Abstract
Purpose
Although the link between mutant TP53 and human cancer is unequivocal, a significant knowledge gap exists in clinically actionable molecular targets in Li-Fraumeni syndrome (LFS), a highly penetrant cancer predisposition syndrome associated with germline mutations in TP53. This study surveyed the epigenome to identify functionally and clinically relevant novel genes implicated in LFS.
Patients and Methods
We performed genome-wide methylation analyses of peripheral blood leukocyte DNA in germline TP53 mutation carriers (n = 72) and individuals with TP53 wild type in whom histologically comparable malignancies developed (n = 111). Targeted bisulfite pyrosequencing was performed on a validation cohort of 30 TP53 mutation carriers and 46 patients with TP53 wild type, and candidate sites were evaluated in primary tumors from patients with LFS across multiple histologic tumor types.
Results
In 183 patients, distinct DNA methylation signatures were associated with deleterious TP53 mutations in peripheral blood leukocytes. TP53-associated DNA methylation marks occurred in genomic regions that harbored p53 binding sites and in genes encoding p53 pathway proteins. Moreover, loss-of-function TP53 mutations were significantly associated with differential methylation at the locus encoding microRNA miR-34A, a key component of the p53 regulatory network (adjusted P < .001), and validated in an independent patient cohort (n = 76, P < .001). Targeted bisulfite pyrosequencing demonstrated that miR-34A was inactivated by hypermethylation across many histologic types of primary tumors from patients with LFS. Moreover, miR-34A tumor hypermethylation was associated with decreased overall survival in a cohort of 29 patients with choroid plexus carcinomas, a characteristic LFS tumor (P < .05).
Conclusion
Epigenetic dysregulation of miR-34A may comprise an important path in TP53-associated cancer predisposition and represents a therapeutically actionable target with potential clinical relevance.
INTRODUCTION
Mutations in the TP53 tumor suppressor gene are the most common genetic aberrations across all human malignancies.1 Germline TP53 mutations are also the hallmark genetic event in Li-Fraumeni syndrome (LFS; Online Mendelian Inheritance in Man 151623), a highly penetrant cancer susceptibility syndrome that confers a predisposition to develop early-onset breast cancer, leukemias, bone and soft tissue sarcomas, brain tumors of various histologies, adrenocortical carcinomas, and a wide range of other malignancies.2 Although the link between mutant p53 and human cancer is unequivocal, the mechanism by which mutant p53 predisposes humans to cancer remains to be elucidated.
To address this gap, we describe the largest systematic analysis of DNA methylation in patients who harbor germline TP53 mutations and individuals with TP53 wild type. These findings uncover distinct TP53 mutation–specific methylation signatures in peripheral blood leukocytes from patients who harbor TP53 mutations relative to those with TP53 wild type in whom histologically comparable sporadic malignancies develop. In particular, we identify and validate a striking link between TP53 mutations and methylation changes at the promoter of the p53-associated microRNA miR-34A. We further provide evidence that dysregulation of miR-34A occurs in various histologic tumor types, is associated with clinical outcomes in a subset of patients with choroid plexus carcinomas (CPCs), and may comprise a significant path in TP53-associated cancer predisposition.
PATIENTS AND METHODS
Patient Cohort
Patients with confirmed TP53 mutations were identified and consented to participate in the study. Peripheral blood was collected in accordance with The Hospital for Sick Children Research Ethics Board (protocol # 0019910602). Patients were recruited to the study from The Hospital for Sick Children, or peripheral blood samples were sent from collaborators from other institutions. TP53 mutation carriers were initially matched approximately 2:1 with age-, sex-, and diagnosis-matched individuals with TP53 wild type; however, after final sample selection and analysis, the samples were not uniformly matched (Table 1).
Table 1.
Sample Attributes of Patients Who Harbor Germline TP53 Mutations and Those With TP53 Wild Type in the Initial and Validation Cohorts
| Cohort | Mutation Carrier | Wild Type |
|---|---|---|
| Initial | ||
| No. of samples | 72 | 111 |
| Mean age, years (range) | 12.54 (0.5-47.0) | 15.46 (0.03-60) |
| Male:female sex* | 34:33 | 46:59 |
| Diagnosis | ||
| Adrenocortical carcinoma | 22 | 24 |
| Brain tumor | 19 | 20 |
| Choroid plexus carcinoma | 9 | 7 |
| Astrocytoma/glioblastoma | 7 | 10 |
| Primitive neuroectodermal tumor | 3 | 4 |
| Breast | 10 | 10 |
| Sarcoma | 14 | 24 |
| Rhabdomyosarcoma | 12 | 21 |
| Other (myofibroblastic sarcoma, leiomyosarcoma) | 2 | 3 |
| Papillary thyroid carcinoma | 0 | 3 |
| Multiple primary malignancies | 24 | 1 |
| Unaffected | 17 | 30 |
| Validation | ||
| No. of samples | 30 | 46 |
| Mean age, years (range) | 18.03 (2-49.0) | 29 (0.83-60.0) |
| Male:female sex* | 8:16 | 12:33 |
| Tumor validation (choroid plexus carcinomas) | ||
| No. of samples | 24 | 18 |
| Mean age, years (range) | 1.22 (0.39-6.9) | 3.35 (0.01-16.53) |
| Male:female sex | 18:6 | 10:8 |
Where reported.
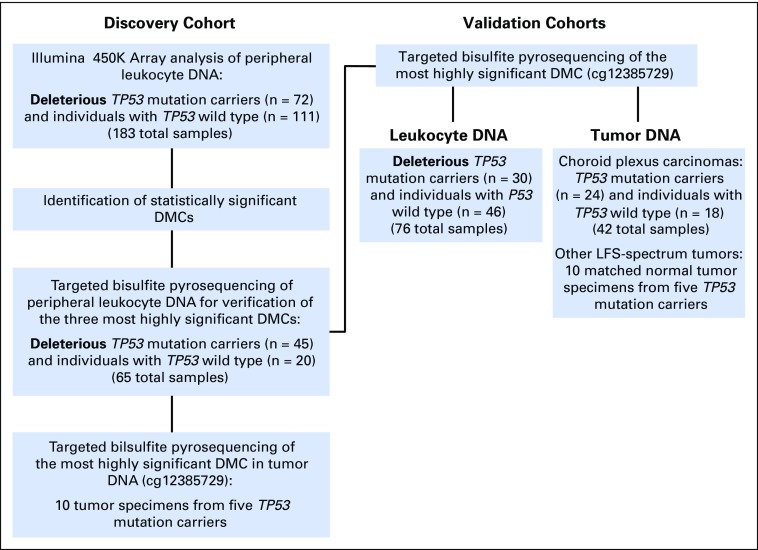
In almost all cases, blood was collected before administration of chemotherapy, but specific information was not available from a number of samples obtained from outside hospitals. In addition, some patients may have undergone treatment of prior malignancies. Because peripheral blood was used as a surrogate nontransformed tissue, patients with hematologic malignancies were excluded. The initial study cohort comprised 206 patients (92 TP53 mutation carriers, 114 individuals with TP53 wild type) for whom leukocyte DNA was used for genome-wide methylation analysis. Among the patients in this cohort, 80 TP53 mutation carriers had at least one other relative in the cohort, which comprised 21 individual families (Data Supplement). The family sizes ranged from two to 11 individuals (Data Supplement). DNA from adjacent normal and matched tumor tissue from patients in the initial cohort as well as patients in the current cohort was obtained where possible (Fig 1).
Fig 1.
Study design, including discovery cohort and validation cohorts for differential methylation analysis. DMC, differentially methylated CpG site.
TP53 Mutation Analysis
TP53 mutation analysis was performed on genomic DNA extracted from peripheral blood leukocytes in the Clinical Laboratory Improvement Amendments-College of American Pathologists molecular diagnostic laboratories at The Hospital for Sick Children and other institutions. By using DNA obtained from whole blood, Sanger sequencing of exons 2 to 11 and flanking intronic regions of the TP53 gene were performed. Multiplex ligation-dependent probe amplification analysis of TP53 copy number was also performed. Mutations in TP53 were classified by their effect on the resultant protein transactivation function as follows: functional, partially functional, or nonfunctional (ie, deleterious mutations) in accordance with the classification scheme provided by the International Agency for Research on Cancer TP53 Database and Sorting Intolerant From Tolerant classifications. Sample numbers on the basis of transactivation functionality in TP53 mutation carriers were as follows: 72 nonfunctional, 15 partially functional, four functional, and 111 nonmutation.
Bisulfite Pyrosequencing of Samples in Initial and Validation Cohorts and Primary Patient Tumors and Normal Tissue
Targeted DNA methylation analysis for the top differential probes from the Illumina 450K Array (San Diego, CA) was performed by using pyrosequencing as previously described.3,4 Verification of leukocyte DNA CpG methylation from the initial cohort was performed on the DNA from 65 patients (45 TP53 mutation carriers, 20 individuals with TP53 wild type). Subsequently, targeted bisulfite pyrosequencing of the cg12385729 CpG site was performed on peripheral leukocyte DNA from 76 patients in an independent validation cohort (30 TP53 mutation carriers, 46 individuals with TP53 wild type). Pyrosequencing assays that contained two polymerase chain reaction primers and one sequencing primer were designed to target CpG sites of interest by using PyroMark assay design software (QIAGEN, Venlo, the Netherlands). Genomic DNA was sodium bisulfite converted in the same manner as for the Illumina microarray and amplified with Hot Start Taq DNA Polymerase (QIAGEN). Amplicons were analyzed on a PyroMark Q24 pyrosequencer (QIAGEN) as specified by the manufacturer. Percent methylation was quantified as the ratio of C to C + T by PyroMark Q24 software. Pyrosequencing primers are provided in the Data Supplement. Forkhead box P2 methylation sites with known methylation levels were used as positive controls. Differential methylation between comparison groups was computed by using a two-tailed t test. The study methods are described in more detail in the Appendix (online only).
Survival Analysis
Survival analysis was performed by using the Kaplan-Meier method. A Mantel-Cox log-rank test was used to compare groups, and curves were generated with GraphPad Prism 6 software (GraphPad Software, La Jolla, CA). Overall survival was time measured from initial diagnosis to death as a result of any cause on last follow-up as of December 1, 2013.
RESULTS
Distinct Methylation Signatures Are Associated With Subclasses of TP53 Mutations
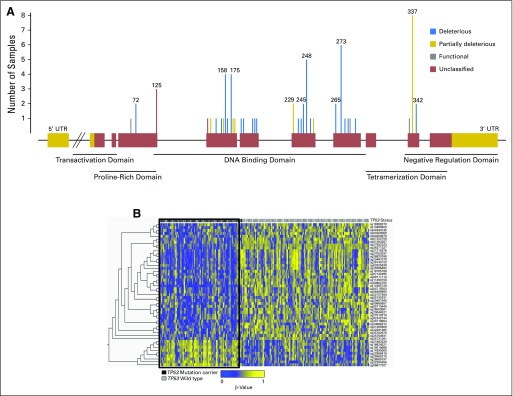
The overall study schema is provided in Figure 1. Differential methylation analyses on peripheral leukocyte DNA from the discovery cohort were first performed to identify TP53 mutation–associated CpG sites. The spectrum of TP53 mutations that occurred in the germline mirrored that of somatic TP53 mutations in sporadic cancers. These mutations were primarily concentrated among the 19 TP53 mutation hot spots in addition to the frequently reported Brazilian founder TP53 p.R337H mutation.5,6 Each individual TP53 mutation in the analysis cohort was classified by its predicted functional impact on the p53 protein (Fig 2A; Data Supplement). Class 1 mutations were defined as those predicted to result in a nonviable or dysfunctional protein product; these include frameshift mutations, early truncating mutations, and deleterious mutations in the p53 DNA-binding domain that affect p53 transactivation function. Class 2 mutations were classified as those that do not lead to abrogation of p53 transactivation function, which primarily occur in the p53 tetramerization domain.
Fig 2.
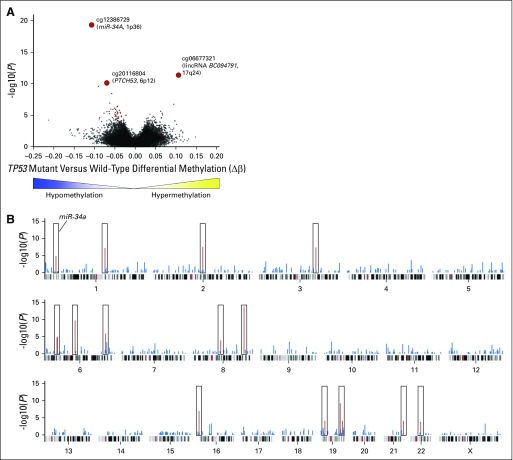
(A) Spectrum of TP53 mutations in the initial patient cohort. The TP53 gene structure is represented by the linearized horizontal projection, and red boxes denote coding exons, whereas gold regions represent noncoding exons. One mutation per family is depicted. Gray lines indicate mutations that are deleterious but retain functional transactivation. Gold lines indicate mutations that lead to partially functional transactivation mutations, and blue lines indicate mutations that are deleterious and result in nonfunctional transactivation. Red lines denote unclassified mutations. Corresponding protein domains of coding regions are indicated below the gene structure. Codon positions are indicated for mutations that occurred in two or more individuals. (B) Heatmap of the top differentially methylated CpG sites in TP53 mutation carriers versus noncarriers. Individual CpG sites are represented by rows, with row names corresponding to CpG IDs. Columns depict individual patients. Bars above each column indicate TP53 mutation status (black, mutation carrier [n = 72]; gray, noncarrier [n = 111]). The 49 most statistically significant differentially methylated sites determined by the Mann-Whitney U test, after multiple hypothesis testing correction by Benjamini-Hochberg method (q < .05), are shown. A black box delineates the methylation signatures of the prominent TP53 mutation carrier cluster with a preponderance of hypomethylated sites (blue) relative to hypermethylated sites (yellow). UTR, untranslated region.
Genome-wide methylation analyses were then performed on DNA from peripheral blood of TP53 mutation carriers relative to individuals with TP53 wild type. Compared with individuals with TP53 wild type, no significant differentially methylated CpG sites (DMCs) in patients with class 2 mutations in TP53 (n = 12; Data Supplement) were found. By contrast, after correction for multiple hypothesis testing, 49 significant DMCs were found between patients who harbored class 1 mutations relative to those with wild type (Appendix Fig A1A, online only). This analysis cohort of patients who harbored class 1 TP53 mutations relative to wild-type comprised 183 individuals (72 mutation, 111 wild type). In these patients, characteristic pediatric and adult-onset LFS malignancies developed, including breast cancer, brain tumors, soft tissue cancers, adrenocortical carcinomas, and other cancers (Table 1). The TP53 class 1 mutation group and TP53 wild-type group were initially matched for age, sex, and cancer diagnosis (Table 1). Both groups also comprised patients in whom a malignancy had not developed at the time of sample collection (17 mutation, 30 wild type). Heat map visualization of the 49 statistically significant DMCs (adjusted P < .05) reveals divergent patterns of hyper- and hypomethylated regions in class 1 TP53 mutation carriers compared with noncarriers, with a greater proportion of hypomethylated DMCs in mutation carriers (73% hypomethylated DMCs; Fig 2B; Table 2).
Table 2.
Statistically Significant Differentially Methylated CpG Sites Between TP53 Mutation Carriers and Noncarriers
| Probe ID | Δβ* | P† | q‡ | CHR | MAPINFO | UCSC Gene Name |
|---|---|---|---|---|---|---|
| cg12385729 | −0.1045 | < .001 | 3.09E-15 | 1 | 9243679 | miR-34A (promoter) |
| cg06677321 | 0.1081 | < .001 | 1.33E-07 | 17 | 67603831 | |
| cg20116804 | −0.0649 | < .001 | 1.73E-06 | 6 | 48036212 | C6orf138 (PTCH53) |
| cg26371327 | −0.09 | < .001 | 5.17E-06 | 1 | 16473143 | EPHA2 |
| cg27582323 | −0.0576 | < .001 | 5.96E-05 | 1 | 16472728 | EPHA2 |
| cg26820168 | −0.0623 | < .001 | 1.59E-03 | 8 | 124217896 | FAM83A |
| cg19518666 | 0.095 | < .001 | 2.09E-03 | 4 | 157692845 | PDGFC |
| cg02242734 | −0.043 | < .001 | 3.44E-03 | 22 | 18064224 | SLC25A18 |
| cg00865857 | −0.0445 | < .001 | 4.13E-03 | 8 | 145926462 | |
| cg20048521 | −0.0503 | < .001 | 5.93E-03 | 6 | 74072242 | C6orf221 |
| cg27053337 | −0.0595 | < .001 | 6.95E-03 | 8 | 124217698 | FAM83A |
| cg21215576 | −0.0548 | < .001 | 6.95E-03 | 8 | 124217760 | FAM83A |
| cg04306069 | −0.048 | < .001 | 6.95E-03 | 5 | 98175255 | |
| cg25731261 | −0.0388 | < .001 | 9.25E-03 | 19 | 47736018 | BBC3 (PUMA) |
| cg06489965 | −0.0452 | < .001 | 1.08E-02 | 16 | 3062975 | CLDN9 |
| cg14395060 | 0.0628 | < .001 | 1.14E-02 | 22 | 32441170 | SLC5A1 |
| cg26140120 | −0.0639 | < .001 | 1.14E-02 | 8 | 124219575 | FAM83A |
| cg23467079 | −0.0692 | < .001 | 1.18E-02 | 8 | 124219425 | FAM83A |
| cg27612019 | −0.0434 | < .001 | 1.29E-02 | 22 | 18064052 | SLC25A18 |
| cg18165186 | −0.0369 | < .001 | 1.30E-02 | 3 | 50314180 | SEMA3B |
| cg00962755 | −0.0486 | < .001 | 1.33E-02 | 1 | 201439001 | PHLDA3 |
| cg07250758 | −0.0422 | < .001 | 1.55E-02 | 11 | 61049251 | VWCE |
| cg23252259 | −0.0764 | < .001 | 1.55E-02 | 6 | 31148612 | |
| cg07142285 | −0.0357 | < .001 | 1.55E-02 | 20 | 57042703 | APCDD1L |
| cg14731462 | 0.0664 | < .001 | 1.57E-02 | 10 | 129789429 | PTPRE |
| cg24869272 | −0.0632 | < .001 | 1.67E-02 | 11 | 850296 | TSPAN4 |
| cg22157494 | −0.0444 | < .001 | 1.88E-02 | 1 | 3774827 | KIAA0562 |
| cg17892917 | 0.0364 | < .001 | 1.88E-02 | 3 | 107823424 | |
| cg26550861 | −0.0505 | < .001 | 1.88E-02 | 6 | 74072376 | C6orf221 |
| cg18419358 | 0.0676 | < .001 | 2.32E-02 | 6 | 158384009 | |
| cg20296493 | −0.0476 | < .001 | 2.48E-02 | 8 | 124219725 | FAM83A |
| cg27500148 | −0.0228 | < .001 | 2.50E-02 | 17 | 37011491 | RPL23 |
| cg05192831 | −0.0338 | < .001 | 3.05E-02 | 16 | 29913007 | ASPHD1 |
| cg04284678 | −0.0451 | < .001 | 3.47E-02 | 14 | 105995251 | TMEM121 |
| cg06508738 | −0.0489 | < .001 | 3.47E-02 | 3 | 138763852 | PRR23C |
| cg04645534 | 0.0444 | < .001 | 3.49E-02 | 8 | 23712550 | STC1 |
| cg02625638 | −0.0338 | < .001 | 3.78E-02 | 8 | 124218648 | FAM83A |
| cg11498156 | −0.0403 | < .001 | 3.78E-02 | 10 | 102890145 | TLX1 |
| cg22989419 | 0.0482 | < .001 | 4.19E-02 | 20 | 9340396 | PLCB4 |
| cg21922810 | 0.0418 | < .001 | 4.30E-02 | 1 | 6669887 | |
| cg21385909 | −0.0555 | < .001 | 4.30E-02 | 15 | 31621843 | KLF13 |
| cg04715503 | −0.0873 | < .001 | 4.39E-02 | 16 | 3062795 | CLDN9 |
| cg22384356 | 0.0646 | < .001 | 4.63E-02 | 8 | 124195192 | FAM83A |
| cg00468228 | 0.0317 | < .001 | 4.79E-02 | 8 | 67092562 | |
| cg09658497 | 0.064 | < .001 | 4.80E-02 | 7 | 2847517 | GNA12 |
| cg13360823 | −0.0428 | < .001 | 4.82E-02 | 19 | 54496353 | CACNG6 |
| cg25962015 | 0.0314 | < .001 | 4.82E-02 | 7 | 26579291 | KIAA0087 |
| cg20401945 | −0.0409 | < .001 | 4.82E-02 | 16 | 29912460 | ASPHD1 |
| cg22291365 | −0.0446 | < .001 | 4.82E-02 | 15 | 74891439 |
Abbreviations: CHR, chromosome; MAPINFO, chromosomal coordinates on human genome assembly GRCh37/hg19; UCSC, University of California Santa Cruz.
Differential methylation level (Δβ) between TP53 mutation carriers relative to wild type.
P value determined by Mann-Whitney U test at each CpG site.
Adjusted P value after Benjamini-Hochberg correction for false discovery rate.
Many of the most significant DMCs overlapped with genes that encode known proteins in the p53 pathway, including EPHA2 and PUMA (BBC3; Data Supplement). Moreover, eight CpG probes mapped to exonic and intronic CpG sites within FAM83A, a putative oncogene7,8 (Data Supplement). Genomic regions highly enriched for DMCs in TP53 mutation carriers include 1p36, 8q24, 16p13, and 19q13 (Appendix Fig A1B). Of note, among the 49 most highly statistically significant DMCs (adjusted P < .05), 14.3% corresponded to known p53 transcription factor binding sites in cancer cells and nontransformed cells9,10 (Data Supplement). This pattern of enrichment of regional methylation differences at p53 binding sites was not observed in the methylation signatures from patients who harbored TP53 class 2 mutations relative to those with TP53 wild type (Data Supplement).
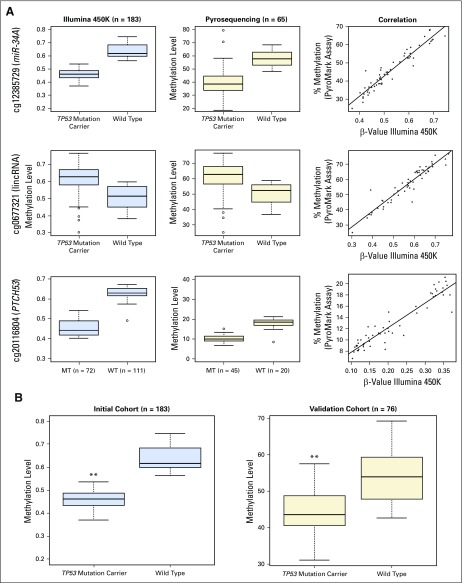
The three most statistically significant DMCs mapped to the promoter region of miR-34A (hypomethylation of cg12385729; adjusted P < .001); a long intergenic noncoding RNA on 17q24, BC094791 (hypermethylation of cg06677321; adjusted P < .001); and a protein-coding gene, PTCH53 (C6orf138), that is transcriptionally regulated by p5311 (hypomethylation of cg20116804; adjusted P < .001; Data Supplement). These findings were further validated by bisulfite pyrosequencing, an orthogonal method of measuring DNA methylation, to verify methylation values on a subset of the same samples from this initial patient cohort (n = 65). The methylation levels were highly correlated between the array-based and bisulfite pyrosequencing, with Spearman and Pearson correlations > 0.95 for all three CpG sites (Appendix Fig A2A, online only).
Diminished miR-34A Promoter Methylation Is a Hallmark of TP53 Mutation Status in Nontransformed Cells
The most statistically significant DMC in TP53 mutation carriers relative to noncarriers (cg12385729) resided in a CpG island in the 5′ promoter and overlapping transcription start site of the microRNA-encoding gene miR-34A (adjusted P < .001; Appendix Fig A1A). Of note, miR-34A is a direct target of the p53 proapoptotic network. This finding led to the inquiry of whether other sites in the miR-34A promoter CpG island are also differentially methylated in TP53 mutation carriers compared with noncarriers. A follow-up analysis of the miR-34A promoter CpG island sites demonstrated that cg24167422, the site directly downstream of cg12385729 on this CpG island, also is significantly hypomethylated in TP53 mutation carriers compared with noncarriers (P < .001). This CpG site was not detected in the initial analysis because the standard deviation of methylation across all samples fell slightly below the cutoff of 0.05 used in the differential methylation analysis (standard deviation, 0.04; Methods).
Statistical approaches were used to confirm that TP53 mutation–associated methylation differences were not confounded by patient attributes such as age and sex, which are known to affect methylation patterns in peripheral blood. The association between methylation at cg12385729 and TP53 mutation status remained in the generalized estimating equation model (unadjusted P < .001). TP53 mutation–associated methylation at the miR-34A CpG island, therefore, was not associated with age and sex because neither was a significant predictor of methylation (P = .98 and .27 for sex and age, respectively). Furthermore, to verify that relatedness of individuals is not a confounding variable, we estimated the difference in cg12385729 methylation in a subset of unrelated patients from the initial cohort and found that this association remained significant (unadjusted P < .001; n = 141).
To further confirm the relative hypomethylation of the miR-34A promoter in nontransformed cells from TP53 mutation carriers, methylation levels at cg12385729 in peripheral blood leukocytes were assessed in an independent cohort of 76 patients (30 TP53 mutation carriers, 46 nonmutation carriers; Appendix Fig A2B; Data Supplement; Table 1). Hypomethylation at the miR-34A promoter CpG site was again strongly observed in this cohort (P < .001; Appendix Fig A2B). Moreover, methylation levels at this CpG site in individuals with TP53 wild type was similar to the levels reported in peripheral blood of a cohort of 850 healthy control subjects.12 As expected, differential methylation at the miR-34A promoter is significantly more pronounced in individuals who harbor class 1 mutations relative to those who harbor class 2 mutations (P < .001; Data Supplement).
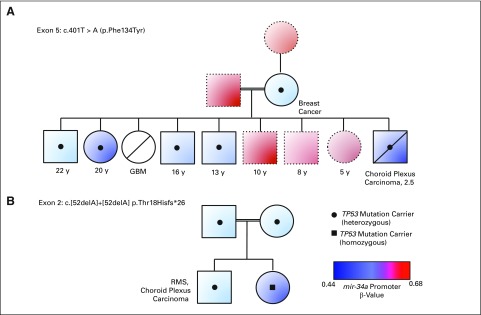
Furthermore, relative loss of miR-34A promoter methylation is consistently associated with TP53 mutation status in two families with LFS (Figs 3A and 3B). Germline miR-34A promoter methylation levels of < 43.9% that correlate with TP53 mutation with 95% accuracy, as demonstrated by bisulfite pyrosequencing of this site in the complete validation cohort (Data Supplement). Taken together, these results suggest that miR-34A promoter methylation levels at this CpG site alone show discriminatory accuracy of TP53 mutation status.
Fig 3.
Correlation of miR-34A promoter CpG site methylation and germline TP53 mutation in families with Li-Fraumeni syndrome. Levels of each pedigree represent a generation. Circles depict females and squares depict males. Direct horizontal linkages indicate maternal and paternal individuals, and direct vertical linkages indicate progeny. Indirect horizontal linkages denote siblings. Horizontal linkages with double lines indicate consanguinity. Color of each circle/square depicts the peripheral blood miR-34A promoter CpG methylation and Illumina 450K Array β-values from the respective individual, whereas black dots/squares indicate TP53 mutation carriers. Age and diagnoses are listed below each individual, where reported. Diagonal lines across a shape indicate that the individual is deceased. (A) Family harbors a familial mutation in exon 5. (B) Family harbors the familial mutation in exon 2. GBM, glioblastoma; RMS, rhabdomyosarcoma.
miR-34A Is Hypermethylated in Tumors From Germline TP53 Mutations Carriers and Is Associated With Poor Clinical Outcomes
To characterize miR-34A promoter methylation in LFS tumors, archived DNA from primary tumor specimens from the same individuals with matched peripheral blood were analyzed. Methylation at the miR-34A promoter in tissue from a patient with LFS with adrenocortical carcinoma and liver metastases and matched normal liver tissue from the same patient demonstrated hypermethylation in the tumor relative to histologically normal tissue (Data Supplement). Moreover, hypermethylation of miR-34A in this tumor tissue was associated with decreased relative miR-34A transcript expression (Data Supplement). Similarly, primary primitive neuroectodermal tumors of the brain from two patients with LFS showed miR-34A promoter hypermethylation relative to normal adjacent cortex (Data Supplement).
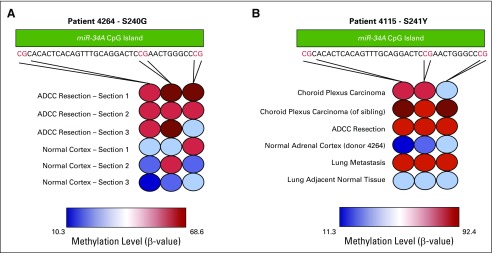
To further investigate this finding, miR-34A promoter methylation was measured in DNA from multiple histologic sections of fresh frozen tumor and normal tissue from both germline TP53 mutation carriers in whom diverse LFS-spectrum cancers developed. Distinct hypermethylation of the miR-34A promoter in tumor but not in histologically normal adjacent tissue was detected in tumor tissue from patients with germline TP53 mutations (Appendix Figs A3A and A3B, online only). This was again confirmed in four of five tumor specimens from germline TP53 mutation carriers (Data Supplement).
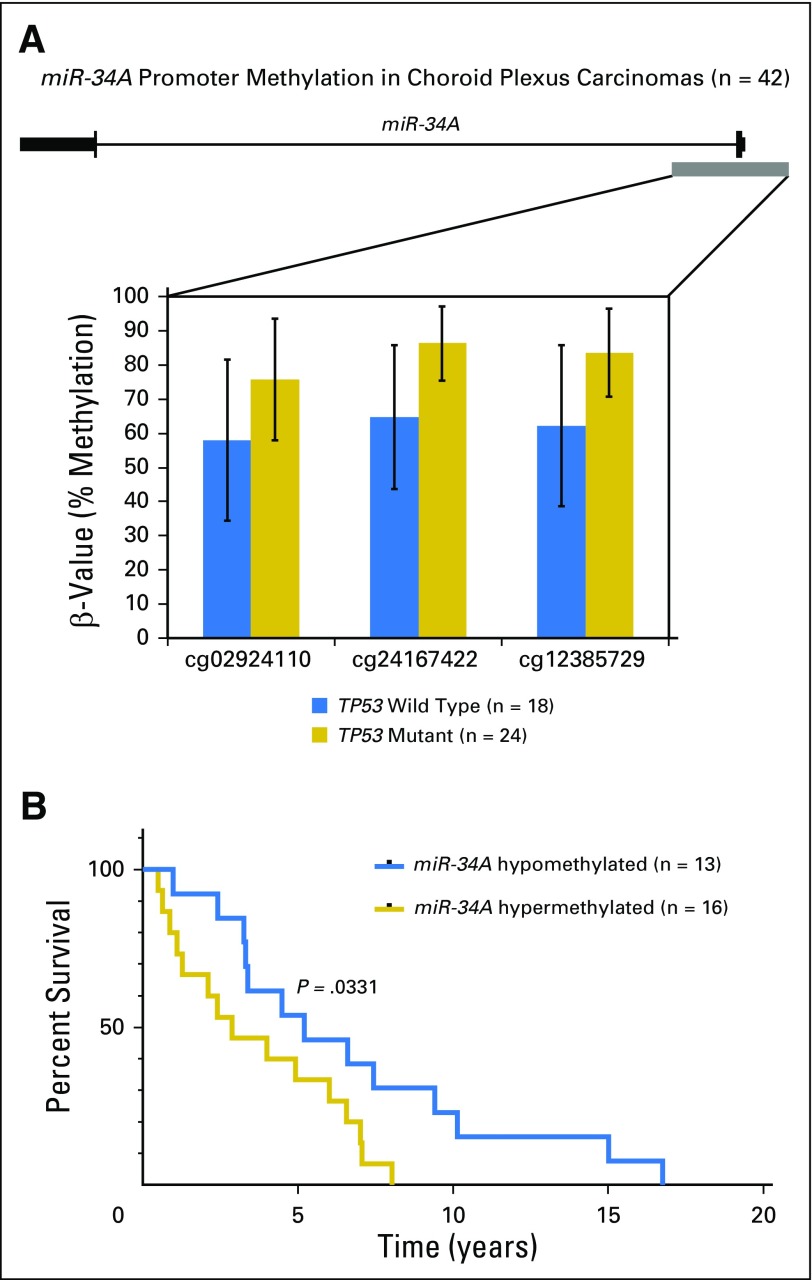
miR-34A promoter hypermethylation was further established in a cohort of 42 CPCs, a characteristic tumor in the clinical context of LFS. Compared with TP53 wild-type CPCs, TP53 mutant CPCs demonstrated statistically significant hypermethylation across three CpG sites of the miR-34A promoter (Fig 4A; Data Supplement). Moreover, when stratified by miR-34A methylation status, which used a > 80% threshold for hypermethylation, patients with tumors that harbored miR-34A promoter hypermethylation had a median survival of 2.88 years after diagnosis, whereas patients whose tumors harbored a hypomethylated miR-34A promoter had a median survival of 5.18 years (Mantel-Cox P = .0331; Fig 4B).
Fig 4.
(A) miR-34A promoter methylation in choroid plexus carcinomas (n = 42). The miR-34A gene structure is depicted, with the promoter CpG island as the gray block. Methylation levels are measured by bisulfite pyrosequencing at three adjacent CpG sites. (B) Kaplan-Meier curve of overall survival in a patient with choroid plexus carcinoma; survival stratified by miR-34A tumor methylation levels. Clinical data were available for 29 patients, and overall survival is shown for patients with miR-34A hypermethylated tumors (n = 16) and miR-34A hypomethylated tumors (n = 13). P = .0331 between the two groups by log-rank (Mantel-Cox) test.
Finally, to assess the potential applicability of these results in a somatic context, the association of miR-34A hypermethylation and TP53 mutation status in tumors was again identified in four tumor types in the publically available The Cancer Genome Atlas data set13 (Data Supplement), which demonstrates that these findings may have implications for cancer beyond the scope of LFS.
DISCUSSION
This study refines the role of epigenetics in a cancer predisposition syndrome and is the first, to our knowledge, to implicate miR-34A in human cancer susceptibility. This work details the largest comprehensive analysis of differential methylation to our knowledge by revealing distinct methylation patterns associated with TP53 mutations and provides a repository of genomic regions of deregulated methylation in the context of dysfunctional TP53. Moreover, there is a clear dynamic interplay between the genes that are aberrantly methylated in the presence of deleterious mutations that affect the p53 DNA binding domain and p53 regulatory regions and signaling networks. These findings suggest that deregulated DNA methylation at defined genomic loci is an important hallmark of TP53 mutation–mediated cancer susceptibility.
The most striking finding from this study is the relative miR-34A promoter hypomethylation at two adjacent CpG sites in peripheral blood from TP53 mutation carriers, which was confirmed in two independent cohorts and shown to cosegregate with TP53 mutations in families with LFS. This result is remarkable because miR-34A is a central microRNA in the p53 network and the first microRNA identified as a direct proapoptotic target of the p53 pathway.14-16 The detection of miR-34A hypomethylation in TP53 mutant nontransformed cells supports a putative model whereby in nontransformed cells that do not harbor mutations in TP53, wild-type p53 is recruited to the miR-34A locus and sustains hypermethylation. In the setting of loss-of-function or deleterious mutations in TP53, mutant p53 may not be able to maintain hypermethylation of the miR-34A promoter, which would lead to upregulation of miR-34A.
Because of the known redundant cellular roles of p53 and miR-34A, upregulation of miR-34A may supplement the necessary basal tumor-suppressive function that is lost when p53 is mutated. This mechanism may guard against mutant p53, even when the wild-type allele remains. Accordingly, miR-34A promoter hypermethylation may be characteristic of TP53 mutant tumors that lack wild-type p53 because this microRNA plays a critical role in cell maintenance, and its loss may cooperate with other genetic and/or epigenetic events to drive malignancy. It is therefore not surprising that, akin to p53, somatic miR-34A dysregulation is pervasive in human cancer and miR-34A inactivation by focal loss of 1p36 or promoter hypermethylation has been reported in a multitude of human malignancies.17-19 The precise mechanisms by which the miR-34A promoter undergoes somatic epimutation in tissues remain to be elucidated, and likely, various pathways may converge to yield this outcome in various tissues.
miR-34A tumor methylation may be of use in clinical prognostication of patients with CPCs, and more work is needed to validate the clinical relevance in patients in whom other tumor types develop. Given the strong association of miR-34A hypermethylation with TP53 mutation status, this finding is consistent with previous reports that TP53 mutations in CPCs are associated with a more clinically aggressive course, with poorer overall and event-free survival.20
Some limitations are inherent to our study design that should be noted. In particular, the analysis was limited to samples collected at our institution and referred to us from collaborators. We initially endeavored to match the TP53 mutation carrier and wild-type cohorts, but due to limitations in sample availability, the analysis cohort was not entirely matched. Moreover, because of the cross-sectional nature of the study, missing data in samples cannot be inferred. Finally, the study did not actively gauge the temporal changes in methylation values during patients’ disease course and relied on the assumption that the methylation values were relatively stable over time. Future studies may be aimed at determining the correlation between miR-34A promoter methylation levels and disease burden in patients with LFS.
As evidenced by the relationship between miR-34A hypermethylation and TP53 mutation status in other tumor types in The Cancer Genome Atlas data set,13 the findings from this study are broadly applicable to other cancer contexts. Given the pervasiveness of TP53 mutations in human malignancies, this has strong implications for targeting miR-34A in cancer. Studies have demonstrated in vivo the utility of miR-34A–based therapies in cancer, including intratumor or systemic delivery of lipid-formulated synthetic miR-34A.21,22 Systemic delivery of miR-34A mimics have already entered phase I clinical trials for numerous malignancies, including hepatocellular carcinoma, melanoma, and hematologic cancers (NCT01829971).
Collectively, this study provides strong support for the impact of TP53 mutations on epigenetic dysregulation in human cancer susceptibility. The study also demonstrates that miR-34A may be important in the pathogenesis of TP53-mediated cancer susceptibility, a putative novel therapeutic target and a marker for clinical prognostication.
Acknowledgment
We thank the patients with LFS and their families who have generously donated specimens for this research. We also thank Faiyaz Notta, PhD, and Sunit Das, MD, PhD, for critical review of the manuscript and Kevin Kelly, PhD, for technical advice.
Appendix
Supplementary Methods
Array-based DNA methylation analysis of initial cohort.
DNA extracted from peripheral blood leukocytes (1 μg) was used for bisulfite treatment (EpiTect Plus Bisulfite Kit; QIAGEN, Venlo, the Netherlands) with the use of DNA protection buffer. For the discovery cohort, bisulfite-treated DNA was then quantified by spectrophotometry (NanoDrop; Thermo Fisher Scientific, Waltham, MA), and > 500 ng was sent to Genome Quebec for hybridization to the Illumina 450K methylation arrays (Infinium; Illumina, San Diego, CA). Similarly, for the family with Li-Fraumeni syndrome (LFS) in Figure 3A and Appendix Figure A2B, bisulfite-treated DNA was quantified by spectrophotometry (NanoDrop), and > 500 ng was analyzed on Illumina 450K methylation arrays in The Centre for Applied Genomics (Toronto, Ontario, Canada). The amount of bisulfite-converted DNA and completeness of bisulfite conversion were assessed with a panel of MethylLight-based quality control reactions.
Plate maps for each 12-sample array were carefully planned to minimize potential batch effects as follows: TP53 mutant and wild-type samples were equally allocated on each array, including a comparable number of samples from patients with the same diagnoses. Equal representation of males and females per array also was ensured. The following quality control measures were embedded in the study design: DNA samples from the same patient, taken at different time points, were hybridized to different arrays, and samples from the same DNA isolation were hybridized to various arrays. This process comprised 11 samples for which duplicates were embedded in the array design and the normalized β-values showed a correlation > 0.99 for all sample pairs. Moreover, six peripheral leukocyte DNA samples from patients with colorectal cancer, previously used in a separate study by M.L. from the Hudson Laboratory, were tested and demonstrated concordance with the previously determined values.
Array preprocessing was performed by using the following functions from the R Bioconductor package: Background subtraction adjustment was applied using NOOB (normal-exponential using out-of-band probes) followed by color adjustments by using Illumina normalization probes and algorithms. Methylation values were then exported as β-values (estimates of CpG methylation levels ranging from 0 to 1) calculated as [M / (M + U)], which represents the ratio of the methylated probe intensity to the overall intensity at each CpG locus. The β-mixture quantile normalization method was applied to the set of β-values. Probes that overlapped with single nucleotide polymorphisms, which mapped to the Y chromosome, and/or Illumina control probes and cross-reactive probes were excluded from the analysis (Koestler DC, et al: Epigenetics 8:816-826, 2013).
Identification of differentially methylated CpG sites in the discovery cohort.
The primary analysis focused on characterization of differentially methylated CpG sites (DMCs) in germline peripheral blood leukocyte DNA from carriers of deleterious, transactivation function–deficient TP53 mutations (class 1) compared with nonmutation carriers with malignancies and healthy control subjects for a total cohort of 183 patients (72 mutation, 111 wild type). Characteristics such as age and sex were similar between the two groups to account for age- or sex-related methylation changes.
By using the imported β-values from the array-based DNA methylation analysis described in the previous section, all samples were subjected to principle component analysis in the R statistical package to identify outlier samples, with median centering of the data and missing value imputation to the 10 nearest neighbors. Four samples with substantial variability in the first three principle components were further excluded from the analysis. For the remaining samples (n = 202), 2,984 cross-reactive probes were excluded.3 Because small standard deviations at individual CpG sites frequently arise by chance and can result in highly significant, but often spurious results, an indiscriminant standard deviation cutoff of 0.05 was implemented to eliminate CpG sites with small standard deviations (Bock C: Nat Rev Genet 13:705-719, 2012). The remaining 80,720 sites were used for downstream statistical analysis. A nonparametric Mann-Whitney U test was used to identify CpGs that were differentially methylated between comparison groups. P values for the differentially methylated CpG sites were corrected for multiple hypothesis testing by the Benjamini-Hochberg procedure, and q = .05 was used as a threshold for statistical significance. β-Values were imported into Qlucore Omics Explorer version 2.2 (Qlucore, New York, NY) for visualization. For unsupervised clustering analyses, the top 1,000 differentially methylated probes between comparison groups were used, and representative heat maps were generated. Statistical significance of target DMCs among various comparison groups was set to false discovery rate q < .05.
Because the analysis cohort comprised both unrelated individuals and related family data, the genome-wide association analysis was also performed by using generalized estimating equations with an exchangeable correlation structure to account for familial correlation. The original or normal quantile-transformed β-values were modeled as the response variable, and domain function was included as a predictor. Age and sex were also assessed as potentially confounding predictors of methylation level (quantile-transformed β-value approximate age or gender using the generalized estimating equation). To address the possibility that differentially methylated regions may be due to differential composition of blood cells, a previously published set of CpGs (Koestler DC, et al: Epigenetics 8:816-826, 2013) was used to assess whether any differences were observed between comparison groups by principle component analysis. No differences were observed in blood composition between TP53 mutation carriers and noncarriers (Data Supplement). Finally, to exclude the possibility that methylation patterns may co-occur at sites with other genetic aberrations, such as copy number changes, low-resolution copy number variations were detected from the Illumina 450K methylation arrays by using the sum of both methylated and unmethylated signals and a similar method as described in Sturm et al (Sturm D, et al: Cancer Cell 22:425-437, 2012). Global germline copy number changes were not observed in any sample through this low-resolution imputation of copy number from methylation array data (data not shown).
Genomic distribution of differential CpG sites and functional interaction analysis.
To assess the genomic distribution of the most-enriched differentially methylated sites, we used the top 1,000 DMCs, identified all the CpG sites tested on the Illumina 450K Array within 1 Mb, and applied the hypergeometric test in R to ascertain the statistical significance of detecting these CpG sites. Briefly, the number of CpG sites in the top 1,000 was compared with the number of total sites (80,720) on the array that were tested to determine the probability of observing that number of sites in that genomic region. P values were then subjected to false discovery rate correction by the Benjamini-Hochberg method. Functional protein interaction analysis was performed by using the genes that mapped to the 49 statistically significant DMCs between TP53 mutation carriers and individuals with TP53 wild type. Visualization of genes in this cohort that are part of the p53 interaction network was performed by using the Cytoscape Reactome Functional Interaction network (Reactome FI Cytoscape Plugin 4).
Statistical analysis for miR-34A correlation with TP53 mutation status within the discovery cohort.
The receiver operating characteristic (ROC) curve was used to evaluate the threshold value of methylation at cg12385729, which resides in the 5′ promoter of the miR-34A gene, in differentiating germline TP53 mutation carriers from noncarriers. Each point on the curve represents the true-positive rate and false-positive rate associated with a particular test value. The area under the curve (AUC) and partial AUC were calculated to indicate the utility of this marker in predicting TP53 mutation status, where AUC values approaching 1 indicate an excellent discriminatory accuracy. This analysis was performed by using the pROC package in the R. Through this approach, the ROC yielded optimal sensitivity and specificity to distinguish TP53 mutation carriers from noncarriers on the basis of miR-34A methylation status.
Quantitative reverse transcriptase-polymerase chain reaction of miR-34A in tumor tissue.
To evaluate expression levels of miR-34A, total RNA (200 ng) was reverse transcribed to cDNA by using a custom reverse transcriptase primer pool for each of miR-34A and U6 small nuclear RNA endogenous control (Thermo Fisher Scientific). The resultant cDNA was also subjected to an additional preamplification step, per manufacturer protocol. Polymerase chain reaction levels of the preamplification product were quantified by using the corresponding TaqMan microRNA assays (Thermo Fisher Scientific). Thermocycling conditions were 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. The amount of miR-34A in TP53 mutation carriers relative to noncarriers was calculated by using the 2-ΔΔCt method and normalized to the U6 endogenous control. Results are fold change or relative quantification relative to U6 small nuclear RNA and are shown as mean ± SEM. Each reaction was performed in triplicate, and the entire procedure was performed in two independent experiments of the same RNA samples.
Fig A1.
(A) Volcano plot of differentially methylated CpG sites (DMCs) in deleterious TP53 mutation carriers relative to noncarriers. Differential methylation in absolute methylation levels (Δβ) is depicted on the x-axis, and the y-axis depicts increasing statistical significance determined by the Mann-Whitney U test (−log10 unadjusted P values). Points in red represent CpG sites that reached statistical significance after multiple hypothesis testing correction by the Benjamini-Hochberg method (q < .05). The three most highly significant DMCs are labeled. (B) Modified Manhattan plot of distribution of top DMCs across the genome. To assess the genomic distribution of the most enriched differentially methylated sites, the top 1,000 DMCs were used, and all CpG sites tested within 1 Mb were identified. The hypergeometric statistical test was applied to ascertain the significance of detecting these CpG sites at these loci relative to the total number of CpGs tested at the specified locus. Each CpG site is projected along its chromosomal location, and red peaks denote statistically significant differentially methylated regions (q < .05). lincRNA, long intergenic noncoding RNA.
Fig A2.
(A) Verification of methylation levels of top three DMCs. Box plots of methylation levels derived from Illumina 450K Array analysis in initial cohort (n = 183; blue box plots) and methylation levels derived from bisulfite pyrosequencing of a subset of the initial cohort (n = 65; beige box plots). The correlation between these values is represented as a line graph with Illumina 450K Array β-values on the x-axes and bisulfite pyrosequencing results on the y-axes. Data are shown for miR-34A promoter (cg12385729), BC097941 lincRNA (cg0667321), and PTCH53 (C6orf138) (cg2016804). (B) Validation of methylation levels of the top miR-34A promoter CpG site (cg12385729) in an independent patient cohort. Comparison between methylation levels at the miR-34A promoter CpG site cg12385729 is shown in an initial cohort (n = 183) on the basis of the Illumina 450K Array platform (blue box plots) and in an independent validation cohort (n = 76) on the basis of bisulfite pyrosequencing of this CpG site (orange box plots). P values were calculated with univariable two-tailed t tests (initial cohort q = 3.1 × 10−15; validation cohort P < .001). **P < .001. lincRNA, long intergenic noncoding RNA; MT, mutation; WT, wild type.
Fig A3.
miR-34A promoter methylation across multiple histologic sections of tumor and normal tissue in patients who harbor germline mutations in TP53. (A) Methylation levels at three CpG sites on the miR-34A promoter CpG island labeled in red in the sequence provided across multiple histologic sections of tissue from patient donor 4264. The first CpG site corresponds to cg12385729. (B) miR-34A promoter methylation levels as in (A) for tissue from patient donor 4115 as well as choroid plexus carcinoma tumor DNA from the patient's male sibling.
Footnotes
Supported in part through grants from the Canadian Institutes for Health Research, Terry Fox Research Institute, and SickKids Foundation. T.J.H. is supported by a Senior Investigator Award of the Ontario Institute for Cancer Research, with funds provided by the Ontario Ministry of Research and Innovation. Establishment and running of the Children's Cancer Centre Tissue Bank at the Royal Children's Hospital Melbourne is made possible through generous support by Cancer in Kids, Leukaemia Auxiliary, the Murdoch Childrens Research Institute, and the Royal Children's Hospital Foundation.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Nardin Samuel, Thomas J. Hudson, David Malkin
Provision of study materials: Lynn Meister, Maria Isabel W. Achatz
Collection and assembly of data: Nardin Samuel, Diana Merino, Ana Novokmet, Jonathan L. Finlay, Christine Elser, Lynn Meister, Michael D. Taylor, Uri Tabori, Meredith Irwin, Jordan R. Hansford, Maria Isabel W. Achatz, David Malkin
Data analysis and interpretation: Nardin Samuel, Gavin Wilson, Mathieu Lemire, Badr Id Said, Youliang Lou, Weili Li, James Tran, Kim E. Nichols, Sanaa Choufani, Marc Remke, Vijay Ramaswamy, Florence M.G. Cavalli, Rosanna Weksberg, Jonathan D. Wasserman, Andrew D. Paterson, Thomas J. Hudson, David Malkin
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Genome-Wide DNA Methylation Analysis Reveals Epigenetic Dysregulation of MicroRNA-34A in TP53-Associated Cancer Susceptibility
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Nardin Samuel
No relationship to disclose
Gavin Wilson
No relationship to disclose
Mathieu Lemire
No relationship to disclose
Badr Id Said
No relationship to disclose
Youliang Lou
No relationship to disclose
Weili Li
No relationship to disclose
Diana Merino
No relationship to disclose
Ana Novokmet
No relationship to disclose
James Tran
No relationship to disclose
Kim E. Nichols
No relationship to disclose
Jonathan L. Finlay
No relationship to disclose
Sanaa Choufani
No relationship to disclose
Marc Remke
No relationship to disclose
Vijay Ramaswamy
No relationship to disclose
Florence M.G. Cavalli
No relationship to disclose
Christine Elser
Honoraria: Pfizer
Consulting or Advisory Role: Pfizer
Lynn Meister
No relationship to disclose
Michael D. Taylor
No relationship to disclose
Uri Tabori
No relationship to disclose
Meredith Irwin
Consulting or Advisory Role: United Therapeutics
Rosanna Weksberg
No relationship to disclose
Jonathan D. Wasserman
No relationship to disclose
Andrew D. Paterson
No relationship to disclose
Jordan R. Hansford
No relationship to disclose
Maria Isabel W. Achatz
Travel, Accommodations, Expenses: AstraZeneca
Thomas J. Hudson
Stock or Other Ownership: AbbVie
David Malkin
No relationship to disclose
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ: Surfing the p53 network Nature 408:307–310,2000 [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez KD Noltner KA Buzin CH, etal: Beyond Li Fraumeni syndrome: Clinical characteristics of families with p53 germline mutations J Clin Oncol 27:1250–1256,2009 [DOI] [PubMed] [Google Scholar]
- 3.Chen YA Lemire M Choufani S, etal: Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray Epigenetics 8:203–209,2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tost J, Gut IG: DNA methylation analysis by pyrosequencing Nat Protoc 2:2265–2275,2007 [DOI] [PubMed] [Google Scholar]
- 5. International Agency for Research on Cancer. IARC TP53 Database. Geneva, Switzerland, World Health Organization, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achatz MI Olivier M Le Calvez F, etal: The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families Cancer Lett 245:96–102,2007 [DOI] [PubMed] [Google Scholar]
- 7.Cipriano R Miskimen KL Bryson BL, etal: Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer Mol Cancer Res 12:1156–1165,2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SY Meier R Furuta S, etal: FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice J Clin Invest 122:3211–3220,2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botcheva K McCorkle SR McCombie WR, etal: Distinct p53 genomic binding patterns in normal and cancer-derived human cells Cell Cycle 10:4237–4249,2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei CL Wu Q Vega VB, etal: A global map of p53 transcription-factor binding sites in the human genome Cell 124:207–219,2006 [DOI] [PubMed] [Google Scholar]
- 11.Chung JH Larsen AR Chen E, etal: A PTCH1 homolog transcriptionally activated by p53 suppresses Hedgehog signaling J Biol Chem 289:33020–33031,2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemire M Zaidi SH Ban M, etal: Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci Nat Commun 6:6326,2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. National Cancer Institute, National Human Genome Research Institute: The Cancer Genome Atlas.
- 14.Raver-Shapira N Marciano E Meiri E, etal: Transcriptional activation of miR-34a contributes to p53-mediated apoptosis Mol Cell 26:731–743,2007 [DOI] [PubMed] [Google Scholar]
- 15.Chang TC Wentzel EA Kent OA, etal: Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis Mol Cell 26:745–752,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L He X Lim LP, etal: A microRNA component of the p53 tumour suppressor network Nature 447:1130–1134,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodygin D Tarasov V Epanchintsev A, etal: Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer Cell Cycle 7:2591–2600,2008 [DOI] [PubMed] [Google Scholar]
- 18.Bader AG: miR-34 - A microRNA replacement therapy is headed to the clinic Front Genet 3:120,2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermeking H: MicroRNAs in the p53 network: Micromanagement of tumour suppression Nat Rev Cancer 12:613–626,2012 [DOI] [PubMed] [Google Scholar]
- 20.Merino DM Shlien A Villani A, etal: Molecular characterization of choroid plexus tumors reveals novel clinically relevant subgroups Clin Cancer Res 21:184–192,2015 [DOI] [PubMed] [Google Scholar]
- 21.Trang P Wiggins JF Daige CL, etal: Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice Mol Ther 19:1116–1122,2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Martino MT Leone E Amodio N, etal: Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: In vitro and in vivo evidence Clin Cancer Res 18:6260–6270,2012 [DOI] [PMC free article] [PubMed] [Google Scholar]