Abstract
β-Type platelet-derived growth factor receptor (PDGFRB) is a typical tyrosine kinase, as a candidate gene associated with reproduction. Its main roles include regulation of gonocytes (migration and proliferation) and of the cell cycle. The objectives of this study were to identify mRNA expression of the goat PDGFRB gene, as well as insertion/deletion (indel) variants and their association with litter size in 1122 healthy Shaanbei white cashmere goats. The results revealed that PDGFRB was widely expressed in all tested tissues, and the expression levels in testes at different developmental stages indicated a potential association with the mitosis-to-meiosis transition. Furthermore, the expression of PDGFRB was relatively higher in the ovary tissue of mothers of two lambs compared with mothers of single lamb. These results implied that PDGFRB was related to goat fertility. Meanwhile, two intronic indels, 5 bp (n = 501) and 10 bp (n = 1122), were identified. Statistical analysis revealed that only the 10 bp indel was associated with first-born litter size (n = 1122, p = 6.030 × 10−5), and that individuals of the genotype insertion/deletion had larger litter sizes than those of genotype insertion/insertion. Overall, these results indicated that the 10 bp indel of PDGFRB could be used in marker-assisted selection during goat genetic breeding.
Keywords: cashmere goat, PDGFRB gene, expression profile, insertion/deletion (indel), litter size
1. Introduction
Since the beginning of the twenty-first century, the goat industry in China has developed rapidly and its share in the national economy has also increased year by year. During this time, litter size has been a low heritability trait that has received widespread attention. Currently, when compared with traditional breeding methods, marker-assisted selection (MAS) based on genetic variation has proved to be more efficient for improving economically relevant traits of low heritability [1,2]. However, crucial genetic variants in the genome that lead to excellent phenotypic traits need to be validated.
At present, numerous potential genetic variations that were associated with phenotypic traits have been revealed by using genome-wide sequencing and genome-wide association study (GWAS) [3–5]. Although the analysis of large amounts of data has produced multiple potential gene variants, only a few relevant experimental need to be verified [4–6]. In order to select the dominant gene mutations that affected phenotypic traits, a method of combining GWAS and MAS has been established [6,7]. In 2017, using GWAS, Liu et al. [8] and Qin et al. [9] found that a single-nucleotide polymorphism (SNP) of the β-type platelet-derived growth factor receptor (PDGFRB) gene was significantly associated with semen production traits in Chinese Holstein bulls, which has since been validated by a larger study. These results implied the feasibility of screening crucial genetic variations by combining different methods.
The PDGFRB gene encodes a receptor tyrosine kinase (PDGFRB), a transmembrane protein belonging to class III receptor tyrosine kinases (RTKs) [10,11]. PDGFRB plays a dominant role in the proliferation and migration of gonocytes. Furthermore, the inhibition of mice PDGFRB tyrosine kinase activity leads to a decrease in testicular size, delayed spermatogenesis and a drastic reduction in epididymal sperm count [12]. Alternatively, the combination of PDGFRB and its ligand is important for the activation of primordial follicles as they transition to the primary stage, and mutations in PDGFRB in mice are lethal prior to follicle development in the ovary [13–15]. Moreover, PDGFRB is not only involved in the synthesis of steroid hormones of mice, but also controlled the development of steroidogenic cells [16]. Alterations in the steroid hormone levels were associated with many types of infertility in both males and females, such as hypogonadism and polycystic ovary syndrome (PCOS) [16]. Meanwhile, as a growth factor, PDGFRB regulates the cell cycle and thus affects cell proliferation [17]. All in all, based on the above studies, these results strongly demonstrated that the PDGFRB gene is associated with mammalian reproduction.
So far, there have been no relevant reports regarding the function of the PDGFRB gene in Shaanbei white cashmere goats (SBWC) reproduction. Therefore, in this study, expression profiles of the PGDFRB gene in SBWC were initially assessed in different tissues (heart, liver, spleen, lung, kidney, testis, brain, skin and muscle), and at different developmental stages in the testis and the ovary of ewes of different litter size. Meanwhile, two intronic insertion/deletion (indel) variants of the intron of PDGFRB gene were identified, namely the 5 bp indel and the 10 bp indel. Importantly, sequencing found a novel 36 bp indel downstream of the 5 bp indel locus, which was separated by a 49 bp sequence. The relationship between these loci and litter size was evaluated in large groups of SBWC. These results not only extend the knowledge of goat PDGFRB genetic variation, but also provide the basis for MAS of goat molecular breeding.
2. Material and methods
2.1. Animal and sample collection
For RNA experiments, we harvested nine tissues (heart, liver, spleen, lung, kidney, testis, brain, skin and muscle) from three-week-old ram (n = 3 per group). Moreover, a total of 16 testis tissues at 0, 3 days, one, two, three, four, six and eight weeks, and ovaries from seven ewes that had different litter size were also sampled. All tissues were stored at −80°C until used for analysis. For DNA experimentation, 1122 healthy female SBWC were randomly sampled and their first-born litter size was recorded. The feeding conditions were similar for all goats.
2.2. Total RNA isolation and cDNA synthesis
According to the manufacturer's instructions, total RNA was extracted from collected tissue samples using Trizol Reagent (Takara, Dalian, China). The purity and concentration of RNA were checked with a Nanodrop 2000 Spectrophotometer and 1% agarose gel electrophoresis. Samples could be used for reverse transcription when the OD260 nm/OD280 nm ratio of RNA was in the range of 1.8–2.0. Subsequently, a Prime Script™ RT Reagent kit (Takara) and universal primers were used in reverse transcription, and negative controls were prepared under the same conditions. The concentration of PrimeScript Reverse Transcriptase was 200 U µl−1, and the reaction volume and amount of RNA were 20 µl and 1 µg, respectively. The resultant cDNA was preserved at −20°C.
2.3. PDGFRB mRNA expression profiles analysis
According to the information on goat PDGFRB mRNA (Ref-seq accession XM_018050163.1 and XM_018050162.1), quantitative real-time PCR (qRT-PCR) primers were designed in Primer-BLAST of the NCBI (table 1). However, the primers needed to cover different exons to ensure cDNA amplification. The expression profiles of PDGFRB were analysed in a StepOnePlus™ Real-Time PCR System (Applied Biosystems, MA, USA) using cDNA from the three-week-old tissues. In addition, the expression profiles of testis tissue at different times (0, 3 days, one, two, three, four, six and eight weeks) and ovarian tissue of ewes of different litter size (MSL, mothers of single lamb; MTL, mothers of two lambs) were also examined.
Table 1.
The qRT-PCR and amplification PCR primer sequences of the goat PDGFRB gene. Note: TD, touch-down PCR.
| primer name | primer sequences (5′-3′) | sizes (bp) | detection methods |
|---|---|---|---|
| PDGFRB-F | GAGTCGGTGGACTATGTGCC | 189 | qRT-PCR |
| PDGFRB-R | CTGGTAGCTGAAGCCCACAA | ||
| Stra8-F | TCTCACACTCCTCCGTCACT | 192 | qRT-PCR |
| Stra8-R | ATGCCTGCAAGAGGATGGTC | ||
| GAPDH-F | AAAGTGGACATCGTCGCCAT | 116 | internal control |
| GAPDH-R | CCGTTCTCTGCCTTGACTGT | ||
| P1(PDGFRB-1F) | CCAGCTCAGGATGGGTCT | 296 | TD |
| P1(PDGFRB-1R) | CCAGCATGGGCACATAGTC | ||
| P2(PDGFRB-2F) | ACCTGAATCTGTCTGGTGTGT | 222 | TD |
| P2(PDGFRB-2R) | CAGAAAGGGAAAGGGACATGC | ||
| P3(PDGFRB-3F) | GCTGGGTGAGGGCTACAAAA | 120 | TD |
| P3(PDGFRB-3R) | AAACACCAGTGCGTCACAGT |
The 13 µl qRT-PCR reaction contained 5 µl cDNA (1/100 dilution), 0.5 µl of each primer, 6.75 µl 2 × SYBR® Premix Ex Taq™ II and 0.25 µl ROX-dye (Takara). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an endogenous control for the normalization of expression levels of PDGFRB gene mRNA (table 1), and three replicates were performed. The amplification cycle was as follows: initial denaturation and activation of the polymerase for 5 min at 95°C for one cycle, followed by 40 cycles at 94°C for 30 s, 60°C for 30 s and then 72°C for 30 s. The final extension was for 10 min at 72°C. And, the 2−ΔΔCt method was used to transform Ct values into normalized relative expression levels of mRNA.
2.4. Genomic DNA isolation and DNA pool construction
Blood samples were obtained from ear tissue of all 1122 SBWC. Genomic DNA was isolated as described by Lan et al. [18] and stored at −20°C. Genomic DNA concentration was quantified by Nanodrop 2000, and the working solution of each DNA sample was brought to 10 ng µl−1. Fifty DNA samples were randomly selected to construct genomic DNA pools for exploring the genetic variation of the PDGFRB gene in SBWC [19].
2.5. Indel identification and sequencing
Based on the goat (Capra hircus) gene sequence (GenBank accession NC_030814.1) and the NCBI SNP-database (https://www.ncbi.nlm.nih.gov/snp), three pairs of primers (table 1, P1–P3) were designed to amplify genomic DNA pools to explored genetic variation in the goat PDGFRB gene. PCR reactions were performed with touch-down (TD) PCR in a 13 µl volume, which contained 6.5 µl 2× mix, 0.5 µl of each primer, 0.5 µl 10 ng µl−1 genomic DNA and 5 µl ddH2O. The TD PCR reaction procedure was as follows: initial denaturation for 5 min at 95°C; followed by 18 cycles of denaturation for 30 s at 94°C; annealing for 30 s at 68°C (with a decrease of 1°C per cycle); extension for 30 s at 72°C; another 23 cycles of 30 s at 94°C, 30 s at 50°C and 2 min at 72°C; and a final extension for 10 min at 72°C, with subsequent cooling to 4°C [20]. The PCR products were detected by means of 3.5% agarose gel electrophoresis that stained with ethidium bromide and SensiCapture 12.3 (Peiqing JS-2012 auto-focus Gel Image Analyzing System, Shanghai, China). The specificity of PCR products was confirmed by sequencing when each pair of primers produced a single objective band.
2.6. Statistical analysis
Genotypic frequencies, allelic frequencies and linkage disequilibrium (LD) were calculated using the SHEsis program (http://analysis.bio-x.cn) [21]. The χ2-test was used to evaluate the Hardy–Weinberg equilibrium (HWE) [22]. Polymorphism information content (PIC) was calculated using online software (http://www.msrcall.com/Gdicall.aspx) [20]. Distribution differences for genotypic and allelic frequencies were analysed using the χ2-test or Fisher exact tests (when the minimum theoretical frequency was less than 5) in SPSS (version 19.0).
Association between the indel loci and first-born litter size was analysed with a general linear model: Yijklm = μ + Si + HYSj + Pk + Gl+eijklm, where Yijklm was the phenotypic value of litter size, μ was the overall population mean, Si was the effect of lambing year, HYSj was the mean of the population, Pk was the fixed effect of the parity, Gl was the fixed effect of the genotype and eijklm was the random error [23,24]. The lambing year and parity were not considered in the model, because the data of litter size used in this study were first-born litter size. Further analysis was performed with SPSS 19.0 software using Student's t-test (t-test), the data were rejected when n < 5. And, p < 0.05 was considered statistically significant.
3. Results
3.1. Tissue expression profiles of the goat PDGFRB gene
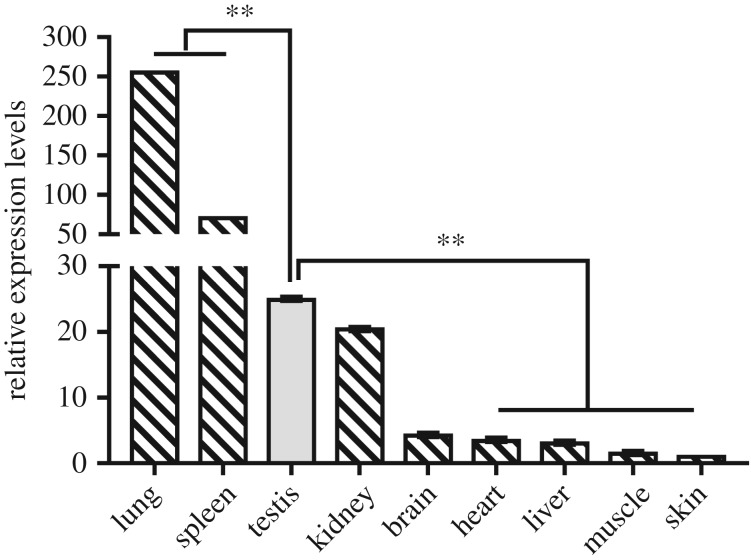
Expression profiles of the PGDFRB gene from different tissues at three weeks old were investigated. The results revealed that the PDGFRB gene was expressed in all tested tissues (heart, liver, spleen, lung, kidney, muscle, brain, skin and testis). Expression level was extremely high in the lung, which was followed by the spleen, and then the testis. Interestingly, the expression levels were significantly different between the testis and skin, muscle, liver, heart, spleen and lung (p < 0.01), but not for the brain or kidney (p > 0.05) (figure 1).
Figure 1.
SBWC PDGFRB mRNA expression patterns detected by qRT-PCR. Different tissues of three-week-old SBWC. Data represent means ± s.e. (n = three samples of each tissues). *p < 0.05; **p < 0.01.
3.2. Goat PDGFRB gene expression profiles of gonads
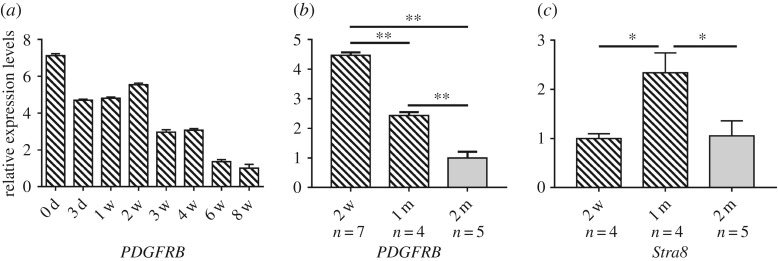
We examined the expression of the goat PDGFRB gene in testis collected at different developmental stages and the ovaries of ewes of different litter size. Results showed that the expression of PDGFRB was highest at 0 day, and the overall expression revealed a downward trend with the extension of time. Moreover, the mRNA expression of PDGFRB at six and eight weeks old was lower than that at other stages (figure 2a).
Figure 2.
SBWC PDGFRB and Stra8 mRNA expression patterns detected by qRT-PCR. (a) The mRNA expression profiles in goat testis at different developmental stages. (b) Expression of PDGFRB mRNA at mitosis (two weeks and one month, n = 7 and n = 4, respectively) and meiosis (two months, n = 5) in testis tissue. (c) Expression of Stra8 mRNA at mitosis (two weeks and one month, n = 4 and n = 4, respectively) and meiosis (two months, n = 5) in testis. Data represent means ± s.e. *p < 0.05; **p < 0.01.
Studies have shown that in the Liaoning cashmere goat (the male parent of SBWC) enter the mitotic stage of spermatogenesis after birth, and the primary spermatocytes that initiate meiosis first occur in one month or so [25]. So, we compared the expression levels of PDGFRB gene in the testis during the mitotic periods of zero to two weeks and one month (n = 7 and n = 4, respectively) and the meiotic period of one to two months (n = 5) [26,27]. Compared to the meiotic stage, the expression of PDGFRB was significantly downregulated as mitosis shifted to meiosis (p < 0.01) (figure 2b). The expression level of Stra8 (stimulated by retinoic acid 8), a marker gene for mammalian germ cell transition from mitosis to meiosis [28], was significantly upregulated during anaphase of mitosis (one month) (p < 0.05) (figure 2c). In short, these results indicated that the PDGFRB gene plays an important role in male reproduction.
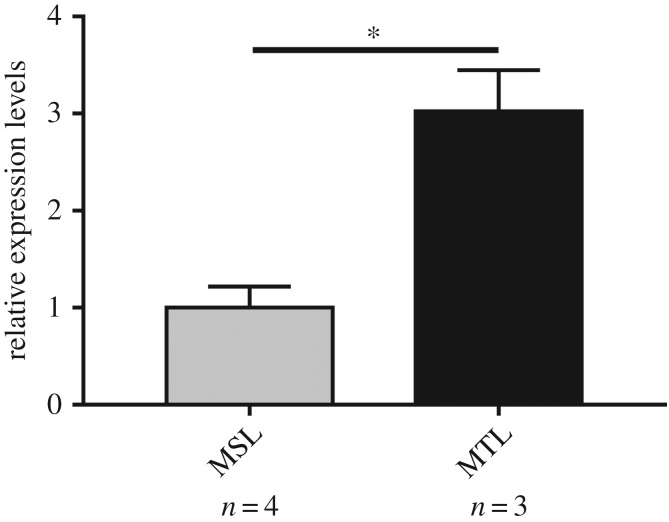
The expression of PDGFRB gene in goat ovaries of ewes of different litter size showed that expression of the PDGFRB gene in MTL was significantly higher than that in MSL (p < 0.05) (figure 3). This proved that the goat PDGFRB gene was also essential for female reproduction.
Figure 3.
The mRNA expression profile of PDGFRB in SBWC ovary. Data represent means ± s.e. *p < 0.05; **p < 0.01.
3.3. Identification indel of the PDGFRB gene
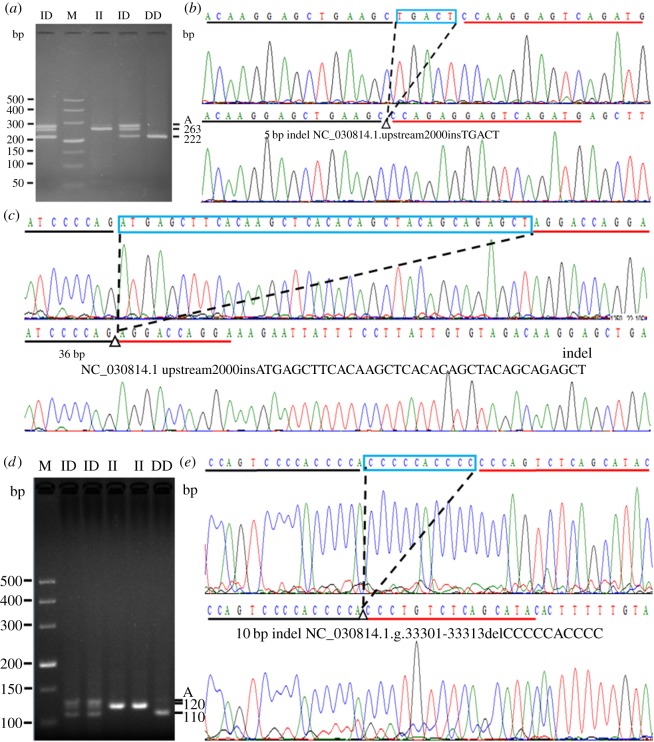
In order to explore potential DNA markers for improving goat fertility, we next carried out an identification of indel variants. In this study, there were two pairs of primers that had genetic variation (table 1, P2–P3). Primer P3 amplified a fragment with an expected size, named 10 bp indel (NC_030814.1. g.33301-33313delCCCCCACCCC) (figure 4d,e). Reciprocally, the DNA product amplified by primer P2 did not match the fragment with an expected size (227/222 bp), and the individuals of 14 insertion/insertion (II) genotypes and 31 DD genotypes were randomly selected and sequenced. The results showed a novel 36 bp indel (NC_030814.1.upstream.2000insATGAGCTTCACAAGCTCACACAGCTACAGCAGAGCT) downstream of the expected 5 bp indel (NC_030814.1.upstream.2000insTGACT), which was separated by a 49 bp sequence (figure 4a–c). Both the 5 and 36 bp indels were inserted or absent at the same time from the sequencing diagram. These results suggested that the two indel loci may be linked.
Figure 4.
The electrophoresis diagrams and sequence diagrams of goat PDGFRB gene indel loci. (a,b) The 5 bp indel locus, (a,c) the 36 bp indel locus and (d,e) 10 bp indel locus. The letter A indicated a heteroduplex and the triangle symbol (Δ) indicated the location of the insertion/deletion (indel). The underlined nucleotides at the beginning of the sequences represent similarities between the ‘ins’ variant and ‘del’ variant.
3.4. Linkage disequilibrium analysis of the PDGFRB gene
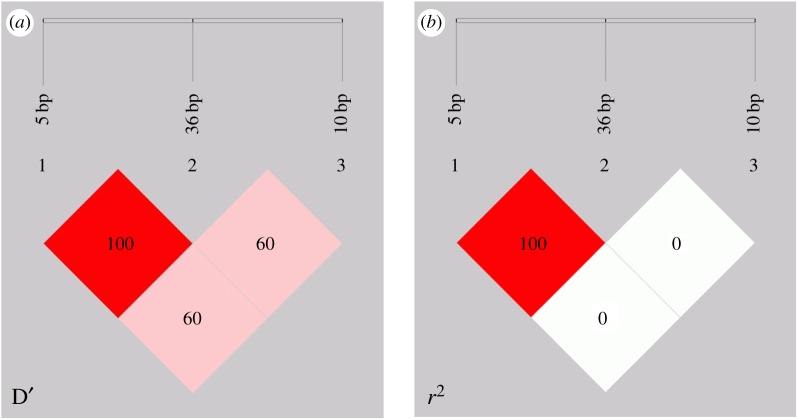
LD of the 5, 36 and 10 bp indel in SBWC was analysed. As shown in figure 5 and table 2, for the 5 and 36 bp indel, the values of D′ and r2 were 1.00 and 1.00, respectively (table 2 and figure 5). The r2 value was used as a pairwise measure of LD, which implied that the 5 bp indel and 36 bp indel were completely linked. In relative terms, there was no LD between these two indel and the 10 bp indel (r2 = 0.007) (table 2 and figure 5).
Figure 5.
LD plot of the PDGFRB gene three indel loci in goat, 5 bp indel, 36 bp indel and 10 bp indel. (a) D′ value, (b) r2 value.
Table 2.
D′ and r2 values of pairwise LD of the PDGFRB gene in SBWC.
|
D' |
r2 |
|||
|---|---|---|---|---|
| loci (bp) | 36 | 10 | 36 | 10 |
| 5 | 1.00 | 0.606 | 1.000 | 0.007 |
| 36 | 0.606 | 0.007 | ||
3.5. Genetic parameter analysis of the indel variants
Next, the genotypes and allele frequencies, as well as the polymorphisms associated with the indel locus in the PDGFRB gene, were calculated. The distribution of genotypes was determined in SBWC by HWE analysis. For the 5 and 36 bp indel, the frequency of allele ‘D’ was highest (0.795), and the frequency of genotypes II, insertion/deletion (ID) and DD were 0.058, 0.293 and 0.649, respectively, in 501 individuals (table 3). At the 10 bp indel, only 3 ‘DD’ genotypes were detected in 1122 individuals, and the frequency of the allele ‘I’ was greatest (0.908) (table 3).
Table 3.
Genotypes, alleles, He, Ne, PIC and HWE analysis the indel within the PDGFRB gene in SBWC. Note: P2, 5 bp and 36 bp indel; P3, 10 bp indel; SBWC, Shaanbei white cashmere goat; HWE, Hardy–Weinberg equilibrium; Ho, homozygosity; He, heterozygosity; Ne, effective allele numbers; PIC, polymorphism information content.
| population parameters |
||||||||
|---|---|---|---|---|---|---|---|---|
| loci | sizes (N) | genotype frequencies | allele frequencies | HWE p-values | Ho | He | Ne | PIC |
| P2 | 501 | II (29, 0.058) | I (0.205) | p > 0.05 | 0.674 | 0.326 | 1.484 | 0.273 |
| ID (147, 0.293) | D (0.795) | |||||||
| DD (325, 0.649) | ||||||||
| P3 | 1122 | II (919, 0.819) | I (0.908) | p > 0.05 | 0.833 | 0.167 | 1.201 | 0.153 |
| ID (200, 0.178) | D (0.092) | |||||||
| DD (3, 0.003) | ||||||||
The χ2-test also showed that the genotype frequencies of these loci (5, 36 and 10 bp) in the SBWC population were in accordance with the HWE (p > 0.05) (table 3). Additionally, gene homozygosity (Ho), heterozygosity (He), effective allele numbers (Ne) and PIC of these indel loci of PDGFRB gene were calculated. The values of PIC at these loci were 0.273 and 0.153, respectively, indicating that they were polymorphic in SBWC (table 3). Among them, the 10 bp indel was close to low genetic diversity (PIC = 0.153) (table 3).
3.6. Analysis of association with first-born litter size in SBWC
In the SBWC population, we analysed the relationship between the indel loci of PDGFRB gene and reproduction (first-born litter size) in ewes. There was no significant correlation between the 5 and 36 bp indel and first-born litter size among 0–501 samples that were selected randomly from the whole population (χ2-test, p > 0.05) (table 4). Interestingly, the 10 bp indel was consistently associated with the first-born litter size in 1122 individuals (Fisher's exact test, p < 0.05), except for 100 individuals at first (table 5). Moreover, genotype distributions were significantly different between the different lambing groups of goats (p < 0.01) (table 6). Association analysis also revealed that individuals of genotype ID outperformed genotype II individuals (p < 0.01) (table 7).
Table 4.
Genotype distribution between mothers of litter size of 5 and 36 bp indel in SBWC. Note: χ2 test.
| mothers of single lamb |
mothers of multi-lamb (≥2) |
||||||
|---|---|---|---|---|---|---|---|
| numbers | II | ID | DD | II | ID | DD | p-values |
| 100 | 3 | 17 | 30 | 4 | 12 | 34 | 0.534 |
| 200 | 7 | 29 | 62 | 7 | 27 | 68 | 0.874 |
| 300 | 13 | 37 | 98 | 13 | 49 | 90 | 0.375 |
| 400 | 15 | 52 | 135 | 14 | 68 | 116 | 0.168 |
| 501 | 15 | 70 | 167 | 14 | 77 | 158 | 0.741 |
Table 5.
Genotype distribution between mothers of litter size of 10 bp indel in SBWC. Note: Fisher exact test. Italics indicate p < 0.05.
| mothers of single lamb |
mothers of multi-lamb (≥2) |
||||||
|---|---|---|---|---|---|---|---|
| numbers | II | ID | DD | II | ID | DD | p-values |
| 100 | 43 | 7 | 0 | 35 | 15 | 0 | 0.09 |
| 200 | 93 | 7 | 0 | 78 | 22 | 0 | 0.004 |
| 300 | 139 | 13 | 0 | 119 | 29 | 0 | 0.007 |
| 400 | 184 | 18 | 0 | 165 | 33 | 0 | 0.024 |
| 500 | 231 | 21 | 0 | 212 | 36 | 0 | 0.035 |
| 600 | 271 | 31 | 0 | 231 | 6 7 | 0 | 3.580 × 10−5 |
| 800 | 352 | 51 | 1 | 313 | 81 | 1 | 0.004 |
| 1000 | 445 | 57 | 1 | 380 | 115 | 2 | 7.699 × 10−7 |
| 1122 | 532 | 84 | 1 | 387 | 116 | 2 | 6.030 × 10−5 |
Table 6.
Genotype distribution between mothers of single lamb, two lambs and three lambs of 10 bp indel in SBWC. Note: Fisher exact test. Italics indicate p < 0.05.
| genotypes |
genotype frequencies |
|||||||
|---|---|---|---|---|---|---|---|---|
| types | sample sizes | II | ID | DD | II | ID | DD | Fisher value p-value |
| mothers of single lamb | 617 | 532 | 84 | 1 | 0.862 | 0.136 | 0.002 |
Fisher = 19.423 p = 4.447 × 10−4 |
| mothers of two lambs | 492 | 376 | 114 | 2 | 0.764 | 0.232 | 0.004 | |
| mothers of three lambs | 13 | 11 | 2 | 0 | 0.846 | 0.154 | 0 | |
Table 7.
Relationship between the 10 bp indel of PDGFRB gene and litter size of first born in SBWC (LSMa ± s.e.) (p < 0.05/p < 0.01). Note: t-test, genotype DD (n = 3) less than 5, we rejected these data. Cells with different letters (a, b/A, B) differed significantly (p < 0.05/p < 0.01). Italics indicate p < 0.05.
| observed genotypes (LSMa ± s.e.) |
|||
|---|---|---|---|
| Traits | II (922) | ID (204) | p-values |
| litter size | 1.43B ± 0.02 | 1.59A ± 0.04 | 1.103 × 10−4 |
4. Discussion
To date, numerous studies have shown that PDGFRB not only participates in testicular development and affects the differentiation of gonocytes, but is also involved in the development of ovarian follicles [12–14]. These observations suggested that the PDGFRB gene could affect fertility. Previous studies have not reported on the expression profile of PDGFRB gene or the association between the PDGFRB indel locus and first-born litter size in SBWC.
Initially, the expression profile of the PDGFRB gene of SBWC was determined in this study, and the results showed it to be widely expressed in various organs (figure 1). It is important to note, however, that PDGFRB is reported to be associated with gonocytes proliferation and migration [12]. The expression of PDGFRB in testis tissues at different developmental stages was also detected, and we found this decreased progressively with development (figure 2a). The expression level of PDGFRB was highest at 0 days, and it was lowest at six and eight weeks (figure 2a). The spermatogonia mature from gonocytes that migrate to the basal membrane, a process which establishes and maintains spermatogenesis in the mature murine testis [12]. Furthermore, the number of PDGFRB-positive cells located in the centre of the seminiferous tubules decreases progressively after birth, they are found on the basal membrane at postnatal 5 (P5), and PDGFRB activity was drastically reduced after P5 [29]. At about a month of age, most of goat gonocytes have differentiated into spermatogonia and are attached to the basal membrane [30]. In Liaoning cashmere goats, spermatogonia gradually proliferate through mitosis since birth, and meiosis is initiated at one month [25]. We, therefore, speculated similar characteristics for this period in SBWC, and combined individuals of 0, 3 days, one, two, three and four weeks as the mitotic period (two weeks and one month), similarly the data of six and eight weeks were considered as the meiotic period (two months). Our results illustrated that the expression level of PDGFRB in SBWC during meiosis was significantly lower than that in the mitotic period (p < 0.01) (figure 2b). Moreover, we also tested the expression level of Stra8 (stimulated by retinoic acid 8) at the same time points as PDGFRB. Retinoic acid (RA) regulates mouse spermatogonia cell differentiation as well as meiotic initiation through the target gene Stra8, and Stra8 was also a marker gene for the germ cell transition from mitosis to meiosis in other mammals, such as rats, humans and goats, etc. [28,31,32]. As shown in figure 2c, Stra8 was significantly upregulated at anaphase of mitosis (one month, p < 0.05) (figure 2c). Overall, these results revealed that the PDGFRB gene was essential for the production of spermatogonia and was associated with goat male reproduction.
Previous studies have also found that PDGFRB had a crucial role in female reproduction. Schmahl et al. [16] showed that mouse PDGFRB was involved in controlling the synthesis of steroid hormones in the ovary. Moreover, Brito et al. [15] identified the expression of PDGFRB in goat ovarian follicles, which is associated with follicular development. From our current study, figure 3 shows that the expression of the PDGFRB gene in MTL was significantly higher than that in MSL (p < 0.05). These findings highlight the role of PDGFRB in female reproduction. This was important because, with the development of society and the improvement of breeding techniques, female livestock commonly outnumber males. The purpose of our study was to screen genetic variation, which could help to implement MAS in goat breeding. The indel variants could be used as a type of genetic variation with important functions, compared with the SNPs, which had the advantages of convenient detection and remarkable effects [20]. Therefore, we studied and analysed the indel variants of PDGFRB in 1122 SBWC.
In this study, we found that there were three insertion/deletions (indels) in the upstream 2000 bp and the 22nd intron regions of the PDGFRB gene in goats, which were the 5 bp indel, 36 bp indel and 10 bp indel, respectively. These indel loci were consistent with the prediction by NCBI, expected for the 36 bp indel. Not only that, but the 36 bp indel was located downstream of the 5 bp indel, and they may have complete LD (table 2 and figure 4). Both indel loci had three genotypes (II, ID and DD), and were consistent with HWE (p > 0.05) (table 3).
Subsequently, the relationship between indel loci and first-born litter size was analysed. In order to make the result more reliable, we adopted a new strategy. In the whole population, 100 individuals were randomly taken as a subset, and different subsets were randomly selected and accumulated for analysis. Interestingly, the 10 bp indel was always associated with first-born litter size in 1122 individuals (p < 0.01), except for 100 individuals (table 5). Meanwhile, Fisher's exact test showed a significant correlation of genotype distribution and litter size within the different lambing groups of goats (p = 4.447 × 10−4) (table 6). Importantly, there were only three individuals with genotype DD (n < 5), so we rejected these data and compared the relationship between individuals with genotype II and ID by t-test. There was a significant difference in first-born litter size between genotype II and ID (p < 0.01), with the mutant (ID) having larger litters than the wild-type (II) (table 7). These results implied that ‘D’ was a dominant allele in the SBWC population, and that this 10 bp indel had great potential for application in breeding. Unfortunately, the 5 bp indel and 36 bp indel show no association between 0 and 501 individuals and first-born litter size (p > 0.05) (table 4).
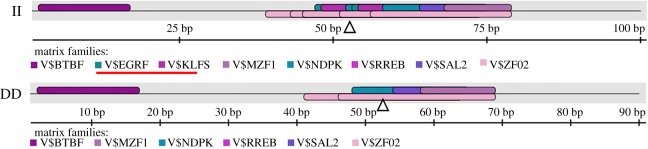
Although the 10 bp indel locus was located in intron 22, several studies have found that the intronic variations of the gene could affect transcription through transcription factors [26,27,33]. Van Laere et al. [34] found that the variation in intron 3 of IGF2 impacted the expression in postnatal muscle, which controlled phenotypic variation, such as muscle growth and fat deposition. Therefore, in order to investigate why the 10 bp indel affected first-born litter size, we predicted the transcription factor binding sites in the regions where indel occurred using the online software Genomatix MatInspector (http://www.genomatix.de/) [35]. The results revealed that there were different transcription factors between genotypes II and DD as follows, namely KLFs (Kruppel-like factors) and EGRF (epidermal growth factor receptor) (figure 6). This discovery provided a possibility that two factors influence goat litter size. Among them, KLF was involved in cellular proliferation, survival and differentiation. Several KLFs participate in oestradiol signalling in the normal reproductive system, thereby regulating reproductive development and function [36]. Meanwhile, EGRF also regulates cell cycle progression, thereby affecting cell proliferation [37]. This suggests that KLFs and EGRF may be associated with reproduction and affected litter size.
Figure 6.
The prediction of transcription factor binding sites in intron of PDGFRB. The triangle symbol (Δ) indicated location of the indel. The red line indicated different transcription factor.
Alternatively, some intronic variations may have LD to mutations with known phenotypes [38]. Such as, Gansmo et al. [39] found that the 40 bp indel in the mouse MDM2 (mouse double minute 2 homologue) gene promoter P1 was completely LD with an SNP, and this indel locus had a positive correlation with a reduced risk of endometrial cancer. Following these findings, the further study was needed regarding the way in which the 10 bp intronic of the goat PDGFRB gene affects the phenotype (first-born litter size).
5. Conclusion
To summarize, PDGFRB gene mRNA was expressed in all tested tissues (heart, liver, spleen, lung, kidney, testis, brain, skin and muscle) and the expression levels in the testis decreased with developmental age. In addition, the expression of PDGFRB was relatively higher in the ovary of MTL compared with those of MSL. Two novel intronic indels, 5 bp and 10 bp, were identified and the latter was significantly associated with first-born litter size (p < 0.001). These results provide the basis for MAS of goat molecular breeding.
Supplementary Material
Supplementary Material
Acknowledgements
We greatly thanked the staffs of Shaanbei white cashmere goat breeding farm, Shaanxi province, PR China, for their collecting samples.
Ethics
The use of all experimental animals completely complied with the local standards for the care and welfare of animals. Furthermore, all animals in this study were approved by Institutional Animal Care and Use Committee (IACUC) of Northwest A&F University, China (NWAFU-314020038).
Data accessibility
The data supporting the present study have been uploaded as part of the manuscript and electronic supplementary material.
Authors' contributions
W.Y., H.Y., X.L. and C.P. carried out the molecular laboratory work and carried out the statistical analyses; K.W. and Y.C. participated in data analysis; T.Z., H.X., H.Z., J.L. and L.Q. collected field data; C.P. and E.Z. conceived of the study, designed the study; W.Y., X.L. and C.P. drafted the manuscript; all authors gave final approval for publication.
Competing interests
We have no competing interests
Funding
This work was funded by the National Key Research and Development Program of China (E.Z., 2016YFD0500508), National Natural Science Foundation of China (H.Y. and C.P., no. 31760650; X.L., no. 31172184) and Provincial Key Projects of Shaanxi (L.Q., 2014KTDZ02-01).
References
- 1.Ma Y, et al. 2015. Bovine HSD17B8 gene and its relationship with growth and meat quality traits. Sci. Bull. 60, 1617–1621. ( 10.1007/s11434-015-0882-0) [DOI] [Google Scholar]
- 2.Yang M, Wei JG, Li PF, Wei SN, Huang YH, Qin QW. 2016. MHC polymorphism and disease resistance to Singapore grouper iridovirus (SGIV) in the orange-spotted grouper, Epinephelus coioides. Sci. Bull. 61, 693–699. ( 10.1007/s11434-016-1055-5) [DOI] [Google Scholar]
- 3.Edwards SL, Beesley J, French JD, Dunning AM. 2013. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797. ( 10.1016/j.ajhg.2013.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidel GE. 2010. Brief introduction to whole-genome selection in cattle using single nucleotide polymorphisms. Reprod. Fertil. Dev. 22, 138–144. ( 10.1071/RD09220) [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Li Z, Cai Y, Xu H, Yang R, Lan X. 2018. Genetic variants in fat- and short-tailed sheep from high-throughput RNA-sequencing data. Anim. Genet. 49, 483–487. ( 10.1111/age.12699) [DOI] [PubMed] [Google Scholar]
- 6.Pausch H, et al. 2014. A nonsense mutation in TMEM95 encoding a nondescript transmembrane protein causes idiopathic male subfertility in cattle. PLoS Genet. 10, e1004044 ( 10.1371/journal.pgen) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin P, et al. 2017. A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci. Rep. 7, 1872 ( 10.1038/s41598-017-02052-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Yin H, Li C, Qin C, Cai W, Cao M, Zhang S. 2017. Genetic effects of PDGFRB and MARCH1 identified in GWAS revealing strong associations with semen production traits in Chinese Holstein bulls. BMC Genet. 18, 63 ( 10.1186/s12863-017-0527-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin C, Yin H, Zhang X, Sun D, Zhang Q, Liu J, Ding X, Zhang Y, Zhang S. 2017. Genome-wide association study for semen traits of the bulls in Chinese Holstein. Anim. Genet. 48, 80–84. ( 10.1111/age.12433) [DOI] [PubMed] [Google Scholar]
- 10.Fredriksson L, Li H, Eriksson U. 2015. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 15, 197–204. ( 10.1016/j.cytogfr.2004.03.007) [DOI] [PubMed] [Google Scholar]
- 11.Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. 2010. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc. Natl Acad. Sci. USA 107, 11 307–11 312. ( 10.1073/pnas.1000806107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basciani S, De Luca G, Dolci S, Brama M, Arizzi M, Mariani S, Rosano G, Spera G, Gnessi L. 2008. Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology 149, 6226–6235. ( 10.1210/en.2008-0349) [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. 1994. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 8, 1888–1896. ( 10.1101/gad.8.16.1888) [DOI] [PubMed] [Google Scholar]
- 14.Pinkas H, Fisch B, Rozansky G, Felz C, Kessler-Icekson G, Krissi H, Nitke S, Ao A, Abir R. 2008. Platelet-derived growth factors (PDGF-A and -B) and their receptors in human fetal and adult ovaries. Mol. Hum. Reprod. 14, 199–206. ( 10.1093/molehr/gan011) [DOI] [PubMed] [Google Scholar]
- 15.Brito IR, et al. 2012. Expression levels of mRNA-encoding PDGF receptors in goat ovaries and the influence of PDGF on the in vitro development of caprine pre-antral follicles. Reprod. Domest. Anim. 47, 695–703. ( 10.1111/j.1439-0531.2011.01946.x) [DOI] [PubMed] [Google Scholar]
- 16.Schmahl J, Rizzolo K, Soriano P. 2008. The PDGF signaling pathway controls multiple steroid-producing lineages. Genes Dev. 22, 3255–3267. ( 10.1101/gad.1723908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D, et al. 2014. miR-34a regulates mesangial cell proliferation via the PDGFR-β/Ras-MAPK signaling pathway. Cell. Mol. Life Sci. 71, 4027–4042. ( 10.1007/s00018-014-1599-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan XY, Pan CY, Chen H, Zhang CL, Li JY, Zhao M, Lei CZ, Zhang AL, Zhang L. 2007. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res. 73, 8–12. ( 10.1016/j.smallrumres.2006.10.009) [DOI] [Google Scholar]
- 19.Lan XY, Zhao HY, Li ZJ, Li AM, Lei CZ, Chen H, Pan CY. 2012. A novel 28-bp insertion-deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds. Genome 55, 547–552. ( 10.1139/G2012-040) [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, et al. 2016. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14-bp insertion/deletion (indel) within the bovine PRNP gene. Prion 10, 409–419. ( 10.1080/19336896.2016.1211593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, He L, Shi Y. 2009. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 19, 519–523. ( 10.1038/cr.2009.33) [DOI] [PubMed] [Google Scholar]
- 22.Chen F, Shi J, Luo YQ, Sun SY, Pu M. 2013. Genetic characterization of the gypsy moth from China (Lepidoptera, Lymantriidae) using inter simple sequence repeats markers. PLoS ONE 8, e73017 ( 10.1371/journal.pone.0073017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Yang Q, Wang K, Zhang S, Pan C, Chen H, Qu L, Yan H, Lan X. 2017. A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats. Anim. Genet. 48, 735–736. ( 10.1111/age.12617) [DOI] [PubMed] [Google Scholar]
- 24.Yang Q, Yan H, Li J, Xu H, Wang K, Zhu H, Chen H, Qu L, Lan X. 2017. A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size. Anim. Genet. 48, 499–500. ( 10.1111/age.12551) [DOI] [PubMed] [Google Scholar]
- 25.Zhan W. 2015. The study of spermatogenesis and seminiferous performance of Liaoning cashmere goat ram. Master's thesis, Jilin Agricultural University, Changchun, People's Republic of China. [Google Scholar]
- 26.Cui Y, et al. 2018. Insertion/deletion within the KDM6A gene is significantly associated with litter size in goat. Front. Genet. 9, 91 ( 10.3389/fgene.2018.00091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Yan H, Wang K, Zhou T, Chen M, Zhu H, Pan C, Zhang E. 2018. Goat CTNNB1: mRNA expression profile of alternative splicing in testis and association analysis with litter size. Gene 679, 297–304. ( 10.1016/j.gene.2018.08.061) [DOI] [PubMed] [Google Scholar]
- 28.Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. 2015. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl Acad. Sci. USA 112, E2347–E2356. ( 10.1073/pnas.1505683112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basciani S, Mariani S, Spera G, Gnessi L. 2010. Role of platelet-derived growth factors in the testis. Endocr. Rev. 31, 916–939. ( 10.1210/er.2010-0004) [DOI] [PubMed] [Google Scholar]
- 30.Han C. 2017. The development of spermatogenic cells and the analysis of lncRNA differential expression in goats. Master's thesis, Northwest A&F University, Yangling, Shaanxi, People's Republic of China. [Google Scholar]
- 31.Frydman N, et al. 2017. Human foetal ovary shares meiotic preventing factors with the developing testis. Hum. Reprod. 32, 631–642. ( 10.1093/humrep/dew343) [DOI] [PubMed] [Google Scholar]
- 32.Yao X, Tang F, Yu M, Zhu H, Chu Z, Li M, Liu W, Hua J, Peng S. 2014. Expression profile of Nanos2 gene in dairy goat and its inhibitory effect on Stra8 during meiosis. Cell Prolif. 47, 396–405. ( 10.1111/cpr.12128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soldner F, et al. 2016. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99. ( 10.1038/nature17939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Laere AS, et al. 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–836. ( 10.1038/nature02064) [DOI] [PubMed] [Google Scholar]
- 35.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. ( 10.1093/bioinformatics/bti473) [DOI] [PubMed] [Google Scholar]
- 36.Knoedler JR, Denver RJ. 2014. Krüppel-like factors are effectors of nuclear receptor signaling. Gen. Comp. Endocrinol. 203, 49–59. ( 10.1016/j.ygcen.2014.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbst RS, Shin DM. 2002. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer 9, 1593–1611. ( 10.1002/cncr.10372) [DOI] [PubMed] [Google Scholar]
- 38.Nakaoka H, et al. 2016. Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLoS Genet. 12, e1005893 ( 10.1371/journal.pgen.1005893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gansmo LB, et al. 2017. MDM2 promoter polymorphism del1518 (rs3730485) and its impact on endometrial and ovarian cancer risk. BMC Cancer 17, 97 ( 10.1186/s12885-017-3094-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the present study have been uploaded as part of the manuscript and electronic supplementary material.