Abstract
Genetic segregation of airway disease traits despite redundancy of calcium-activated chloride channel family members. Physiol Genomics 25: 502–513, 2006. First published March 28, 2006; doi:10.1152/physiolgenomics.00321.2005.—Complex airway diseases such as asthma and chronic obstructive pulmonary disease exhibit stereotyped traits (especially airway hyperreactivity and mucous cell metaplasia) that are variably expressed in each patient. Here, we used a mouse model for virus-induced long-term expression of these traits to determine whether individual traits can be genetically segregated and thereby linked to separate determinants. We showed that an F2 intercross population derived from susceptible and nonsusceptible mouse strains can manifest individual phenotypic extremes that exhibit one or the other disease trait. Functional genomic analysis of these extremes further indicated that a member of the calcium-activated chloride channel (CLCA) gene family designated mClca3 was inducible with mucous cell metaplasia but not airway hyperreactivity. In confirmation of this finding, we found that mClca3 gene transfer to mouse airway epithelium was sufficient to induce mucous cell metaplasia but not airway hyperreactivity. However, newly developed mClca3−/− mice exhibited the same degree of mucous cell metaplasia and airway hyperreactivity as wild-type mice. Bioinformatic analysis of the Clca locus led to the identification of mClca5, and gene transfer indicated that mClca5 also selectively drives mucous cell metaplasia. Thus, in addition to the capacity of CLCA family members to exhibit diverse functional activities, there is also preserved function so that more than one family member mediates mucous cell metaplasia. Nonetheless, Clca expression appears to be a selective determinant of mucous cell metaplasia so that shared homologies between CLCA family members may still represent a useful target for focused therapeutic intervention in hypersecretory airway disease.
Keywords: airway hyperreactivity, mucous (goblet) cell metaplasia, viral bronchiolitis, asthma, chronic obstructive pulmonary disease
COMPLEX AIRWAY DISEASES such as asthma and chronic obstructive pulmonary disease (COPD) represent some of the world’s most common disorders (53). Each of these conditions is marked by the development of disease traits, chiefly, airway hyperreactivity and mucous cell metaplasia, which exhibit variable expression in affected individuals. Despite the capacity to precisely quantify these traits, the basis for their variability in different individuals remains uncertain. In that regard, there is considerable evidence that the manifestation of airway disease traits may be influenced by genetic background (14, 39). Approaches to define this issue have included linkage analysis in patient populations as well as mouse models that aim to replicate the disease condition (28, 46). The most common model has been the use of mice in studies of the susceptibility to either allergen- or cigarette smoke-induced disease traits. In general, however, these studies have not directly addressed the possibility that disease traits might be genetically segregated and so have not led to defining a more specific pathway responsible for each individual trait.
In the present work, we aimed to segregate airway disease traits and thereby identify a pathway that might be associated with a single trait. We especially focused on defining the role of genes responsible for chronic airway hyperreactivity versus mucous cell metaplasia. We took advantage of our earlier study (52) indicating that both of these traits were inducible on a long-term basis after viral bronchiolitis in the C57BL/6J strain of inbred mice. We identified the Balb/cJ strain as one that responds similarly during the acute infection but then fails to develop any chronic alteration in airway behavior. We took advantage of this difference in genetic susceptibility to develop an F2 intercross population with phenotypic extremes that exhibited one or the other disease trait, and we analyzed these extremes with the use of an oligonucleotide microarray for gene expression. This combined genetic and genomic strategy provides evidence of a selective association between the expression of a member of the calcium-activated chloride channel (Clca) gene family, i.e., mouse (m)Clca3, with the development of mucous cell metaplasia but not airway hyperreactivity. When mucous cell metaplasia persisted despite the loss of mClca3 function in newly developed mClca3−/− mice, we then defined the role of another Clca family member, i.e., mClca5, as being capable of shared biological function. The findings thereby provide a useful model for segregating and defining complex disease traits in general and airway disease traits in particular. Moreover, the results help to clarify the specific role of the CLCA gene family in the development of mucous cell metaplasia in airway disease and thereby point to homologies in this family as a target for selective therapeutic intervention.
MATERIALS AND METHODS
Mice generation and handling.
Wild-type C57BL/6J, C57BL/10.D2, C57BL/10SnJ, Balb/cJ, Balb.B10, C3H, A/J, and F1 hybrid CB6F1/J mice were obtained from Jackson Laboratory. F1 hybrid mice were bred to generate an F2 intercross population (CB6F2/J). mClca3−/− mice were generated by homologous recombination with a mutant mClca3 gene with deletion of the 3′ half of exon 7, exons 8–11, and the intervening introns (Supplementary Fig. 1; available at the Physiological Genomics web site).1 The targeting construct was prepared with 1) a 5′ homology region of 3,946 bp extending from 961 nt upstream of exon 5 to 96 nt into exon7 and 2) a 3′ homology region of 5,227 bp from 90 bp after the splice donor site of exon 11 to 1,109 bp downstream of exon 14. The intervening region was substituted by a neomycin resistance cassette of 1.2 kb in reverse orientation. The targeting construct also included a Diptheria toxin cassette adjacent to the 3′ homology arm to permit negative selection of unspecific insertion events. The KspI linearized targeting vector was transfected into C57BL/6 embryonic stem cells (Eurogentec), and selected clones were screened for the null allele by PCR. Embryonic stem cell clones were injected into Balb/c blastocysts. Chimeric males were mated with C57BL/6 females, and heterozygotes were crossed to generate homozygous mClca3−/− mice. With the use of a real-time PCR assay directed against sequences in mClca3 exons 8 and 9, no detectable mClca3 mRNA was found in mClca3−/− mice. Mice were maintained under pathogen-free conditions, and the Animal Studies Committee approved all mouse study protocols. Viral inoculation with Sendai virus (SeV) and allergen challenge with ovalbumin (Ova) were performed as described previously (49, 52). For the present experiments, mice were inoculated intranasally with SeV (2 × 105 plaque-forming units) or an equivalent amount of UV-inactivated SeV (SeV-UV). Viral expression level in lung tissue was monitored by viral plaque assay and real-time quantitative RT-PCR as described previously (52).
Measurements of mRNA levels.
RNA was purified from homogenized lung tissue using the RNeasy kit (Qiagen). Target mRNA levels were quantified with real-time PCR using fluorogenic probe-primer combinations specific for mClca3, mClca5, and mucin 5AC (Muc5ac) mRNA and the TaqMan One-Step system (Applied Biosystems; Foster City, CA). Sequences of the forward primers, reverse primers, and probes were as follows: 5′-TGTGAAATTTGTGATGCTATTGCTTT-3′, 5′-ATGAATGCAATTGTTGTTGTTAACTTG-3′, and 5′-TTTGTAACCATTATAAGCTGCAAT-3′ for simian virus 40 (SV40); 5′-ACCGGCTGCCGCTAAAGAGCTTGAG-3′, 5′-AGACCATTTTCTGAAGCTGATCCGAAG-3′, and 5′-AGCTGTCCAAAATGACAGGAGGCCTGCAGACATA-3 for mClca3; 5′-CTGGCCTTTCGGACAGCCAGCCTTAAGA-3′, 5′-AATGGTGGGTGTTGTTCAGCGTGTAAGT-3′, and 5′-AGTGCCCATGCTTAGCTGTCCCTG-3′ for mClca5; and 5′-TACCACTCCCTGCTTCTGCAGCGTGTCA-3′, 5′-ATAGTAACAGTGGCCATCAAGGTCTGTCT-3′, and 5′-TATACCCCTTGGGA-TCCATCATCTACA-3′ for Muc5ac, respectively. All probes were designed to span an intron and did not react with genomic DNA. All real-time PCR data were normalized to the Gapdh level using the Taqman GAPDH control kit (Applied Biosystems). RNA standards of a known copy number were used to calculate mRNA levels. All quantitative PCR results are expressed as mRNA copy number per 107 GAPDH mRNA copies. For the gene transfer experiments, mRNA copy number was also normalized to SV40 polyA mRNA copies as described below.
Western blot analysis and immunostaining.
For Western blot analysis, whole lung lysate was separated by electrophoresis on a 7.5% TrisHCl gel (Bio-Rad) and transferred to polyvinylidene difluoride membranes, which were blocked with 5% dry milk-Tris-buffered saline-Tween and probed first with rabbit anti-human (h)CLCA1/mClca3 Ab (obtained from R. Levitt, Genera) and anti-rabbit secondary Ab and then with mouse anti-chicken β-actin mAb (Chemicon). For immunostaining with bright-field microscopy, mouse lungs were fixed, embedded, cut into sections, dewaxed, rehydrated, and incubated with mouse anti-MUC5AC mAb (clone 45M1, Neomarkers) or control Ab as described previously (51, 52). Photomicrography and quantification of reporter by cell counting (cells/mm basement membrane) using Image J and NIH Image software were performed as described previously (44, 51). For immunofluorescence, sections were incubated with 50 mM glycine (pH 9.6) at 90°C for 10 min for antigen retrieval and then with anti-hCLCA1/mClca3 Ab (10 μg/ml) and anti-MUC5AC mAb (2 μg/ml) in 70 mM NaCl, 30 mM HEPES, and 2 mM CaCl2 (pH 7.4) with 1% BSA, 1% goat serum, and 0.1% cold water fish gelatin (Electron Microscopy Sciences) followed by 10 μg/ml Alexa 488-conjugated goat anti-rabbit IgG (Molecular Probes) and 10 μg/ml rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) in the same buffer for 4 h at 25°C. In some experiments, tissue sections were blocked with 2% fish gel, incubated with biotinylated anti-MUC5AC mAb and Alexa 555-conjugated streptavidin or anti-hCLCA1/mClca3 Ab and Alexa 633-conjugated goat anti-rabbit secondary Ab (Molecular Probes), counterstained with Sytox green (Molecular Probes), and then imaged by confocal microscopy using a Zeiss laser scanning system with LSM-510 software.
Airway reactivity measurements.
In screening experiments, airway reactivity to nebulized methacholine was determined in unrestrained mice using a whole body plethysmograph and BioSystem XA version 2.7.9 software (Buxco Electronic) to derive values for enhanced pause (Penh) as described previously (23, 52). Mice were placed in the plethysmograph for a 5-min acclimatization interval, followed by 3-min acquisition intervals before (baseline Penh) and after a 48-s exposure to nebulized vehicle (PBS) or doubling concentrations of methacholine (5–80 mg/ml) delivered from a Collison 6 jet nebulizer (BGI). In validation experiments, airway reactivity was also determined by measurements of total lung resistance and dynamic compliance as described previously (35). In this case, mice were anesthetized, ventilated through a tracheostomy with a Harvard Apparatus model 687 at 6–10 ml/kg and positive end-expiratory pressure of 2–4 cmH2O, and monitored for intrapleural pressure using an oroesophageal tube. Methacholine was delivered at 3-min intervals using an in-line nebulizer (Aerogen Laboratory, 2.4–4 μm particle size). Between deliveries, the respiratory flow signal was obtained with a pneumotach (SenSym SCXL004, Buxco Electronics) and was integrated to calculate lung volume. Intraesophageal and airway pressure were measured via pressure transducers (Validyne DP45, Buxco Electronics) directly connected to the respective catheters.
Genetic segregation and microarray analysis.
From the group of CB6F2/J mice (n = 486), we identified extreme phenotypes as those with values for mucous cell metaplasia or airway reactivity that were above the 90th percentile or below the 10th percentile as described previously (31). Within this extreme population, we then identified four extreme cohorts: 1) high levels of mucous cell metaplasia and airway reactivity (H/H), 2) high mucous cell metaplasia and low airway reactivity (H/L), 3) low mucous cell metaplasia and high airway reactivity (L/H), or 4) low mucous cell metaplasia and low airway reactivity (L/L). Within each cohort, we randomly selected three mice for analysis of gene expression levels using oligonucleotide microarrays (Affymetrix Murine Genome U74v2 Set) hybridized in triplicate to labeled cRNA as described previously (43). Differential expression was analyzed by GeneChip 3.1 software (Affymetrix) and was assessed by pairwise comparisons of samples from the four extreme cohorts of CB6F2/J mice. Criteria for differential expression required increases or decreases in 12 of the 16 possible pairwise comparisons. Subsequently, microarrays were normalized using the GC-RMA algorithm as implemented in Bioconductor (17, 54). Mean log2 expression values for each of the extreme CB6F2/J cohorts were plotted using Spotfire Decisionsite for Functional Genomics. Microarray data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus under Series Accession No. GSE3925.
Adenoviral-associated virus 5 vectors.
Cellular RNA was purified from IL-13-treated mouse tracheal epithelial cell cultures (48) using the Micro RNA Isolation Kit (Stratagene) and reverse transcribed with the Superscript II Kit (Invitrogen). The product served as the template to clone full-length mClca3 and mClca5 cDNAs using PCR. mClca3 or mClca5 cDNA was ligated to the 3xFlag reporter sequence, and these constructs or 3xFlag sequence alone was inserted into the pTRUF2 adenoviral-associated virus 5 (AAV5) shuttle vector. High-titer AAV5 viruses were generated at the University of North Carolina Gene Therapy Center Vector Core Lab as described previously (55). For gene transfer experiments, purified AAV5 (2 × 1011 particles in 50 μl) was delivered intranasally in the same manner as for SeV inoculations. The expression of recombinant AAV5 was monitored by real-time PCR for SV40 polyA (which is present only in recombinant genes) using the Two-Step TaqMan system. Total RNA was reverse transcribed using the Archival cDNA kit (Applied Biosystems), and the cDNA was used as a template. Sequences for the forward primer, reverse primer, and probe were as follows: 5′-TGTGAAATTTGTGATGCTATTGCTTT-3′, 5-ATGAATGCAATTGTTGTTG-TTAACTGG-3′, and 5′-TTTGTAACCATTATAAGCTGCAAT-3′.
Statistical analysis.
Values for Penh and histochemistry of mouse tissues were analyzed using ANOVA for a factorial experimental design. If significance was achieved by one-way analysis, post-ANOVA comparison of means was performed using Scheffe`’s F-test. PCR data were compared by an unpaired Student’s t-test. If variances were unequal, Welch’s correction was applied. The significance level for all analyses was 0.05.
RESULTS
Independent genetic control of airway disease traits.
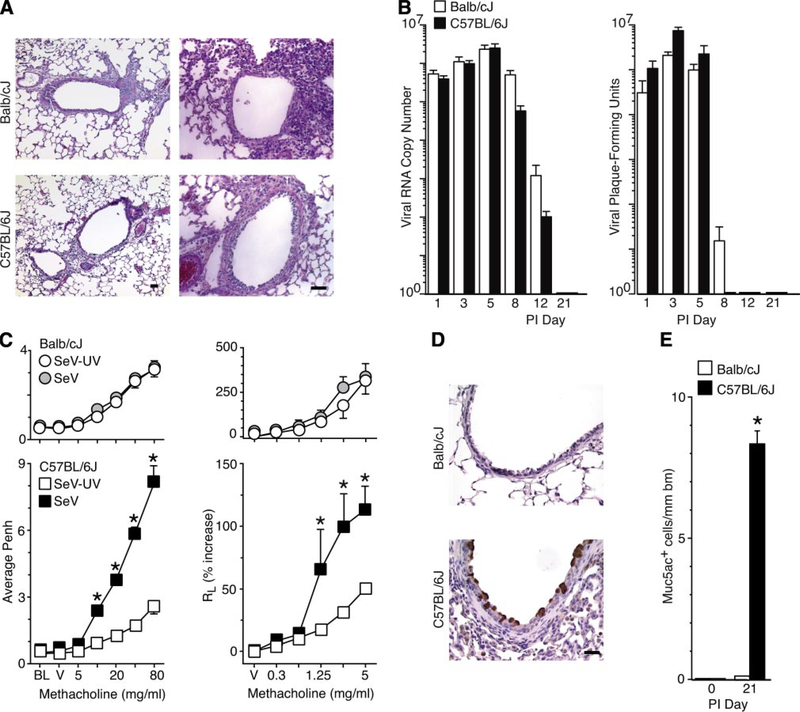
The initial experiments were aimed at identifying strains of mice with different susceptibility to virus-induced airway disease traits. Previous work indicated that C57BL/6J mice manifest acute bronchiolitis followed by persistent airway hyperreactivity and mucous cell metaplasia after SeV infection (48, 52). In the present experiments, we found that Balb/cJ mice exhibited a similar bronchiolitis but failed to develop long-term airway hyperreactivity or mucous cell metaplasia (Fig. 1). Thus C57BL/6J and Balb/cJ strains were closely matched for the acute response but were divergent in the chronic response to viral infection and thereby offered an ideal substrate to define genetic susceptibility to developing chronic airway disease traits. A/J and C3H/HeJ mice, which are often used for studies of genetic susceptibility to asthma, developed a severe alveolitis after SeV inoculation and so were not suitable for comparison (data not shown).
Fig. 1.
Susceptibility to virus-induced airway disease traits in inbred mouse strains. Balb/cJ and C57BL/6J mice were inoculated with Sendai virus (SeV) or an equivalent amount of UV-inactivated SeV (SeV-UV) and analyzed as follows. A: sections were subjected to hematoxylin-eosin staining at 8 days postinoculation (PI). B: lungs were used to quantify levels of viral RNA by real-time PCR and replicating virus by plaque-forming assay. C: airway reactivity to inhaled methacholine (MCH) was monitored using enhanced pause (Penh) or total lung resistance (RL) at 21 days after inoculation. Measurements are made at baseline (BL) and after exposure to vehicle (V) or MCH (5–80 mg/ml or 0.3–5 mg/ml). D: lung sections were subjected to immunostaining for mucin 5AC (Muc5ac) at 21 days after inoculation. E: quantitative analysis of immunostaining from D. All values represent means ± SE. *Significant difference between strains in B or from SeV-UV controls for C and E. Bars 20 μm.
We next questioned whether the major histocompatibility complex (MHC) haplotype might be responsible for the observed differences in C57BL/6J versus Balb/cJ behavior. However, B10.D2nSnJ congenic mice, which share the Balb/cJ H-2d haplotype but are maintained on a C57BL/10SnJ background (7), showed levels of SeV-induced airway hyperreactivity and mucous cell metaplasia equal to those of the C57BL/6J and parental C57BL/10SnJ strain (Supplementary Fig. 2). Similarly, Balb.B10 congenic mice, which share the C57BL/6J H-2b haplotype but are maintained on a Balb/cJ background (2), manifested no induction of airway hyperreactivity or mucous cell metaplasia, consistent with findings in the parental Balb/cJ strain. Thus MHC haplotype restriction is not responsible for the difference between C57BL/6J versus Balb/cJ susceptibility to viral induction of airway hyperreactivity and mucous cell metaplasia.
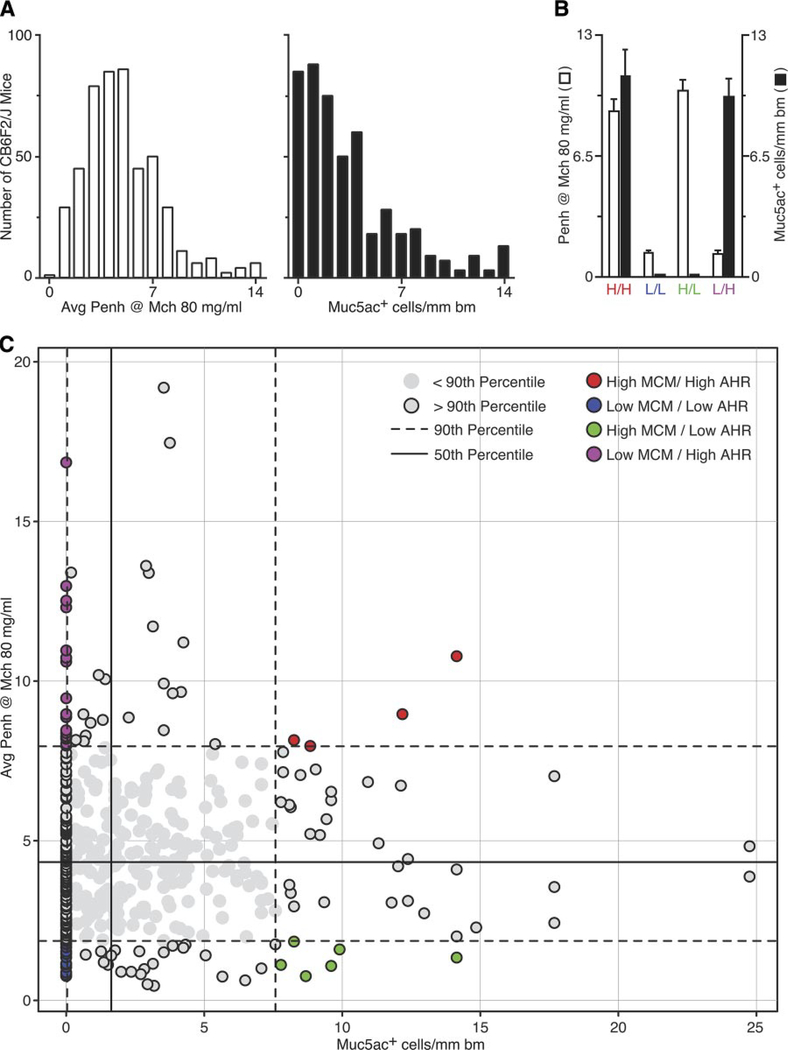
To determine whether individual airway disease traits can be genetically segregated and thereby linked to separate determinants, we next pursued an intercross analysis of susceptible C57BL/6J and nonsusceptible Balb/cJ strains. Characterization of the corresponding F1 hybrid mice (designated CB6F1/J) derived from a parental cross (C57BL/6J × Balb/cJ) yielded an intermediate phenotype (data not shown), so we developed an expanded F2 hybrid population (designated CB6F2/J) derived from an F1 intercross (CB6F1/J × CB6F1/J). Characterization of a CB6F2/J population of 486 male mice revealed a broad range of airway reactivity and mucous cell metaplasia after SeV infection (Fig. 2). This diversity of responsiveness thereby provided for a genetically informative population of phenotypic extremes. Within these extremes (the upper or lower 10% of the population), we identified four distinct CB6F2/J cohorts that showed either 1) H/H (analogous to the C57BL/6J parental strain), 2) L/L (analogous to the Balb/cJ parental strain), 3) H/L, or 4) L/H (Fig. 2, B and C). These findings indicate that these two airway disease traits can be genetically segregated in individual mice and therefore have independent genetic control elements.
Fig. 2.
Distribution and segregation of airway disease traits in an F2 hybrid population. A: F2 mice (CB6F2/J) derived from an F1 intercross (Balb/cJ × C57BL/6J) were inoculated with SeV and assessed for airway hyperreactivity and mucous cell metaplasia 21 days later, as described in Fig. 1. Values represent distributions for a population of 486 male mice. B: segregation analysis of phenotype data from A. Quadrants are set at the 50th percentile for each trait. Within each quadrant, the extreme upper (for high traits) or lower (for low traits) 10% of the population is indicated by shaded circles with a black border that lie outside of the 90th percentile (indicated by dashed lines). Within this population, we identified four extreme cohorts (identified with colored circles) that exhibited either high mucous cell metaplasia (MCM) and high airway hyperreactivity (AHR) (H/H; n = 4), low MCM and low AHR (L/L; n = 14), high MCM and low AHR (H/L; n = 6), and low MCM and high AHR (L/H; n = 18). C: phenotype data for each extreme cohort from B. The left y-axis indicates values for of AHR, and the right y-axis indicates values of MCM. Values represent means ± SE for all mice in each cohort.
Genomic linkage to mClca3 gene expression.
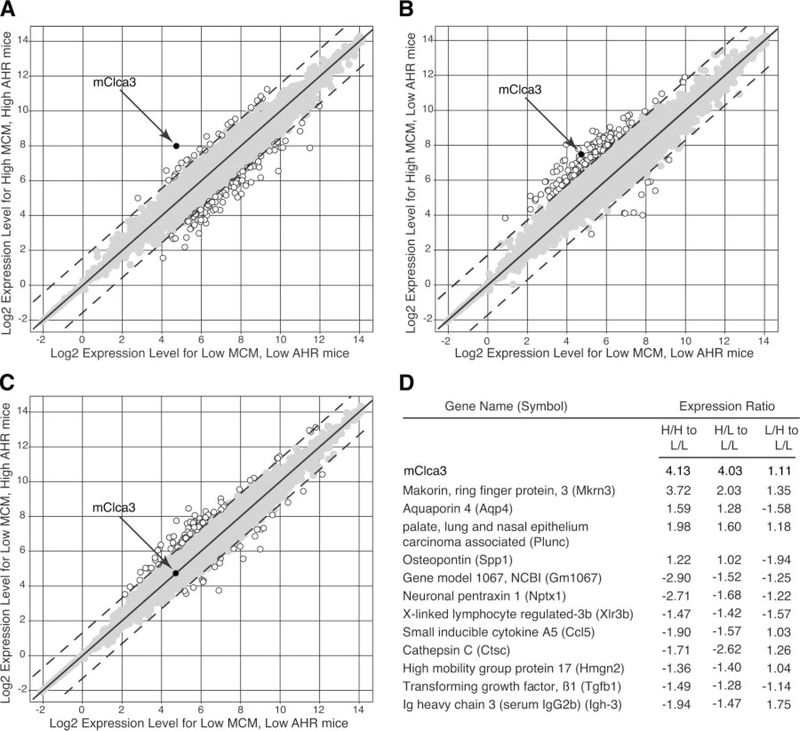
We next took advantage of this matrix of phenotypes to better define the genomic profiles associated with SeV-induced airway hyperreactivity or mucous cell metaplasia. By oligonucleotide microarray analysis, we found that 2,275 genes exhibited differential expression when we compared postviral lung RNA from one parental strain versus the other (data not shown). However, when we performed the same analysis in a comparison of the four extreme CB6F2/J cohorts, the list of candidate genes was decreased by >90%. In a comparison of cohorts that exhibited mucous metaplasia compared with ones that did not, mClca3 was identified as a gene that was consistently upregulated (Fig. 3). However, we noted that Muc5ac and other Clca family members were not represented on the microarray set.
Fig. 3.
Genomic linkage of mouse calcium-activated chloride channel (mClca3) gene expression to MCM but not AHR in F2 hybrid mice. A–C: from each of the four F2 extreme cohorts described in Fig. 2, lung mRNA was analyzed by an oligonucleotide microarray (n = 3 randomly selected mice/cohort). Scatterplots depict normalized gene expression in L/L mice (x-axis) versus each of the other 3 cohorts (H/H, H/L, and L/H; y-axis). Open circles indicate genes with differential expression ≥6 SD from the mean (dashed lines); shaded circles indicate genes with values < 6 SD from the mean. D: using the matrix from A–C, the ratio of gene expression was calculated for each cohort comparison; values are presented for genes with significantly different expression for high versus low MCM conditions (i.e., H/H vs. L/L and H/L vs. L/L).
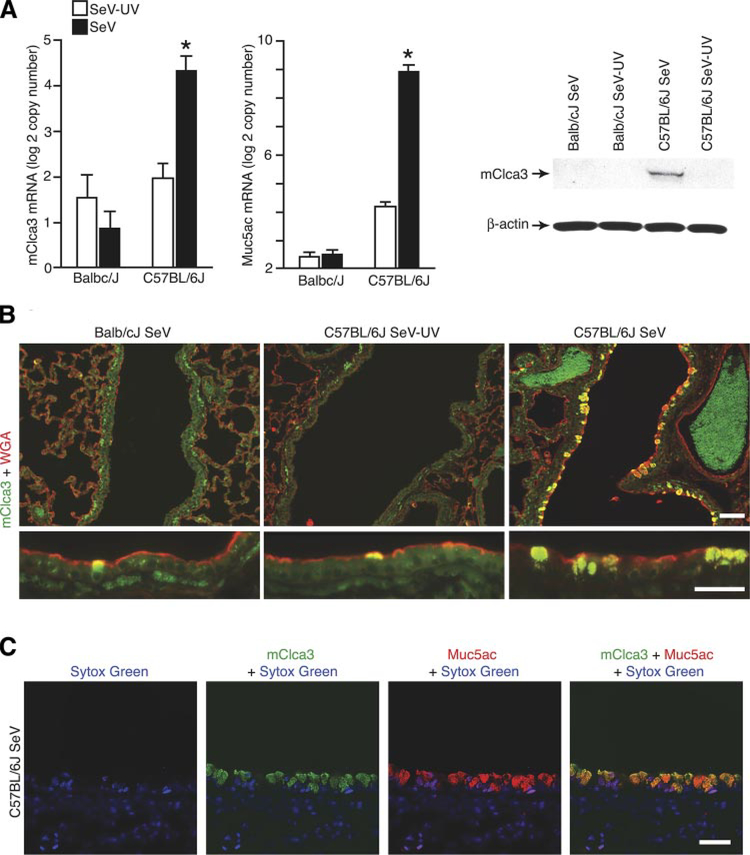
To verify the genomic analysis, we next assessed mClca3 expression in response to viral infection in susceptible and nonsusceptible parental strains. Here, we found a fourfold induction of mClca3 mRNA levels in concert with Muc5ac levels in the lungs of C57BL/6 mice but no induction of mClca3 in Balb/cJ mice after SeV infection (Fig. 4A). Western blot analysis of lung lysates and immunostaining of lung tissue sections confirmed coordinate increases in the levels of mClca3 protein after SeV infection (Fig. 4B). Specificity for mucous cell expression of mClca3 was further established by colocalization with wheat germ agglutinin as a marker for glycoprotein accumulation in mucus-producing cells (Fig. 4C). In addition, mClca3 and Muc5ac immunostaining were colocalized by confocal microscopy (Fig. 4D).
Fig. 4.
Detection of mClca3 gene expression in concert with mucous cell metaplasia in F0 parental mice. Balb/cJ and C57BL/6J mice were inoculated with SeV or SeV-UV and then analyzed as follows. A: lung RNA from 21 days after inoculation was subjected to real-time PCR for mRNA levels of mCcla3 and Muc5ac normalized to the Gapdh control. Values represent means ± SE for 4–6 mice. *Significant difference from SeV-UV. B: corresponding lung lysates from C57BL/6J mice were subjected to Western blot analysis with anti-human (h)CLCA1/ mClca3 Ab and control anti-β-actin mAb. C: corresponding lung sections from C57BL/6J mice were immunostained with anti-hClca1/mClca3 Ab and FITC-conjugated secondary Ab as well as anti-wheat germ agglutinin (WGA) Ab and CY3-conjugated secondary Ab. D: corresponding lung sections were immunostained with anti-hCLCA1/mClca3 Ab and Alexa 633-conjugated secondary Ab (green) as well as biotinylated anti-Muc5ac Ab and Alexa 555-streptavidin (red) and counterstained with Sytox green (blue). Bars = 20 μm.
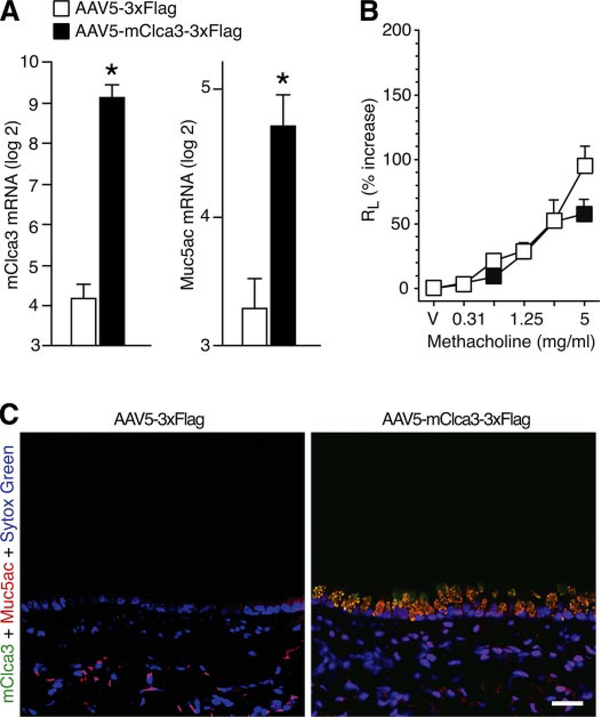
Recognizing that endogenous mClca3 expression coincided with the development of mucous cell metaplasia after viral infection, we next determined whether gene transfer of mClca3 was sufficient to cause this trait. Here, we used an AAV5 gene transfer vector that is able to enter polarized epithelial cells through the apical cell surface and is less likely to trigger an airway inflammatory response than adenovirus vectors used in previous studies of CLCA function (34, 56). We found that mice inoculated with AAV5 encoding mClca3 tagged with 3xFlag (AAV5-mClca3–3xFlag) developed a significant increase in the expression of mClca3 that was accompanied by a coordinate increase in Muc5ac gene expression (Fig. 5A). This induction of mClca3 mRNA was not found in mice that were inoculated with a control vector encoding only for 3xFlag mRNA. In addition, increased mClca3 mRNA coincided with a significant increase in mClca3 and Muc5ac protein expression, as detected by immunostaining and confocal microscopy (Fig. 5B). Consistent with the genomic analysis of the F2 cohort, mClca3 expression caused no change in airway reactivity despite levels of expression that were comparable to those after SeV infection (Fig. 5C). This same pattern of selective action of mClca3 gene transfer on mucous cell metaplasia found in C57BL/6J mice was also found in Balb/cJ mice (data not shown), indicating that events upstream of mClca3 expression were responsible for the strain differences in susceptibility to viral induction of mucous cell metaplasia.
Fig. 5.
Effect of mClca3 gene transfer on MCM. C57BL/6J mice were treated with adenoviral-associated virus 5 (AAV5) encoding mClca3–3xFlag or 3xFlag reporter alone and then studied 21 days later as follows. A: lung RNA was subjected to real-time PCR for mRNA levels of mClca3 and Muc5ac normalized to the Gapdh control and the simian virus (SV40) polyA level of expression. Values represent means ± SE for 3–5 mice. *Significant difference from treatment with AAV5–3xFlag. B: corresponding lung sections from C57BL/6J mice were immunostained with anti-hCLCA1/mClca3 Ab and Alexa 633-conjugated secondary Ab (green) as well as biotinylated anti-Muc5ac Ab and Alexa 555-streptavidin (red) and counterstained with Sytox green (blue). Bar = 20 μm. C: AHR to MCH was assessed using RL measurements as described in Fig. 1.
Persistent airway disease traits despite mClca3 deficiency.
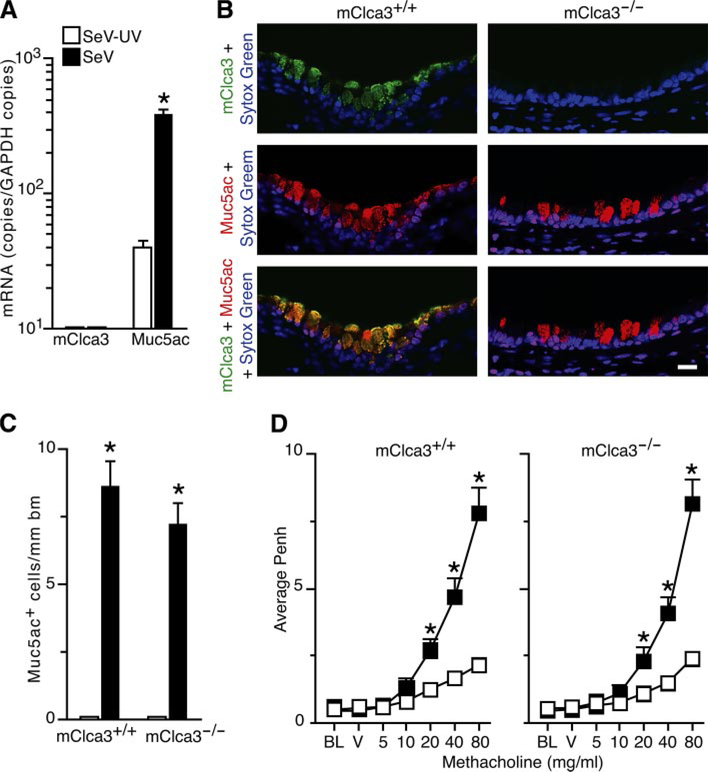
To further define whether mClca3 is necessary for virus-induced airway disease traits, we next developed mClca3−/− mice in the C57BL/6J background. These mice bred and developed normally and exhibited similar degrees of acute illness based on behavior and weight change after SeV inoculation (data not shown). Despite this mClca3 deficiency, mClca3−/− mice exhibited the same degree of mucous cell metaplasia as wild-type control mice (Fig. 6, A–C). In addition, mClca3−/− mice exhibited the same baseline airway reactivity and the same level of induction of airway hyperreactivity to inhaled methacholine (Fig. 6D). Each of these traits developed precisely as expected despite the loss of mClca3 gene expression.
Fig. 6.
Development of virus-induced MCM and AHR in mClca3−/− mice. Both mClca3−/− and wild-type control mice were inoculated with SeV or SeV-UV and analyzed 21 days later follows. A: lung RNA was subjected to real-time PCR for mRNA levels of mClca3 and Muc5ac normalized to the Gapdh control. Values represent means ± SE for 3–5 mice. B: corresponding lung sections were subjected to immunostaining with anti-hCLCA1/mClca3 Ab and Alexa 633-conjugated secondary Ab (green) as well as biotinylated anti-Muc5ac Ab and Alexa 555-streptavidin (red) and counterstained with Sytox green (blue). Bar = 20 μm. C: quantitative analysis of immunostaining from B. D: AHR to inhaled MCH was assessed using Penh measurements as described in Fig. 1. Values represent means ± SE for 8 mice. *Significant differences from SeV-UV in A, C, and D.
These findings indicated that mClca3 expression was not necessary for the development of mucous cell metaplasia or airway hyperreactivity after viral infection. Previous work by others had indicated that mClca3 was necessary and sufficient for allergen (Ova)-induced goblet cell metaplasia and airway hyperreactivity in Balb/cJ mice based on adenovirus-mediated gene transfer of sense and antisense mClca3 constructs (38). Thus, in comparative experiments, we also took advantage of the Ova sensitization/challenge model, which causes transient increases in goblet cell metaplasia and airway hyperreactivity in C57BL/6J mice (52). In this case, similar to the observations after viral infection, we found no difference in the degree of mucous cell metaplasia or airway hyperreactivity induced by allergen in mClca3−/− versus wild-type control mice (Supplementary Fig. 3). Thus, in both viral and allergic settings, mClca3 gene expression did not appear to be necessary for the development of mucous cell metaplasia or airway hyperreactivity.
mClca5 (like mClca3) causes mucous cell metaplasia without airway hyperreactivity.
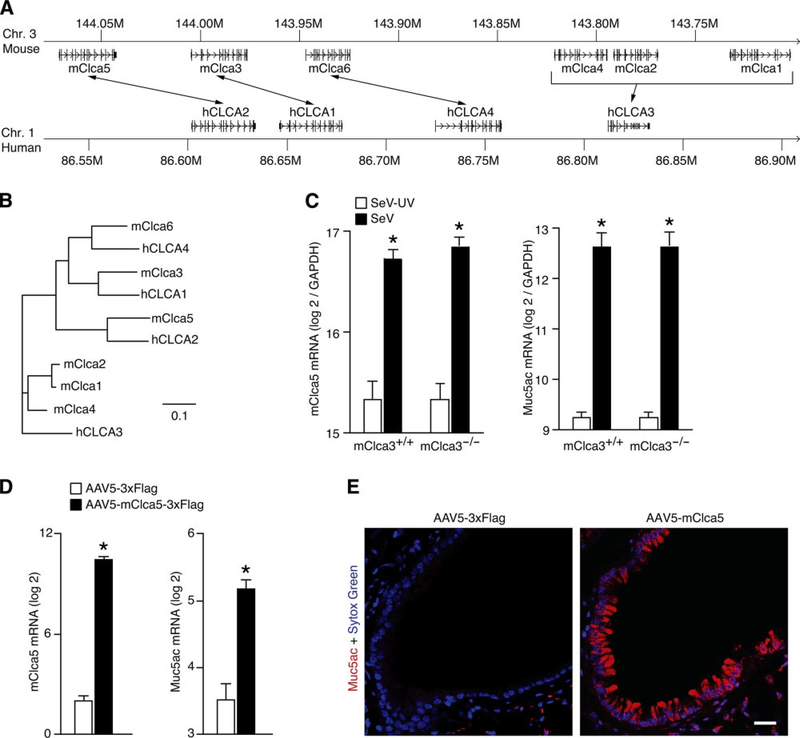
Because mClca3 gene expression was sufficient but not necessary for the development of mucous cell metaplasia, we reasoned that homologs of the Clca gene might be capable of mediating the chronic change in airway behavior. Using the BLAST search program (3), we uncovered two additional genes flanking mClca3 on mouse chromosome 3 (Fig. 7A). We also found that these genes were homologous to two of the hCLCA genes for which no mouse homolog had been identified (Fig. 7B). We thus tentatively named these genes mClca5 and mClca6, and, subsequently, these genes were cloned and given the same designations (15). Others have reported that mClca5 (as well as the human homolog hCLCA2) are both expressed in airway tissue, whereas mClca6 has not been found in the mouse lung or airway (15, 21). Thus we proceeded to evaluate mClca5 gene expression as a potential compensatory mechanism allowing for the development of mucous cell metaplasia in the setting of mClca3 deficiency.
Fig. 7.
Identification of mClca5 as a regulator of MCM. A: syntenic map of mouse Clca and human CLCA loci with homologous genes connected by arrows, generated with University of California-Southern California Genome Browser (27). B: corresponding homology tree for genes annotated in A generated from ClustalX alignment (11) of GenBank sequences (5) in TreeView (40). Bar = 10% sequence diversity. C: wild-type C57BL/6J and mClca3−/− mice were inoculated with SeV or SeV-UV, and, 21 days later, lung RNA was subjected to real-time RT-PCR for mRNA levels of mClca5 and Muc5ac normalized to the Gapdh control. D: wild-type C57BL/6J mice were inoculated with AAV5-mClca5–3xFlag or AAV5–3xFlag, and, 21 days later, lung RNA was subjected to real-time RT-PCR for mRNA levels of SV40 as a marker of viral expression as well as mClca5 and Muc5ac normalized to the Gapdh control and SV40 level of expression. For C and D, values represent means ± SE for 3–5 mice. *Significant differences from SeV-UV or AAV5–3xFlag controls. E: corresponding lung sections from D were subjected to immunostaining for Muc5ac (red) and counterstained with Sytox green (blue). Bar = 20 μm.
To evaluate the potential compensatory role of mClca5 in the development of mucous cell metaplasia, we first quantified the levels of endogenous mClca5 gene expression in mClca3−/− and wild-type control mice during virus-induced mucous cell metaplasia. In both cases, we found a significant increase in lung levels of mClca5 in concert with Muc5ac gene expression after inoculation with SeV (Fig. 7C). To next determine whether mClca5 gene expression is sufficient for the induction of mucous cell metaplasia, we took the same gene transfer approach as for mClca3 function. In this case, we found that mice inoculated with AAV5-mClca5–3xFlag demonstrated a significant increase in mClca5 and concomitant Muc5ac gene expression compared with mice inoculated with AAV5–3xFlag (Fig. 7D). Moreover, the levels of mClca5 gene expression were similar to those found during SeV-induced mucous cell metaplasia. Furthermore, the induction of mClca5 and Muc5ac gene expression was associated with concomitant increases in Muc5ac-positive mucous cells (Fig. 7e). Similar to the case for mClca3, we found that mClca5 gene expression at comparable levels to viral induction did not cause airway hyperreactivity (data not shown). Together, these findings indicate that mClca3 or mClca5 expression may both selectively drive the development of mucous cell metaplasia and so may act as functional substitutes for one another during induction by viral infection.
DISCUSSION
The present study establishes several new insights into the pathogenesis of complex airway diseases. In particular, we developed an experimental model for segregating airway disease traits and thereby defined independent genetic susceptibilities for developing the individual traits of mucous cell metaplasia versus airway hyperreactivity. We also took advantage of segregation into phenotypic extremes and genomic analysis to identify mClca3 gene expression as being linked to one trait (mucous cell metaplasia) but not the other (airway hyperreactivity). In addition, we developed a mClca3−/− mouse and Clca gene transfer system to show that mClca3 is sufficient but not necessary for mucous cell metaplasia. This finding, in turn, led to the identification of mClca5 as a functional homolog and was thereby capable of substituting for mClca3 in the development of this disease trait. The study thereby establishes the principles of genetic segregation and redundancy in the development of airway disease. Because of the clinical overlap and variability among individual patients with inflammatory airway diseases such as asthma and COPD, this principle is critical for understanding how these conditions develop and for developing more specific biomarkers and therapies for them.
Our study comes in the context of others that aimed to define the genetic influence on the development of complex airway diseases. In that regard, inbred mouse strains have been used to define genetic differences in native airway reactivity as well as increases in reactivity after exposure to inhaled irritants (12, 32, 33, 37, 57). Others have examined differences between mouse strains or among populations of hyperreactive to hyporeactive crosses to establish determinants of airway hyperreactivity after allergen challenge (8, 13, 16, 50). In the usual case, these previous studies were directed at defining a genetic locus for a single quantitative trait rather than segregating one trait from another in a complex phenotype. We submit that the present results will encourage a reductionist approach that isolates one trait within a complex phenotype and thereby allows for more precise definition of molecular mechanism for the trait under study.
In that regard, our results should also help to clarify the findings from previous studies of CLCA function in mice and humans. In an initial study (38), mClca3 was found to be sufficient to cause airway hyperreactivity and goblet cell metaplasia when expressed in the mouse airway using adenoviral gene transfer. Furthermore, expression of an antisense construct for mClca3 suppressed hyperreactivity and mucus production induced by allergen challenge in mice. On the basis of this information, mClca3 was proposed as both necessary and sufficient for allergen-induced airway hyperreactivity and goblet cell metaplasia. However, others recently reported that allergen-induced mucous cell metaplasia is no different in mClca3−/− mice (developed in a 129/C57BL/6J chimeric background) compared with wild-type control mice, but these investigators did not examine airway reactivity (42). In addition, they analyzed only mClca1, mClca2, and mClca4 gene expression, and, when these genes showed no allergen induction, they concluded that other Clca family members could not compensate for the loss of mClca3 in the development of mucous cell metaplasia. The present analysis indicates that mClca5 is a suitable candidate for compensatory function.
In that context, we note that the airway system may be designed for redundancy in the pathways leading to mucous cell metaplasia and hyperreactivity. There are currently at least 20 mucin genes and 6 CLCA family members, and many of each of these appear to be expressed in airway tissue (10, 22, 26, 30, 41). The genes for CLCA family members are clustered on the short arm of chromosome 1 in human (syntenic for chromosome 3 in the mouse), suggesting the presence of a large family derived from gene duplication. Airway mucin genes (including Muc5ac) likely arose from a single ancestral gene on chromosome 11 as well (9). Moreover, mucous cell mucins and CLCA proteins may regulate cell adhesion as well as apoptosis in addition to other possible roles in host defense, repair, and ion movement (20). Thus redundant pathways for preserving the CLCA-mucin axis and the consequent goblet cell lineage would likely confer a useful advantage to the host. An obvious corollary of this circumstance is that hCLCA1/mClca3 inhibition achieved by niflumic acid (a chloride channel blocker) may be effective by producing a more widespread inhibition of CLCA activities than a targeted gene knockout (4, 19, 45, 58). Similarly, for antisense strategies, it is possible that the inhibitory effect on goblet cell metaplasia found in previous studies was due to cross-reactivity with other closely related and homologous targets, including other CLCAs (30, 41). The present study suggests that loss of either mClca3 or mClca5 alone would not be sufficient to block mucous cell metaplasia in vivo.
Precisely how mClca3 or mClca5 cause mucous cell metaplasia is uncertain. Structure-function studies indicate that expression of CLCA proteins results in a chloride current that is induced by the calcium ionophore ionomycin and inhibited by niflumic acid (24), but a connection between ion channel function and mucin gene expression remains uncertain. Possibilities include regulation of ATP-mediated mucin exocytosis as well as adenosine induction of mucin synthesis via EGF receptor transactivation because these processes are also inhibited by niflumic acid (6, 36). However, other studies have suggested that CLCA proteins may function not as ion channels but instead as signaling molecules (18). For example, all CLCA family members contain a VWFa domain with signaling capabilities. Indeed, mClca1 binding to β4-integrin activates focal adhesion kinase and downstream ERK with the potential to regulate mucin synthesis (1). This work and most other previous studies were performed in cell lines that may exhibit differences from primary culture cells in the regulation of mucin gene expression (Y. Alevy, A. C. Patel, and M. J. Holtzman, unpublished observations). Thus additional work is needed to determine the role of ion channel activity or proteinprotein interactions in regulating mucin synthesis, mucin secretion/exocytosis, and mucous cell metaplasia. The present results suggest that mClca3 and mClca5 may share structural determinants that regulate mucous cell metaplasia and thereby provide a basis for further definition of structure-function relationships for this family of proteins.
The present study was not designed to determine the upstream events that regulate CLCA expression or the downstream events that mediate CLCA function. However, we note that the 5-regulatory region of the mClca3 gene contains a putative binding site for the Stat6 transcription factor that mediates IL-13 receptor signal transduction and that IL-13 stimulates mClca3 (and mClca5) gene expression in cultured airway epithelial cells (A. C. Patel, J. D. Morton, and M. J. Holtzman, unpublished observations). Moreover, IL-13 expression is inducible in the mouse model of viral infection as well as the one for allergen challenge (48). Thus it appears likely that mClca3 expression is driven at least in part by IL-13 in these settings.
In summary, the present report provides the first model for genetic segregation of airway disease traits of mucous cell metaplasia and airway hyperreactivity and the first evidence that mClca3 gene expression is associated with one trait but not the other. The results further indicate that other Clca family members, i.e., mClca5, may also mediate mucous cell metaplasia in the setting of an isolated deficiency in the mClca3-dependent pathway responsible for mucous cell formation. The mouse system appears to be directly relevant to human airway disease because the human homologs for mClca3 and mClca5, i.e., hCLCA1 and hCLCA2, are both overexpressed in airway tissue of asthmatic and cystic fibrosis subjects and are again colocalized to mucous cells (25, 29, 47). The results thereby highlight the complex nature of chronic airway disease as well as individual asthma traits and provide a rational basis to specifically and effectively inhibit mucous cell metaplasia by globally inhibiting CLCA family members that may mediate this trait.
Supplementary Material
Acknowledgments
GRANTS
This research was supported by grants from the National Institutes of Health, Martin Schaeffer Fund, and Alan A. and Edith L. Wolff Charitable Trust.
Footnotes
The Supplementary Material (Supplementary Figs. 1–3) for this article is available online at http://physiolgenomics.physiology.org/cgi/content/full/00321.2005/DC1.
REFERENCES
- 1.Abdel-Ghany M, Cheng HC, Elble RC, and Pauli BU. Focal adhesion kinase activated by beta4 integrin ligation to mCLCA1 mediates early metastatic growth. J Biol Chem 277: 34391–34400, 2002. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R, Byrne JA, and Oldstone MB. Virus specificity of cytotoxic T lymphocytes generated during acute lymphocytic choriomeningitis virus infection: role of the H-2 region in determining cross-reactivity for different lymphocytic choriomeningitis virus strains. J Virol 51: 34–41, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990. [DOI] [PubMed] [Google Scholar]
- 4.Barnett J, Chow J, Ives D, Chiou M, Mackenzie R, Osen E, Nguyen B, Tsing S, Bach C, and Freire J. Purification, characterization, and selective inhibition of human prostaglandin G/H synthase 1 and 2 expressed in the baculovirus system. Biochim Biophys Acta 1209: 130–139, 1994. [DOI] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, and Wheeler DL. GenBank. Nucleic Acids Res 34: D16–D20, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand CA, Danahay H, Poll CT, Laboisse C, Hopfer U, and Bridges RJ. Niflumic acid inhibits ATP-stimulated exocytosis in a mucin secreting epithelial cell line. Am J Physiol Cell Physiol 286: C247–C255, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Bevan MJ. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J Exp Med 143: 1283–1288, 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer JP, Kisselgof AB, and Martin TR. Genetic variability in pulmonary physiological, cellular, and antibody responses to antigen in mice. Am J Respir Crit Care Med 160: 1150–1156, 1999. [DOI] [PubMed] [Google Scholar]
- 9.Buisine MP, Desseyn JL, Porchet N, Degand P, Laine A, and Aubert JP. Genomic organization of the 3-region of the human MUC5AC mucin gene: additional evidence for a common ancestral gene for the 11p15.5 mucin gene family. Biochem J 332: 729–738, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Zhao YH, Kalaslavadi TJ, Hamati E, Nehrke K, Le AD, Ann DK, and Wu R. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am J Respir Cell Mol Biol 30: 155–165, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, and Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba Y, Yanagisawa R, and Sagai M. Strain and route differences in airway responsiveness to acetylcholine in mice. Res Commun Mol Pathol Pharmacol 90: 169–172, 1995. [PubMed] [Google Scholar]
- 13.De Sanctis GT, Singer JB, Jiao A, Yandava CN, Lee YH, Haynes TC, Lander ES, Beier DR, and Drazen JM. Quantitative trait locus mapping of airway responsiveness to chromosomes 6 and 7 in inbred mice. Am J Physiol Lung Cell Mol Physiol 277: L1118–L1123, 1999. [DOI] [PubMed] [Google Scholar]
- 14.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, and Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 170: 1294–1301, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Evans SR, Thoreson WB, and Beck CL. Molecular and functional analyses of two new calcium-activated chloride channel family members from mouse eye and intestine. J Biol Chem 279: 41792–41800, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, Peltz G, and Wills-Karp M. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol 23: 537–545, 2000. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, and Zhang JM. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, and Thompson N. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J Biol Chem 280: 27205–27212, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Gruber AD, Elble RC, Ji HL, Schreur KD, Fuller CM, and Pauli BU. Genomic cloning, molecular characterization, and functional analysis of human CLCA1, the first human member of the family of Ca2+ -activated Cl− channel proteins. Genomics 54: 200–214, 1998. [DOI] [PubMed] [Google Scholar]
- 20.Gruber AD and Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+ -activated Cl− channel CLCA2. Cancer Res 59: 5488–5491, 1999. [PubMed] [Google Scholar]
- 21.Gruber AD, Schreur KD, Ji HL, Fuller CM, and Pauli BU. Molecular cloning and transmembrane structure of hCLCA2 from human lung, trachea, and mammary gland. Am J Physiol Cell Physiol 276: C1261–C1270, 1999. [DOI] [PubMed] [Google Scholar]
- 22.Gustincich s Batalov S, Beisel KW, Bono H, Carninci P, Fletcher CF, Grimmond S, Hirokawa N, Jarvis ED, Jegla T, Kawasawa Y, Le-Mieux J, Miki H, Raviola E, Teasdale RD, Tominaga N, Yagi K, Zimmer A, Hayashizaki Y, and Okazaki Y. Analysis of the mouse transcriptosome for genes involved in the function of the nervous system. Genome Res 13: 1395–1401, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamelmann E, Schwarze J, Takeda K, Oshiba A, Larsen GL, Irvin CG, and Gelfand EW. Measurement of airway responsiveness in allergic mice using barometric plethsmography. Am J Respir Crit Care Med 156: 766–775, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Hartzell C, Putzier I, and Arreola J. Calcium-activated chloride channels. Annu Rev Physiol 67: 719–758, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Hauber HP, Manoukian JJ, Nguyen LH, Sobol SE, Levitt RC, Holroyd KJ, McElvaney NG, Griffin S, and Hamid Q. Increased expression of interleukin-9, interleukin-9 receptor, and the calcium-activated chloride channel hCLCA1 in the upper airways of patients with cystic fibrosis. Laryngoscope 113: 1037–1042, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Higuchi T, Orita T, Nakanishi S, Katsuya K, Watanabe H, Yamasaki Y, Waga I, Nanayama T, Yamamoto Y, Munger W, Sun HW, Falk RJ, Jennette JC, Alcorta DA, Li H, Yamamoto T, Saito Y, and Nakamura M. Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J Biol Chem 279: 1968–1979, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, and Kent WJ. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtzman MJ, Kim EY, and Morton JD. Genetic and genomic approaches to complex lung diseases using mouse models In: Computational Genetics and Genomics: Tools for Understanding Disease, edited by Totawa Peltz G., NJ: Humana, 2005, p. 99–141. [Google Scholar]
- 29.Hoshino M, Morita S, Iwashita H, Sagiya Y, Nagi T, Nakanishi A, Ashida Y, Nishimura O, Fujisawa Y, and Fujino M. Increased expression of the human Ca2+ -activated Cl− channel 1 (CaCC1) gene in the asthmatic airway. Am J Respir Crit Care Med 165: 1132–1136, 2002. [DOI] [PubMed] [Google Scholar]
- 30.Jentsch TJ, Stein V, Weinreich F, and Zdebik AA. Molecular structure and physiological function of chloride channels. Physiol Rev 82: 503–568, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, Aud D, Peltz G, and Wills-Karp M. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immun 1: 221–226, 2000. [DOI] [PubMed] [Google Scholar]
- 32.Konno S, Adachi M, Matsuura T, Sunouchi K, Hoshino H, Okazawa A, Kobayashi H, and Takahashi T. Bronchial reactivity to methacholine and serotonin in six inbred mouse strains. Arerugi 42: 42–47, 1993. [PubMed] [Google Scholar]
- 33.Levitt RC and Mitzner W. Autosomal recessive inheritance of airway hyperreactivity to 5-hydroxytryptamine. J Appl Physiol 67: 1125–1132, 1989. [DOI] [PubMed] [Google Scholar]
- 34.Look DC, Roswit WT, Frick AG, Gris-Alevy Y, Dickhaus DM, Walter MJ, and Holtzman MJ. Direct suppression of Stat1 function during adenoviral infection. Immunity 9: 871–880, 1998. [DOI] [PubMed] [Google Scholar]
- 35.Martin TR, Gerard NP, Galli SJ, and Drazen JM. Pulmonary responses to bronchoconstrictor agonists in the mouse. J Appl Physiol 64: 2318–2323, 1988. [DOI] [PubMed] [Google Scholar]
- 36.McNamara N, Gallup M, Khong A, Sucher A, Maltseva I, Fahy J, and Basbaum C. Adenosine up-regulation of the mucine gene, MUC2, in asthma. FASEB J 18: 1770–1772, 2004. [DOI] [PubMed] [Google Scholar]
- 37.Miyabara Y, Yanagisawa R, Shimojo N, Takano H, Lim HB, Ichinose T, and Sagai M. Murine strain differences in airway inflammation caused by diesel exhaust particles. Eur Respir J 11: 291–298, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, and Fujino M. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA 98: 5175–5180, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ober C HLA-G: an asthma gene on chromosome 6p. Immunol Allergy Clin North Am 25: 669–679, 2005. [DOI] [PubMed] [Google Scholar]
- 40.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358, 1996. [DOI] [PubMed] [Google Scholar]
- 41.Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, and Elble RC. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol 27: 901–905, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Robichaud A, Tuck S, Kargman S, Tam J, Wong E, Abramovitz M, Mortimer J, Burston H, Masson P, Hirota J, Slipetz D, Kennedy B, O’Neill G, and Xanthoudakis S. Gob-5 is not essential for mucus overproduction in preclinial murine models of allergic asthma. Am J Respir Cell Mol Biol 33: 303–314, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, and Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity 15: 435–443, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Sampath D, Castro M, Look DC, and Holtzman MJ. Constitutive activation of an epithelial signal transducer and activator of transcription (Stat1) pathway in asthma. J Clin Invest 103: 1353–1361, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott-Ward TS, Li H, Schmidt A, Cai Z, and Sheppard DN. Direct block of the cystic fibrosis transmembrane conductance regulator Cl− channel by niflumic acid. Mol Membr Biol 21: 27–38, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro SD, DeMeo DL, and Silverman EK. Smoke and mirrors: mouse models as a reflection of human chronic obstructive pulmonary disease. Am J Respir Crit Care Med 170: 929–931, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Toda M, Tulie MK, Levitt RC, and Hamid Q. A calcium-activated chloride channel (hCLCA1) is strongly related to IL-9 expression and mucus production in bronchial epithelium of patients with asthma. Am J Respir Crit Care Med 109: 246–250, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, Spoor MS, You Y, Brody SL, and Holtzman MJ. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest 116: 309–321, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O’Sullivan MP, Walter MJ, Schwendener RA, Cook DN, Danoff TM, and Holtzman MJ. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nat Med 11: 1180–1187, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Oosterhout AJ, Jeurink PV, Groot PC, Hofman GA, Nijkamp FP, and Demant P. Genetic analysis of antigen-induced airway manifestations of asthma using recombinant congenic mouse strains. Chest 121: 13S, 2002. [DOI] [PubMed] [Google Scholar]
- 51.Walter MJ, Kajiwara N, Karanja P, Castro M, and Holtzman MJ. IL-12 p40 production by barrier epithelial cells during airway inflammation. J Exp Med 193: 339–352, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter MJ, Morton JD, Kajiwara N, Agapov E, and Holtzman MJ. Viral induction of a chronic asthma phenotype and genetic segregation from the acute response. J Clin Invest 110: 165–175, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. Global strategy for asthma management and prevention. NHLBI/WHO Workshop Report In: National Heart Lung and Blood Institute. Bethesda, MD: National Institutes of Health, 2002, p. 1–16. [Google Scholar]
- 54.Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, and Spencer F. A model based background adjustment for oligonucleotide expression arrays In: Johns Hopkins University, Dept. of Biostatistics Working Papers. Baltimore, MD: Johns Hopkins Univ., 2004. [Google Scholar]
- 55.Xiao X, Li J, and Samulski RJ. Production of high titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72: 2224–2232, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zabner J, Seiler M, Walters R, Kotin RM, Fulgeras W, Davidson BL, and Chiorini JA. Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J Virol 74: 3852–3858, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang LY, Levitt RC, and Kleeberger SR. Differential susceptibility to ozone-induced airways hyperreactivity in inbred strains of mice. Exp Lung Res 21: 503–518, 1995. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, Shapiro M, Dong Q, Louahed J, Weiss C, Wan S, Chen Q, Dragwa CR, Savio D, Huang M, Fuller C, Tomer Y, Nicolaides NC, McLane MP, and Levitt RC. A calcium-activated chloride channel blocker inhibits goblet cell metaplasia and mucus overproduction In: Novartis Foundation Symposium 248 on Mucus Hypersecretion in Respiratory Disease. Hoboken, NJ: Wiley, 2002, p. 150–165. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.