A number of genes with conserved roles in reproduction were first discovered in yeast, flies, and worms using a forward genetics approach, such as a mutagenesis screen. Aside from occasional rare mutations spontaneously arising in mice, rats, sheep, and dogs, identification of reproduction-related genes in mammals has depended historically upon a reverse genetic approach, targeting candidate genes for disruption in the mouse, based on similarity to simpler model organisms or by identifying interesting expression or functional characteristics. Because the majority of male infertility is expected to arise spontaneously, mouse models that disrupt genes with male reproductive roles by point mutations may provide a more bona fide model of human male infertility than large gene deletions; the AZF deletions on the Y chromosome represent an exception to this general rule. The most significant forward genetics screen to identify mammal-specific fertility genes is the Reproductive Genomics ethylnitrosourea mutagenesis screen initiated at The Jackson Laboratory. To date this effort has identified roughly 30 mouse lines with various gonadal and extragonadal defects in both the male and female reproductive systems, with male germ cell mutant phenotypes ranging from sperm motility defects to Sertoli cell-only testes [1, 2]. The causative mutations for six of the repro mutants were previously identified, including repro4/Mtap2, repro5/Brwd1, repro8/Eif4g3, repro9/Mybl1, repro32/Capza3, and repro34/Stx2 [3–8]. In this issue of Biology of Reproduction, the link has been made between repro42 and Spata22 by La Salle and colleagues [9]. The phenotype of several other repro mutants has been characterized, but the causative mutations are still unknown [10–12]. Collectively these mutations have identified multiple autosomal fertility genes in a variety of pathways (Table 1 and Supplemental Table S1 [available online at www.biolreprod.org]).
Table 1.
A partial list of reproduction-defective (repro) mouse lines affecting germ cells with identified mutations and divided into early and late categories consistent with IVF classification of male infertility.a
| Lineb | Map position | Gene | Mutation | Spermatogenesis Phenotype | Reference |
|---|---|---|---|---|---|
| Early | |||||
| repro42 | chr11:73143243–73159546 | Spata22 | T>A; Y275X (nonsense) | Meiotic arrest, zygonemac | [9] |
| repro9 | chr1:9658916–9690290 | Mybl1 | C>A; A213E (missense) | Meiotic arrest, pachynema | [6] |
| repro8 | chr4:137549385–137762386 | Eif4g3 | G>C; A1572P (missense) | Meiotic arrest, diplonema | [5] |
| repro4 | chr1:66368572–66489157 | Mtap2 | T>A (intronic) | Meiotic arrest, diplonema-MI division | [8] |
| Late | |||||
| repro34 | chr5:129490436–129514439 | Stx2 | C>T; R41X (nonsense) | Spermiogenesis arrest, azoospermic | [7] |
| repro5 | chr16:96213699–96304035 | Brwd1 | C>T (splicing defect) | Spermiogenesis defect, oligoasthenoteratospermicc | [3] |
| repro32 | chr6:139990046–139991307 | Capza3 | T>A; M44K (missense) | Spermiogenesis defect, oligoasthenoteratospermic | [4] |
See Supplemental Table S1 (available online at www.biolreprod.org) for more mouse lines. A complete list and detailed description of mutant phenotypes and current map intervals can be found on the Reproductive Genomics website (http://reproductivegenomics.jax.org).
Within the early and late categories, mutants are listed with increasing attainment of maturation during spermatogenesis.
Mutants that also show female infertility.
Vertebrate genomes typically display increasing spatiotemporal compartmentalization of function compared to their fungal and invertebrate counterparts. For example, SPO11 has a broadly conserved role in meiotic recombination initiation, but among its yeast cofactors only MEI4 has a mammalian ortholog [13] (n.b., forkhead transcription factor mei4 in Schizosaccharomyces pombe [14] and the mei4 allele of CCNB1IP1 [15] are unrelated to mouse Mei4). Similarly, a subset of proteins in the Fanconi anemia complex, including FANCD2 and FANCI and their monoubiquinase UBE2T-FANCL, is broadly conserved with invertebrate orthologs and functions in response to DNA cross-linking agents and during meiosis. Yet mutations in multiple vertebrate-specific members of the Fanconi complex also result in defects in primordial, mitotic, and meiotic germ cells [16–22]. Moreover, reduction of BRCA2 (Fanconi protein J) in the germline causes meiotic arrest in the mouse, contrasting to the elevation of breast cancer risk associated with missense mutations in the same gene in humans [23]. This illustrates that human diseases may be specific to the type of mutation induced and to whether the mutant allele is dominant or recessive—a caveat to predicting the nature of mutations to expect in human male infertility based solely on mouse models of deleted alleles. Therefore, forward genetics combined with targeted knock-in of mutated alleles may be critical to the identification of other meiotic proteins essential in vertebrates.
The phenotype of the repro42 mutant allele indicates a potential role for SPATA22 in meiotic DNA repair or recombination. Recent findings indicate a role for ataxia telangiectasia mutated (ATM) kinase in fine-tuning the number of meiotic crossovers by inhibiting the number of SPO11-induced double-strand breaks [24]. Another component of the Fanconi anemia complex, BTBD12/SLX4 (BTB/POZ domain containing 12/structure-specific endonuclease subunit homolog 4), is stabilized by ATM phosphorylation and has been recently shown to be required for male and female fertility, blocking meiotic progression at the diplotene stage during spermatogenesis with some cells displaying relatively normal synapsis [25]. So it is intriguing that SPATA22 has a putative site for phosphorylation by ATM/ATR kinases, yet its phenotype is arrested earlier in meiotic prophase with synaptic defects. These findings imply that these two proteins have distinct actions during meiotic prophase. Many of the Fanconi complex components, including BTBD12/SLX4, share a common null phenotype—ocular deficits, anemia, and germ cell defects—in contrast to targeted inactivation of mismatch repair (MMR) processes, homologous recombination (HR) pathways, or SPATA22. Disruption of the HR pathway (e.g., Mre11 and Nbn) causes sex-specific reproductive defects while also altering other somatic tissues [26–28]. With the exception of Pms2, which has no effect on female fertility, deletions of most MMR proteins (e.g., Msh4, Msh5, Mlh1, and Mlh3) affect gametes in both sexes, with arrest points in spermatogenesis ranging from the zygonema to the meiotic division stage [29–34]. Synaptonemal complex (SC) protein deficits (Sycp1-3, Syce1-3, Tex11, Tex12, and Fkbp6) cause infertility in both sexes but in a sexually dimorphic fashion—arrest of meiotic prophase in spermatocytes but defects in the first meiotic division in oocytes [35–43]. Although SPATA22 could affect recombination or synapsis indirectly, such as by altering meiotic chromatin, putative phosphorylation sites in SPATA22 recognized by the BRCT (BRCA1 C-terminal) interaction domain offer the intriguing possibility that the protein could act in concert with BRCA1 (Fig. 1). TEX15 has been proposed to interact with BRCA1, and its loss prevents synapsis, but, in contrast to SPATA22, it is required only for male fertility, and the Tex15 mutant arrest is at pachynema [44]. BRCT domain-containing Mei1 in Arabidopsis is required for male pollen development [45], and its mouse ortholog Topbp1 localizes to meiotic chromatin, but its somatic requirement for DNA repair confers embryonic lethality upon its mutant [46]. However, mouse MEI1 (which is unrelated to the Arabidopsis gene) lacks these domains that could allow direct binding to SPATA22, and Mei1 mutant mice have a phenotype similar to that described above for SC mutants [47]. Collectively there is an impression that assembly of the SC and processing of meiotic recombination intermediates is an admixture of sex-specific processes and checkpoints superimposed upon common components. This may account for some reported sex-specific differences in the distribution of crossover events. Two criteria—the lack of a sex bias and the arrest point of the repro42 phenotype—are insufficient alone for a strong correlation between this mutant and specific classes of other meiotic mutants affecting synapsis and recombination.
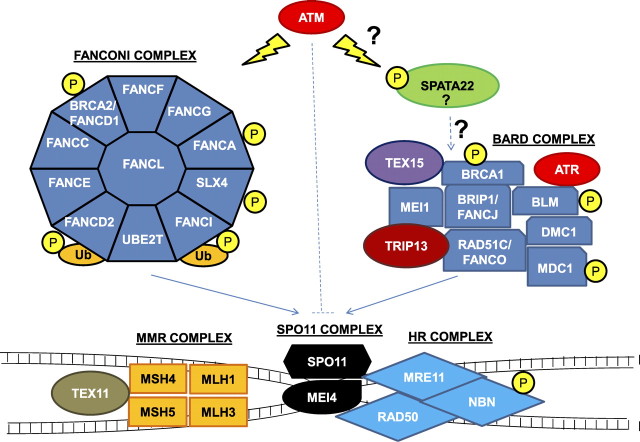
Fig. 1.
A speculative model for pathways converging on mammalian meiotic recombination. ATM, through multiple phosphorylation events, targets the Fanconi complex to SPO11 positive double-strand breaks through the action of UBE2T (E2) and FANCL (E3) monoubiquitination of FANCD2/FANCI. Multiple components of complexes involved in HR, MMR, and BRCA1 (BARD) that affect processing of SPO11-induced double strand breaks are also ATM targets. The precise role for SPATA22 in meiotic recombination is unclear but appears distinct from BTBD12/SLX4, despite potential mutual targeting by ATM phosphorylation. In addition to stimulating DNA break processing, ATM also has a distinct inhibitory action in limiting the number of new breaks by an unknown mechanism inhibiting the SPO11 complex. The substructure of the complexes shown is diagrammatic and not intended to represent the known direct protein-protein interactions. This biochemical model is temporo-spatially compressed to emphasize interactions between complexes and common posttranslational modifications, simplifying the subunit complexity likely present at distinct sites on DNA.
Identification of novel meiotic mutants in the mouse provides tools to determine the stepwise processes within meiotic recombination. Spermatocyte-specific TEX11 appears to promote crossover processing of Holliday junctions in males [48], whereas the more broadly expressed TRIP13 acts to favor a noncrossover outcome [49]. Significant recent progress has been made in identifying the collaborations between repair and SC proteins in mammals necessary to create a milieu that biases strand invasion or resolution favoring greater interhomolog versus intersister exchange during meiosis than in somatic cells [50]. Future efforts aimed at determining the mechanism of SPATA22 action are likely to advance our understanding of the current model of mammalian meiosis, as will identification of mutations in other repro mutant lines with meiotic arrests.
Supplementary Material
References
- 1. Lessard C, Pendola JK, Hartford SA, Schimenti JC, Handel MA, Eppig JJ. New mouse genetic models for human contraceptive development. Cytogenet Genome Res 2004; 105:222–227. [DOI] [PubMed] [Google Scholar]
- 2. Handel MA, Lessard C, Reinholdt L, Schimenti J, Eppig JJ. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol Cell Endocrinol 2006; 250:201–205. [DOI] [PubMed] [Google Scholar]
- 3. Sun F, Handel MAA. Mutation in Mtap2 is associated with arrest of mammalian spermatocytes before the first meiotic division. Genes 2011; 2:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, Handel M, Eppig JJ, Schimenti JC. The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol 2008; 317:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geyer CB, Inselman AL, Sunman JA, Bornstein S, Handel MA, Eddy EM. A missense mutation in the Capza3 gene and disruption of F-actin organization in spermatids of repro32 infertile male mice. Dev Biol 2009; 330:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development 2010; 137:1699–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, Eppig JJ, Handel MA, Shen L, Schimenti JC. A-MYB. (MYBL1) transcription factor is a master regulator of male meiosis. Development 2011; 138:3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akiyama K, Akimaru S, Asano Y, Khalaj M, Kiyosu C, Masoudi AA, Takahashi S, Katayama K, Tsuji T, Noguchi J, Kunieda T. A new ENU-induced mutant mouse with defective spermatogenesis caused by a nonsense mutation of the syntaxin 2/epimorphin (Stx2/Epim) gene. J Reprod Dev 2008; 54:122–128. [DOI] [PubMed] [Google Scholar]
- 9. La Salle S, Palmer K, O'Brien M, Schimenti J, Eppig J, Handel MA. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol Reprod 2012; 86:45,1–12. 10.1095/biolreprod.111.095752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lessard C, Lothrop H, Schimenti JC, Handel MA. Mutagenesis-generated mouse models of human infertility with abnormal sperm. Hum Reprod 2007; 22:159–166. [DOI] [PubMed] [Google Scholar]
- 11. Khalaj M, Abbasi AR, Nishimura R, Akiyama K, Tsuji T, Noguchi J, Okuda K, Kunieda T. Leydig cell hyperplasia in an ENU-induced mutant mouse with germ cell depletion. J Reprod Dev 2008; 54:225–228. [DOI] [PubMed] [Google Scholar]
- 12. Asano Y, Akiyama K, Tsuji T, Takahashi S, Noguchi J, Kunieda T. Characterization and linkage mapping of an ENU-induced mutant mouse with defective spermatogenesis. Exp Anim 2009; 58:525–532. [DOI] [PubMed] [Google Scholar]
- 13. Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 2010; 24:1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol 1998; 18:2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, Langlais KK, Backus VL, Schimenti KJ, O'Brien MJ, Eppig JJ, Schimenti JC. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet 2007; 3:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whitney MA, Royle G, Low MJ, Kelly MA, Axthelm MK, Reifsteck C, Olson S, Braun RE, Heinrich MC, Rathbun RK, Bagby GC, Grompe M. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood 1996; 88:49–58. [PubMed] [Google Scholar]
- 17. Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, Joenje H, Berns A, Arwert F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet 2000; 9:1805–1811. [DOI] [PubMed] [Google Scholar]
- 18. Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, Lu N, Seed B, D'Andrea AD. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood 2001; 98:3435–3440. [DOI] [PubMed] [Google Scholar]
- 19. Koomen M, Cheng NC, van de Vrugt HJ, Godthelp BC, van der Valk MA, Oostra AB, Zdzienicka MZ, Joenje H, Arwert F. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum Mol Genet 2002; 11:273–281. [DOI] [PubMed] [Google Scholar]
- 20. Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet 2003; 12:2063–2076. [DOI] [PubMed] [Google Scholar]
- 21. Zhao QG, Lu BS, Huang PT. Functions of FANCL in primordial germ cell formation and Fanconi anemia [in Chinese]. Yi Chuan Xue Bao 2005; 32:993–1000. [PubMed] [Google Scholar]
- 22. Bakker ST, van de Vrugt HJ, Visser JA, Delzenne-Goette E, van der Wal A, Berns MA, van de Ven M, Oostra AB, Vries SD, Kramer P, Arwert F, van der Valk M et al. . Fancf-deficient mice are prone to develop ovarian tumors. J Pathol 2012; 226:28–39. [DOI] [PubMed] [Google Scholar]
- 23. Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, Flaws JA, Handel MA. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 2004; 131:131–142. [DOI] [PubMed] [Google Scholar]
- 24. Lange J, Pan J, Cole F, Thelen M, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature 2011; 479:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 2011; 7:e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell 2003; 12:1511–1523. [DOI] [PubMed] [Google Scholar]
- 27. Kang J, Bronson RT, Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J 2002; 21:1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cherry SM, Adelman CA, Theunissen JW, Hassold TJ, Hunt PA, Petrini JH. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr Biol 2007; 17:373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 2000; 14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 30. Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW, Kucherlapati R. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 1999; 21:123–127. [DOI] [PubMed] [Google Scholar]
- 31. de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, te Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev 1999; 13:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, Pollard JW, Kolodner RD et al. . Meiotic pachytene arrest in MLH1-deficient mice. Cell 1996; 85:1125–1134. [DOI] [PubMed] [Google Scholar]
- 33. Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, Schwartzberg P, Collins FS et al. . Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 2002; 31:385–390. [DOI] [PubMed] [Google Scholar]
- 34. Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N, Flavell RA, Liskay RM. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 1995; 82:309–319. [DOI] [PubMed] [Google Scholar]
- 35. de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 2005; 19:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kouznetsova A, Novak I, Jessberger R, Hoog C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J Cell Sci 2005; 118:2271–2278. [DOI] [PubMed] [Google Scholar]
- 37. Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 2006; 173:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 2000; 5:73–83. [DOI] [PubMed] [Google Scholar]
- 39. Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet 2009; 5:e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG, Cooke HJ. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 2007; 176:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Hoog C, Cooke HJ, Alsheimer M, Benavente R. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet 2011; 7:e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, Hoog C. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci 2008; 121:2445–2451. [DOI] [PubMed] [Google Scholar]
- 43. Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, Mo R, Hui CC et al. . Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 2003; 300:1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 2008; 180:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grelon M, Gendrot G, Vezon D, Pelletier G. The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J 2003; 35:465–475. [DOI] [PubMed] [Google Scholar]
- 46. Jeon Y, Ko E, Lee KY, Ko MJ, Park SY, Kang J, Jeon CH, Lee H, Hwang DS. TopBP1 deficiency causes an early embryonic lethality and induces cellular senescence in primary cells. J Biol Chem 2011; 286:5414–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Libby BJ, De La Fuente R, O'Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 2002; 242:174–187. [DOI] [PubMed] [Google Scholar]
- 48. Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 2008; 22:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet 2007; 3:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li XC, Bolcun-Filas E, Schimenti JC. Genetic evidence that synaptonemal complex axial elements govern recombination pathway choice in mice. Genetics 2011; 189:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.