Abstract
Lim-domain only 4 (LMO4) plays a critical role in mediating the ototoxic side-effects of cisplatin, a highly effective anti-cancer drug. However, the signaling mechanism by which cochlear LMO4 mediates otopathology is yet to be fully understood. Knockout cell culture models are useful tools for investigating the functional roles of novel genes and delineating associated signaling pathways. Therefore, LMO4 knockout organ of Corti cells were generated by using the CRISPR (clustered regularly interspersed short palindromic repeats)/Cas9 (CRISPR-associated protein 9) system. Successful knockout of LMO4 in UB/OC1 cells was verified by the absence of LMO4 protein bands in immunoblots. Though the Knockout of LMO4 retarded the growth rate and the migratory potential of the cells it did not inhibit their long-term viability as the LMO4 knockout UB/OC1 cells were able to survive, proliferate, and form colonies. In addition, the knockout of LMO4 did not alter the expression of myosin VIIa, a biomarker of hair cells, suggesting that the knockout cells retain important characteristic features of cochlear sensory receptor cells. Thus, the findings of this study indicate that CRISPR/Cas9 system is a simple and versatile method for knocking out genes of interest in organ of Corti cells and that LMO4 knockout UB/OC1 cells are viable experimental models for studying the functional role of LMO4 in ototoxicity.
Keywords: cisplatin, cochlea, LMO4, hearing loss, ototoxicity
1. INTRODUCTION
LMO4 is a transcriptional regulator that serves as a scaffold for complexes of DNA-binding factors and many regulatory proteins.1–3 It is heavily expressed in epithelial cells4,5 including the sensory receptor cells of the inner ear.6 One of the important functions of LMO4 is the regulation of cell survival and cell death signaling.7–10 LMO4 promotes the proliferation of neuronal cells, inhibits differentiation of mammary epithelial cells, and regulates the function of progenitor cells.3,11 In the inner ear, deficiency of LMO4 leads to developmental abnormalities in the organ of Corti.12 Consistent with other models, silencing of LMO4 in organ of Corti explants facilitated apoptosis13 while overexpression of LMO4 in UB/OC1 cell cultures enhanced cell proliferation.14 A critical role of LMO4 in protecting the cochlea from oxidative stress induced damage was suggested by the recent findings in cisplatin-mediated ototoxicity. Treatment with cisplatin induced nitration and downregulation of cochlear LMO4 whereas inhibition of nitration attenuated cisplatin-induced downregulation of LMO4 and prevented associated hearing loss.13–15 The mechanism by which nitration of cochlear LMO4 leads to ototoxicity could be further clarified by using genetically modified organ of Corti cells in which LMO4 is either permanently deleted or altered.
Genetic manipulation of target genes has been successfully employed to elucidate the functional role of specific genes in auditory physiology as well as pathology. For instance, the otoprotective function of Akt2 and Akt3 in gentamicin-induced hair cell loss was determined by using Akt knockout mice.16 The role of prestin in enhancing the susceptibility of outer hair cells to cell death induced by cholesterol chelator 2-hydroxypropyl-β-cyclodextrine was determined by using prestin knockout mice.17 The requirement of Neuroplastin 65 for synaptogenesis in inner hair cells was detected by using Neuroplastin knockout mice.18 Alternatively, genetically modified organ of Corti cell culture models are used for determining the functions of manipulated genes. For example, the role of copper transporter Ctr1 in facilitating the uptake of cisplatin was determined by Knockdown of Ctr1 in HEI-OC1 cells19 while the involvement of c-Myb in increasing the sensitivity of these cells to neomycin-induced cytotoxicity was evaluated by knocking down c-Myb.20 Silencing of TRPV1 or NOX3 by using short interfering RNAs reduced cisplatin-induced apoptosis21 while transient knockdown of LMO4 exacerbated cisplatin-induced cell death in UB/OC1 cells.14 Though the later study suggested a central role of LMO4 in cisplatin-induced ototoxicity, stable LMO4 knockout organ of Corti cell culture models are not currently available to further investigate the underlying mechanism.
UB/OC1 cells are immortal cell lines derived from the mouse organ of Corti.22,23 They are amenable to genetic manipulations and have been used to study signaling mechanisms that regulate auditory function.25–27 These cells express LMO4 in both differentiation and proliferation stages, and have been widely used as a model to study ototoxicity.14,23,28,29 LMO4 mutants and knockouts have been used in other models to elucidate the function of LMO4 in these systems. For example, the link between LMO4 and mammary hyperplasia was elucidated by using breast epithelial cells in which LMO4 was ablated.24 However, LMO4 knockout organ of Corti cells have not been generated so far. Therefore, the aim of the present study is to establish a stable LMO4 knockout UB/OC1 cell culture model and characterize the growth and differentiation of knockout UB/OC1 cells in order to facilitate the use of this cell culture model to further elucidate LMO4 signaling in ototoxicity.
2. MATERIALS AND METHODS
2.1. Cell culture
UB/OC1 cells were provided by Dr. Mathew C Holley (University of Sheffield, UK) and were initially cultured in minimum essential medium (GlutaMAX, catalog no. 41090–036, Thermo Fisher Scientific, Rockford, IL) containing 10% fetal bovine serum (Gibco BRL, Gaithersburg, MD) and 50 U/mL γ-interferon (catalog no. 315–05, PeproTech, Rocky Hill, NJ), in a humidified atmosphere containing 10% CO2. To facilitate differentiation the cells were cultured for a week at 37°C without γ-interferon. Differentiation was verified by assessing the expression of myosin VIIa and fully differentiated cells were used for all experiments.14
2.2. Chemicals and reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO), unless noted otherwise.
2.3. CRISPR/Cas9 knockout of LMO4
UB/OC1 cells were plated at 50% confluence and cultured for 24 h. Then the cells were transfected with LMO4 CRISPR/Cas9 knockout plasmid (sc-401636; Santa Cruz Biotechnology Inc., Santa Cruz, CA), which is tagged with green fluorescent protein (GFP). Although human plasmids were used the mouse LMO4 shares the target sequences of the guide RNAs in the plasmid. Lipofectamine-3000 (L3000008, Life Technologies, Carlsbad, CA) was used for the transfection, following the manufacturer’s protocol. After 48 h, the transfection efficiency was determined by assessing the expression of GFP using flowcytometry. GFP positive cells were isolated and cultured individually in a 96 well plate for 3 weeks. Then the cell colonies were trypsinized, plated on 60 mm plates, and propagated for further analysis. Control cells were transfected with plasmids without LMO4 specific guide RNAs and were subjected to the same protocol as that of the knockout cells.
2.4. Cisplatin treatment
LMO4 knockout, control, and wild-type UB/OC1 cells were treated with 5 μm of cisplatin mixed with culture medium for 24 h. Cell viability was assessed by counting the number of viable cells after trypan blue staining.
2.5. Immunoblotting
Proteins were extracted from the cells by homogenizing in radio-immunoprecipitation assay buffer containing 5 mm ethyl-enediaminetetraacetic acid, protease, and phosphatase inhibitors (Thermo Fisher Scientific). The protein concentration was determined by Bradford protein assay and 30 of protein was loaded on 4–20% Mini-Protean TGX gel (456–1093, Bio-Rad Laboratories, Inc., Hercules, CA). The separated proteins were transferred on to apolyvinylidene difluoride membrane, blocked with 5% fat-free milk in tris-buffered saline containing 0.05% Tween 20, and incubated overnight with rabbit anti-LMO4 (1:1000, ab39383, Abcam, Cambridge, MA). After incubation with peroxidase-conjugated anti-rabbit secondary antibody (32460, Thermo Fisher Scientific), the blots were developed using chemiluminescence detection reagent (34076, Thermo Fisher Scientific), and visualized using FluorChem E System (ProteinSimple, San Jose, CA). Background-corrected bands (ImageJ, NIH, Bethesda, MD) were normalized against actin (sc-1615, Santa Cruz Biotechnology Inc.).
2.6. Sequencing
The LMO4 knockout cells were propagated and mRNA was extracted by using RNeasy Microarray Tissue Mini Kit (Catalog no. 73304, Qiagen, Valencia, CA) and transcribed to cDNA (Catalog no.1038960, Qiagen) following the manufacturer’s protocols. The target locus (377 base pair fragment) was amplified by using primers flanking the guide RNAs (Forward GGACCGCTTTCTGCTCTATG; Reverse GGGCTGTGGGTCTATCATGT). The amplified PCR product was sequenced by Genwiz (South Plainfield, NJ) by employing the sequence primer TGTAGTGAAACCGA TCTCCCG.
2.7. Cell growth assay
The reduction of 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyl- tetrazolium bromide (MTT) by metabolically active cells was used as a measure of cell growth. MTT Cell Growth Assay Kit (CT02, EMD Millipore, EMD Millipore Corporation, Temecula, CA) was used to measure cell growth from day 1 to day 5, following the manufacturer’s protocol. Briefly, UB/OC1-vector controls and UB/OC1-LMO4 Knockout cells were plated on 96 well plates (500 cells/well), cultured at 37°C (5% CO2) for the specified number of days, and then incubated with 10 μL of MTT solution (5 mg/mL in phosphate-buffered saline) for an additional 4 h (37°C and 5% CO2). The formazan crystals, formed by the reduction of MTT by viable cells, were dissolved in 0.04 N hydrochloric acid in isopropanol and the absorbance at 570 nm was measured using a microplate reader.
2.8. Colony-forming assay
LMO4 knockout cells (200 cells) were plated on a six well plate and cultured for 2 weeks at 37°C. Then the cells were washed with ice cold phosphate-buffered saline and fixed with ice-cold methanol for 20 min. The fixed cells were stained with 0.5% crystal violet solution (in 25% methanol) and incubated for 10 min at room temperature. Finally, the cells were rinsed with water and the colonies were counted using an inverted microscope.
2.9. Immunocytochemistry
UB/OC1-vector controls and UB/OC1-LMO4 Knockout cells were plated on two-well chamber slides (Nunc Lab-Tek II Chamber Slide system, 154461, Fisher Scientific, Pittsburgh, PA) and cultured for 24 h at 37°C. Then the cells were fixed with pre-cold methanol/acetone (1:1, v/v) mixture for 15 min, permeabilized with 0.2% (v/v) Triton X-100 in phosphate-buffered saline for 30 min, and blocked with goat serum (5% v/v in phosphate-buffered saline) for 1 h. The cells were then incubated at room temperature with anti-myosin VIIa (sc-74516, Santa Cruz Biotechnology Inc.) for 1 h followed by Alexa Fluor 568 donkey anti-mouse secondary antibody (A10037, Life Technologies) and fluorescein phalloidin (F432, Life Technologies) for 1 h. The stained cells were mounted on glass slides with ProLong Gold antifade reagent containing DAPI (P36935, Life Technologies) and examined using the Carl Zeiss Laser Scanning Systems (Zeiss LSM 780, Jena, Germany).
2.10. Wound healing assay
UB/OC1-vector controls and UB/OC1-LMO4 Knockout cells were plated in 60 mm cell culture plates and cultured to establish a monolayer of cells. After 24 h a scratch was made in the monolayers using a sterile 200 μL pipette tip and the image of the wound was captured at regular intervals using an inverted microscope. The width of the scratch was measured at the beginning and after 6, 12, and 24 h and wound closure was determined using image J software.
3. RESULTS
3.1. LMO4 is knocked out in UB/OC1 cells using CRISPR/Cas9 system
Generation of LMO4 knockout UB/OC1 cells is important to fully delineate the role of LMO4 in ototoxicity. CRISPR/Cas9 system is increasingly used for genome editing and has been successfully applied for generating knockout cell models to study the function of target genes. We employed a pool of three plasmids encoding Cas9 nuclease and target-specific guide RNAs to cause double-strand break within the LMO4 gene (Figure 1). Transfection of LMO4 CRISPR/Cas9 knockout plasmid into the UB/OC1 cells was verified by the detection of GFP in the transfected cells (Figure 2A). LMO4 protein was not detected in the immunoblots of transfected cells (Figure 2B) suggesting the knockout of LMO4 in UB/OC1 cells using the CRISPR/Cas9 system. In addition, knockout of LMO4 was verified by a six base pair deletion in the target locus (Figure 2C). Off-target mutation analysis was performed by using web-based tool Cas-OFFinder (http://www.rgenome.net). This analysis indicated that there are no off-target matching sequences in the mouse genome for the guide RNAs.
FIGURE 1.
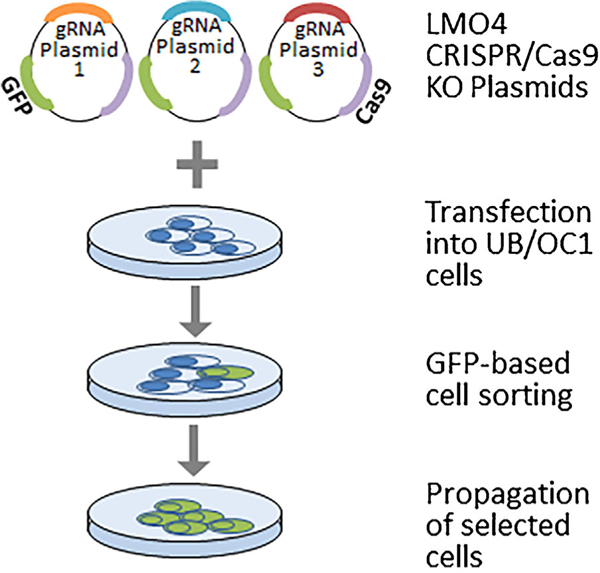
Schematic of the experimental design for generating LMO4 knockout UB/OC1 cells. UB/OC1 cells were transfected with LMO4 CRISPR/Cas9 knockout plasmids containing target-specific guide RNAs and GFP. Successful transfection of the plasmid was verified by the expression of GFP. The transfected cells containing GFP were isolated by cell sorting and propagated for further analysis.
FIGURE 2.
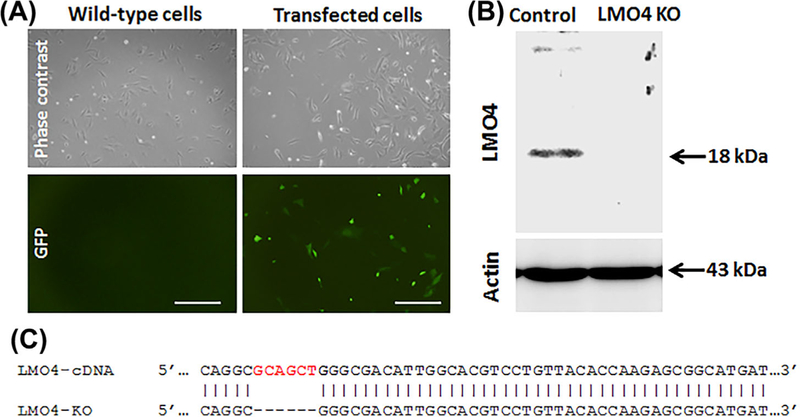
Knockout of LMO4 in UB/OC1 cells. A, Green fluorescent cells indicate the presence of GFP in UB/OC1 cells transfected with CRISPR/Cas9 knockout plasmid. GFP is not detected in the wild-type cells. The images are representative of three biological replicates. Scale bar represents 200 μm. B, Immunoblots indicate that LMO4 protein is not detected in the knockout cells. Actin is used for normalization. The images are representative of three biological replicates. C, Modified DNA sequence of target locus in the knockout cells indicates a 6 base pair deletion.
3.2. Knockout of LMO4 slows down the proliferation of UB/OC1 cells
The effect of LMO4 knockout on the growth of UB/OC1 cells needs to be ascertained in order to determine the suitability of this model for ototoxicity studies. Cell growth was assessed by monitoring the conversion of MTT to formazan. The growth rates of the LMO4 knockout cells were similar to that of the vector controls during the first 4 days (Figure 3A). However, the growth rate was much slower on the 5th day. In addition, colony-forming assay was used to determine the ability of the LMO4 knockout UB/OC 1 cells to survive, proliferate and form colonies (Figure 3B). The plating efficiency of LMO4 knockout cells, which indicates the percentage of cells seeded into a dish that finally grow to form a colony, was 22 ± 3 (n = 3). These findings collectively suggest that though the knockout of LMO4 retarded the proliferation of UB/OC 1 cells they are viable models that are capable of forming colonies.
FIGURE 3.
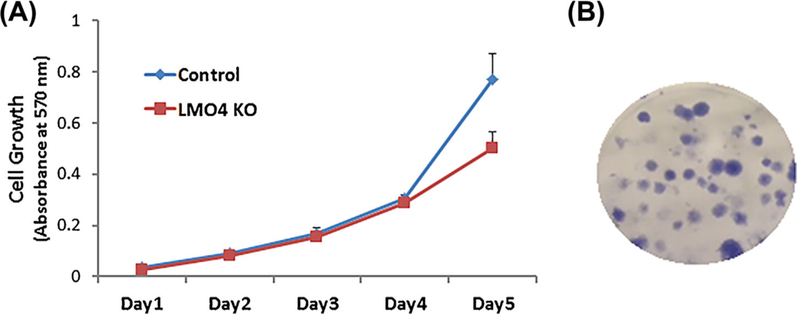
Growth pattern of LMO4 knockout UB/OC1 cells. A, Growth curve of the cells, determined by using MTT calorimetric assay, indicated that the growth rate of the LMO4 knockout cells is similar to that of the control cells during the initial 4 days and is relatively slower on day 5. The results are expressed as mean ± standard deviation, n = 6. B, Representative image of LMO4 knockout UB/OC1 cell colonies, 2 weeks after plating, indicates the survival potential of the cells and their ability to form colonies. The image is representative of three biological replicates.
3.3. Differentiation of UB/OC1 cells is not affected by knockout of LMO4
Expression of myosin VIIa was used to verify whether the knockout of LMO4 could alter the differentiation of UB/OC1 cells. Immunoreactivity to anti-myosin VIIa, indicated by red staining, suggested that myosin VIIa is expressed in both the vector control and LMO4 knockout UB/OC1 cells (Figure 4). The specificity of the immunoreaction of anti-myosin VIIa was previously verified by the presence of a protein band at the expected molecular weight and the absence of non-specific bands in immunoblots. These results suggest that knockout of LMO4 did not alter the differentiation of UB/OC1 cells.
FIGURE 4.
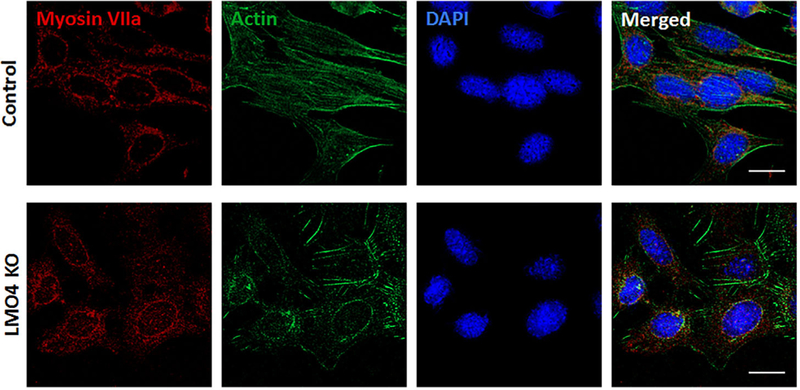
Differentiation of LMO4 knockout UB/OC1 cells. Myosin Vila, a biomarker of hair cells, is detected in both the wild-type and knockout cells suggesting that the knockout of LMO4 does not alter the differentiation of UB/OC1 cells. Red staining indicates immunoreactivity to antimyosin Vila, green indicates actin staining with phalloidin, while blue indicates nuclear staining with DAPI. The images are representative of three biological replicates. Scale bar represents 20 μm.
3.4. Migratory potential is decreased by knockout of LMO4
The migration of LMO4 knockout UB/OC1 cells was assessed by wound healing assay. The results indicated that the LMO4 knockout cells were able to close the wound, 24 h after the scratch. However, compared to the vector controls the LMO4 knockout UB/OC1 cells took a longer time to completely close the wound (Figure 5). Measurement of the wound width 12 h after the scratch indicated that only 38% of the wound was closed by the LMO4 knockout UB/OC1 cells whereas 78% of the wound was closed by the vector control cells. This suggested that knockout of LMO4 decreases the migratory potential of the UB/OC1 cells.
FIGURE 5.
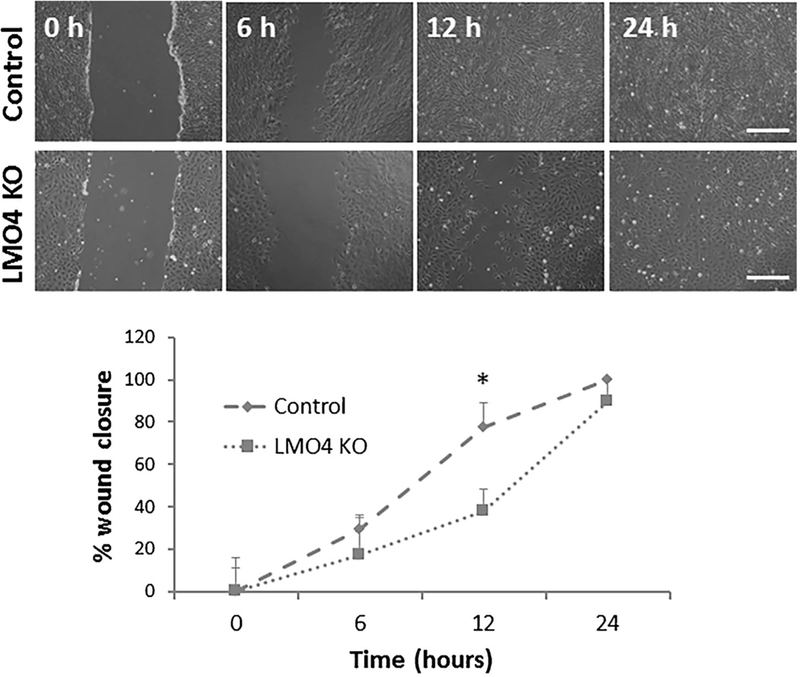
Migratory potential of LMO4 knockout UB/OC1 cells. Wound healing assay suggests that the migratory potential of LMO4 knockout cells is relatively slower than that of the control cells. The wound closure in control cells after 24 h is considered as 100%. The results are expressed as mean ± standard deviation, n = 3. Significant differences between the groups were determined by one-tailed t-tests, * indicates P < 0.05. Scale bar represents 200 μm.
3.5. Knockout of LMO4 enhances the susceptibility of UB/OC1 cells to cisplatin-induced ototoxicity
Viability of UB/OC1 cells post cisplatin treatment was assessed by using trypan blue staining. Treatment of LMO4 knockout UB/OC1 cells with 5 μm cisplatin significantly decreased the number of viable cells when compared to both control and wild-type cells (Figure 6). Cisplatin treatment induced a 52% and 54% decrease in cell viability in wild-type and control cells, respectively, whereas it induced a 70% decrease in cell viability in the LMO4 knockout cells. This enhanced vulnerability of LMO4 knockout UB/OC1 cells indicates that the knockout cells could be employed to study the role of LMO4 in ototoxicity.
FIGURE 6.
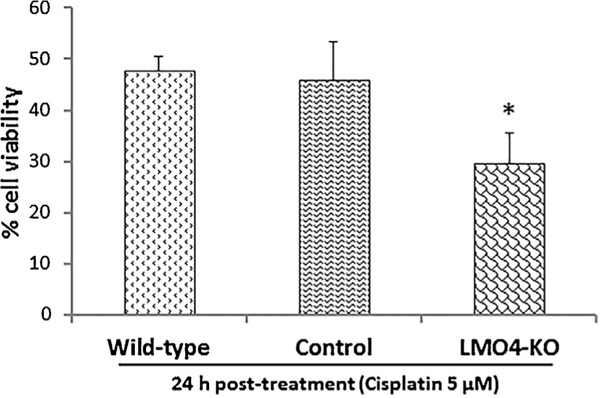
Viability of LMO4 knockout cells after cisplatin treatment. Cell counts indicate that cisplatin treatment decreases the viability of UB/OC1 cells. However, the number of viable cells was much lower in the LMO4 knockout cells relative to the control and wild-type cells. The results are expressed as mean ± standard deviation, n = 3. Significant differences between the groups were determined by one-tailed i-tests, * indicates P < 0.05, relative to control.
4. DISCUSSION
Nitration of LMO4 plays a role in cisplatin ototoxicity.13,14 However, in vivo evaluation of its functional significance in the auditory system by using global knockout models is not feasible because of phenotypic abnormalities and prenatal death of LMO4 knockout mice.30 Therefore, generation of genetically engineered cell culture models is critical for delineating the function of LMO4 in acquired hearing loss. In this study, LMO4 knockout organ of Corti cells were generated for the first time, by using the CRISPR/Cas9 system. Although knockout of LMO4 retarded the growth rate of UB/OC1 cells, the knockout cells were able to survive and form colonies when cultured for 2 weeks. Moreover, knockout of LMO4 did not alter the differentiation of UB/OC1 cells. This suggested that these knockout cells are viable models that can be used for investigating the functional role of LMO4 in ototoxicity.
UB/OC1 cells were chosen for genetically manipulating LMO4 and establishing a LMO4 deficient organ of Corti cell culture model because previous studies have demonstrated that cisplatin-induced nitration and downregulation of LMO4 mediates apoptosis in UB/OC1 cells.14,15 Moreover, transient repression of LMO4 in UB/OC1 cells by using short interfering RNAs did not have lethal consequences.14 In this study CRISPR/Cas9 system was used to establish stable LMO4 knockout organ of Corti cells because of the simplicity and efficiency of this gene editing tool.31 Moreover, this method facilitated the verification of the transfection of LMO4 CRISPR/Cas9 knockout plasmid into the UB/OC1 cells in 48 h, by using the GFP tag included in the plasmid. The guide RNAs in the plasmid, which is designed to provide high target specificity, targeted the LMO4 gene in the UB/OC1 cells for disruption. This was indicated by the absence of LMO4 protein bands in the immunoblots of LMO4 knockout UB/OC1 cells whereas the actin protein bands were detected in both the control and knockout cells. These results suggested that CRISPR/Cas9 system is an effective tool for generating stable LMO4 knockout organ of Corti cell lines.
In order to employ this cell culture model for investigating the role of LMO4 in auditory dysfunction it is essential to determine the impact of the knockout of LMO4 on the growth and differentiation of UB/OC1 cells. The results of this study indicated that the growth pattern of LMO4 knockout UB/OC1 cells was similar to that of the vector controls during the first 4 days. Although the growth rate was relatively slower on day 5, the knockout cells were able to survive for longer periods and form colonies. Moreover, myosin Vila, a biomarker of hair cells, was detected in the knockout cells suggesting the suitability of this model for studying the role of LMO4 in mediating the toxic effects of ototoxicants that target the sensory epithelial cells of the inner ear. Finally, the knockout cells were able to migrate, though at a slower pace, suggesting that the knockout of LMO4 does not completely inhibit the migratory potential of UB/OC1 cells. Collectively, the growth, differentiation, and migration of the knockout cells suggested that they can be useful models for investigating the role of LMO4 in ototoxicity.
The LMO4 knockout UB/OC1 cells could be used for several applications. For instance, LMO4 is a key regulator of apoptosis,32,33 an important cell death mechanism by which many ototoxicants cause hearing loss.34–37 Consistent with its anti-apoptotic role, the knockout of LMO4 enhanced the vulnerability of UB/OC1 cells to cisplatin-induced cytotoxicity. Therefore, this knockout cell culture model could be used for evaluating the apoptotic signaling mechanisms that regulate cochlear cell death after exposure to ototoxic drugs, organic solvents, and heavy metals. In addition, the role of cochlear LMO4 in auditory dysfunction could be analyzed after reintroduction of LMO4 into the knockout cells, under different experimental conditions.
Furthermore, these knockout cells could be used to generate LMO4 mutant cells. Specific mutations that target different sites of posttranslational modifications could be induced by transfecting corresponding plasmids into the LMO4 knockout UB/OC1 cells. This in turn will facilitate the investigation of the functional importance of posttranslational modifications of LMO4, such as nitration of LMO4 in cisplatin-induced ototoxicity. Thus, LMO4 knockout organ of Corti cells could be a valuable tool for investigating the ototoxic pathways regulated by cochlear LMO4.
5. CONCLUSION
The generation of LMO4 knockout UB/OC1 cells by using the CRISPR/Cas9 system suggests that this gene editing tool could be employed for knockout of other genes of interest in organ of Corti cells. The ability of the LMO4 knockout UB/OC 1 cells to grow, form colonies, and migrate suggests that they are viable models that could be used for delineating the role of LMO4 in ototoxicity. Furthermore, the expression of myosin Vila in the knockout cells suggests that they can be considered as an experimental model for sensory receptor cells of the cochlea. Taken together, the establishment of LMO4 knockout organ of Corti cells and the characterization of its growth and differentiation suggest that this novel cell culture model could be a useful tool for investigating the cochlear LMO4 signaling mechanism and its implications in auditory dysfunction.
ACKNOWLEDGMENTS
This research was supported by start-up funds to SJ from Wayne State University and P30 grant (P30 ES020957) to Center for Urban Responses to Environmental Stressors (CURES) from NIEHS. The Microscopy, Imaging and Cytometry Resources Core is supported, in part, by NIH Center grant P30 CA022453.
Funding information
Wayne State University, Grant number: P30 ES020957
Abbreviations:
- CRISPR/Cas9
clustered regularly interspersed short palindromic repeats/CRISPR-associated protein 9
- GFP
green fluorescent protein
- LMO4
Lim-domain only 4
- MTT
3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromid.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflict of interest to declare.
How to cite this article: Rathinam R, Rosati R, Jamesdaniel S. CRISPR/Cas9-mediated knockout of Lim-domain only four retards organ of Corti cell growth. J Cell Biochem. 2017;1–9. https://doi.org/10.1002/Jcb.26529
REFERENCES
- 1.Grutz G, Forster A, Rabbitts TH. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/ NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene. 1998;17:2799–2703. [DOI] [PubMed] [Google Scholar]
- 2.Visvader JE, Venter D, Hahm K, et al. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2001;98:14452–14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sum EY, Peng B, Yu X, et al. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. [DOI] [PubMed] [Google Scholar]
- 4.Sum EY, O’Reilly LA, Jonas N, Lindeman G, Visvader JE, The LIM domain protein Lmo4 is highly expressed in proliferating mouse epithelial tissues. J Histochem Cytochem. 2005;53: 475–486. [DOI] [PubMed] [Google Scholar]
- 5.Lu Z, Lam K, Wang N, Xu X, Cortes M, Andersen B. LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene. 2006;25: 2920–2930. [DOI] [PubMed] [Google Scholar]
- 6.Jamesdaniel S Downstream targets of Lmo4 are modulated by cisplatin in the inner ear of Wistar rats. PLoS ONE. 2014;9: e115263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novotny-Diermayr V, Lin B, Gu L, Cao X. Modulation of the interleukin-6 receptor subunit glycoprotein 130 complex and its signaling by LMO4 interaction. J Biol Chem. 2005;280: 12747–12757. [DOI] [PubMed] [Google Scholar]
- 8.Schock SC, Xu J, Duquette PM, et al. Rescue of neurons from ischemic injury by peroxisome proliferator-activated receptor- gamma requires a novel essential cofactor LMO4. J Neurosci. 2008;28:12433–12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setogawa T, Shinozaki-Yabana S, Masuda T, Matsuura K, Akiyama T. The tumor suppressor LKB1 induces p21 expression in collaboration with LMO4, GATA-6, and Ldb1. Biochem Biophys Res Commun. 2006;343:1186–1190. [DOI] [PubMed] [Google Scholar]
- 10.Wang N, Lin KK, Lu Z, et al. The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene 2007;26:6431–41. [DOI] [PubMed] [Google Scholar]
- 11.Ochoa SD, Salvador S, LaBonne C. The LIM adaptor protein LMO4 is an essential regulator of neural crest development. Dev Biol. 2012;26:313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng M, Luo XJ, Pan L, et al. LMO4 functions as a negative regulator of sensory organ formation in the mammalian cochlea. J Neurosci. 2014;34:10072–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamesdaniel S, Coling D, Hinduja S, et al. Cisplatin-induced ototoxicity is mediated by nitroxidative modification of cochlear proteins characterized by nitration of Lmo4. J Biol Chem. 2012;287:18674–18686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamesdaniel S, Rathinam R, Neumann WL. Targeting nitrative stress for attenuating cisplatin-induced downregulation of cochlear LIM domain only 4 and ototoxicity. Redox Biol. 2016;10:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathinam R, Ghosh S, Neumann WL, Jamesdaniel S. Cisplatin- induced apoptosis in auditory, renal, and neuronal cells is associated with nitration and downregulation of LMO4. Cell Death Discov. 2015;1:pii15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand Y, Levano S, Radojevic V, et al. All Akt isoforms (Akt1, Akt2, Akt3) are involved in normal hearing, but only Akt2 and Akt3 are involved in auditory hair cell survival in the mammalian inner ear. PLoS ONE. 2015;10:e0121599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi S, Homma K, Zhou Y, et al. Susceptibility of outer hair cells to cholesterol chelator 2-hydroxypropyl-beta-cyclodextrine is prestin-dependent. Sci Rep. 2016;6:21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrott L, Bowl MR, Aguilar C, et al. Absence of Neuro-plastin-65 Affects Synaptogenesis in Mouse Inner Hair Cells and Causes Profound Hearing Loss. J Neurosci. 2016;36: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.More SS, Akil O, Ianculescu AG, Geier EG, Lustig LR, Giacomini KM, Role of the copper transporter, CTR1, in platinum-induced ototoxicity. J Neurosci. 2010;30:9500–9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Liu W, Fan Z, et al. c-Myb knockdown increases the neomycin-induced damage to hair-cell-like HEI-OC1 cells in vitro. Sci Rep. 2017;7:41094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjea D, Jajoo S, Whitworth C, et al. Short interfering RNA against transient receptor potential vanilloid 1 attenuates cisplatin-induced hearing loss in the rat. J Neurosci. 2008;28: 13056–13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivolta MN, Grix N, Lawlor P, Ashmore JF, Jagger DJ, Holley MC. Auditory hair cell precursors immortalized from the mammalian inner ear. Proc Biol Sci. 1998;265:1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivolta MN, Halsall A, Johnson CM, Tones MA, Holley MC. Transcript profiling of functionally related groups of genes during conditional differentiation of a mammalian cochlear hair cell line. Genome Res. 2002;12:1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sum EY, Segara D, Duscio B, et al. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci USA. 2005;102: 7659–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagger DJ, Griesinger CB, Rivolta MN, Holley MC, Ashmore JF. Calcium signalling mediated by the 9 acetylcholine receptor in a cochlear cell line from the Immortomouse. J Physiol. 2000;527: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccardi S, Bergling S, SigoillotF, et al. MiR-210promotes sensory hair cell formation in the organ of corti. BMC Genomics. 2016;17:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zallocchi M, Meehan DT, Delimont D, et al. Localization and expression of clarin-1, the Clrn1 gene product, in auditory hair cells and photoreceptors. Hear Res. 2009;255:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur T, Borse V, Sheth S, et al. Adenosine A1 Receptor Protects Against Cisplatin Ototoxicity by Suppressing the NOX3/STAT1 Inflammatory Pathway in the Cochlea. J Neurosci. 2016;36: 3962–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JS, Jou I, Park SM. Attenuation of noise-induced hearing loss using methylene blue. Cell Death Dis. 2014;5:e1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol. 2004;24:2074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen HH, Schock SC, Xu J, Safarpour F, Thompson CS, Stewart AF. Extracellular ATP-dependent upregulation of the transcription cofactor LMO4 promotes neuron survival from hypoxia. Exp Cell Res. 2007;313:3106–3116. [DOI] [PubMed] [Google Scholar]
- 33.Tian Y, Wang N, Lu Z. Repression of Lim only protein 4-activated transcription inhibits proliferation and induces apoptosis of normal mammary epithelial cells and breast cancer cells. Clin Exp Metastasis. 2010;27: 455–463. [DOI] [PubMed] [Google Scholar]
- 34.Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 27 2006; 1–19. [DOI] [PubMed] [Google Scholar]
- 35.Huang T, Cheng AG, Stupak H, et al. Oxidative stress-induced apoptosis of cochlear sensory cells: otoprotective strategies Int J Dev Neurosci. 2000;18:259–270. [DOI] [PubMed] [Google Scholar]
- 36.Op de Beeck K, Schacht J, Van Camp G. Apoptosis in acquired and genetic hearing impairment: the programmed death of the hair cell. Hear Res. 2011;281:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybak LP, Whitworth CA, Mukheijea D, Ramkumar V. Mechanisms of cisplatin-induced ototoxicity and prevention. Hear Res. 2007;226: 157–167. [DOI] [PubMed] [Google Scholar]


