The Problem of, and the Need for Accessing Meiotic Events in Human Fetal Ovaries
Meiosis is the characteristic feature of sexual reproduction; its molecular regulation has been preserved throughout eukaryotic evolution. The defining stage of meiosis is prophase I (Fig. 1), in which homologous chromosomes pair and remain tethered until the first meiotic division, when they must segregate equally into daughter cells that then enter meiosis II. The importance of this stage is underscored by the fact that approximately 50% of all spontaneous miscarriages are due to nondisjunction errors at the first meiotic division [1, 2]. Although a large body of literature exists to confirm the molecular conservation of these processes across eukaryotes, one of the more startling observations from mammalian meiosis is that there are differences between the sexes in their meiotic progression [3] and, more specifically, in the source of meiotic errors in humans, because approximately 90% of chromosomally aneuploid human fetuses arise as a result of errors in maternal meiosis I [2, 5].
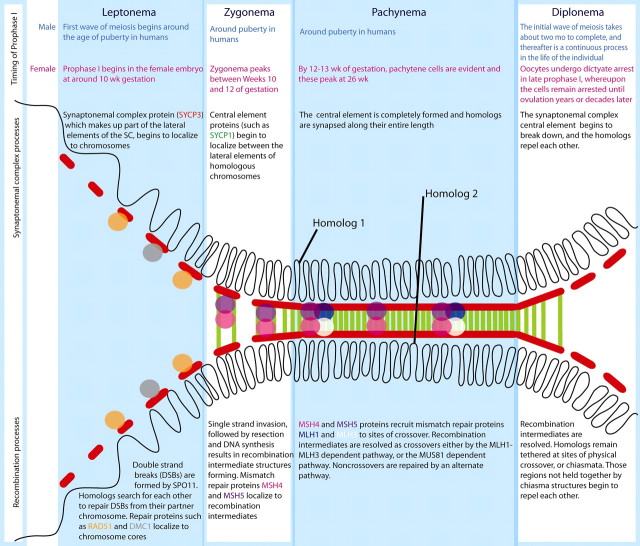
Fig. 1.
Cartoon summary of prophase I events in humans. The first four stages of prophase I are depicted (leptonema, zygonema, pachynema, and diplonema), together with the progression of synapsis events (shown by the localization of synaptonemal complex protein 3 [SYCP3; red], and SYCP1 [green]) and the appearance of major markers for recombination (colored circles). Homologous chromosomes, consisting of paired sister chromatids, are depicted as single looping lines radiating from the synaptonemal complex core. The timing of prophase I stages for male and female meiosis (in humans) is shown above the cartoon image, underscoring the temporal differences in prophase I progression between male and female germ cells. The complexity of synapsis and recombination events is greatly understated in this summary image, but some of the important genes are described here, all of which are components of key gene cliques within the HFOnet.
The timing and progression of meiosis also vary between the sexes. In females, oogonia enter meiosis during fetal development, arresting toward the end of prophase I in a prolonged state of diplotene known as dictyate arrest (Fig. 1). Meiosis resumes after puberty, when cohorts of oocytes are stimulated to undergo the first meiotic division with each estrous cycle and only complete the second meiotic division upon fertilization. Thus, oogenesis begins during fetal life, but it may take months (in rodents) or years (in primates) to complete. By contrast, male meiosis is not interrupted by arrest periods, and it occurs in a continuum from around the time of (or just prior to) puberty, after which spermatogonia continue to enter prophase I in waves throughout the life of the individual.
What becomes obvious from these temporal differences is the inherent difficulty in studying meiotic events in females. The availability of female meiotic material is hampered, not only by the fact that one must retrieve such tissue from fetuses, but also because of the extremely limited amount of ovarian tissue available at these stages. Even in the mouse, where animal numbers may not be limiting, the use of female meiotic tissues for high-throughput biochemical, proteomics, or genomics research is hindered because of the small size of the fetal ovary.
These issues are exacerbated in humans, with the result that very few studies have focused on meiotic events in human fetal ovaries [6–12]. Most of the published reports focus on confirmation in human oocytes of the molecular pathways involved in synapsis and recombination derived from mouse data [6, 13]. Several groups have attempted to address whether and how events during fetal meiosis in human oocytes may be a causative factor in human nondisjunction [6, 8, 10, 14]. These studies have mainly used surface spread chromosome preparations (e.g., Fig. 2), and although substantial data have been accumulated, little new information has been obtained concerning the details of genetic regulation that is unique/specific to humans. An exciting report from Zheng et al. [15] in this issue of Biology of Reproduction attempts to overcome these difficulties by providing the first functional gene network for human fetal germ cells, HFOnet. This network provides a tool to predict and assess the meiotic role of genes from multiple pathways in a tissue (the human fetal ovary) that is poorly accessible through traditional laboratory sources.
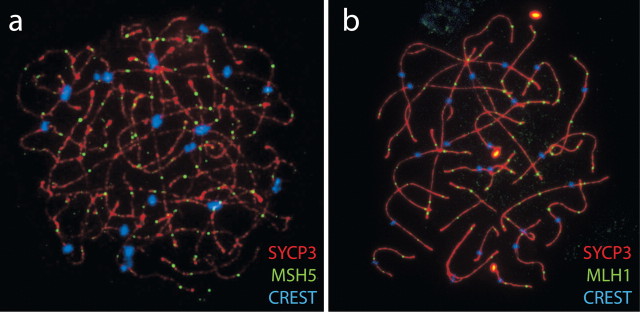
Fig. 2.
Examples of human oocyte chromosome spreads. Chromosome spreads were prepared from 21-wk-old fetal ovaries and immunostained against MutS protein homolog 5 (MSH5; a) and MutL homolog 1, colon cancer, nonpolyposis type 2 (MLH1; b), both in green, to reveal sites of recombination processing. In both images, the synaptonemal complex is highlighted in red by the use of anti-SYCP3 antibodies, whereas the centromere is stained blue with CREST (calcinosis, Raynaud phenomenon, esophageal dysfunction, sclerodactyly, and telangiectasia) human autoimmune serum. Methods are similar to those described previously [6]. In the absence of the HFOnet, such immunofluorescent techniques are one of only a few techniques available for studying regulators of meiotic function in fetal oocytes. This technique requires prior knowledge of the proteins of interest but can be used to validate and confirm information obtained from the HFOnet. Original magnification ×240.
Creating a Probabilistic Network of Genetic Interactions in Human Fetal Oocytes
High-throughput proteomic and genomic screening efforts during the past 10 yr have provided a staggering amount of information on various cellular and disease processes. The issue now is to integrate this mass of information with data from wet-bench studies to delineate networks of regulatory systems that define these different biological events. One such approach is to create a Bayesian network, which is defined as “a structured directed graph representation of relationships between variables. The nodes represent the random variables … and the edges represent the influence of one variable on another” [16]. The network developed by Zheng et al. [15] integrates datasets that describe a number of variables, including gene expression, protein-protein interactions, disease phenotype, protein domain, phylogeny, and gene ontology. The sheer number of datasets used allows for identification of highly significant gene linkages and clustering of functional modules that, in turn, increase dramatically the prediction accuracy of this network.
Zheng et al. [15] base their network on protein-protein interaction data, with the rationale that the most relevant genetic interactions are likely to be observed between genes whose protein products interact physically. Using five different databases to identify potentially interacting gene pairs, the authors then selected pairs of genes that are both expressed specifically in human fetal ovarian germ cells. They obtained approximately 8000 genes and identified from them approximately 20 000 physically interacting pairs, or “positive gold standards.” These interacting pairs were compared to “negative gold standards,” which represent pairs of unrelated genes whose constituents are either not expressed in the fetal ovary, or whose protein products reside in different cellular compartments.
With >20 000 positive and >4 500 000 negative gene pairs in hand, Zheng et al. [15] calculated the likelihood, termed the joint likelihood ratio (LR), of their interactions using a Bayesian approach to develop a naïve functional network assembled from six defined genomic features. These included gene expression, gene ontology, phylogeny, protein domain, disease phenotype, and essential genes. The LR scores for gene pairs above a defined threshold were defined as those that involve functionally linked genes. For example, it stands to reason that a functional relationship (and thus a higher LR score) would be ascribed to a given pair of genes that are present together in similar groups of organisms (phylogeny), that are expressed in similar temporal and spatial patterns (gene expression), mutations in which result in the same disease phenotypes, and whose gene products function in similar biological and molecular processes (ontology). On the other hand, a low LR score would be predicted for a gene pair whose components, although expressed in similar temporal and spatial patterns in human, are not colocalized in other organisms and/or do not function in similar disease processes. Thus, if gene pairs are linked to other gene pairs on the basis of one common component (one gene of the pair becoming “the node” for such interactions), and if all of these pairs provide similar LRs based on one or more of the six genomic features, one can begin to accumulate a clustering effect that gives rise to a complex topological view of the biological process under investigation. Each of the six genomic features may then be considered to be predictive of this clustering and topology, based on the individual LR score generated for that feature based on the negative and positive gold standards.
To assess the validity of their approach, the authors examined the coverage and precision of their functional associations, based on the proportion and total number of positive gold standards with LR scores above their defined threshold. As expected, the integrated Bayesian prediction was consistently more precise than that derived from individual genomic features, although some specific features are more predictive than others.
Interpreting the HFOnet
The HFOnet provides a comprehensive and integrated view of genetic interactions in human oocytes during the critical stages of meiotic induction and prophase I progression. The interconnected gene pairs exhibit a joint LR above a defined threshold, resulting in the creation of a physical network map that provides a visual representation of the functional integration of genes during oocyte meiosis. Although the network in its most simple form can be used for assessing the strength of particular gene interactions and for identifying so-called cliques (in which highly interactive protein groups are found to interact with each and every other member of the clique), the strength of this probabilistic model lies in the ability to make predictions about unknown genetic interactions. As proof of principle, the genes that encode the meiosis-specific DNA mismatch repair (MMR) proteins, MutS protein homologs 4 and 5 (MSH4 and MSH5), form a clique that involves other non-meiosis-specific MMR genes, including MSH3. The gene encoding Ankyrin repeat domain-containing protein 17 (ANKRD17) is also found in this clique, although this protein has never been implicated in meiotic processes. Interestingly, however, the gene is expressed highly in fetal ovary and in adult testis, suggesting a possible involvement in meiotic processes. In this way, genes that are known to be meiosis specific, MSH4 and MSH5, are linked functionally to genes that are not meiosis specific, such as MSH3, as well as to genes whose function in meiosis is yet to be determined/demonstrated (ANKRD17).
Novel meiotic genes could almost certainly be procured using alternative methods, such as comparative approaches that consider meiosis in other organisms or in males. Indeed, as in the case of ANKRD17, simple gene expression analysis could pinpoint yet more meiotic gene candidates. The benefits of HFOnet over these other strategies, however, lies in the heterogeneity of its dataset, because of its having been obtained through a multifactorial approach. Thus, for ANKRD17, the HFOnet is able to verify germ cell specificity and ascribe this gene to a functional network, giving researchers a “heads up” as to where to focus research efforts. Thus, unlike the high-throughput microarray methods used routinely, the HFOnet can distinguish oocyte gene pairs from somatic cell gene pairs in a way not otherwise possible with the limited tissue availability in human fetal ovaries. In addition, through the use of hubs (genes that connect to more than one other gene) and cliques, it is possible to validate and reinforce the role of any given gene in a specific meiotic process and to ascribe a functional relationship to a gene that might ordinarily have not been determined without detailed wet-bench analysis.
References
- 1. Hunt PA, Hassold TJ. Human female meiosis: what makes a good egg go bad? Trends Genet 2008; 24:86–93. [DOI] [PubMed] [Google Scholar]
- 2. Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2:280–291. [DOI] [PubMed] [Google Scholar]
- 3. Morelli MA, Cohen PE. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction 2005; 130:761–781. [DOI] [PubMed] [Google Scholar]
- 4. Hunt PA, Hassold TJ. Sex matters in meiosis. Science 2002; 296:2181–2183. [DOI] [PubMed] [Google Scholar]
- 5. Martin RH, Ko E, Rademaker A. Distribution of aneuploidy in human gametes: comparison between human sperm and oocytes. Am J Med Genet 1991; 39:321–331. [DOI] [PubMed] [Google Scholar]
- 6. Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am J Hum Genet 2005; 76:112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tease C, Hartshorne GM, Hulten MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 2002; 70:1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng EY, Hunt PA, Naluai-Cecchini TA, Fligner CL, Fujimoto VY, Pasternack TL, Schwartz JM, Steinauer JE, Woodruff TJ, Cherry SM, Hansen TA, Vallente RU et al. Meiotic recombination in human oocytes. PLoS Genet 2009; 5:e1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Cruz R, Casanovas A, Brieno-Enriquez M, Robles P, Roig I, Pujol A, Cabero L, Durban M, Garcia Caldes M. Cytogenetic analyses of human oocytes provide new data on non-disjunction mechanisms and the origin of trisomy 16. Hum Reprod 2010; 25:179–191. [DOI] [PubMed] [Google Scholar]
- 10. Robles P, Roig I, Garcia R, Brieno M, Martin M, Barbero JL, Cabero LI, Garcia-Caldes M. Analysis of recombination along chromosome 21 during human female pachytene stage. Reprod Biomed Online 2009; 18:784–794. [DOI] [PubMed] [Google Scholar]
- 11. Hartshorne GM, Barlow AL, Child TJ, Barlow DH, Hulten MA. Immunocytogenetic detection of normal and abnormal oocytes in human fetal ovarian tissue in culture. Hum Reprod 1999; 14:172–182. [DOI] [PubMed] [Google Scholar]
- 12. Wallace BM, Hulten MA. Meiotic chromosome pairing in the normal human female. Ann Hum Genet 1985; 49(pt 3):215–226. [DOI] [PubMed] [Google Scholar]
- 13. Roig I, Liebe B, Egozcue J, Cabero L, Garcia M, Scherthan H. Female-specific features of recombinational double-stranded DNA repair in relation to synapsis and telomere dynamics in human oocytes. Chromosoma 2004; 113:22–33. [DOI] [PubMed] [Google Scholar]
- 14. Barlow AL, Tease C, Hulten MA. Meiotic chromosome pairing in fetal oocytes of trisomy 21 human females. Cytogenet Genome Res 2002; 96:45–51. [DOI] [PubMed] [Google Scholar]
- 15. Zheng P, Griswold MD, Hassold TJ, Hunt PA, Small CL, Ye P. Predicting meiotic pathways in human fetal oogenesis. Biol Reprod 2010; 82:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pe'er D. Bayesian network analysis of signaling networks: a primer. Sci STKE 2005; 2005:pl4. [DOI] [PubMed] [Google Scholar]