Abstract
Background
Breast cancer patients under neoadjuvant chemotherapy includes a heterogeneous group of patients who eventually develop distal disease, not detectable by current methods. We propose the use of exosomal miRNAs and circulating tumor cells as diagnostic and predictive biomarkers in these patients.
Methods
Fifty-three breast cancer women initially diagnosed with localized breast cancer under neoadjuvant chemotherapy were prospectively enrolled in this study. However, six of them were later re-evaluated and diagnosed as metastatic breast cancer patients by PET-CT scan. Additionally, eight healthy donors were included. Circulating tumor cells and serum exosomal miRNAs were isolated from blood samples before and at the middle of neoadjuvant therapy and exosomal miRNA levels analyzed by qPCR.
Results
Before neoadjuvant therapy, exosomal miRNA-21 and 105 expression levels were higher in metastatic versus non-metastatic patients and healthy donors. Likewise, higher levels of miRNA-222 were observed in basal-like (p = 0.037) and in luminal B versus luminal A (p = 0.0145) tumor subtypes. Exosomal miRNA-222 levels correlated with clinical and pathological variables such as progesterone receptor status (p = 0.017) and Ki67 (p = 0.05). During neoadjuvant treatment, exosomal miRNA-21 expression levels directly correlated with tumor size (p = 0.039) and inversely with Ki67 expression (p = 0.031). Finally, higher levels of exosomal miRNA-21, miRNA-222, and miRNA-155 were significantly associated with the presence of circulating tumor cells.
Conclusion
Liquid biopsies based on exosomal miRNAs and circulating tumor cells can be a complementary clinical tool for improving breast cancer diagnosis and prognosis.
Electronic supplementary material
The online version of this article (10.1186/s13058-019-1109-0) contains supplementary material, which is available to authorized users.
Keywords: Exosomes, microRNA, Circulating tumor cells, Localized breast cancer, Neoadjuvant chemotherapy, Conservative surgery, Cancer diagnosis, Cancer prognosis
Background
Breast cancer (BC) is the most frequently diagnosed cancer in women and the second most common cancer worldwide [1]. Despite considerable advances in early detection, diagnosis, and treatment, BC is among the leading causes of cancer-related deaths in women due to recurrent metastatic disease [2]. Approximately 20 to 25% of women diagnosed with localized BC (LBC) are subjected to neoadjuvant therapy [3]. These tumors remain a noteworthy clinical problem, as a significative percentage of these patients will develop metastatic disease, despite appropriate treatment [4, 5]. Therefore, early detection of the systemic disease is especially important for improving the clinical outcomes in these patients [6]. Thus, despite the improvement of the imaging techniques and diagnostic biomarkers, they are not yet fully satisfactory, principally due to important limitations in detecting distal disease [7, 8]. In this context, liquid biopsy (LB) emerges as an increasing important tool for early tumor diagnosis, recurrence monitoring, and therapeutic guidance [9]. LB provides a non-invasive alternative to traditional “solid biopsies,” which cannot be consistently performed in certain situations, in “real time,” or as easily under recurring sampling and monitoring. LB also has significant value improving our knowledge about the metastatic processes occurring in blood. Consequently, with further clinical validation, LB could allow a better patient risk stratification and, therefore, a better patient treatment choice [10].
The presence of circulating tumor cells (CTCs) and cell-free nucleic acids (cfNAs) such as DNA, mRNA, and microRNA in blood has been recognized, and their clinical relevance is considered attractive as novel biomarkers [11, 12].
CTCs can be used as markers of disease progression, as early indicators of metastasis, and as mediators of drug resistance in BC [13, 14].
The interest of exosomal microRNAs (EmiRs) in cancer has been vastly intensified [15]. In fact, aberrantly expressed microRNAs in tissues, serum/plasma, and CTCs have been explored in the development of new BC biomarkers [16–18]. However, this has not yet been proved to be successful, mainly due to confounding factors impacting levels of circulating miRNAs and potentially compromising their potential as disease biomarkers [19]. In response to this, exosomes are suggested, as containers of miRNAs, to solve these problems [20].
In this work, we sought to evaluate the role of an EmiR panel in the diagnosis and prediction of treatment response in BC patients under neoadjuvant therapy. We further correlated the CTC findings with specific EmiR profiles and clinical and pathological characteristics of the matching primary tumors.
Methods
Experimental design
BC women (n = 53) initially diagnosed with LBC susceptible to receive neoadjuvant chemotherapy were prospectively enrolled in this study. Forty-seven of them underwent neoadjuvant treatment while the other six, initially diagnosed as LBC, were newly evaluated and considered as metastatic breast cancer (MBC) after PET-CT scan evaluation (Additional file 1). Peripheral blood samples (10 ml in EDTA Vacutainer® tubes for CTCs and 5 ml in BD Vacutainer® SST™ II Advance tubes for serum) were extracted at diagnosis time (Ext1) and after 4 cycles of doxorubicin/cyclophosphamide (Ext2). CTCs and exosomes were isolated following the protocol established by our group [10], and identification and counting were performed in a computerized fluorescence microscope (Zeiss AXIO Imager) (Additional file 1).
Biomarker analysis
miRNAs were extracted from exosomes using the Maxwell® 16 miRNA Tissue kit (Promega, USA). Complementary DNA was synthesized using 10 ng of total miRNA and the TaqMan™ Advanced miRNA cDNA Synthesis Kit (Applied Biosystems, USA) following the manufacturer’s protocol. A panel of five miRNAs was designed based on their association with previously reported regulatory roles in cell proliferation, dissemination, and invasion when overexpressed in BC. In addition, the selection was based on their reported mRNA targets of genes as ESR1, progesterone receptors (PGR), and ERBB2 and oncogenes as FOXO3, using the database starBase v3.0 [21]: miR21-5p (significantly correlated with advanced clinical stage, lymph node metastasis, and poor prognosis) [22], miR222-3p (facilitates growth, metastasis, and invasion of a variety of malignant tumors), miR221-3p (predicting distant metastases and poor prognosis) [23], miR155-5p (closely related to the status of estrogen receptor (ER) and PGR) [24], and miR105-5p (potently inducing migration and proliferation in metastatic breast cancer (MBC) cells) [25]. Expression levels of miRNAs were analyzed using TaqMan™ MicroRNA assay probes and TaqMan™ Universal PCR Master Mix (Applied Biosystems, USA) in a 7900HT Fast Real-Time PCR System.
Statistical methods
The main objective was to investigate the expression of five microRNAs in exosomes of BC patients and correlate their expression levels with clinical and pathological parameters and BC subtypes. The secondary objective was to test the association between the presence of CTC count at baseline and EmiR expression.
The statistical analyses were performed using R and SPSS 14.0 software (SPSS). To test if the microRNA expression was significantly different between patients with different clinicopathological parameters and BC subtypes, a non-parametric Wilcoxon signed-rank test was used when two groups were compared. To compare more than two groups, a non-parametric Kruskal-Wallis test was used instead. To test the correlation between number of CTCs and expression of EmiRs, Spearman’s rank correlation coefficient was used. Logistic binary regression and receiver-operating characteristics (ROC) curves were performed to test the sensibility and specificity of the Emir-21 (Ext1) and clinical biomarkers to identify MBC.
The presence of at least one CTC per 10 ml was considered a positive result according to the reported analytic detection limit of our assay [10]. Data are expressed as means or numbers (%). Two-tailed p < 0.05 values were considered statistically significant. Fisher test was calculated to assess the association between clinicopathological variables and the CTC status.
Results
Exosomal miRNA levels associated with LBC and MBC stages
After fully characterizing exosomes derived from cell lines (Additional file 1: Figure S1), we firstly compared the EmiR panel (21, 222, 221, 105, and 155) between 53 BC patients and 8 healthy donors to investigate the diagnostic role of these EmiRs and, secondly, between different disease stages: LBC and MBC to detect their prognostic implications.
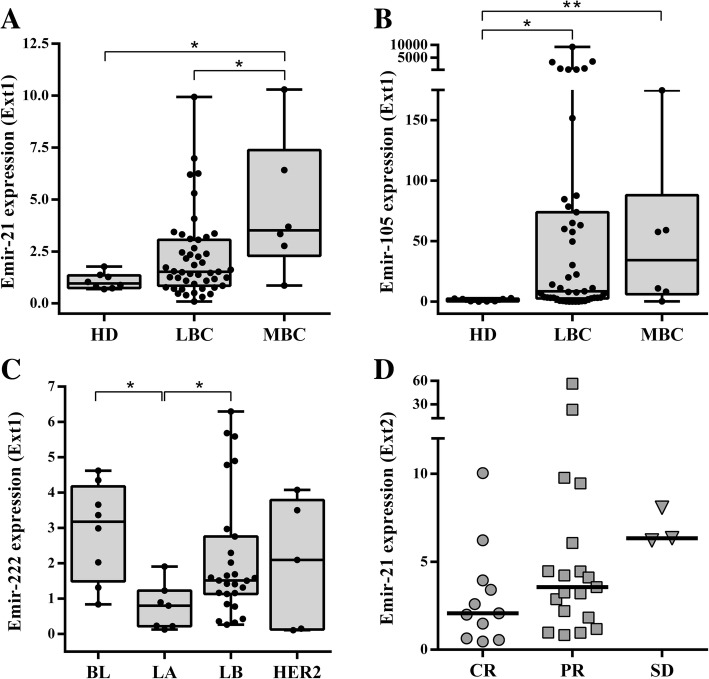
EmiR expression levels were statistically different between groups (healthy donors and LBC and MBC patients). Both EmiR-21 (p = 0.017) and EmiR-105 (p = 0.009) showed statistically significant differences between the three groups (Kruskal-Wallis test). Higher expression levels of EmiR-21 and EmiR-105 (p = 0.013 and p = 0.029 respectively) were found in MBC patients compared to healthy donors (Wilcoxon test). Furthermore, comparisons between LBC and MBC were found significantly different for EmiR-21 (p = 0.027) but not for EmiR-105 (p = 0.71). Emir-21 diagnostic potential of MBC was compared with the current clinical biomarkers carbohydrate antigen 19.9 and carcinoembryonic antigen by logistic binary regression. The Emir-21 was an independent diagnostic biomarker with a HR = 1.404 (95% CI = 1.028–1.918); (p = 0.033) (Additional file 1: Table S1) and the area under the curve (AUC) of the ROC curve was 0.777 (95% CI = 0.566–0.987) (p = 0.029) (Additional file 1: Figure S2). However, none of the clinical biomarkers could significantly identify MBC patients (Additional file 1: Table S1). Finally, EmiR-105 (p = 0.002), but not EmiR-21 (p = 0.09), was able to discriminate between healthy donors and LBC patients (Fig. 1a, b).
Fig. 1.
EmiR expression is associated with clinical stage, cancer subtype, and clinical response to neoadjuvant treatment. Expression of EmiR-21 (a) and EmiR-105 (b) compared between healthy donors (HD), non-metastatic (LBC), and metastatic (MBC) breast cancer patients. EmiR-222 (c) expression according to Perou’s classification in the LBC cohort, divided into four groups: basal-like (BL), luminal A (LA), luminal B (LB), and HER2. EmiR-21 (d) expression at Ext2 in patients with complete response (CR), partial response (PR), or stable disease (SD). Data are presented as a box and whiskers plot (min to max). *p > 0.05 and **p < 0.01. No significant comparisons are not represented
Exosomal miRNAs and clinical characteristics in LBC patients under neoadjuvant chemotherapy
The association between expression levels of the EmiR panel at Ext1 and Ext2 and the clinical and pathological features are respectively summarized in Tables 1 and 2.
Table 1.
Association between EmiR expression at Ext1 and clinicopathological characteristics in LBC patients
| Ext1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EmiR-21 | EmiR-222 | EmiR-221 | EmiR-105 | EmiR-155 | ||||||||
| n | Median | p value | Median | p value | Median | p value | Median | p value | Median | p value | ||
| Age (years) | < 50 | 27 | 1.518 | 0.983 | 1.506 | 0.966 | 1.161 | 0.667 | 11.199 | 0.897 | 1.901 | 0.302 |
| ≥ 50 | 20 | 1.671 | 1.592 | 1.375 | 8.362 | 3.247 | ||||||
| Menopause | Pre | 30 | 1.579 | 0.330 | 1.585 | 0.658 | 1.399 | 0.232 | 9.474 | 0.947 | 1.904 | 0.250 |
| Post | 17 | 1.424 | 1.424 | 1.088 | 8.597 | 3.370 | ||||||
| T | T1–T2 | 27 | 1.265 | 0.039* | 1.506 | 0.813 | 1.173 | 0.780 | 3.851 | 0.067 | 1.906 | 0.093 |
| T3–T4 | 20 | 1.787 | 1.556 | 1.379 | 22.184 | 4.073 | ||||||
| N | 0 | 20 | 1.443 | 1.000 | 1.315 | 0.342 | 1.197 | 0.733 | 8.362 | 0.846 | 2.436 | 0.770 |
| 1–3 | 23 | 1.543 | 1.584 | 1.088 | 30.239 | 3.019 | ||||||
| Estrogen receptor | Negative | 13 | 1.615 | 0.812 | 2.992 | 0.161 | 1.221 | 0.812 | 49.757 | 0.140 | 2.776 | 0.981 |
| Positive | 34 | 1.462 | 1.417 | 1.155 | 7.072 | 2.892 | ||||||
| Progesterone receptor | Negative | 17 | 1.615 | 0.773 | 2.992 | 0.018* | 1.597 | 0.465 | 49.757 | 0.116 | 2.776 | 0.947 |
| Positive | 30 | 1.403 | 1.271 | 1.064 | 7.072 | 2.778 | ||||||
| HER2 | Negative | 36 | 1.531 | 0.466 | 1.465 | 0.880 | 1.149 | 0.563 | 8.362 | 0.744 | 3.117 | 0.421 |
| Positive | 11 | 1.265 | 2.019 | 1.596 | 14.130 | 1.055 | ||||||
| KI67 | < 20% | 8 | 1.157 | 0.458 | 0.850 | 0.050 | 1.279 | 0.573 | 3.380 | 0.103 | 2.219 | 0.373 |
| ≥ 20 | 37 | 1.518 | 1.584 | 1.161 | 14.130 | 3.019 | ||||||
| Perou’s classification | Basal-like | 8 | 1.566 | 0.976 | 3.179 | 0.037* | 1.191 | 0.986 | 39.998 | 0.306 | 4.127 | 0.495 |
| Luminal A | 7 | 1.543 | 0.803 | 1.656 | 3.389 | 1.901 | ||||||
| Luminal B | 27 | 1.424 | 1.513 | 1.139 | 7.750 | 3.215 | ||||||
| HER2neu | 5 | 1.960 | 2.094 | 1.597 | 60.097 | 1.055 | ||||||
Wilcoxon signed-rank test and Kruskal-Wallis test were used. Abbreviations: CTCs circulating tumor cells, Ext1 basal extraction, Ext2 extraction during neoadjuvant treatment, T tumor size, N lymph node status. *p > 0.05
Table 2.
Association between EmiR expression at Ext2 and clinicopathological characteristics in LBC patients
| Ext2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EmiR-21 | EmiR-222 | EmiR-221 | EmiR-105 | EmiR-155 | ||||||||
| n | Median | p value | Median | p value | Median | p value | Median | p value | Median | p value | ||
| Age (years) | < 50 | 19 | 3.975 | 0.585 | 1.851 | 0.536 | 1.705 | 0.771 | 2.534 | 0.166 | 2.173 | 0.536 |
| ≥ 50 | 14 | 3.033 | 2.100 | 1.718 | 11.182 | 1.401 | ||||||
| Menopause | Pre | 20 | 4.159 | 0.094 | 2.250 | 0.461 | 1.890 | 0.338 | 3.916 | 0.224 | 2.177 | 0.768 |
| Post | 13 | 2.192 | 1.551 | 1.422 | 10.603 | 1.507 | ||||||
| T | T1–T2 | 18 | 3.668 | 0.233 | 2.382 | 0.828 | 2.238 | 0.219 | 16.929 | 0.112 | 2.601 | 0.159 |
| T3–T4 | 15 | 3.246 | 1.851 | 1.286 | 3.484 | 1.819 | ||||||
| N | 0 | 11 | 3.975 | 0.621 | 3.037 | 0.621 | 2.490 | 0.006* | 21.588 | 0.445 | 1.344 | 0.928 |
| 1–3 | 18 | 3.477 | 2.250 | 1.095 | 8.885 | 2.177 | ||||||
| Estrogen receptor | Negative | 8 | 2.133 | 0.450 | 1.213 | 0.475 | 1.095 | 0.585 | 5.405 | 0.529 | 0.839 | 0.120 |
| Positive | 25 | 3.933 | 2.649 | 2.000 | 11.760 | 2.173 | ||||||
| Progesterone receptor | Negative | 11 | 2.192 | 0.541 | 1.338 | 0.760 | 1.787 | 0.541 | 7.327 | 0.789 | 3.224 | 0.849 |
| Positive | 22 | 3.668 | 2.250 | 1.686 | 8.528 | 1.899 | ||||||
| HER2 | Negative | 27 | 3.933 | 0.031* | 3.037 | 0.056 | 1.705 | 0.744 | 5.297 | 0.484 | 2.173 | 0.161 |
| Positive | 6 | 0.857 | 0.536 | 41.874 | 46.810 | 0.639 | ||||||
| KI67 | < 20% | 6 | 4.996 | 0.427 | 1.464 | 0.283 | 0.321 | 0.176 | 3.691 | 0.963 | 1.355 | 0.191 |
| ≥ 20 | 27 | 3.246 | 3.037 | 1.787 | 10.444 | 3.201 | ||||||
| Perou’s classification | Basal-like | 5 | 2.192 | 0.655 | 1.338 | 0.666 | 1.286 | 0.666 | 3.484 | 0.510 | 0.983 | 0.232 |
| Luminal A | 5 | 6.058 | 0.729 | 0.334 | 2.085 | 1.367 | ||||||
| Luminal B | 20 | 3.324 | 3.052 | 2.038 | 16.580 | 3.459 | ||||||
| HER2neu | 3 | 2.074 | 0.175 | 0.307 | 10.603 | 0.695 | ||||||
Wilcoxon signed-rank test and Kruskal-Wallis test were used. Abbreviations: CTCs circulating tumor cells, Ext2 extraction during neoadjuvant treatment, T tumor size, N lymph node status. *p > 0.05
At Ext1, 20 of 47 patients (42.55%) with larger tumors (III–IV) showed significantly higher EmiR-21 levels (p = 0.039) than those with smaller tumors.
According to hormone receptor expression at time of diagnosis, PGR-negative patients (17 of 47, 36.2%) showed a positive association with EmiR-222 expression (p = 0.017) at Ext1. Furthermore, 37 of 45 patients (82%) showed association between EmiR-222 at Ext1 and the proliferation marker Ki-67 (p = 0.050), that it was significantly associated with lower EmiR-21 levels at Ext2 (p = 0.030). With respect to HER2, an inverse significant association was found for EmiR-21 at Ext2 (p = 0.031) but not at Ext1 (p = 0.466). At the Ext2, we found lower levels of EmiR-221 Ext2 in lymph node-affected patients (p = 0.006).
BC subtypes and EmiR profile
We correlated our five EmiR panel with the four major distinct molecular BC subtypes, according to Perou’s classification (Table 1) to address the clinical and relevant need of identifying subgroups. At Ext1, EmiR-222 was significantly associated with the different patient subgroups (p = 0.037): it was under-expressed in luminal A tumors vs. basal-like tumors (p = 0.004) and under-expressed in luminal A vs. luminal B (p = 0.015). However, non-significant differences were found between basal-like and luminal A/B, basal-like and HER2, or luminal A/B and HER2 (Fig. 1c).
Predictive role of EmiRs
We analyzed the association of the EmiR panel at Ext2 with clinical response evaluated 3 months after treatment initiation. EmiR-21 did not show significant differences among partial response (PR), stable disease (SD), and complete response (CR) groups (p = 0.062) and neither when comparing CR patients vs. those with worse response to therapy (PR + SD) (p = 0.060) (Fig. 1d).
Association between EmiR expression and CTC presence in LBC
We identified CTCs in 17 of 47 LBC patients (36.17%) at Ext1. Mean number of CTCs present was 1.23 cells per 10 ml of blood (range 0–10). At Ext2, CTCs were identified in 19 of 47 (40.43%) LBC patients, with a mean value of 1.60 cells per 10 ml (range 0–11). No significant differences were found between the presence of CTCs at Ext1 vs. Ext2 (p = 0.301 and p = 0.392 respectively). Correlation between CTC status and the clinical and pathological characteristics is shown in Additional file 1: Table S2.
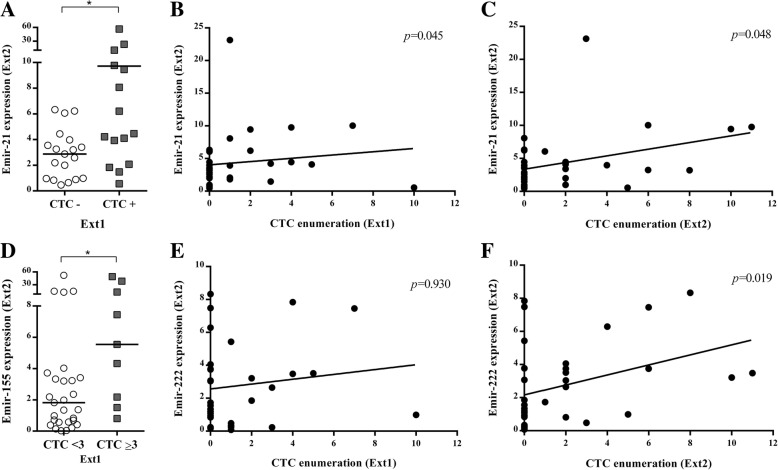
Moreover, EmiR-21, EmiR-222, and EmiR-155 showed a positive correlation with the presence and/or number of CTCs in LBC patients. In fact, higher levels of EmiR-21 at Ext2 were associated with the presence of CTCs at Ext1 (p = 0.032) (Fig. 2a). Besides, according to the number of CTCs, we observed higher EmiR-21 levels at Ext2 in those patients with higher number of CTCs at Ext1 (p = 0.045) and Ext2 (p = 0.048) (Fig. 2b, c). Furthermore, we observed a similar association between CTCs and EmiR-155 levels at Ext2. EmiR-155 was significantly overexpressed in the group of patients with ≥ 3 CTCs/10 ml at Ext1 (p = 0.039) (Fig. 2d). Finally, an association was found between higher levels of EmiR-222 and higher number of CTCs at Ext2 (p = 0.019) but not at Ext1 (Fig. 2f, e).
Fig. 2.
CTC detection correlates with EmiR expression. Comparison of EmiR-21 expression at Ext2 in patients with the presence or absence of CTCs at Ext1 (a), and correlation with the number of CTCs at Ext1 (b) and Ext2 (c). EmiR-155 expression comparison between patients with three or more CTCs per sample and less than three CTCs (d). Correlation of EmiR-222 expression and number of CTCs at Ext1 (e) and Ext2 (F). *p > 0.05
Discussion
In the present study, we analyzed the role of five different exosomal miRNAs as a clinical tool to understand diagnostic, prognostic, and treatment response assessment.
Interestingly, one of the most important results observed in our study was the correlation between EmiR expression and disease stage. This way, EmiR-21 expression levels can distinguish localized from distant diseased patients. We also found differences in EmiR-105 levels between localized BC and healthy donors. These results might have important clinical implications for the correct identification of the disease and subsequent finest treatment choice. This could be particularly important in BC patients undergoing neoadjuvant chemotherapy, as it involves an advanced non-metastatic stage and includes a variety of clinical scenarios. A significative proportion of patients under neoadjuvant chemotherapy will end up developing metastatic disease, despite the administrated treatment. In addition, a percentage of them present distal disease at the time of diagnosis, which the current clinical methods are unable to detect. In consequence, these patients could be misdiagnosed and hence not appropriately treated. Therefore, the ability of LB to differentiate among localized disease, occult systemic disease, or not and healthy donors should be noted. A potential application would lay on the context of BC diagnostic, as despite the generally satisfactory results from mammograms, they have some important limitations, which can involve false negatives (i.e., greater proportion of cancers are detected after a negative mammogram). In fact, younger women (40–49 years) have lower mammographic sensitivity than older ones (≥ 50 years). We suggest that these limitations might be addressed by incorporating EmiR-21 and EmiR-105 analysis, in addition to mammogram tests. In this way, the LB could identify patients with metastatic disease, even those patients who are misdiagnosed as non-metastatic by the current clinical methods.
To assess the diagnostic ability of these EmiRs, we analyzed the correlation between EmiR levels in serum and different clinicopathological characteristics. miRNA-21 has been shown to be overexpressed in numerous types of tumor tissues [26], to be involved in cancer at almost all stages [27], and to be associated with proliferation and therefore growth. In the present study, serum EmiR-21 expression levels were positively correlated with tumor size, indicating that patients with higher serum EmiR-21 levels have larger tumors. Interestingly, we found lower levels of EmiR-221 during neoadjuvant treatment, in those patients with lymph node affection (N1-N3). The expression of miR-221 has been identified as a good prognostic biomarker in breast cancer tissues, and it is associated with ER positivity and lymph node negativity [28]. However, the miR-221 is associated with tamoxifen resistance in breast cancer cells [29]. In addition, Miller et al. [30] reported that miR-221/222 expressions were upregulated in endocrine therapy-resistant luminal-type breast cancer cells, which could explain our results. The association between EmiR expression levels, tumor size, and lymph node status suggests their potential beneficial use as diagnostic biomarkers to improve the stratification of metastasis risk in LBC.
We also observed lower levels of EmiR-21 in HER2-positive patients during neoadjuvant treatment with Trastuzumab (Ext2). Since EmiR-21 levels were not different between HER2-positive and HER2-negative patients at diagnosis time (Ext1), our results are in accordance with conclusions from similar studies [31, 32] suggesting that this low expression of EmiR-21 might be caused by the MAPK (ERK1/2) pathway blockage through HER2/neu. These data highlight the prognostic value of EmiR-21 to predict the treatment response of these patients to Trastuzumab.
Additionally, an inverse association was detected between EmiR-21 and Ki67 at Ext2. This correlation might predict the biological behavior of BC and its treatment response. Similar conclusions can be found in additional studies where the correlation among Ki67, HER2, and treatment was analyzed [33].
A correct BC diagnosis necessarily involves the identification of the different BC subtypes, what is crucial not only for risk stratification, but also for selecting the most appropriate patient treatment. Currently, identification of these BC subtypes depends on traditional pathological assessments with a limited number of biomarkers to define each subtype due to the technical impossibility of molecular profiling each tissue. However, despite the clinical relevance of these histopathological methods, the information provided is limited, with deficient genetic characterization of each patient and tissue. We observed associations between specific EmiRs and different BC subtypes. Our results, in agreement with the study of Sung EH et al. [34] in which miRNAs related to different BC subtypes and their target genes were identified, suggest that EmiRs could act as putative biomarkers valuable to improve classification and diagnosis of different BC subtypes. Therefore, identification of additional EmiRs will help to improve the genetic characterization of these tumor subtypes. Hence, we suggest that EmiRs, on further validation studies, as liquid biopsy biomarkers might become a non-invasive supplementary tool to the current BC classification methods. The use of these liquid biomarkers involves the possibility to obtain genetic information about the status of the disease on different temporal points, since BC is a dynamic disease that changes in the space and over time. These biological dynamics might be responsible for future relapses and resistance to treatment [35]. Therefore, the detection of changes in EmiRNA levels in biological fluids over time, might provide this information. Despite it was not the objective of our work, the EmiRNAs could be used to detect relapses in early stages and to detect early resistance to treatment.
Finally, we found a positive association between the presence of CTCs at baseline status and higher levels of EmiR-21, EmiR-155, and EmiR-222. As these miRNAs have been associated with tumor aggressiveness, promoting the proliferation and migration of tumor cells [22–24], higher levels of these EmiRs could explain more CTC dissemination.
Conclusion
Currently used techniques are not sufficiently sensitive or specific so as to ensure a correct diagnosis of metastatic disease in breast cancer and certainly are unsatisfactory for therapeutic guidance. New approaches are needed to address this problem, requiring further research that involves novel markers and technologies to discover and validate improved biomarkers such as EmiRs. In this way, liquid biopsy based on serum EmiRs can become an additional clinical tool for improving BC diagnosis.
Limitations of the study and interpretation
We would like to remark that our study included a single hospital with a limited cohort of patients; therefore, our results need further validation with a larger patient population. Inclusion of more patients, but also healthy donors, would improve the statistical power of our results. According with these limitations and as a result of the data of the EmiRNA profile established in this work, we are validating these results in different Spanish hospitals, using as central laboratory, the GENyO center.
On the other hand, the clinical utility of EmiRs should include reproducibility and analytic validity. These requirements involve the establishment of standard operating protocols (SOP) through multicenter clinical trials. These SOP not only should include the molecular analyses, but also include how the sample is collected, stored, and transported until the final molecular analyses. For LB to become a clinical reality instead of a simple proposal, the establishment and approval of these SOP by different laboratories and the performance of multicenter clinical trials is needed.
Additional files
Figure S1. Exosome characterization by TEM and Western blot. TEM images of exosomes derived from BC cell lines demonstrated that our methodology was successful in isolating exosomes, observing double-membrane vesicles with a diameter of ~ 150 nm (A). Furthermore, Western blot characterization showed positive expression of Hsp70 and CD9 exosomal proteins in these exosomes but negative expression of GM-130, which was present in the MCF-7 lysate positive control (B). Figure S2. ROC Curve of MBC identification by EmiR-21 (Ext1). Gray line represents EmiR-21 (Ext1) values for sensibility and specificity while black dotted line represents random predictor baseline. Light gray area represents EmiR-21 (Ext1) area under the curve (AUC) = 0.777. Table S1. Univariate logistic binary regression for MBC identification. Abbreviations: CA19.9, Carbohydrate Antigen 19.9; CEA, Carcinoembryonic Antigen; Ext1, basal extraction; HR, Hazard Ratio. Table S2. Association between CTC presence and clinicopathological features. Abbreviations: CTCs, circulating tumor cells; Ext1, basal extraction; Ext2, extraction during neoadjuvant treatment; T, tumor size; N, lymph node status. (ZIP 352 kb)
Acknowledgements
We would like to extend our gratitude to the all the patients and the healthy volunteers who participated in the study and the University of Granada, Biomedicine PhD program.
Funding
This work was supported by “Granada Research of Excellence Initiative on BioHealth (GREIB)”, the PhD grant from the University of Granada and the PhD grant from the Spanish Government (FPU) 2014, REF FPU14/05461.
Availability of data and materials
Not applicable.
Abbreviations
- AUC
Area under the curve
- BC
Breast cancer
- cfNAs
Cell-free nucleic acids
- CR
Complete response
- CTCs
Circulating tumor cells
- EmiRs
Exosomal microRNAs
- ER
Estrogen receptor
- Ext1
Diagnosis time
- Ext2
After 4 cycles of doxorubicin/cyclophosphamide
- LB
Liquid biopsy
- LBC
Localized breast cancer
- MBC
Metastatic breast cancer
- N
Lymph node status
- PGR
Progesterone receptor
- PR
Partial response
- ROC
Receiver-operating characteristics
- SD
Stable disease
- SOP
Standard operating protocols
- T
Tumor size
Authors’ contributions
MJS, JAL, and JLP conceived and designed the study. MJS, AR, DdeM-P, FGO, IR-F, JM-M, and PC-S analyzed and interpreted the data. MJS, CR, and JAL wrote the manuscript. HI and MCG-N provided the critical revisions. HI and JAL approved the final version of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Independent Ethics Committee of San Cecilio Hospital and written consents were provided and signed by all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alba Rodríguez-Martínez, Email: alba.rodriguez@genyo.es.
Diego de Miguel-Pérez, Email: diego.miguel@genyo.es.
Francisco Gabriel Ortega, Email: gabrielfranciscoortega@gmail.com.
José Luis García-Puche, Email: jlpuche@ugr.es.
Inmaculada Robles-Fernández, Email: inmaculada.robles@genyo.es.
José Exposito, Email: jose.exposito.sspa@juntadeandalucia.es.
Jordi Martorell-Marugan, Email: jordi.martorell@genyo.es.
Pedro Carmona-Sáez, Email: pedro.carmona@genyo.es.
María del Carmen Garrido-Navas, Email: carmen.garrido@genyo.es.
Christian Rolfo, Email: Christian.Rolfo@uza.be.
Hugh Ilyine, Email: hugh@destinagenomics.com.
José Antonio Lorente, Email: jlorente@ugr.es.
Marta Legueren, Email: marta.legeren@gmail.com.
María José Serrano, Phone: +34 958 715 500, Email: mjose.serrano@genyo.es.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6:229. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tryfonidis K, Senkus E, Cardoso MJ, Cardoso F. Management of locally advanced breast cancer—perspectives and future directions. Nat Rev Clin Oncol. 2015;12:147–162. doi: 10.1038/nrclinonc.2015.13. [DOI] [PubMed] [Google Scholar]
- 4.Garg PK, Prakash G. Current definition of locally advanced breast cancer. Curr Oncol. 2015;22:409. doi: 10.3747/co.22.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandilaras V, Bouganim N, Spayne J, Dent R, Arnaout A, Boileau JF, et al. Concurrent chemoradiotherapy for locally advanced breast cancer-time for a new paradigm? Curr Oncol. 2015;22:25–32. doi: 10.3747/co.22.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allemani C, Sant M, Weir HK, Richardson LC, Baili P, Storm H, et al. Breast cancer survival in the US and Europe: a CONCORD high-resolution study. Int J Cancer. 2013;132:1170–1181. doi: 10.1002/ijc.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heywang-Köbrunner SH, Hacker A, Sedlacek S. Advantages and disadvantages of mammography screening. Breast Care. 2011;6:199–207. doi: 10.1159/000329005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoon CK, Starker EQ, Wilson AM, Emmert-Buck MR, Libutti SK, Tangrea MA. Current molecular diagnostics of breast cancer and the potential incorporation of microRNA. Expert Rev Mol Diagn. 2009;9:455–466. doi: 10.1586/erm.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadal R, Lorente JA, Rosell R, Serrano MJ. Relevance of molecular characterization of circulating tumor cells in breast cancer in the era of targeted therapies. Expert Rev Mol Diagn. 2013;13:295–307. doi: 10.1586/erm.13.7. [DOI] [PubMed] [Google Scholar]
- 10.Nadal R, Fernandez A, Sanchez-Rovira P, Salido M, Rodríguez M, García-Puche JL, et al. Biomarkers characterization of circulating tumour cells in breast cancer patients. Breast Cancer Res. 2012;14:R71. doi: 10.1186/bcr3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi G, Mu Z, Rademaker AW, Austin LK, Strickland KS, Costa RLB, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast Cancer. Clin Cancer Res. 2018;24:560–568. doi: 10.1158/1078-0432.CCR-17-2092. [DOI] [PubMed] [Google Scholar]
- 13.Serrano MJ, Lorente JA, Delgado Rodríguez M, Fernández A, Fernández M, de la Torre C, et al. Circulating tumour cells in peripheral blood: potential impact on breast cancer outcome. Clin Transl Oncol. 2011;13:204–208. doi: 10.1007/s12094-011-0641-x. [DOI] [PubMed] [Google Scholar]
- 14.Riethdorf S, Müller V, Loibl S, Nekljudova V, Weber K, Huober J, et al. Prognostic impact of circulating tumor cells for breast cpatients treated in the neoadjuvant “Geparquattro” trial. Clin Cancer Res. 2017;23:5384–5393. doi: 10.1158/1078-0432.CCR-17-0255. [DOI] [PubMed] [Google Scholar]
- 15.Jia Y, Chen Y, Wang Q, Jayasinghe U, Luo X, Wei Q, et al. Exosome: emerging biomarker in breast cancer. Oncotarget. 2017;8:41717–41733. doi: 10.18632/oncotarget.16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, et al. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damavandi Z, Torkashvand S, Vasei M, Soltani BM, Tavallaei M, Mowla SJ. Aberrant expression of breast development-related MicroRNAs, miR-22, miR-132, and miR-212, in breast tumor tissues. J Breast Cancer. 2016;19:148–155. doi: 10.4048/jbc.2016.19.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega FG, Lorente JA, Garcia Puche JL, Ruiz MP, Sanchez-Martin RM, de Miguel-Pérez D, et al. miRNA in situ hybridization in circulating tumor cells - MishCTC. Sci Rep. 2015;5:9207. doi: 10.1038/srep09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet. 2017;62:15–24. doi: 10.1038/jhg.2016.89. [DOI] [PubMed] [Google Scholar]
- 23.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375–1380. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 25.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Reports. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanna JA, Wimberly H, Kumar S, Slack F, Agarwal S, Rimm DL. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. Biotechniques. 2012;52:235–245. doi: 10.2144/000113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan R, Yang Y, Yang X, Zhao L, Lu J, Meng QH. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther. 2014;21:290–296. doi: 10.1038/cgt.2014.29. [DOI] [PubMed] [Google Scholar]
- 30.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast Cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–18524. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shokouh TZ, Ezatollah A, Barand P. Interrelationships between Ki67, HER2/neu, p53, ER, and PR status and their associations with tumor grade and lymph node involvement in breast carcinoma subtypes: retrospective-observational analytical study. Medicine (Baltimore) 2015;94:e1359. doi: 10.1097/MD.0000000000001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun EH, Zhou Q, Liu K-S, Wei W, Wang CM, Liu XF, et al. Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray. Eur Rev Med Pharmacol Sci. 2014;18:2783–2788. [PubMed] [Google Scholar]
- 35.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Exosome characterization by TEM and Western blot. TEM images of exosomes derived from BC cell lines demonstrated that our methodology was successful in isolating exosomes, observing double-membrane vesicles with a diameter of ~ 150 nm (A). Furthermore, Western blot characterization showed positive expression of Hsp70 and CD9 exosomal proteins in these exosomes but negative expression of GM-130, which was present in the MCF-7 lysate positive control (B). Figure S2. ROC Curve of MBC identification by EmiR-21 (Ext1). Gray line represents EmiR-21 (Ext1) values for sensibility and specificity while black dotted line represents random predictor baseline. Light gray area represents EmiR-21 (Ext1) area under the curve (AUC) = 0.777. Table S1. Univariate logistic binary regression for MBC identification. Abbreviations: CA19.9, Carbohydrate Antigen 19.9; CEA, Carcinoembryonic Antigen; Ext1, basal extraction; HR, Hazard Ratio. Table S2. Association between CTC presence and clinicopathological features. Abbreviations: CTCs, circulating tumor cells; Ext1, basal extraction; Ext2, extraction during neoadjuvant treatment; T, tumor size; N, lymph node status. (ZIP 352 kb)
Data Availability Statement
Not applicable.