haemocanis; 2 were collected from cats and one had no host species record. Three fleas were positive for both D. caninum and Bartonella spp. One flea was positive for both Bartonella spp. and M. haemofelis or M. haemocanis.
Conclusions
This study highlights the need for ongoing flea control, particularly given the relatively high prevalence of Bartonella spp., which is of concern for both animal welfare and human health. The study demonstrates the ongoing need to educate pet owners about the effects of both flea infestation and also the pathogen risks these fleas present.
Keywords: Companion animal, Disease, Flea-borne, Pathogen, Siphonaptera, Vector, Bartonella
Background
The promiscuous blood-feeding behaviour of both sexes of fleas (Siphonaptera), their mobility, their persistence in the environment and their ability to act as vectors of a range of pathogens all contribute to making them the most clinically important ectoparasites of dogs and cats worldwide [1]. Of particular importance for companion animals are the cat flea, Ctenocephalides felis felis, and the dog flea, C. canis, which are widely distributed globally. These species are generally considered to be host-preferential rather than host-specific and will try to feed on any available animal; C. f. felis has been found on over fifty different host species, which contributes to its persistence in the environment [2]. The prevalence of flea infestation in companion animals is commonly reported as varying between 10–40% [3–6], although in some instances peak infestation rates of more than 70% have been reported [6]. Infestation rates are highly variable from year to year and also depend on location, whether animals are rural or urban, lifestyle - for example outdoor access, whether they live in multi-pet households and the frequency of effective insecticidal treatments. Seasonal variations in infestation prevalence are also commonly seen, with a lower prevalence in winter and an increase from spring to autumn [7]. A knowledge of the prevalence of C. f. felis and C. canis on domestic animals and other wild hosts has important implications for flea control, since it affects the extent to which they may act as reservoirs of fleas, from which treated animals may be continuously reinfested.
Besides the direct effects resulting from blood-feeding, Ctenocephalides species are important as competent vectors for a wide range of pathogens, many of which are zoonotic [8–10]. In particular, these fleas may be vectors of rickettsiae, such as Yersinia pestis, Rickettsia typhi, Rickettsia felis, Rickettsia conorii and Bartonella henselae [11] and are the intermediate hosts for cysticercoid larvae of Dypidilium caninum tapeworms [12, 13]. Amongst the factors that contribute to the highly effective role of fleas as vectors includes the transovarial and transtadial transmission of some pathogens [14, 15]. Dipylidium caninum infection is very widely prevalent and, being dependent on the continuous presence of vectors for its local endemicity, infestations are seen in both neglected animals and owned domestic dogs and cats [16].
Several Bartonella spp. transmitted by fleas can induce clinical disease in both dogs and humans and these pathogens may be particularly prevalent. Bartonellosis is contracted by handling animals infested with fleas, animal owners and veterinarians are at particular risk [17]. Domestic cats are considered to be the natural reservoir for Bartonella henselae and Bartonella clarridgeiae, the causative agents of cat scratch disease, and dogs may also be infected with these pathogens [18, 19]. Ctenocephalides felis felis is assumed to be the main vector for B. henselae and B. clarridgeiae [20]. Several studies have reported the presence of Bartonella spp. DNA in various flea species suggesting their role as potential vectors [2].
Three species of haemotropic mycoplasmas (also known as haemoplasmas), Mycoplasma haemofelis, “Candidatus Mycoplasma haemominutum” and “Candidatus Mycoplasma turicensis”, have been reported in UK cats [21]. Mycoplasma haemofelis is the most pathogenic feline haemoplasma, occasionally causing severe haemolytic anaemia in acute infections [22]. Infections with the other two haemoplasma species may cause a drop in erythrocyte parameters, but these cats do not usually become clinically anaemic unless their health is compromised or they are immunosuppressed [22]. The natural route of transmission of these pathogens between cats is yet to be determined, but the possible role of an arthropod vector is supported by the detection of feline haemoplasma DNA in fleas and ticks collected from cats and/or the environment [23, 24].
The aim of the present study was to undertake a large-scale survey of flea-borne pathogens in fleas collected from cats and dogs in the UK. Increasing levels of pet ownership and increasing urbanisation, along with factors such as climate change and globilisation may, over time, affect the endemicity and prevalance of insect vectors and change the dynamics of pathogen transmission. Therefore, a more detailed understanding of the current prevalence and distribution of flea-borne pathogens is important and should be supported by continued surveillance. This will aid in better education of veterinarians, physicians and public health workers and help develop and implement more effective preventive measures. Such surveillance has been facilitated in recent years by the wide availability of molecular tools, although, given the relatively low prevalances of flea-borne pathogens, large sample numbers are required for statistically meaningful results.
Methods
Flea collection and questionnaire
To collect flea samples for the present study, a nationwide campaign was instigated in March 2018 to recruit veterinary practices. Practices that registered an interest in participating were sent a kit, consisting of an inspection protocol, questionnaires, envelopes, sealable bags and flea combs. The protocol instructed veterinary practitioners to select 5 cats and 5 dogs per week at random for four weeks. The randomisation procedure to be adopted was not specified; however, veterinarians were asked to undertake flea inspections for animals where the infestation status was unknown, for example when giving booster injections, routine operations, or when offering free flea checks or at other routine nurse clinics. Each flea check was done using a dampened flea comb. The pet was combed, focusing attention on the parts of the body most likely to harbour fleas: the lower back, tail-head, and posterior and inner thighs. The dampened comb increased the probability of fleas sticking to the comb-teeth long enough to allow capture. At the end of the grooming process, the entire comb was placed in plastic sample bag and sealed. Veterinarians were asked to complete a questionnaire for each animal regardless of whether fleas were found or not, recording the owner address, pet species, breed, sex, neutered status, presence and abundance of fleas, whether the pet had been abroad in the previous two weeks and its insecticidal treatment history. Veterinarians could print and post the questionnaires or submit them online. All flea samples were sent by standard post to the University of Bristol where they were stored at -20 °C.
Data handling, mapping and flea identification
Questionnaire data were entered into Microsoft Excel and the WGS84 (World Geodetic System) map coordinates of each pet owner’s location was recorded. The geographical program QGIS (Open Source Geospatial Foundation Project. https://qgis.org/en/site/, Version 2.18.2) was used to map the location of samples. Fleas were identified to species level using dichotomous keys [25, 26].
DNA extraction and amplification
After identification, fleas were transferred into individual micro-tubes and all the fleas of the same species collected from a single pet, were pooled together into what will be described as a ‘sample’. DNA extraction was carried out using a high throughput system, DNeasy 96 Blood & Tissue Kit (Qiagen, Manchester, UK). The flea samples were crushed using micro-pestles in their respective tubes and thoroughly mixed in 180 μl Buffer ATL and 20 μl proteinase K by vortexing. The samples were spun down and incubated overnight at 56 °C to ensure complete tissue lysis. After overnight incubation, the lysates were spun down and all the contents from each tube were transferred to individual wells of a 96 deep-well plate, leaving behind the flea exoskeleton. Further extraction steps were carried out as per the manufacturer’s guidelines. To check the efficacy of the DNA extraction, a conventional PCR targeting the 18S rRNA gene of fleas [27] was run before running the PCRs for the detection of pathogens.
Flea DNA in extracted samples was detected with conventional PCR that amplified a 1200 base pair (bp) region of the flea 18S rRNA gene. A master mix was made as follows: 5 μl of 2× GoTaq Hot Start Mastermix (Promega, UK), 0.2 μl of 10 μM each forward (18S-F)/reverse (18S-R) primer mix (Table 1) and 2.8 μl water. Two μl of extracted DNA were then added to 8 μl of master mix in 96 well PCR plates using a high throughput automated pipetting system (epMotion P5073, Eppendorf, Stevenage, UK). Flea DNA and water were used as positive and negative controls, respectively. The thermal cycling protocol consisted of an initial denaturation at 95 °C for 2 min, followed by 40 cycles of 95 °C for 20 s, 56 °C for 20 s and 72 °C for 90 s in a thermal cycler (Biorad T100 thermal cycler, Biorad, Watford, UK). Amplified DNA was subjected to electrophoresis in a 2% agarose gel pre-stained with 0.05 μg/ml ethidium bromide and viewed under ultraviolet light. Positive samples were identified as having a defined band of ~1200 bp on the gel.
Table 1.
Details of the qPCR/PCR assays used in the study for the detection of pathogen DNA in flea samples
| Target species (gene) | PCR primer and probe sequences (5'-3') | Product size (bp) | Reference |
|---|---|---|---|
| Flea (18S rRNA) | 18S-F: GATCGTACCCACATTACTTG | 1200 | [27] |
| 18S-R: AAAGAGCTCTCAATCTGTCA | |||
| Dipyllidium caninum (28S rRNA) | F: GCATGCAAGTCAAAGGGTCCTACG | 653 | [48] |
| R: CACATTCAACGCCCGACTCCTGTAG | |||
| Bartonella spp. (ssrA) | F: GCTATGGTAATAAATGGACAATGAAATAA | 299 | [28]a |
| R: GGCTTCTGTTGCCAGGTG | |||
| FAM-ACCCCGCTTAAACCTGCGACG-BHQ1 | |||
| Mycoplasma haemofelis (16S rRNA) | F: GTGCTACAATGGCGAACACA | 80 | [29] |
| R: TCCTATCCGAACTGAGACGAA | |||
| FAM-TGTGTTGCAAACCAGCGATGGT-BHQ1 | |||
| “Candidatus Mycoplasma haemominutum” (16S rRNA) | F: TGATCTATTGTKAAAGGCACTTGCT | 135 | [29] |
| R: TTAGCCTCYGGTGTTCCTCAA | |||
| FAM-TTCAATGTGTAGCGGTGGAATGCGT-BHQ1 | |||
| “Candidatus Mycoplasma turicensis” (16S rRNA) | F: AGAGGCGAAGGCGAAAACT | 138 | [29] |
| R: ACGTAAGCTACAACGCCGAAA | |||
| FAM-CGTAAACGATGGGTATTAGATGTCGGGAT-BHQ1 |
aThe reverse primer has been modified compared to the one described in the paper
Bartonella spp. quantitative PCRs and DNA sequencing
Bartonella spp. were detected using a quantitative PCR (qPCR) targeting a fragment of the ssrA gene [28] modified as follows: each qPCR consisted of GoTaq Hot Start Mastermix (Promega, Southampton, UK), MgCl2 to a final concentration of 4.5 mM, forward and reverse primers (Table 1) at a final concentration of 500 nM each and TaqMan probe (Table 1) at a final concentration of 100 nM, 5 μl of template DNA and water to a final volume of 25 μl. The thermal cycling protocol consisted of an initial denaturation at 95 °C for 2 min and 40 cycles of 95 °C for 15 s and 60 °C for 30 s (Agilent MX3005P qPCR, Agilent, Stockport, UK). Fluorescence data were collected at 520 nm at the end of each annealing/extension step. A positive control sample (of known copy number) and negative control (water) were included on each plate. All samples positive for Bartonella spp. were prepared using a Nucleospin® 96 PCR Clean-up Core Kit (Macherey-Nagel, Düren, Germany) and submitted to a commercial sequencing laboratory (DNA Sequencing & Services, MRC I PPU, School of Life Sciences, University of Dundee, UK) for DNA sequencing using Applied Biosystems Big-Dye Ver 3.1 chemistry on an Applied Biosystems model 3730 automated capillary DNA sequencer.
Dipyllidium caninum PCR
Conventional PCR was used to amplify a 653 bp region of the 28S rRNA gene of D. caninum in the flea samples. A master mix was made as follows: 5 μl of 2× GoTaq Hot Start Mastermix (Promega, Southampton, UK), 0.2 μl of 10 μM each forward and reverse primer (Table 1) and 2.8 μl water. Two μl of extracted DNA were then added to 8 μl of master mix in 96-well PCR plates using a high throughput automated pipetting system (epMotion P5073, Eppendorf, Stevenage, UK). Dipylidium caninum-positive DNA samples (Clinvet, South Africa, www.clinvet.com) were used as positive controls and water as a negative control. The thermal cycling protocol consisted of an initial denaturation at 95 °C for 2 min, followed by 40 cycles of 95 °C for 20 s, 66 °C for 5 s and 72 °C for 30 s in a thermal cycler (Biorad T100 thermal cycler). Agarose gel electrophoresis was used to visualise target amplicons. Positive samples were identified from a defined band of ~650 bp on the gel.
Haemoplasma species qPCRs
Feline haemoplasma DNA was detected using individual species-specific qPCRs targeting the 16S rRNA gene of M. haemofelis, “Ca. M. haemominutum” and “Ca. M. turicensis” (Table 1), as previously described [29]. The canine haemoplasma M. haemocanis is also detected by the M. haemofelis qPCR. The qPCR assay for each species consisted of GoTaq Hot Start Mastermix (Promega), MgCl2 to a final concentration of 4.5 mM, forward and reverse primers (for each species as shown in Table 1) at a final concentration of 200 nM each, TaqMan probe (for each species as shown in Table 1) at 50 nM, 2 μl of template DNA and water to a final volume of 10 μl. A positive control, M. haemofelis (of known copy number) and negative control (water) were included on each plate. The thermal cycling conditions were identical to those described above for the Bartonella spp. qPCR.
Results
Species abundance and distributions
A total of 326 veterinary practices from across the UK participated between April and June 2018 (Fig. 1); during this time a total of 1627 animals (cases) were examined. For 1475, of these a questionnaire was submitted that was wholly or at least partially completed. Among the 1475 animals with questionnaire returns, 812 were cats and 662 were dogs and in one instance no information about the host species was present on the otherwise completed questionnaire (Table 2). Three hundred and twenty-three of the 1475 cases (21.9% ± 95% CI 2.1%) with wholly or partially completed questionnaire records were infested with fleas. Of the 812 cats examined, 228 (28.1% ± 95% CI 3.09%) were infested with fleas and of the 662 dogs examined, 95 (14.4% ± 95% CI 2.67%) carried fleas (Table 2). Amongst the 228 flea infested cats, 92% carried the cat flea C. f. felis, 1.3% the dog flea C. canis, and 4% the rabbit flea S. cuniculi. Among the 95 flea-infested dogs, 90% were infested by cat fleas, 3% by dog fleas, and 2% by rabbit fleas. Small numbers of both cats and dogs were infested with hedgehog, A. erinacei, or hen, Ceratophyllus spp. fleas (Table 2). The median number of fleas collected per animal was 1, although a maximum number of 89 fleas were reported from a cat and 18 from a dog. Flea samples from two cases were too damaged to be identified. The flea species from different cases were widely distributed although, notably, relatively few cases were reported from northern areas, particularly from Scotland (Fig. 2). An additional 152 flea samples were sent without any questionnaire records at all, so no information about the host species from which they were collected was available (Table 2). These fleas were included in the pathogen analysis but could not be included in the calculation of prevalence by host. The data for these 152 flea samples is reported separately from the other samples.
Fig. 1.
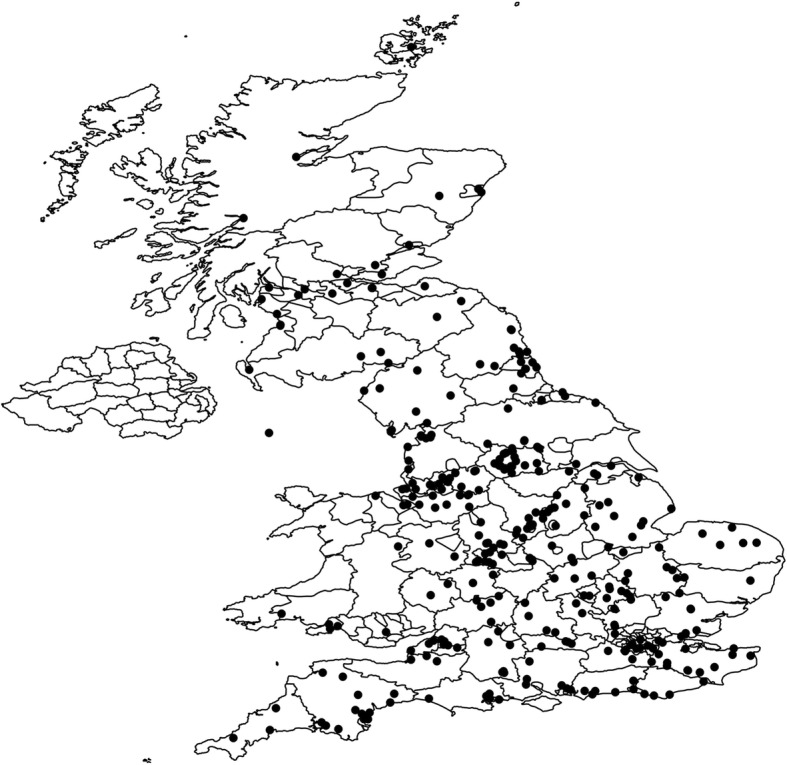
Distribution of 326 veterinary practices that participated in the survey. Black dots indicate the postcode location of the veterinary practice
Table 2.
The number of cats and dogs examined during the survey, the number with or without fleas and number infested with various species of fleas collected from each pet species. In some cases flea identification was not possible (No ID) because the specimens were too damaged, while in others the host species was not recorded (no records) by the veterinary practice
| Pet species | No. of examined animals with records | Fleas absent n (%) | Fleas present n (%) | C. felis felis | C. canis | S. cuniculi | A. erinacei | Ceratophyllus spp. | No ID |
|---|---|---|---|---|---|---|---|---|---|
| Cat | 812 | 584 (71.9) | 228 (28.1) | 210 (25.6) | 3 (0.4) | 9 (1.1) | 2 (0.25) | 3 (0.4) | 1 |
| Dog | 662 | 567 (85.6) | 95 (14.4) | 86 (13.0) | 3 (0.5) | 2 (0.3) | 1 (0.2) | 2 (0.3) | 1 |
| No records | – | – | 152 | 133 | 3 | 8 | 3 | 4 | 0 |
| Total | 1474 | – | 475 | 429 | 9 | 19 | 6 | 9 | 2 |
Fig. 2.
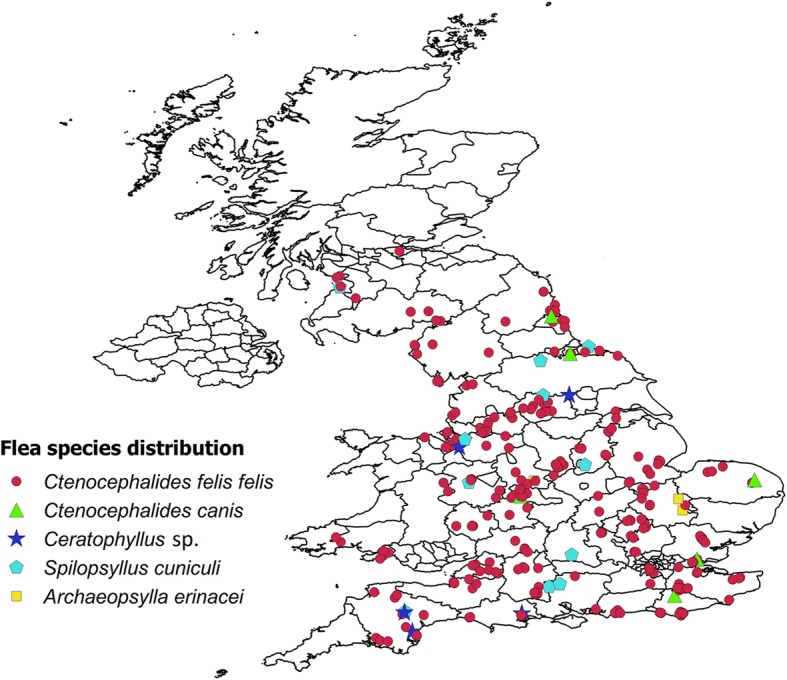
Distribution of different flea species collected from dogs and cats in the UK. The different symbols indicate the postcode location of the pet owner or the veterinary practice
Prevalence and geographical location flea-borne pathogens
DNA samples from 470 pooled flea samples were analysed for D. caninum, Bartonella spp. and haemoplasma species. Of the 470 pooled flea samples, 66 were positive for the DNA of at least one of these pathogens (14% ± 95% CI 3.14%).
Fifty-three of the 470 pooled flea samples (11.3% ± 95% CI 2.85%) were found to be positive for Bartonella spp. DNA. Among these, 35 were collected from cats and 4 from dogs, 14 flea samples did not have any record of their host species. Forty-nine of the Bartonella spp. positive samples were from C. f. felis, 3 were from S. cuniculi and one was from an A. erinacei. Thirty-two of the infected C. f. felis samples were collected from cats, whereas as only 4 samples were collected from dogs; two Bartonella-positive samples from S. cuniculi and one from A. erinacei were also collected from cats. Among the 14 flea samples without any host record, 13 were from C. f. felis and one was from S. cuniculi.
Among the 53 Bartonella spp. qPCR positive samples, 4 could not be sequenced and the remaining 49 samples belonged to four different Bartonella species. Thirty-two were from cats and 3 were from dogs and 14 had no record of the host species from which the fleas were collected (Table 3). Seventeen samples were found to be B. henselae, 27 were B. clarridgeiae, and these belonged to two different strains (Table 3). Four samples were B. alsatica, and 1 was B. grahamii. Fifty of the 53 fleas carrying Bartonella spp. were C. f. felis, two were S. cuniculi and 1 was A. erinacei. The S. cuniculi and A. erinacei flea samples were positive for B. alsatica. Most of the samples positive for Bartonella spp. came from central and southern England (Fig. 3).
Table 3.
Number of different Bartonella species detected in flea samples of different species in different cat or dog hosts. In some cases, the host species was not recorded (no record) by the veterinary practice
| Bartonella spp. | No. of infected flea samples | qPCR Ct values (range) | GenBank ID | Sequence homology (%) | Flea species | Host species |
|---|---|---|---|---|---|---|
| B. grahamii strain as4aup | 1 | 36.94 | HG519007.1 | 99 | C. f. felis | Cat |
| B. alsatica strain IBS 382 | 4 | 22.7–33.17 | JN029776.1 | 95–99 |
S. cuniculi (n = 2); A. erinacei (n = 1); C. f. felis (n = 1) |
Cat (n = 3); no record (n = 1) |
| B. clarridgeiae strain 73 | 24 | 18.1–36.8 | HG519012.1 | 97–99 | C. f. felis | Cat (n = 15); dog (n = 3); no record (n = 6) |
| B. clarridgeiaestrain Rc_AL817-1 | 3 | 35.9–37.5 | KY417894.1 | 96–99 | C. f. felis | Cat (n = 2); no record (n = 1) |
| B. henselae Houston-I | 17 | 21.0–36.3 | CP020742.1 | 94–100 | C. f. felis | Cat (n = 11); no record (n = 6) |
Fig. 3.
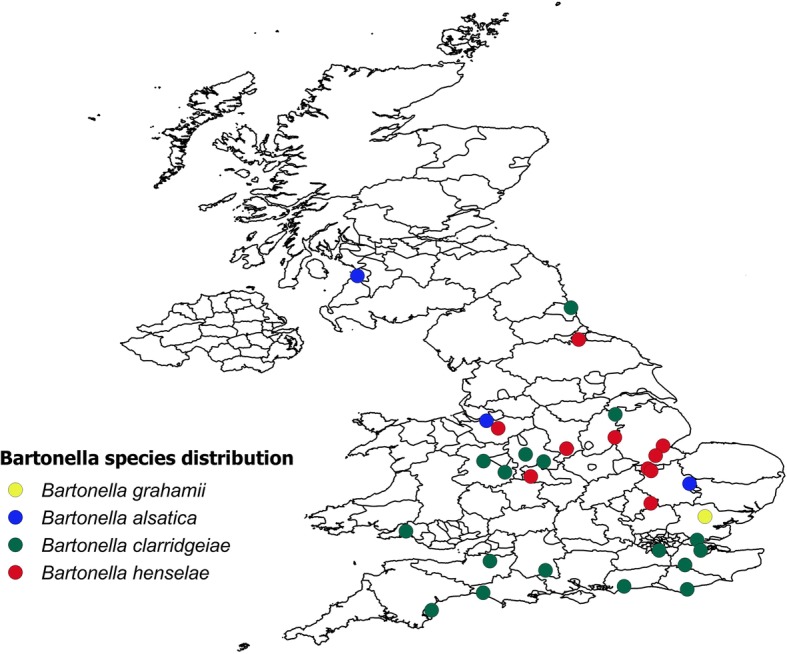
Distribution of Bartonella species detected in fleas collected from dogs and cats in the UK. The circles indicate the postcode location of the pet owner or the veterinary practice
Fourteen of the 470 (3% ± 95% CI 1.53%) pooled flea DNA samples were found to be positive for D. caninum DNA. A positive result was indicated by a single band of ~650 bp by gel electrophoresis and all positive samples had bands of greater than 25 ng per 5 μl loaded. The distribution of the positive samples was widespread, although they were mostly concentrated in southern England and none of the fleas in northern England or Scotland were found to be positive for the DNA of this cestode. Among the 14 positive flea samples, 13 were C.f. felis and one was C. canis. Ten of the infected C. f. felis samples were collected from cats and one from a dog, the other 3 positive flea samples did not have a record of the host species.
The species-specific qPCRs for haemoplasma species found that only 3 flea samples were positive for M. haemofelis or M. haemocanis, all of which were found in C. f. felis fleas, 2 of which were collected from cats and one had no record of the host species. Thus, it would be expected that the infecting species in these flea samples was M. haemofelis. None of the flea samples were found to be positive for either “Ca. M. haemominutum” or “Ca. M. turicensis”.
Three flea samples were positive for both D. caninum and Bartonella spp. One flea sample was positive for both Bartonella spp. and M. haemofelis or M. haemocanis.
Discussion
In the present study, the flea infestation rate was high for both cats (28.1 ± 3.09%) and dogs (14.4 ± 2.67%), but both host species were almost equally likely to carry the cat flea, C. f. felis. Relatively few dog fleas were found, but these did occur more frequently on dogs. Relatively low numbers of rabbit, hedgehog and hen fleas were found, with no Pulex irritans identified. The protocol for recruitment to the study highlighted the need for veterinarians to seach pets brought into the practice for reasons other than flea infestation, suggesting that if known flea-infested animals were included, the prevalence would be even higher. Previous studies have shown that the flea species primarily associated with cats is C. f. felis; however, for dogs the results are more variable. Ctenocephalides felis felis was shown to be the most common species of flea on domestic dogs in the UK [30–32]. High levels of infestation of dogs by C. f. felis have also been reported in North America [33], Denmark [34] and Finland [35]. In contrast, C. canis was found to be more common on dogs in the UK than C. f. felis [36], as was the case in Ireland [37, 38]. In a further study in Ireland of 193 cases, 90% of all infestations on cats were with C. f. felis; only a single cat was found to be infested by C. canis. In contrast, in the dogs 17.5% were infested by C. f. felis and 75.7% by C. canis [39]. A preponderance of C. canis on dogs has also been reported in New Zealand [12], Denmark [40] and the Republic of Korea [41]. In a study of fleas infesting kennel dogs from two localities in Israel, a total of 355 fleas were collected from 107 dogs, of which 74.8% had C. canis, 63.6% had C.f. felis, 14.0% had P. irritans and one animal had Xenopsylla cheopis [42].
Flea borne pathogens in cats and dogs can result in significant levels of clinical disease and are of concern for veterinarians and pet owners [16]. In this study, 11% ± 2.85% of the pooled flea samples of different species and collected from both cats and dogs were found positive for Bartonella spp. Four different Bartonella species were found in the positive flea samples, and these samples were distributed mostly around central and southern UK. This could possibly be because fewer veterinary practices were recruited in northern parts of the UK, but equally may reflect a more southerly distribution of fleas since relatively few cases were submitted from northern England or Scotland; further spatial analysis is required to quantify this trend. The effects of infection by Bartonella spp. may range from asymptomatic to fatal. The most common zoonotic species is B. henselae, for which cats are the major natural reservoir. Fleas, C. f. felis in particular, are the known vectors for B. henselae, B. clarridgeiae and Bartonella koehlerae [17]. The results of this study highlight the anticipated strong association between Bartonella spp. and C. f. felis feeding on cats. Evidence of exposure to Bartonella spp. in cats has been found in many countries, particularly in regions with high humidity [43, 44]. In cats, bartonellosis can result in lymphadenopathy, endocarditis, myocarditis and hyperglobulinemia. However, most cats infected with a Bartonella spp. will show no clinical signs [45]. Since B. henselae survives at least nine days in flea faeces, flea control is imperative to attempt to reduce the risk of infection of other cats, dogs, or people [9, 46]. In this study Bartonella spp. were also found in fleas collected from dogs, which has also been noted in other studies [42] where 7.8% of pooled flea samples from kennel dogs from two localities in Israel were positive for Bartonella DNA. It should be noted that qPCR for Bartonella spp. detection alone may not be the most sensitive approach, which may be achieved more accurately using a combination of conventional and nested PCRs from blood and liquid culture samples [1]. Hence, the prevalence figures reported here may be an underestimate.
Dipylidium caninum is a common intestinal cestode parasite of dogs and cats. The onchospheres are contained in egg packets, each with about 20 eggs, and these are either expelled by the active segment or released by its disintegration. After ingestion by a larval flea intermediate host, the onchospheres travel to its haemocoel, where they develop into cysticercoids. The final host is infected by ingestion of the flea containing the cysticercoids. Occasionally humans have become infected following ingestion of the saliva of infected pets [12]. The potential zoonotic transmission and wide geographical range emphasize the importance of protecting dogs and cats from D. caninum. Routine treatment may be an effective approach to tapeworm management; however, the brief pre-patent period and lack of residual activity of most treatments means that reinfection may occur rapidly. Hence effective and persistent flea control is an important element of any tapeworm management regime. A relatively low prevalence of Dipylidium has been reported in studies using coproscopy. For example, in a study of 2775 dog faecal samples from the Lazio Region of central Italy (1156 from households and 1619 from shelter dogs) only 0.1% were found to be infected [16]. In Greece, in a study of the faeces from 1150 cats, a prevalence of 0.2% was detected [47]. However, the poor sensitivity of coproscopy means that faecal analysis is likely to significantly underestimate infection by Dipylidium. In PCR analyses of flea samples from 435 cats, 4.37% of fleas were found to be infected with D. caninum and in fleas from 396 dogs, 9.1% carried Dipylidium [48]. In the present study, the prevalence of D. caninum was similar, with 14 of the 470 (3 ± 1.53%) flea DNA samples found to be positive for D. caninum DNA but the majority of the D. caninum-positive flea samples were collected from cats and only one was collected from a dog. Of particular interest is the recent finding of two distinct genotypes of D. caninum in dogs and cats, suggesting that two distinct species may be present [49], but this was not investigated in the present study.
Like Bartonella spp., the DNA of haemoplasmas has been amplified from the blood of cats in many regions of the world [44], with M. haemofelis usually considered to be the most pathogenic species [22, 50]. The present study found only 3 flea samples positive for M. haemofelis or M. haemocanis DNA, and it was not possible to differentiate between these two haemoplasma species as their 16S rDNA sequences are near identical [51]. A similarly low prevalence of M. haemofelis DNA was found in ticks collected from pets in the UK [24] and no haemoplasma DNA was found in fleas collected from cats in southern Italy [44]. In contrast in a study involving 1585 cats, found 2.8% cats to be positive for M. haemofelis and 1.7% positive for “Ca. M. turicensis” [29]. The zoonotic importance of feline haemoplasmas is still being questioned [52]. Clinical signs of disease depend on the degree of anaemia, the stage of infection and the immune status of infected cats. Direct transmission may occur with the hemoplasmas, and studies have found some of the agents in saliva [53] and that subcutaneous inoculation of hemoplasma-containing blood resulted in infection transmission [54]. Infection does not necessarily result in clinical disease and in some cases healthy cats can also be positive for haemoplasma DNA in blood [29, 55, 56] and so PCR assay results do not always correlate well with clinical illness.
The methods used to evaluate the prevalence of flea-borne pathogens and the role of fleas as vectors vary in their sensitivity and each is subject to different biases. The use of host blood samples can be problematic. In a study of ectoparasites on cats and vector-borne pathogens in feline blood samples in southern Italy which used qPCR there was little agreement between serological and molecular results in individual cats and the presence of ectoparasites, with the exception of B. clarridgeiae and B. henselae [44]. The authors argued that the ability to detect pathogens in the blood depends strongly on the immunological sensitivity of the host; in addition, the bacteraemia of some pathogens is transient, lasting only a few hours. This makes it difficult to detect pathogens in the blood and requires samples to be taken at very specific time points and the use of highly sensitive molecular tools; the use of serology may therefore underestimate the prevalence of pathogens. The present study investigated the presence of pathogen DNA amplified from fleas but, even though fleas tend to remain on the same host once acquired, the blood in the gut could potentially have come from more than one host, particularly where pets live in close contact in the same household; the presence of pathogen DNA in the gut alone also does not demonstrate vectoral competence [24]. There is a possibility of carryover of the pathogen DNA from the host blood, especially where the Ct values of the qPCR is higher than 36 cycles. The detection of D. caninum DNA from adult fleas collected off a host also requires careful interpretation because, although it is highly indicative, it represents the potential for infection rather than infection itself, since the adult flea would need to be ingested by the grooming host to result in infection. As a result, ideally, a combination of epidemiological indicators is required to establish the true prevalence and the role of arthropod vectors in the transmission of pathogens [57].
Conclusions
The present study indicates that veterinary practices were able to find fleas in a quarter of cats and one sixth of the dogs examined during the study period and the flea samples were found to be positive for a range of infectious agents; in particular the study highlights the relatively high prevalence of Bartonella spp., particularly in central and southern areas, which is of concern for both animal welfare and human health. The study highlights the ongoing need to educate pet owners about the effects of both flea infestation but also the pathogen risks these fleas present.
Acknowledgements
We would like to thank Amanda Melvin and Daniel Guerrero of MSD Animal Health for their help and support throughout the flea collection process, and Megan Barstow, Anna Koi and Marina Metaxaki for laboratory assistance. We are extremely grateful to Clinvet, South Africa, for supplying samples of D. caninum worm segments and D. caninum-infected flea samples for PCR validation. We are grateful to MSD Animal Health for financial support and the Molecular Diagnostic Unit, Diagnostic Laboratories, Langford Vets, for help in performing the PCRs. We would also like to thank all of the participating veterinary practices.
Funding
The study was designed, and the data analysed and interpreted by SA and RW. MSD Animal Health funded the study. The MSD Animal Health sales team also helped to recruit participating veterinary practices. Veterinarians did not receive payment for their participation.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Nucleotide sequence data from this study are available in the GenBank database under the accession numbers MK298151-MK298192.
Abbreviations
- 95% CI
95% confidence interval
- qPCR
Quantitative polymerase chain reaction
- Ct
Cycle threshold
- ng
Nano gram
- μl
Micro litre
- bp
Base pair
- nM
Nano molar
- °C
Degree centigrade
Authors’ contributions
SA was study investigator, coordinated and supervised the sample analysis and undertook statistical analysis. RW was study director and led manuscript preparation. RW and HN assisted in the design and management of the study and interpretation of data. ST and CH provided advice, laboratory support and critically reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval
Ethical approval for this study was given by the University of Bristol’s Animal Welfare and Ethical Review Body with the University Investigation Reference Number, UIN/18/013.
Consent for publication
Not applicable.
Competing interests
SA has no competing interests. RW and CH have had research funded by a range of pharmaceutical companies and animal health charities. RW is director of AgriEnt Ltd. ST is the Chief Medical Officer of Linnaeus Group. HN is an employee of MSD Animal Health.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Swaid Abdullah, Email: swaid.abdullah@uq.edu.au.
Chris Helps, Email: c.r.helps@bristol.ac.uk.
Séverine Tasker, Email: s.tasker@bristol.ac.uk.
Hannah Newbury, Email: hannah.newbury@merck.com.
Richard Wall, Email: richard.wall@bristol.ac.uk.
References
- 1.Drummond MR, Lania BG, de Paiva Diniz PP, Gilioli R, Demolin DM, Scorpio DG, et al. Improvement of Bartonella henselae DNA detection in cat blood samples by combining molecular and culture methods. J Clin Microbiol. 2018;JCM-01732. [DOI] [PMC free article] [PubMed]
- 2.Chomel BB, Kasten RW, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, et al. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck W, Boch K, Mackensen H, Wiegand B, Pfister K. Qualitative and quantitative observations on the flea population dynamics of dogs and cats in several areas of Germany. Vet Parasitol. 2006;137:130–136. doi: 10.1016/j.vetpar.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Bond R, Riddle A, Mottram L, Beugnet F, Stevenson R. Survey of flea infestation in dogs and cats in the United Kingdom during 2005. Vet Rec. 2007;160:503–506. doi: 10.1136/vr.160.15.503. [DOI] [PubMed] [Google Scholar]
- 5.Farkas R, Gyurkovsky M, Solymosi N, Beugnet F. Studies on flea infestation of dogs and cats combined with a questionnaire survey in Hungary. Med Vet Entomol. 2009;23:187–194. doi: 10.1111/j.1365-2915.2009.00798.x. [DOI] [PubMed] [Google Scholar]
- 6.Beugnet F, Franc M. Results of a European multicentric field efficacy study of fipronil-(S) methoprene combination on flea infestation of dogs and cats during 2009 summer. Parasite. 2010;17:337–342. doi: 10.1051/parasite/2010174337. [DOI] [PubMed] [Google Scholar]
- 7.Durden LA, Judy TN, Martin JE, Spedding LS. Fleas parasitizing domestic dogs in Georgia, USA: species composition and seasonal abundance. Vet Parasitol. 2005;130:157–162. doi: 10.1016/j.vetpar.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Beugnet F, Marie JL. Emerging arthropod-borne diseases of companion animals in Europe. Vet Parasitol. 2009;163:298–305. doi: 10.1016/j.vetpar.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Shaw SE, Kenny MJ, Tasker S, Birtles RJ. Pathogen carriage by the cat flea Ctenocephalides felis (Bouché) in the United Kingdom. Vet Microbiol. 2004;102:183–188. doi: 10.1016/j.vetmic.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Lappin MR, Hawley J. Presence of Bartonella species and Rickettsia species DNA in the blood, oral cavity, skin and claw beds of cats in the United States. Vet Dermatol. 2009;20:509–514. doi: 10.1111/j.1365-3164.2009.00800.x. [DOI] [PubMed] [Google Scholar]
- 11.Chandra S, Forsyth M, Lawrence AL, Emery D, Šlapeta J. Cat fleas (Ctenocephalides felis) from cats and dogs in New Zealand: molecular characterisation, presence of Rickettsia felis and Bartonella clarridgeiae and comparison with Australia. Vet Parasitol. 2017;234:25–30. doi: 10.1016/j.vetpar.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Guzman RF. A survey of cats and dogs for fleas: with particular reference to their role as intermediate hosts of Dipylidium caninum. New Zeal Vet J. 1984;32:71–73. doi: 10.1080/00480169.1984.35067. [DOI] [PubMed] [Google Scholar]
- 13.Azad AF, Radulovic S, Higgins JA, Noden BH, Troyer JM. Flea-borne rickettsioses: ecologic considerations. Emerg Infect Dis. 1997;3:319. doi: 10.3201/eid0303.970308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farhang-Azad A, Traub R, Baqar S. Transovarial transmission of murine typhus rickettsiae in Xenopsylla cheopis fleas. Science. 1985;227:543–545. doi: 10.1126/science.3966162. [DOI] [PubMed] [Google Scholar]
- 15.Azad AF, Sacci JB, Nelson WM, Dasch GA, Schmidtmann ET, Carl M. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. Proc Natl Acad Sci USA. 1992;89:43–46. doi: 10.1073/pnas.89.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaramozzino P, Carvelli A, Iacoponi F, De Liberato C. Endoparasites in household and shelter dogs from central Italy. Int J Vet Sci Med. 2018;6:45–47. doi: 10.1016/j.ijvsm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care. 2010;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser PO, Riess T, O’Rourke F, Linke D, Kempf VA. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol. 2011;301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Brenner EC, Chomel BB, Singhasivanon OU, Namekata DY, Kasten RW, Kass PH, et al. Bartonella infection in urban and rural dogs from the tropics: Brazil, Colombia. Sri Lanka and Vietnam. Epidemiol Infect. 2013;141:54–61. doi: 10.1017/S0950268812000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudebouch N, Sarih M, Beaucournu JC, Amarouch H, Hassar M, Raoult D, et al. Bartonella clarridgeiae, B. henselae and Rickettsia felis in fleas from Morocco. Ann Trop Med Parasitol. 2011;105:493–498. doi: 10.1179/1364859411Y.0000000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasker S, Binns SH, Day MJ, Gruffydd-Jones TJ, Harbour DA, Helps CR, et al. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum” in cats in the United Kingdom. Vet Rec. 2003;152:193–198. doi: 10.1136/vr.152.7.193. [DOI] [PubMed] [Google Scholar]
- 22.Tasker S, Peters IR, Day MJ, Willi B, Hofmann-Lehmann R, Gruffydd-Jones TJ, et al. Distribution of Mycoplasma haemofelis in blood and tissues following experimental infection. Microb Pathog. 2009;47:334–340. doi: 10.1016/j.micpath.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornok S, Meli ML, Perreten A, Farkas R, Willi B, Beugnet F, et al. Molecular investigation of hard ticks (Acari: Ixodidae) and fleas (Siphonaptera: Pulicidae) as potential vectors of rickettsial and mycoplasmal agents. Vet Microbiol. 2010;140:98–104. doi: 10.1016/j.vetmic.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Duplan F, Davies S, Filler S, Abdullah S, Keyte S, Newbury H, et al. Anaplasma phagocytophilum, Bartonella spp., haemoplasma species and Hepatozoon spp. in ticks infesting cats: a large-scale survey. Parasit Vectors. 2018;11:201. doi: 10.1186/s13071-018-2789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaucournu JC, Launay H. Les puces (Siphonaptera) de France et du Bassin mediterraneen occidental (Faune de France). Paris, France: Fédération française des sociétés de sciences naturelles; 1990.
- 26.Whitaker, A.P. Siphonaptera. Handbooks for the Identification of British Insects. The Natural History Museum, London. 2007;1:1–178.
- 27.Kaewmongkol G, Kaewmongkol S, McInnes LM, Burmej H, Bennett MD, Adams PJ, et al. Genetic characterization of flea-derived Bartonella species from native animals in Australia suggests host-parasite co-evolution. Infect Genet Evol. 2011;11:1868–1872. doi: 10.1016/j.meegid.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Diaz MH, Bai Y, Malania L, Winchell JM, Kosoy MY. Development of a novel genus-specific real-time PCR assay for detection and differentiation of Bartonella species and genotypes. J Clin Microbiol. 2012;JCM-06621. [DOI] [PMC free article] [PubMed]
- 29.Peters IR, Helps CR, Willi B, Hofmann-Lehmann R, Tasker S. The prevalence of three species of feline haemoplasmas in samples submitted to a diagnostics service as determined by three novel real-time duplex PCR assays. Vet Microbiol. 2008;126:142–150. doi: 10.1016/j.vetmic.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Beresford-Jones WP. Prevalence of fleas on dogs and cats in an area of central London. J Small Anim Pract. 1981;22:27–29. doi: 10.1111/j.1748-5827.1981.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 31.Geary MR. Ectoparasite survey. Vet Dermatol. 1977: Newsletter 2:2-3.
- 32.Chesney CJ. Species of flea found on cats and dogs in south west England: further evidence of their polyxenous state and implications for flea control. Vet Rec. 1995;136:356–358. doi: 10.1136/vr.136.14.356. [DOI] [PubMed] [Google Scholar]
- 33.Harman DW, Halliwell RE, Greiner EC. Flea species from dogs and cats in north-central Florida. Vet Parasitol. 1987;23:135–140. doi: 10.1016/0304-4017(87)90031-8. [DOI] [PubMed] [Google Scholar]
- 34.Kristensen S, Haarløv N, Mourier H. A study of skin diseases in dogs and cats. IV. Patterns of flea infestation in dogs and cats in Denmark. Nord Vet Med. 1978;30:401. [PubMed]
- 35.Saari S, Nikander S. Flea species found on dogs in Finland - a morphological study. Suomen Elanilaakarilehti. 1991;97:362–366. [Google Scholar]
- 36.Edwards FB. Fleas. Vet Rec. 1969;85:665. [Google Scholar]
- 37.Baker KP, Hatch C. The species of fleas found on Dublin dogs. Vet Rec. 1972;91:151–152. doi: 10.1136/vr.91.6.151. [DOI] [PubMed] [Google Scholar]
- 38.Baker KP, Mulcahy R. Fleas on hedgehogs and dogs in the Dublin area. Vet Rec. 1986;119:16–17. doi: 10.1136/vr.119.1.16. [DOI] [PubMed] [Google Scholar]
- 39.Wall R, Shaw SE, Penaliggon J. The prevalence of flea species on cats and dogs in Ireland. Med Vet Entomol. 1997;11:404–406. doi: 10.1111/j.1365-2915.1997.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 40.Haarlov N, Kristensen S. Beiträge zur Dermatologie von Hund und Katze. 3. Flöhe von Hunden und Katzen in Dänemark. Tierärztl Praxis. 1977;5:507–511. [PubMed] [Google Scholar]
- 41.Ahn KS, Huh SE, Seol SW, Kim HJ, Suh KH, Shin S. Ctenocephalides canis is the dominant flea species of dogs in the Republic of Korea. Parasit Vectors. 2018;11:196. doi: 10.1186/s13071-018-2769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sofer S, Gutiérrez R, Morick D, Mumcuoglu KY, Harrus S. Molecular detection of zoonotic bartonellae (B. henselae, B. elizabethae and B. rochalimae) in fleas collected from dogs in Israel. Med Vet Entomol. 2015;29:344–348. doi: 10.1111/mve.12116. [DOI] [PubMed] [Google Scholar]
- 43.Solano-Gallego L, Hegarty B, Espada Y, Llull J, Breitschwerdt E. Serological and molecular evidence of exposure to arthropod-borne organisms in cats from northeastern Spain. Vet Microbiol. 2006;118:274–277. doi: 10.1016/j.vetmic.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Persichetti MF, Solano-Gallego L, Serrano L, Altet L, Reale S, Masucci M, et al. Detection of vector-borne pathogens in cats and their ectoparasites in southern Italy. Parasit Vectors. 2016;9:247. doi: 10.1186/s13071-016-1534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whittemore JC, Hawley JR, Radecki SV, Steinberg JD, Lappin MR. Bartonella species antibodies and hyperglobulinemia in privately owned cats. J Vet Intern Med. 2012;26:639–644. doi: 10.1111/j.1939-1676.2012.00925.x. [DOI] [PubMed] [Google Scholar]
- 46.Šlapeta J, Lawrence A, Reichel MP. Cat fleas (Ctenocephalides felis) carrying Rickettsia felis and Bartonella species in Hong Kong. Parasitol Int. 2018;67:209–212. doi: 10.1016/j.parint.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Symeonidou I, Gelasakis AI, Arsenopoulos K, Angelou A, Beugnet F, Papadopoulos E. Feline gastrointestinal parasitism in Greece: emergent zoonotic species and associated risk factors. Parasit Vectors. 2018;11:227. doi: 10.1186/s13071-018-2812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beugnet F, Labuschagne M, Fourie J, Jacques G, Farkas R, Cozma V, et al. Occurrence of Dipylidium caninum in fleas from client-owned cats and dogs in Europe using a new PCR detection assay. Vet Parasitol. 2014;205:300–306. doi: 10.1016/j.vetpar.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Beugnet F, Labuschagne M, de Vos C, Crafford D, Fourie J. Analysis of Dipylidium caninum tapeworms from dogs and cats, or their respective fleas - Part 2. Distinct canine and feline host association with two different Dipylidium caninum genotypes. Parasite. 2018;25:31. [DOI] [PMC free article] [PubMed]
- 50.Westfall DS, Jensen WA, Reagan WJ, Radecki SV, Lappin MR. Inoculation of two genotypes of Hemobartonella felis (California and Ohio variants) to induce infection in cats and the response to treatment with azithromycin. Am J Vet Res. 2001;62:687–691. doi: 10.2460/ajvr.2001.62.687. [DOI] [PubMed] [Google Scholar]
- 51.Brinson JJ, Messick JB. Use of a polymerase chain reaction assay for detection of Haemobartonella canis in a dog. J Am Vet Med Assoc. 2001;218:1943–1945. doi: 10.2460/javma.2001.218.1943. [DOI] [PubMed] [Google Scholar]
- 52.dos Santos AP, dos Santos RP, Biondo AW, Dora JM, Goldani LZ, De Oliveira ST, et al. Hemoplasma infection in HIV-positive patient, Brazil. Emerg Infect Dis. 2008;14:1922. [DOI] [PMC free article] [PubMed]
- 53.Dean RS, Helps CR, Jones TJ, Tasker S. Use of real-time PCR to detect Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum” in the saliva and salivary glands of haemoplasma-infected cats. J Feline Med Surgery. 2008;10:413–417. doi: 10.1016/j.jfms.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Museux K, Boretti FS, Willi B, Riond B, Hoelzle K, Hoelzle LE, et al. In vivo transmission studies of “Candidatus Mycoplasma turicensis” in the domestic cat. Vet Res. 2009;40:1–4. doi: 10.1051/vetres/2009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willi B, Boretti FS, Baumgartner C, Tasker S, Wenger B, Cattori V, et al. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland. J Clin Microbiol. 2006;44:961–969. doi: 10.1128/JCM.44.3.961-969.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roura X, Peters IR, Altet L, Tabar MD, Barker EN, Planellas M, et al. Prevalence of hemotropic mycoplasmas in healthy and unhealthy cats and dogs in Spain. J Vet Diagn Invest. 2010;22:270–274. doi: 10.1177/104063871002200219. [DOI] [PubMed] [Google Scholar]
- 57.Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB. Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J Clin Microbiol. 2010;48:3782–3785. doi: 10.1128/JCM.01029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article. Nucleotide sequence data from this study are available in the GenBank database under the accession numbers MK298151-MK298192.


