Abstract
Background
The NAC (NAM, ATAF1/2, and CUC2) transcription factor family represents a group of large plant-specific transcriptional regulators, participating in plant development and response to external stress. However, there is no comprehensive study on the NAC genes of Tartary buckwheat (Fagopyrum tataricum), a large group of extensively cultivated medicinal and edible plants. The recently published Tartary buckwheat genome permits us to explore all the FtNAC genes on a genome-wide basis.
Results
In the present study, 80 NAC (FtNAC) genes of Tartary buckwheat were obtained and named uniformly according to their distribution on chromosomes. Phylogenetic analysis of NAC proteins in both Tartary buckwheat and Arabidopsis showed that the FtNAC proteins are widely distributed in 15 subgroups with one subgroup unclassified. Gene structure analysis found that multitudinous FtNAC genes contained three exons, indicating that the structural diversity in Tartary buckwheat NAC genes is relatively low. Some duplication genes of FtNAC have a conserved structure that was different from others, indicating that these genes may have a variety of functions. By observing gene expression, we found that FtNAC genes showed abundant differences in expression levels in various tissues and at different stages of fruit development.
Conclusions
In this research, 80 NAC genes were identified in Tartary buckwheat, and their phylogenetic relationships, gene structures, duplication, global expression and potential roles in Tartary buckwheat development were studied. Comprehensive analysis will be useful for a follow-up study of functional characteristics of FtNAC genes and for the development of high-quality Tartary buckwheat varieties.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5500-0) contains supplementary material, which is available to authorized users.
Keywords: Tartary buckwheat, NAC family, Genome-wide, Fruit development, Expression profile, Phylogenetics
Background
Transcription factors (TFs) play an important role in controlling multitudinous vital growth and development processes such as signal transduction, cellular morphogenesis, and response to environmental stressors during the growth and development of plants [1, 2]. TFs activate or inhibit the transcription rate of target genes by binding to specific cis-acting promoter elements to regulate gene expression [3, 4]. The NAC domain protein family is a larger plant-specific TF family. The family name is based on the three proteins: no apical meristem (NAM), ATAF1–2, and cup-shaped cotyledon (CUC), which all possess a similar DNA-binding domain [5, 6]. Typically, the structure of NAC proteins can be divided into conserved N-terminal DNA-binding domains and highly dispersed C-terminal transcriptional regulatory regions [7, 8]. NAC domains comprise five subdomains, A through E, located at the N-terminal, and usually contain approximately 150 amino acid residues, which are related to DNA binding, dimer formation and localization [9–11].
Since the NAC gene was first found to influence pattern formation in embryos and flowers of petunias [5], numerous studies have shown that the NAC protein is important for plant growth and environmental adaptation. [11, 12]. NAC genes affect many important biological processes. In Arabidopsis, NAP is involved in floral morphogenesis [13]. AtNAC1 and AtNAC2 are involved in the development of lateral roots [14, 15]. CUC1 is the second gene indispensable for the formation of cup-shaped cotyledons [16]. ATAF1/2 and CUC2 participate in cell division [17]. RD26 and ANAC036 are involved in hormone signal pathways [18, 19]. NAM-B1 is involved in the deposition of nutrients in wheat grains [20, 21]. OsNAC5 may participate in nutrient substance remobilization from green tissues to seeds [22].
NAC genes are also important regulators of plant abiotic stress response [11]. In Arabidopsis, overexpressing ANAC019/055/072 enhanced tolerance to drought by stress-induced gene expression [23]. In rice, the overexpression of OsNAC6/SNAC2 enhanced seedling tolerance to drought, salt, and cold stress [24, 25]. In Arabidopsis, the heteroexpression of active membrane-bound GmNTL1/GmNLT11 proteins was found to result in improved tolerance of abiotic stressors [26]. In Capsicum, silencing CaNAC2 increased the sensitivity of seedlings to cold stress [27]. In Tartary buckwheat, FtNAC4/6/7/8/9 was involved in responses to drought, high salt, cold, SA and JA stress [28]. In addition, NAC transcription factors play an important role in secondary cell wall (SCW) biosynthesis in plants [29, 30]. In Arabidopsis, the dominant repression of VND6/7 (vascular-related NAC domain) causes an inhibition of secondary wall thickening in vessels [31]. NST1 and NST3 (NAC secondary wall thickening promoting factor) regulated secondary wall biosynthesis, including production of xylary and interfascicular fibers and pod shattering, in a functionally redundant manner [32–34]. AtXND1 (xylem NAC domain 1) negatively regulated terminal secondary wall biosynthesis [35]. In Medicago sativa, an MtNST1 gene mutant exhibited a lack of lignification in interfascicular fibers [36]. In cotton, transgenic experiments indicate that GhFSN1 positively regulates SCW thickness [37].
Tartary buckwheat is a widely cultivated dual-purpose medicinal and food grain crop that contains abundant proteins with a well-balanced composition of essential amino acids, and its content of amino acids is higher than that of primary grain crops. Tartary buckwheat is a rich source of vitamins B1, B2 and B6, dietary fiber and a variety of minerals, such as niacin, magnesium, manganese and phosphorus (adopted from USDA Food Composition Databases: https://ndb.nal.usda.gov/ndb/). Importantly, Tartary buckwheat is a rich source of beneficial phytochemicals. Flavonoids, especially rutin, have anti-fatigue properties and anti-inflammatory activity and can be used to treat microangiopathy [38–41]. Although NAC genes affect cell division and expansion [17], lateral root development [14, 15], maintenance of the shoot apical meristem [5], flower formation [13] and secondary cell wall biosynthesis [29, 30], there are still many problems to be solved. The NAC gene family has been widely studied in many species, such as Arabidopsis thaliana [7], rice [42], wheat [43], soybean [44], maize [45], potato [46], cassava [47], Chinese cabbage [48], pepper [49], melon [50] and apple [51]. However, there is no systematic study of the NAC family in Tartary buckwheat. Due to the fact that NAC genes are crucial to many developmental processes and responses to abiotic stress, it is of great significance to comprehensively study the NAC family of Tartary buckwheat. The recent completion of Tartary buckwheat genome sequencing allowed us to study the NAC gene family of Tartary buckwheat at the genome level [52]. To date, only the ARF and MADS gene families have been systematically analyzed in Tartary buckwheat [53, 54]. In this article, we report the gene structure, chromosomal location, duplication, global expression and potential developmental roles of Tartary buckwheat NAC genes. In addition, a homologous analysis between NAC genes in Arabidopsis thaliana, beet, soybean, tomato, sunflower, rice, grape and Tartary buckwheat was also conducted. The expression of FtNAC genes during plant development, especially fruit development, was comprehensively analyzed. This study is helpful as a follow-up study of the functional characteristics of FtNAC genes in the development of Tartary buckwheat.
Results
Identification of FtNAC genes in Tartary buckwheat
Eighty NAC genes were identified by the two BLAST methods, and the redundant forms of the same gene were removed simultaneously. In this study, FtNACs were named FtNAC 1 to FtNAC 80 according to their chromosome location (Additional file 1: Table S1).
We introduced the basic information of 80 NAC genes in detail, including isoelectric point (PI), molecular weight (MW) and coding sequence length (CDS). Of the 80 FtNAC proteins, the smallest protein was FtNAC 37 with 91 amino acids, and the largest protein was FtNAC 24 (621 aa) (Additional file 1: Table S1). The MWs of the proteins ranged from 10.59 to 69.92 KDa, and the pIs ranged from 4.36 (FtNAC 7) to 11.46 (FtNAC 58). From the location information, we can see that 69 FtNACs were located in the nucleus, 4 FtNACs were located in the cytoplasm, 3 FtNACs were located in the chloroplast, 3 FtNACs were located in the mitochondria, and 1 FtNAC was located in the plasma membrane (Additional file 1: Table S1).
Phylogenetic analysis and classification of FtNAC genes
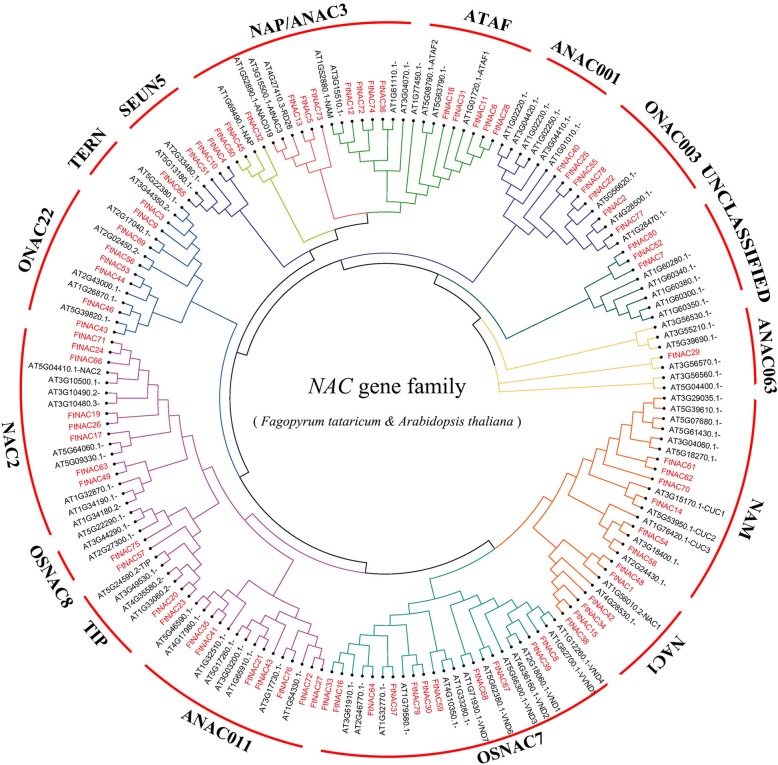
We used amino acid sequences of FtNAC and AtNAC proteins to construct an unrooted phylogenetic tree to explore their evolutionary relationship. According to their homology with NAC proteins in Arabidopsis, 80 FtNAC genes were divided into 14 subgroups (Fig. 1). In our analysis, we found that the NAC proteins in Tartary buckwheat had representatives from the ANAC063, NAM, NAC1, OsNAC7, ANAC011, TIP, OsNAC8, NAC2, ONAC22, TERN, SENU5, NAP/ANAC3, ATAF, and ONAC003 subgroups. However, in Tartary buckwheat, no NAC members from the ANAC001 subgroup were identified. The subgroups OSNAC8 and ANAC063 each contained only one FtNAC protein, whereas the subgroup OSNAC7 contained the greatest number (11) of FtNAC proteins. The ATNAC3 and NAP members represented a single clade (Figs. 1 and 2a). In an earlier analysis, these two classes were reported to be sister taxa, suggestive of their coevolution [47, 55]. These results indicate that the functions of FtNAC genes in Tartary buckwheat are diverse, which is consistent with the reports in Arabidopsis and rice.
Fig. 1.
Unrooted phylogenetic tree representing relationships among the NAC proteins of Tartary buckwheat and Arabidopsis. The tree divided the FtNAC proteins into 16 subgroups represented by different colored clusters within the tree. A phylogenetic tree was constructed from the NAC protein sequence of Tartary buckwheat and Arabidopsis thaliana. The phylogenetic trees were derived using the neighbor-joining (NJ) method in Geneious R11. The parameters used included a Blosum62 cost matrix, the Jukes-Cantor model, and global alignment with free end gaps and bootstrap value of 1000
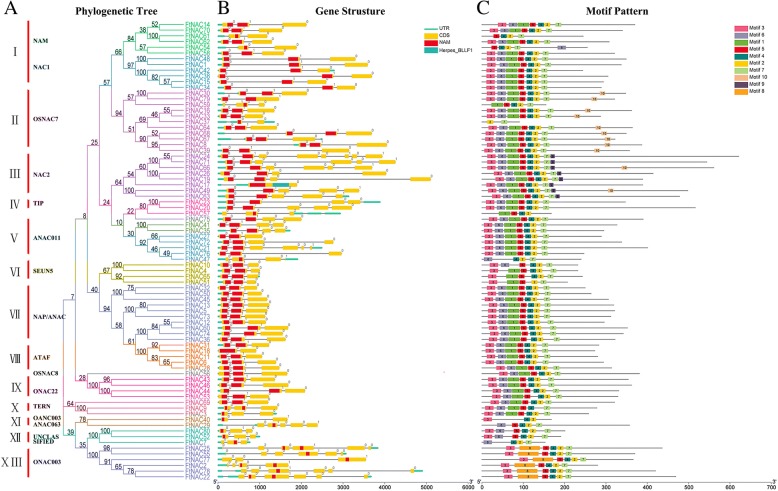
Fig. 2.
Phylogenetic relationships, gene structure and architecture of conserved protein motifs in NAC genes from Tartary buckwheat. a The phylogenetic tree was constructed based on the full-length sequences of Tartary buckwheat NAC proteins using Geneious R11 software. b Exon-intron structure of Tartary buckwheat NAC genes. Green boxes indicate untranslated 5′- and 3′-regions; yellow boxes indicate exons; black lines indicate introns. The NAM domains are highlighted by red boxes. The number indicates the phases of the corresponding introns. c The motif composition of Tartary buckwheat NAC proteins. The motifs, numbered 1–10, are displayed in different colored boxes. The sequence information for each motif is provided in Additional file 2. The length of the protein can be estimated using the scale at the bottom
Gene structure and motif composition of the FtNAC gene family
A phylogenetic tree was constructed from 80 amino acid sequences of FtNAC, and the gene family was divided into thirteen subgroups, named I-XIII, according to bootstrap values and motif composition (Fig. 2a). The largest, subgroup I, had 12 members, whereas the subgroups V and X had only two members. Two NAC genes, FtNAC75 and FtNAC56, could not be assigned to any subgroup because they had low bootstrap values. To analyze the structure of FtNAC genes, we compared the genomic DNA sequences to analyze the composition of introns and exons (Fig. 2b). The coding sequences of the entire FtNAC family were interrupted by introns, with the number of exons ranging from 1 to 8 (Fig. 2b). FtNAC24 had 7 introns, whereas FtNAC17 contained no introns in the open reading frame (ORF). The gene structure is relatively conserved within the NAC gene family; among its members, the majority of genes have 3 exons (Fig. 2b). In general, the closest members from the same subgroups had a similar exon/intron structure with regard to intron number, intron phase, and exon length.
To further study the characteristic regions of FtNAC proteins, the motifs of 80 FtNAC proteins were analyzed online using MEME. Ten conserved motifs were predicted, and the specific amino acid sequences of each motif were also provided (Fig. 2c, Additional file 2: Table S2). We found that most of the closely related members in the phylogenetic tree showed common motifs with the same alignment and position, which indicates that the members of NAC that clustered in the same subgroup may have similar biological functions. Most of the FtNAC proteins contained motif 3 (subdomain A), motif 6 (subdomain B), motif 1 and motif 5 (subdomain C), motif 4 and motif 2 (subdomain D), and motif 7 (subdomain E) in succession. Notably, some conserved motifs in specific subgroups were identified. All of the FtNAC proteins in subgroup XIII lacked motif 1 and motif 6 and contained motif 8, and all of the members of subgroup III contained motif 9. Additionally, all of the FtNAC proteins in subgroup II contained motif 10, except for FtNAC37 and FtNAC74, which contained only 2 motifs. This pattern suggests that the specific motifs may be related to specific functions of different subgroups. The motif composition and gene structure of the members from each subgroup obtained from phylogenetic analysis were similar, which indicates that the classification was more reliable.
Evolutionary analysis of FtNAC genes and several different species
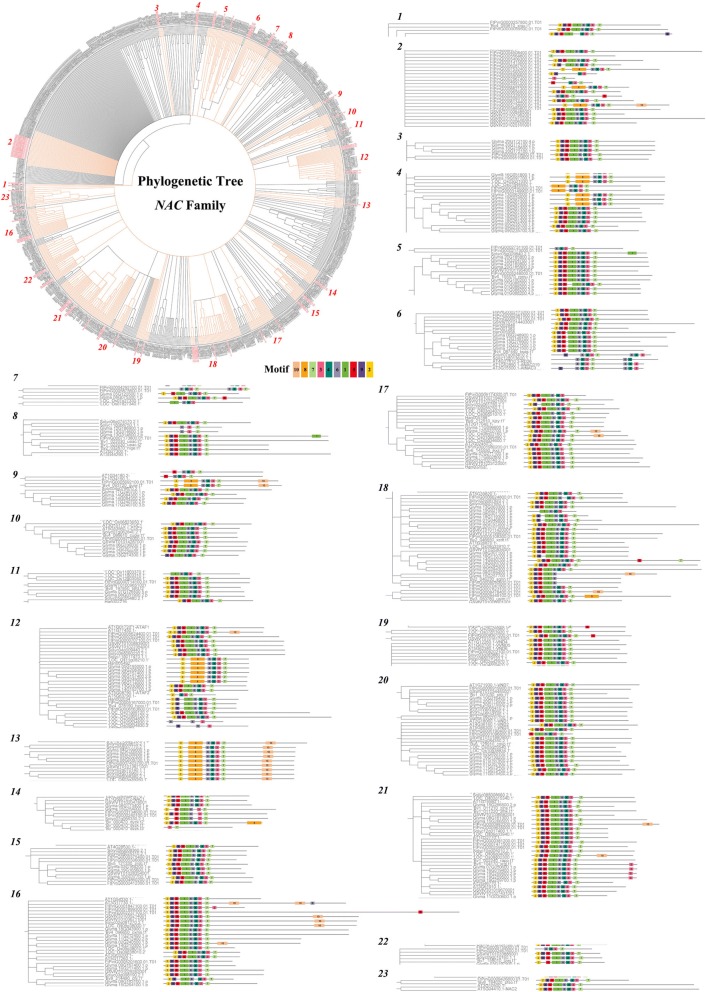
In comparison with the existing NAC genes of Tartary buckwheat, we also studied the evolutionary diversification of NAC genes. A phylogenetic tree was constructed containing the NAC protein sequences from one monocotyledonous plant (rice) and seven dicotyledonous plants (Arabidopsis thaliana, Tartary buckwheat, beet, soybean, tomato, sunflower and grape) using Geneious R11. All NAC proteins were divided into twenty-three subclades within the phylogenetic tree. However, a number of sequence relationships were not well defined (Fig. 3).
Fig. 3.
Phylogenetic relationships and motif compositions of NAC proteins from 8 different plant species. Left panel: an unrooted phylogenetic tree constructed using the neighbor-joining method in Geneious R11. Right panel: distribution of conserved motifs in NAC proteins. The different colored boxes represent different motifs and their positions in each NAC protein sequence
Using MEME, ten different conserved motifs were found, among which motif 2 and motif 9, motif 5, motif 1 and motif 6, motif 4 and motif 3, and motif 7 encoded NAC subdomains A-E, respectively. (Fig. 3). Notably, all of the NACs without motifs 9, 5 and 1 had motif 8 in the N-terminal but not in the C-terminal (Fig. 3). Additionally, some NACs contained at least one of the motifs 1, 3, 4, 5, 6, 7, or 9 in the C-terminal and N-terminal simultaneously (Fig. 3). The majority of NAC proteins in the same clade, particularly the most closely related proteins, typically shared common motifs (for example, FtNAC49 and Glyma.08G009700.1.p), indicating a similar potential function between these NAC proteins.
Chromosomal location and synteny analysis of FtNAC genes
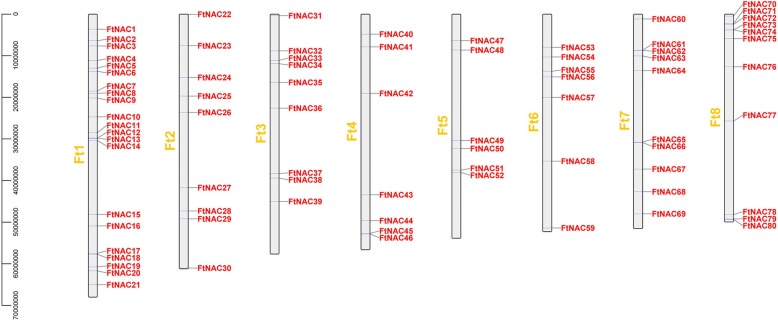
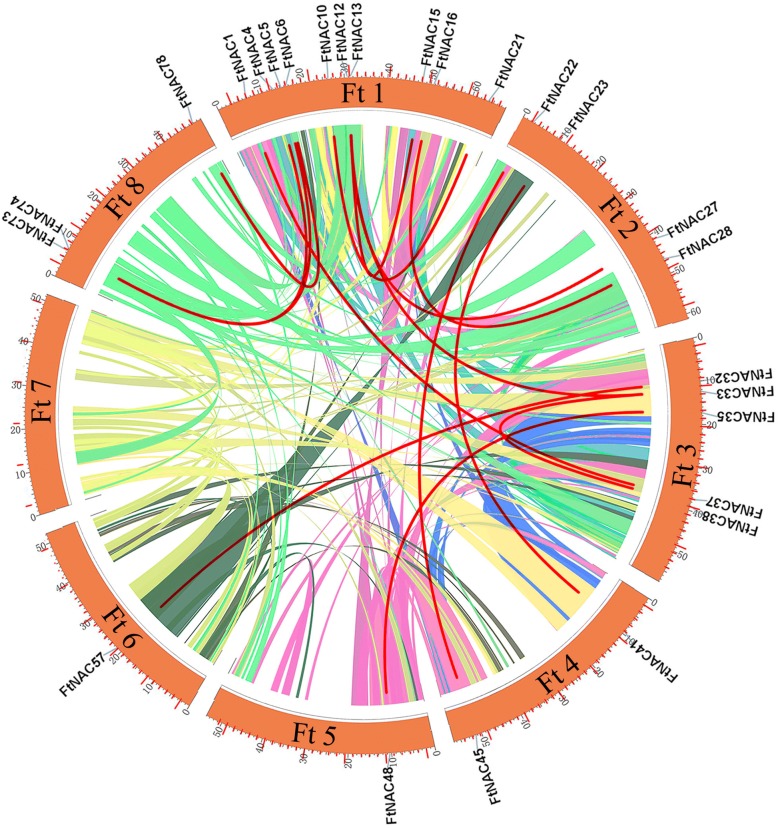
A total of 80 FtNAC genes were unevenly distributed on 8 chromosomes of Tartary buckwheat (Fig. 4). The largest, chromosome 1, had the most FtNAC genes (21, ~ 25.93%), followed by the smallest, chromosome 8 (11, ~ 13.58%) and chromosome 7 (10, ~ 12.35%), whereas there were only six genes in chromosome 5 (~ 1.31%) (Fig. 4). To identify the duplication events in FtNAC genes, a collinearity analysis was performed using MCScanX software. Fourteen pairs of fragment duplication genes were found in 80 FtNAC genes (Fig. 5). Fragment duplication genes were the most common on chromosome 1, followed by chromosome 3, whereas there were no fragment duplication gene pairs on chromosome 7 (Fig. 5).
Fig. 4.
Distribution of FtNAC genes among 8 chromosomes. Vertical bars represent the chromosomes of Tartary buckwheat. The chromosome number is to the left of each chromosome. The scale on the left represents chromosome length
Fig. 5.
Schematic representations of the interchromosomal relationships of Tartary buckwheat NAC genes. Colored lines indicate all synteny blocks in the Tartary buckwheat genome, and the red lines indicate duplicated NAC gene pairs. The chromosome number is indicated below each chromosome
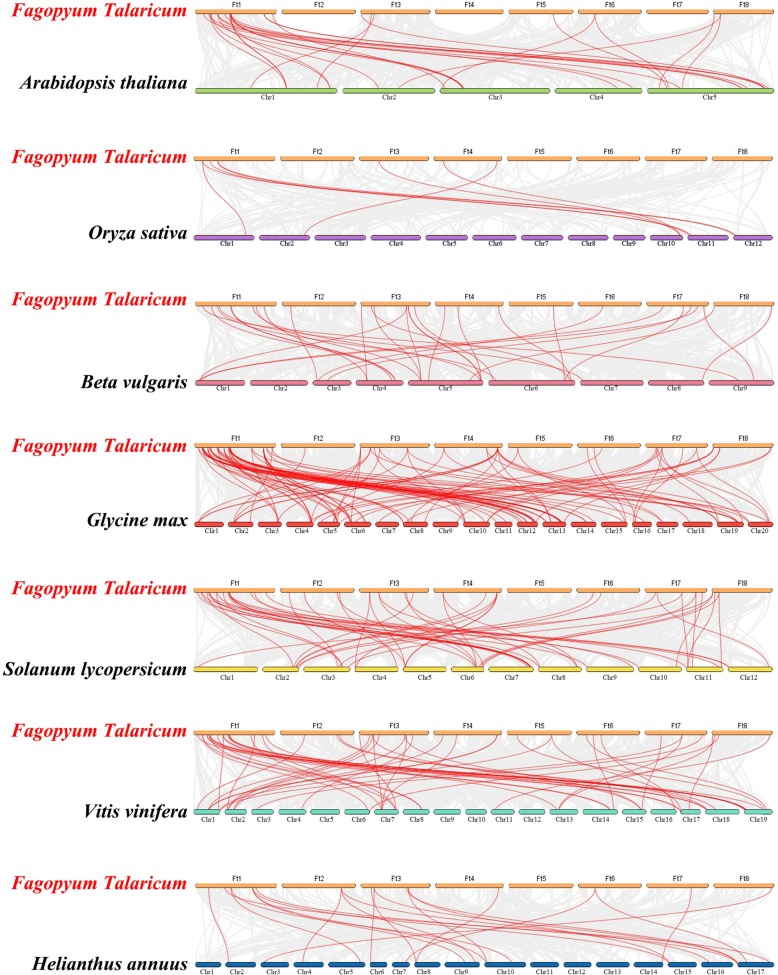
To further explore the evolutionary relationship of the Tartary buckwheat NAC family, we constructed a phylogenetic tree consisting of six dicotyledonous plants (Arabidopsis, beet, tomato, soybean, sunflower and grape) and a monocotyledonous plant (rice) (Fig. 6). The order of genome size was as follows: sunflower (3.6 Gb) [56], soybean (1.025 Gb) [57], tomato (828.349 Mb) [58], beet (714–758 Mb) [59], grape (490 Mb) [60], rice (466 Mb) [61] and Arabidopsis thaliana (125 Mb) [62]. In total, 43 FtNAC genes displayed a syntenic relationship with grape, followed by soybean (40), tomato (40), beet (29), Arabidopsis (20), sunflower (15) and rice (6). The number of homologous pairs in the other seven species (soybean, tomato, Arabidopsis thaliana, grape, beet, sunflower and rice) were 73, 36, 28, 24, 21, 17 and 6, respectively (Additional file 3: Table S3). Tartary buckwheat chromosome 1 had the most homologous gene pairs with other species. In addition, the number of gene pairs between Tartary buckwheat and other species was not related to the genome size of the other species.
Fig. 6.
Synteny analysis of NAC genes between Tartary buckwheat and seven representative plant species. Gray lines in the background indicate the collinear blocks within the Tartary buckwheat and other plant genomes, whereas the red lines highlight the syntenic NAC gene pairs
Some FtNAC genes were formed in at least six syntenic gene pairs, particularly those of Tartary buckwheat and soybean, such as FtNAC3 and FtNAC46, which may play a vital role in the evolution of the NAC gene family. Tartary buckwheat and rice shared less than 10 colinear gene pairs, which may be associated with the phylogenetic relationship between these species.
Expression patterns of FtNAC genes in different plant tissues
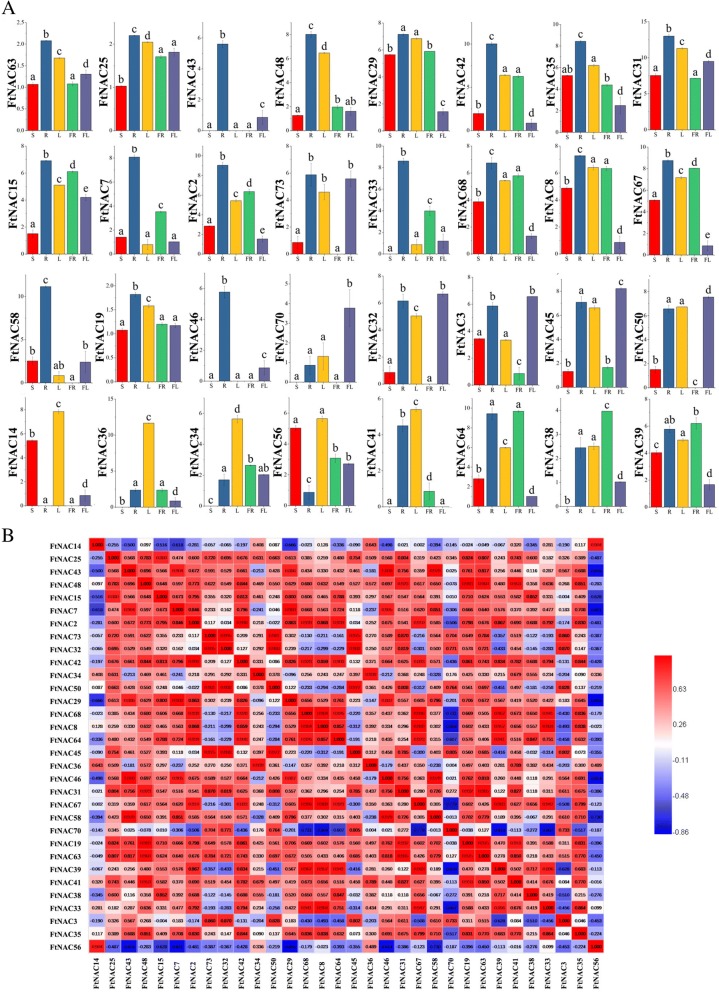
To explore the physiological role of the FtNAC genes, we randomly selected 32 genes distributed among the branches (Fig. 2, Additional file 4: Table S4) and performed qPCR. The expression levels of 32 FtNAC genes in stem, root, leaf, fruit and flower tissues were evaluated (Fig. 7a). As shown in Fig. 7, the various expression patterns of each gene in different tissues indicated that FtNAC genes perform different functions in Tartary buckwheat.
Fig. 7.
Tissue-specific gene expression of 32 Tartary buckwheat NAC genes and the correlation between the gene expression of FtNACs. a The expression patterns of 32 Tartary buckwheat NAC genes in flower, leaf, root, stem and fruit tissues were examined using a qPCR assay. b The correlation between the gene expression of FtNACs. Red: positively correlated; Blue: negatively correlated. Error bars were obtained from three measurements. Small letter(s) above the bars indicate significant differences (α = 0.05, LSD) among the treatments
Three FtNAC genes (FtNAC43, FtNAC46 and FtNAC58) were more highly expressed in roots than in other organs. (Fig. 7a). FtNAC70 was highly expressed in flower tissue, and FtNAC36 was highly expressed in leaf tissue (Fig. 7a). FtNAC14 was highly expressed in both leaf and stem tissue (Fig. 7a). FtNAC31 was highly expressed in all organs of Tartary buckwheat and may play crucial roles in adaptation to the environment during plant growth and development. In addition, the expression patterns of some FtNAC genes in the same subgroup were similar in Tartary buckwheat, such as four genes in subgroup VII (FtNAC73, FtNAC32, FtNAC50 and FtNAC45) and three genes in subgroup II (FtNAC68, FtNAC67 and FtNAC8) (Figs. 2 and 7a). Furthermore, FtNAC32 and FtNAC45 were duplicated FtNAC members. However, some duplicated FtNAC genes (FtNAC15 and FtNAC38, FtNAC35 and FtNAC41, etc.) in the same subgroup showed diffe rent expression patterns (Figs. 2, 5 and 7a), indicating that the functions of these genes may have changed during the evolution of Tartary buckwheat. In general, understanding the expression patterns of FtNAC genes in different tissues can lay a foundation for identifying functional genes in Tartary buckwheat.
We also studied the correlation of expression patterns for 32 FtNAC genes in Tartary buckwheat (Fig. 7b). Most of the FtNAC genes were positively correlated, and all of the FtNAC genes that were significantly correlated (FtNAC43 and FtNAC58/FtNAC46/ FtNAC33/FtNAC7, FtNAC14 and FtNAC56, FtNAC42 and FtNAC68/FtNAC64/FtNAC67/FtNAC2, etc.) were found to be positively correlated (Fig. 7b).
Differential expression of FtNAC genes during fruit development in Tartary buckwheat
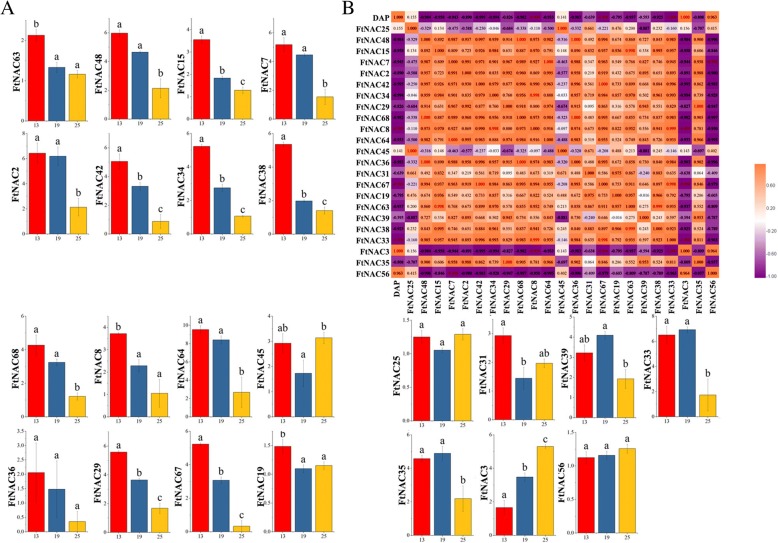
NAC transcription factors have been studied in many plants in relation to plant development and biological stress, but only a few studies have evaluated their role in fruit development. Tartary buckwheat is a nutritional and medicinal crop, and as with other cereal crops, the fruit of the Tartary buckwheat is the most important part and of the greatest concern. The expression levels of 23 FtNAC genes in three fruit development stages were determined by qPCR to explore the function of these genes. (Fig. 8a). The expression of three FtNAC genes (FtNAC42, FtNAC34 and FtNAC15) showed a consistent decreasing trend across the three fruit development stages, whereas FtNAC3 showed an increasing trend (Fig. 8a). The expression of FtNAC45 initially decreased and then increased across the three fruit development stages. FtNAC56 was expressed at a similar level at all three stages.
Fig. 8.
Gene expression of 23 Tartary buckwheat NAC genes during fruit development and the correlation between the gene expression of FtNACs. a The expression patterns of 23 Tartary buckwheat NAC genes at three fruit development stages were examined using a qPCR assay. b The correlation between the gene expression of FtNACs. Orange: positively correlated; Purple: negatively correlated. Error bars were obtained from three measurements. Small letter(s) above the bars indicate significant differences (α = 0.05, LSD) among the treatments
We also studied the correlation of expression patterns for 23 FtNAC genes at three fruit development stages in Tartary buckwheat (Fig. 8b). Based on the correlation results, we observed that most FtNAC genes were positively correlated and that the FtNAC genes were significantly correlated (FtNAC29 and FtNAC67/FtNAC8, FtNAC6 and FtNAC38/ FtNAC15, etc.) were found to be positively correlated (Fig. 8b).
Discussion
FtNAC gene identification and evolutionary analysis in Tartary buckwheat
The NAC domain protein family is a larger plant-specific TF family. In recent years, 117 proteins have been identified in Arabidopsis thaliana [7], 151 in rice [42], 167 in cassava [47], 104 in pepper [49], and 180 in apple [51], but there are few studies on the NAC family in Tartary buckwheat. However, in previous studies, eight FtNAC genes were cloned and found to respond to at least one treatment, among which FtNAC4(FtNAC5) and FtNAC7(FtNAC31) showed extremely significant responses to salt, drought, ABA, and SA treatments [28]. In the current study, we identified 80 members of the FtNAC family (Fig. 1), which were designated FtNAC1 to FtNAC80 on the basis of their chromosomal locations (Fig. 4, Additional file 1: Table S1). Clearly, the NAC gene family in Tartary buckwheat is by far the smallest compared with that in other plant species. This size difference can be attributed to the genome of Tartary buckwheat containing fewer NAC genes than other plants, probably because more duplication events have occurred in other species after differentiation from their earliest ancestors. The duplication of genes can amplify the number of genes; for example, four large duplication events have occurred in the Arabidopsis genomes [63, 64]. The ρ genome duplication (WGD) event is inferred to have occurred after different lineages, while the σWGD event occurred in all Poales [65]. Hence, the low number of FtNAC genes may be due to the absence of a ρWGD event during the evolution of Tartary buckwheat. The genome size of each species was very different (such as Tartary buckwheat, 489.3 Mb [52]; Arabidopsis thaliana, 125 Mb [62]; rice, 466 Mb [61]; soybean, 1.025 Gb [57]), indicating that the NAC gene family was stable in different species during evolution. Although the length, MW and pI of NAC genes vary greatly, the structure of the genes were relatively conservative; among these FtNACs, the majority of genes have 3 exons (Fig. 3b), while the number of exons ranged from 1 to 8 [43]. The gene structure of NAC genes in Tartary buckwheat was similar to that of NAC genes in rice and cassava [42, 47]. A collinearity analysis of the data in this study showed that there were 14 pairs of fragment replication genes without tandem replication events (Fig. 5). Therefore, chromosome fragment replication events drive the evolution of FtNAC genes, which greatly facilitates the expansion of the NAC gene family in plants with smaller genomes. The duplicated FtNAC genes may have functional redundancy. Among the 14 duplicated gene pairs, FtNAC32 and FtNAC45 have a similar, consistent pattern of expression. In the current theory, duplicated genes may pass through 4 selection modes [66], which may result in differences in expression patterns. There is evidence of this from the expression patterns of FtNAC35 and FtNAC41. FtNAC35 had the highest expression level in roots, followed by stems, while FtNAC41 had no expression in stems and the highest expression in leaves. In our study, the NAC genes in the phylogenetic tree of related species were relatively similar, and many branches contained the NAC genes from dicotyledons and monocotyledons, which indicates that the diversity of NAC genes appeared before the differentiation of monocotyledons and dicotyledons (Fig. 3) [67]. Collinearity analysis showed that there were only 10 pairs of homologous genes in Tartary buckwheat and rice, whereas more orthologous gene pairs were found in six of the dicotyledonous leaves, including those of soybean and tomato (Fig. 6). Consistent with the phylogenetic relationship between Tartary buckwheat and the other seven plants, this analysis suggests that the NAC genes in dicotyledonous leaves are more homologous and consistent.
Functional analysis of special structures of FtNAC genes in Tartary buckwheat
The domains and motifs of transcription factors are often related to protein interaction, transcriptional activity and DNA binding [68]. The conserved motifs in the N-terminus of the NAC genes have highly conserved DNA-binding abilities (Fig. 2c), which indicates that these motifs are very important for the function of NAC genes [7]. Motif analysis showed that most of the NAC genes of Tartary buckwheat contained motif 1 to motif 7 (Fig. 2c). However, among the FtNAC members in subgroup XIII, motif 8 (DEFIPTLDGEDGICYTHPEKLPGVTKDGLARHFFHRPSKAYTTGTRKRRK) replaced motifs 1 (part of subdomain C, binds to DNA) and 6 (subdomain B, has distinctive functions) (Fig. 2c), which might confer a distinctive function to this subgroup [7, 11]. In an evolutionary analysis of NAC genes in several different plants, it was also found that motif 8 (EFIPTJEGEBGICYTHPEKLPGVTKDGLSRHFFHRPSKAYTTGTRKRRKI) replaced motifs 5 (part of subdomain A, promotes functional dimerization), 9 (subdomain B) and 1 (part of subdomain C), although they were not all present together (Fig. 3). The genes of subgroup XIII cluster with the ONAC003 subgroup containing Arabidopsis AT4G28500, which has been found to function in the formation of secondary cell wall fibers [69]. FtNAC25 and FtNAC2, the genes of subgroup XIII, were expressed in all tissues. We speculate that most of the genes in subgroup XIII have similar expression patterns due to their common conserved motifs and thus play a role in secondary wall formation.
Tartary buckwheat NAC genes may play an important role in plant development, particularly in fruits
Valuable insights regarding the roles of FtNAC genes involved in specific Tartary buckwheat physiological processes were obtained from the different expression patterns of FtNAC genes. The biological function of new proteins is typically assigned through alignment with homologous sequences in Arabidopsis. Several studies have assigned a wider range of functions to NAC proteins using this approach, such as temperature stress in Chinese cabbage [48] and dehydration stress in soybean [44]. In a multispecies phylogenetic tree, genes in one branch often have the same function. Therefore, the function of NAC in Tartary buckwheat was predicted by clustering with Arabidopsis NAC protein sequences in the phylogenetic tree. Gene expression is a basic form of evidence for gene function. qPCR analysis revealed that FtNAC70 may be involved in the development of flowers. FtNAC43, FtNAC46 and FtNAC58 may play a role in the roots because they exhibit a higher expression level in roots. FtPinG0009426800.01 was the most highly expressed in leaves and may play a role in leaf development. The expression pattern of FtNAC15 was different from that of FtNAC38, although the two genes are duplicated genes within the same subgroup. The NAM groups may regulate cell division and leaf development [16, 17, 70–73]. The NAM subgroup gene FtNAC14 is most highly expressed in the leaf followed by the stem and is orthologous with AT5G53950.1(CUC2), indicating that FtNAC14 may function in leaf development through expression in both the leaf and stem (Fig. 7a). The multispecies evolutionary analysis showed that they fall in the same subgroup and have the same motif composition (Fig. 3). The proteins of a given subgroup contain the same motif, indicating that the NAC genes of the same subgroup may perform similar functions [7]. The expression of FtNAC56 was significantly positively correlated with the expression of FtNAC14 (Fig. 7b). The overexpression of homologous genes AT2G02450.1 (LOV1) in switchgrass alters the leaf angle, cell wall composition, and flowering time [56]. FtNAC56 was also most highly expressed in the leaf followed by the stem, suggesting that these two genes have synergistic functions in leaves and stems. Although they belong to different subgroups, they have the same motif composition (Figs. 2 and 7a). Thus, FtNAM may participate in leaf development. These results suggest that FtNAC genes are extensively involved in the root, stem and leaf development of Tartary buckwheat.
Among the 32 FtNAC genes, most were expressed in fruits, although 8 FtNAC genes were not expressed (Fig. 7a). Therefore, we explored the expression pattern of 23 FtNAC genes at three developmental stages of Tartary buckwheat fruit. Fruit development is an essential process of cell division, differentiation and expansion, which determines fruit quality. In Tartary buckwheat, the most noticeable changes were fruit size and texture, which were related to the activity of different genes [74, 75]. In previous studies, the development of Tartary buckwheat fruit was divided into five stages [76]. We selected three representative stages (13DAP, 19DAP and 25DAP) corresponding to the initial filling stage, milk stage and initial maturity stage to study the role of FtNAC genes in fruit development. FtNAC45, which belongs to the NAP and AtNAC3 subgroups, initially decreased and then increased during the three fruit development stages and may play a role in the fruit development of Tartary buckwheat. FtNAC45 and FtNAC31 showed similar expression patterns at 13 DAP and 19 DAP but exhibited differences at 25 DAP. The overexpression of SlNAC1 alters fruit pigmentation and softening [77], whereas the suppression of tomato SlNAC1 delays fruit ripening [78]. ATAF2 is orthologous to SlNAC1 [79] and is involved in auxin biosynthesis [80], which influences the fruit size of Tartary buckwheat [74]. There was a positive correlation between auxin concentrations and cell division or final seed size in a comparison of two kinds of Tartary buckwheat from 7 to 19 DAP [74]. The size of the fruit is mainly determined by the early development of the embryo, not by the later stage [81, 82]. FtNAC31 is orthologous to AT5G08790.1 (ATAF2) (Fig. 3) and is highly expressed at 13 DAP and may function in the early stages fruit development in Tartary buckwheat. Both FtNAC67 and FtNAC68 are highly expressed at 13 DAP and 19 DAP. These two genes cluster to OSNAC7. Genes in this subgroup have been reported many times to function in secondary wall biosynthesis [29–34, 36, 37, 83]. This result suggests that the FtOSNAC7 genes function in the secondary wall biosynthesis of Tartary buckwheat fruit, with the development from 13 DAP~ 19 DAP, Tartary buckwheat fruit gradually developed completely [76]. According to the expression patterns of FtNAC genes, it can be speculated that these genes may be involved in similar regulatory networks, but the specific function of FtNAC genes in Tartary buckwheat fruit needs to be further verified.
Conclusion
In our study, a comprehensive analysis including gene structure, chromosomal location, sequence homology synteny and expression patterns of the NAC gene family in Tartary buckwheat was first performed. We identified a total of 80 FtNAC genes in Tartary buckwheat, which will provide basic information for the functional characterization of FtNAC genes. Furthermore, expression analysis of FtNAC genes in various tissue and developmental stages of fruit implied that FtNACs may participate in the development of Tartary buckwheat. These data are useful for further exploration of NAC gene-mediated physiological processes in Tartary buckwheat. This systematic analysis of the NAC gene in Tartary buckwheat is helpful for the follow-up study of the functional characteristics of FtNAC genes and the cultivation of high-quality Tartary buckwheat varieties.
Methods
Identification of FtNAC genes in Tartary buckwheat
We downloaded the genome of Tartary buckwheat from the TBGP (Tartary Buckwheat Genome Project; http://www.mbkbase.org/Pinku1/). The FtNAC genes were obtained using two BLASTP methods. First, all AtNACs were used to search possible FtNACs on the TBGP website, and further identified using BLASTP [84] (score value ≥100 and e-value ≤1e− 10) [53]. Second, we downloaded the hidden Markov model (HMM) file of the NAC domain (PF 02365) through Pfam (http://pfam.xfam.org/). All genes were retrieved using HMMER 3.0 (default parameters) with a cutoff of 0.01 [85]. The possible FtNACs were confirmed with PFAM and SMART programs [86, 87], and the HMMERs were used to further verify the results. Finally, all NAC gene models were used to analyze the CDS length, MW, PI and subcellular localization using tools on the ExPasy website.
Analysis of gene structure
To explore the structure of FtNAC genes, ClustalW (default parameters) was used [88]. We used the GSDS (Gene Structure Display Server) analysis to analyze the constituents of the exons/introns of the FtNAC genes. We used MEME to analyze the motifs of FtNAC proteins, (http://meme-suite.org/tools/meme), with the parameters as follows: motif width set to 6–200 and number set to 10 [53, 89].
Chromosomal location, gene duplication and evolutionary analysis of FtNAC genes
Circos were used to locate FtNAC genes on the chromosomes of Tartary buckwheat. We used MCScanX (default parameters) to examine duplication genes [90]. We analyzed the homology of the NAC gene between Tartary buckwheat and the other seven plants using Dual Synteny Plotter (https://github.com/CJ-Chen/TBtools).
Phylogenetic analysis and classification of the FtNAC gene family
According to the classification of AtNAC, all the identified FtNAC genes were divided into different groups. NAC protein sequences (from Arabidopsis, rice, beet, soybean, tomato, sunflower and grape) were obtained by UniProt https://www.uniprot.org/). The NJ phylogenetic trees were constructed using Geneious R11 with Blosum62 cost matrix, the Jukes-Cantor model, global alignment with free end gaps and bootstrap value of 1000.
Plant growth
Xiqiao is produced by physical mutagenesis and is characterized by higher rutin and crude protein contents. It has many advantages, such as a high seed setting rate, strong anti-falling ability, high 1000-grain weight and many grains per plant [94]. Xiqiao was planted in a field of the Sichuan Agricultural University (29°97’ N, 102°97′ E; elevation, 580 m), Ya’an, Sichuan, China and in a consistent growth environment for five consecutive years since 2013. The flower, fruit from three different fruit developmental stages (13, 19, and 25 DAP) [76], stem, root, and leaf from the maturity period were collected from three different Tartary buckwheat plants with identical developmental statuses. Samples were quickly frozen in liquid nitrogen after collection and stored at − 80°C before use.
Expression analysis of FtNAC genes using qPCR
The selected gene expression analysis used qPCR, and primers were designed by Primer3 (Additional file 4: Table S4). We used the FtH3 gene, which was stably expressed at each growth stage in almost all tissues, as an internal control [53]. The qPCR with SYBR Premix Ex Taq II (TaKaRa) was repeated at least three times and the data were analyzed using the 2−(∆∆Ct) method [91].
The data were analyzed using Origin Pro 2018b, and the means were compared using LSD at the 0.05 and 0.01 levels of significance.
Additional files
Table S1. List of the 80 FtNAC genes identified in this study. (XLS 184 kb)
Table S2. Analysis and distribution of conserved motifs in Tartary buckwheat NAC proteins. (XLS 31 kb)
Table S3. One-to-one orthologous relationships between Tartary buckwheat and other plants. (XLS 88 kb)
Table S4. The primer sequences of qRT-PCR. (XLS 32 kb)
Acknowledgements
We thank all colleagues in our laboratory for helpful discussions and technical assistance.
Funding
This research was supported by the National Natural Science Foundation of China (31500289) and the National Key R&D Program of China (2018YFD1000706).
Availability of data and materials
The datasets supporting the conclusions of this article are included in the article and in its additional files.
Abbreviations
- ABA
Abscisic acid
- At
Arabidopsis thaliana
- BLASTP
Basic Local Alignment Search Tool-Protein
- CDS
Coding sequence
- CUC
Cup-shaped cotyledon
- DAP
Days after pollination
- ERFs
Ethylene response factors
- Ft
Fagopyrum tataricum
- HMM
Hidden Markov model
- JA
Jasmonic acid
- MW
Molecular weight
- NAM
No apical meristem
- NAP
NAC-like, activated by AP3/PI
- NST
NAC secondary wall thickening promoting factor
- PI
Isoelectric point
- qPCR
Quantitative real-time PCR
- SA
Salicylic acid
- SCW
Secondary cell wall
- TFs
Transcription factors
- VND
Vascular-related NAC domain
- XND
Xylem NAC Domain
Authors’ contributions
M.-Y.L. planned and designed the research and analyzed the data. Z.-T.M. wrote the manuscript. W.-J.S., L.H., Q.W., Z.-Z.T., T.-L.B., and C.-L.L. performed the experiments. H.C. supervised the research. M.-Y.L. and Z.-T.M. contributed equally. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Moyang Liu, Email: lmyyunxi@163.com.
Zhaotang Ma, Email: WaohUncle_Ma@163.com.
Wenjun Sun, Email: sunnan82475@163.com.
Li Huang, Email: 18428385443@163.com.
Qi Wu, Email: wuqi@sicau.edu.cn.
Zizhong Tang, Email: 3530279123456789@163.com.
Tongliang Bu, Email: tlbu@163.com.
Chenglei Li, Email: gezhucao1998@yahoo.com.
Hui Chen, Phone: (+86) 18981604486, Email: chenhui@sicau.edu.cn.
References
- 1.Slobodan R, Diego RP, Ingo D, Bernd MR. PlnTFDB: an integrative plant transcription factor database. Bmc Bioinformatics. 2007;8(1):42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Z, Jinpu J, Liang T, Yi Z, Xiaocheng G, Ge G, Jingchu L. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39(Database issue):D1114. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riechmann JL, Heard J, Martin G, Reuber L, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 4.Wray G, Hahn MW, Abouheif E, Balhoff J, Pizer M, Rockman MV, Romano LA. The Evolution of Transcriptional Regulation in Eukaryotes[J]. Mol Biol Evol. 2003;20(9):1377-419. [DOI] [PubMed]
- 5.Souer E, Houwelingen A, van Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–170. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 6.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9(6):841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY. Molecular analysis of the NAC gene family in rice. Mol Gen Genet MGG. 2000;262(6):1047–1051. doi: 10.1007/PL00008647. [DOI] [PubMed] [Google Scholar]
- 9.Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O'Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Plant Signal Behav. 2010;426(7):183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 10.Olsen NA, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C. 2010 Papers Collection of National Symposium on Plant Biology 2010. 2010. Dual Function of Arabidopsis ATAF1 in Abiotic and Biotic Stress Responses; pp. 1279–1290. [Google Scholar]
- 13.Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92(1):93–103. doi: 10.1016/S0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 14.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, Mu R, Cao W, Zhang Z, Zhang J, Chen S. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2010;44(6):903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 16.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128(7):1127. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 17.Youn-Sung K, Sang-Gyu K, Jung-Eun P, Hye-Young P, Mi-Hye L, Nam-Hai C, Chung-Mo P. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18(11):3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2010;39(6):863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 19.Hiroaki K, Taizo M, Yoshibumi K, Tamao S, Atsushi K. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J Plant Physiol. 2010;167(7):571–577. doi: 10.1016/j.jplph.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Cristobal U, Assaf D, Tzion F, Ann B, Jorge D. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314(5803):1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters BM, Uauy C, Dubcovsky J, Grusak MA. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J Exp Bot. 2009;60(15):4263–4274. doi: 10.1093/jxb/erp257. [DOI] [PubMed] [Google Scholar]
- 22.Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, Grusak MA, Fett JP. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta. 2009;230(5):985–1002. doi: 10.1007/s00425-009-1000-9. [DOI] [PubMed] [Google Scholar]
- 23.Lam-Son Phan T, Kazuo N, Yoh S, Simpson SD, Yasunari F, Kyonoshin M, Miki F, Motoaki S, Kazuo S, Kazuko YS. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazuo N, Tran LSP, Dong VN, Miki F, Kyonoshin M, Daisuke T, Yusuke I, Nagao H, Kazuo S, Kazuko YS. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2010;51(4):617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 25.Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Erratum to: characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2010;72(4–5):567–568. doi: 10.1007/s11103-010-9598-3. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Wang N, Ji D, Xue Z, Yu Y, Jiang Y, Liu J, Liu Z, Xiang F. Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol. 2016;172(3):1804. doi: 10.1104/pp.16.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo WL, Wang SB, Chen RG, Chen BH, Du XH, Yin YX, Gong ZH, Zhang YY. Characterization and expression profile of CaNAC2 pepper gene. Front Plant Sci. 2015;6(6):755. doi: 10.3389/fpls.2015.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng R, Zhao H, Xiao Y, Huang Y, Yao P, Lei Y, Li C, Chen H, Wu Q. Cloning, characterization, and expression analysis of eight stress-related NAC genes in Tartary buckwheat. Crop Sci. 2018;58:1–14. doi: 10.2135/cropsci2017.02.0079. [DOI] [Google Scholar]
- 29.Zhou J, Zhong R, Ye ZH. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One. 2014;9(8):e105726. doi: 10.1371/journal.pone.0105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci. 2015;6(288):288. doi: 10.3389/fpls.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minoru K, Makiko U, Nobuyuki N, Gorou H, Masatoshi Y, Jun I, Tetsuro M, Hiroo F, Taku D. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19(16):1855. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiqin Z, Richardson EA, Zheng-Hua Y. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225(6):1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- 33.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, OhmeTakagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in Woody tissues of Arabidopsis. Plant Cell. 2007;19(1):270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2010;56(5):768–778. doi: 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- 35.Chengsong Z, Utku A, Grant EH, Haigler CH, Beers EP. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 2010;53(3):425–436. doi: 10.1111/j.1365-313X.2007.03350.x. [DOI] [PubMed] [Google Scholar]
- 36.Qiao Z, Lina GG, Huanzhong W, Yining Z, Shi-You D, Fang C, Dixon RA. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 2010;63(1):100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, Hu S, Li XB. The cotton ( Gossypium hirsutum ) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2017;217. [DOI] [PubMed]
- 38.Jin HM, Wei P. Anti-fatigue properties of tartary buckwheat extracts in mice. Int J Mol Sci. 2011;12(8):4770–4780. doi: 10.3390/ijms12084770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karki R, Park CH, Kim DW. Extract of buckwheat sprouts scavenges oxidation and inhibits pro-inflammatory mediators in lipopolysaccharide-stimulated macrophages (RAW264.7) J Integr Med. 2013;11(4):246–252. doi: 10.3736/jintegrmed2013036. [DOI] [PubMed] [Google Scholar]
- 40.Tsai H, Deng H, Tsai S, Hsu Y. Bioactivity comparison of extracts from various parts of common and tartary buckwheats: evaluation of the antioxidant- and angiotensin-converting enzyme inhibitory activities. Chem Cent J. 2012;6(1):78. doi: 10.1186/1752-153X-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosoyama H, Mitomi A, Sagesaka Y, Kakuda T. Endothelium-dependent vasorelaxation effect of rutin-free tartary buckwheat extract in isolated rat thoracic aorta. J Nutr Biochem. 2008;19(10):700–707. doi: 10.1016/j.jnutbio.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Borrill P, Harrington SA, Uauy C. Genome-Wide Sequence and Expression Analysis of the NAC Transcription Factor Family in Polyploid Wheat. G3 Genesgenetics. 2017;7(9):3019–3029. doi: 10.1534/g3.117.043679. [DOI] [Google Scholar]
- 44.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchishinozaki K, Shinozaki K, Tran LSP. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18(4):263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiriga K, Sharma R, Kumar K, Yadav SK, Hossain F, Thirunavukkarasu N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene. 2014;2(1):407–417. doi: 10.1016/j.mgene.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.) DNA Res. 2013;20(4):403–423. doi: 10.1093/dnares/dst019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu W, Wei Y, Xia Z, Yan Y, Hou X, Zou M, Lu C, Wang W, Peng M. Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS One. 2015;10(8):e0136993. doi: 10.1371/journal.pone.0136993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Wang F, Li MY, Jiang Q, Tan GF, Xiong AS. Genome wide analysis of the NAC transcription factor family in Chinese cabbage to elucidate responses to temperature stress ☆. Sci Hortic. 2014;165(3):82–90. doi: 10.1016/j.scienta.2013.11.005. [DOI] [Google Scholar]
- 49.Diao W, Snyder JC, Wang S, Liu J, Pan B, Guo G, Ge W, Dawood M. Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. Int J Mol Sci. 2018;19(4):1048–1061. doi: 10.3390/ijms19041028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei S, Gao L, Zhang Y, Zhang F, Yang X, Huang D. Genome-wide investigation of the NAC transcription factor familyin melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016;35(9):1827–1839. doi: 10.1007/s00299-016-1997-8. [DOI] [PubMed] [Google Scholar]
- 51.Su H, Zhang S, Yuan X, Chen C, Wang XF, Hao YJ. Genome-wide analysis and identification of stress-responsive genes of the NAM–ATAF1,2–CUC2 transcription factor family in apple. Plant Physiol Biochem Ppb. 2013;71(2):11–21. doi: 10.1016/j.plaphy.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Li X, Ma B, Gao Q, Du H, Han Y, Li Y, Cao Y, Qi M, Zhu Y. The Tartary buckwheat genome provides insights into Rutin biosynthesis and abiotic stress tolerance. Mol Plant. 2017;10(9):1224–1237. doi: 10.1016/j.molp.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Liu M, Ma Z, Wang A, Zheng T, Huang L, Sun W, Zhang Y, Jin W, Zhan J, Cai Y, et al. Genome-wide investigation of the auxin response factor gene family in Tartary buckwheat (Fagopyrum tataricum) Int J Mol Sci. 2018;19(11):3526. doi: 10.3390/ijms19113526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M, Fu Q, Ma Z, Sun W, Huang L, Wu Q, Tang Z, Bu T, Li C, Chen H. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum) Planta. 2019;1:1–18. doi: 10.1007/s00425-019-03089-3. [DOI] [PubMed] [Google Scholar]
- 55.Baranwal VK, Khurana P. Genome-wide analysis, expression dynamics and varietal comparison of NAC gene family at various developmental stages in Morus notabilis. Mol Genet Genomics. 2016;291(3):1305–1317. doi: 10.1007/s00438-016-1186-z. [DOI] [PubMed] [Google Scholar]
- 56.Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546(7656):148. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 57.Shen Y, Liu J, Geng H, Zhang J, Liu Y, Zhang H, Xing S, Du J, Ma S, Tian Z. De novo assembly of a Chinese soybean genome. Sci China Life Sci. 2018;61(8):1–14. doi: 10.1007/s11427-018-9360-0. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, van der Hoeven RS, Nielsen R, Mueller LA, Tanksley SD. Characteristics of the tomato nuclear genome as determined by sequencing undermethylated EcoRI digested fragments. Theor Appl Genet. 2005;112(1):72. doi: 10.1007/s00122-005-0107-z. [DOI] [PubMed] [Google Scholar]
- 59.Dohm JC, Minoche AE, Daniela HW, Salvador CG, Falk Z, Hakim T, Oliver R, Thomas Rosleff SR, Ralf S, Richard R. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) Nature. 2014;505(7484):546. doi: 10.1038/nature12817. [DOI] [PubMed] [Google Scholar]
- 60.Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 61.Goff SA, Darrell R, Tien-Hung L, Gernot P, Ronglin W, Molly D, Jane G, Allen S, Paul O, Hemant V. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the Arabidopsis Genome[J]. Genome Res. 2003;13(2):137. [DOI] [PMC free article] [PubMed]
- 62.Korbinian S, Stephan O, Felix O, Klein JD, Xi W, Christa L, Smith LM, Joffrey F, Norman W. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci U S A. 2011;108(25):10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 64.Blanc G, Hokamp K, Wolfe KH. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the. 2003;13. [DOI] [PMC free article] [PubMed]
- 65.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prince VE, Bryan PF. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 67.Cenci A, Guignon V, Roux N, Rouard M. Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol Biol. 2014;85(1–2):63–80. doi: 10.1007/s11103-013-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu L, White MJ, Macrae TH. Transcription factors and their genes in higher plants functional domains, evolution and regulation. FEBS J. 2010;262(2):247–257. doi: 10.1046/j.1432-1327.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- 69.Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011;11(1):173. doi: 10.1186/1471-2229-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duval M, Hsieh TF, Kim SY, Thomas TL. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol. 2002;50(2):237–248. doi: 10.1023/A:1016028530943. [DOI] [PubMed] [Google Scholar]
- 71.Krisztina N, Thomas B, Alexis P, Tetsuya I, Halima M, Mitsuhiro A, Patrick L. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18(11):2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ken-Ichiro H, Md Rezaul K, Shinobu T, Ken-Ichiro T, Masahiko F, Mitsuhiro A, Masao T. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18(11):2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agata B, Magdalena RS, Dorota BWT, Agnieszka P, Mitsuhiro A, Dorota K. The CUP-SHAPED COTYLEDON2 and 3 genes have a post-meristematic effect on Arabidopsis thaliana phyllotaxis. Ann Bot. 2015;115(5):807–820. doi: 10.1093/aob/mcv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu M, Ma Z, Zheng T, Wang J, Huang L, Sun W, Zhang Y, Jin W, Zhan J, Cai Y, et al. The potential role of auxin and abscisic acid balance and FtARF2 in the final size determination of Tartary buckwheat fruit. Int J Mol Sci. 2018;19(9):2755. doi: 10.3390/ijms19092755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo X, Zhao H, Yao P, Li Q, Huang Y, Li C, Chen H, Wu Q. An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and Proanthocyanidins from Tartary buckwheat. J Plant Growth Regul. 2017;37(1):1–9. [Google Scholar]
- 76.Liu M, Ma Z, Zheng T, Sun W, Zhang Y, Jin W, Zhan J, Cai Y, Tang Y, Wu Q. Insights into the correlation between Physiological changes in and seed development of tartary buckwheat (Fagopyrum tataricum Gaertn.) Bmc Genomics. 2018;19(1):648. doi: 10.1186/s12864-018-5036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma N, Feng H, Xia M, Dong L, Yang D, Wu C, Meng Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014;14(1):351. doi: 10.1186/s12870-014-0351-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen M, Yang D, Ma X, Zhao W, Liang X, Ma N, Meng Q. Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J Plant Physiol. 2016;193:88–96. doi: 10.1016/j.jplph.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 79.Selth LA, Dogra SC, Saif RM, Helen H, Randles JW, Ali RM. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell. 2005;17(1):311–325. doi: 10.1105/tpc.104.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huh SU, Lee SB, Kim HH, Paek KH. ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol Cells. 2012;34(3):305–313. doi: 10.1007/s10059-012-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Venkatesan S. Control of seed size in plants. Proc Natl Acad Sci U S A. 2005;102(50):17887–17888. doi: 10.1073/pnas.0509021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber H, Borisjuk L, Wobus U. Molecular Physiology of Legume Seed Development [J]. Annu Rev Plant Biol. 2005;56(1):253-79. [DOI] [PubMed]
- 83.Mitsuda N, Seki M, Shinozaki K, Ohmetakagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17(11):2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finn RD, Jody C, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28(1):263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(Database issue):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson JD, Gibson TJ, Higgins DG: Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinforma 2002;Chapter 2(Unit 2):Unit 2.3. [DOI] [PubMed]
- 89.Bailey TL, Mikael B, Buske FA, Martin F, Grant CE, Luca C, Jingyuan R, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yupeng W, Haibao T, Debarry JD, Xu T, Jingping L, Xiyin W, Tae-Ho L, Huizhe J, Barry M, Hui G. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔ C T method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of the 80 FtNAC genes identified in this study. (XLS 184 kb)
Table S2. Analysis and distribution of conserved motifs in Tartary buckwheat NAC proteins. (XLS 31 kb)
Table S3. One-to-one orthologous relationships between Tartary buckwheat and other plants. (XLS 88 kb)
Table S4. The primer sequences of qRT-PCR. (XLS 32 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included in the article and in its additional files.