Abstract
The adaptive fasting response is invoked as a promising cardiometabolic and neurodegenerative therapeutic pathway. We and others have defined the carbohydrate transporter glucose transporter 8 (GLUT8) as a critical regulator of hepatic and whole-organism metabolic homeostasis in the overfed and diabetic states. However, the functions of this critical transporter in the physiological fasting response remain poorly understood. Here, we tested the hypothesis that GLUT8 modulates the adaptive hepatic fasting response. We demonstrate that mice with targeted Slc2a8 disruption exhibit enhanced thermogenesis, ketogenesis, and peripheral lipid mobilization during fasting. These metabolic enhancements were observed in the context of mildly impaired hepatic mitochondrial oxidative metabolism in vivo and in vitro. Mechanistically, we show that hepatic peroxisome proliferator–activated receptor α (PPARα) and its transcriptional fasting response target hepatokine, fibroblast growth factor (FGF)21, are cell-autonomously hyperactivated in GLUT8-deficient liver and in isolated primary murine hepatocytes during nutrient depletion. Hepatic PPARα knockdown in GLUT8-deficient mice normalized the enhanced ketogenic and FGF21 secretory responses and decreased mitochondrial respiratory function without altering the hyperthermic response to fasting. Our data demonstrate that hepatocyte GLUT8 regulates adaptive fasting in part through regulation of the PPARα signaling cascade. Moreover, the ketotic and thermic responses to fasting are differentially encoded within the GLUT8-PPARα communication axis. GLUT8 therefore represents a therapeutic target that can be leveraged against cardiometabolic disease.
The carbohydrate transporter GLUT8 (Slc2a8) regulates fasting responses including lipid mobilization, ketogenesis, FGF21 secretion, and thermogenesis. This is in part mediated by hepatic PPARα.
Increasing evidence demonstrates the adaptive effects of physiological fasting in promoting healthspan and in the treatment and prevention of metabolic disease. Indeed, rodent models demonstrate prevention and reversal of all aspects of the metabolic syndrome with intermittent fasting (1), and diabetes, obesity, and heart disease are each mitigated by subjecting mice and humans to variations of fasting or to drugs that mimic the fasting state (2–4). Leveraging the adaptive hepatic fasting response and developing pharmacological fasting mimetics targeting hepatic glucose transport are thus gaining traction as credible therapeutic goals (4–9).
Prolonged fasting necessitates a shift from using diet-derived substrate toward mobilizing cellular nutrient stores via peripheral lipolysis and nutrient redistribution by the liver (10, 11). This process is largely coordinated through hepatic peroxisome proliferator–activated receptor α (PPARα), a lipid ligand–activated transcription factor that upregulates peroxisomal and mitochondrial biogenesis to oxidize newly mobilized lipids and ultimately to generate ketone bodies (12). One key transcriptional target of PPARα is the fasting-induced hepatokine fibroblast growth factor (FGF)21. In addition to promoting the pro-oxidative and ketogenic effects of PPARα within the hepatocyte, FGF21 enhances peripheral lipid mobilization and thermogenesis (13–21), although the precise function and specific contexts for those functions can vary (22). Although PPARα activation via the fibrate class of synthetic agonists has been used clinically for decades (23), the potential utility of FGF21 in the pharmacological treatment of metabolic disease (24) warrants continued investigation of the hepatic PPARα-FGF21 signaling axis.
We recently demonstrated that the hepatic carbohydrate transporter, glucose transporter 8 (GLUT8), which is encoded by the Slc2a8 gene, mediates key aspects of the hepatic fasting signaling cascade, including autophagy, AMPK/mTORC1 signaling, and peripheral thermogenesis, induced by the disaccharide GLUT inhibitor trehalose (4, 7, 8, 25, 26). We therefore examined the extent to which GLUT8 modulates hepatic fasting signaling pathways and extrahepatic energy homeostasis during physiological fasting. Here, we demonstrate that GLUT8 cell autonomously modulates hepatic PPARα-FGF21 signaling during fasting. Mice with targeted GLUT8 gene disruption exhibit enhanced thermogenesis, ketogenesis, and peripheral lipid mobilization during fasting. These metabolic enhancements were observed in the context of impaired hepatic mitochondrial oxidative metabolism in vivo and in vitro. Mechanistically, we show that hepatic PPARα and its transcriptional fasting response target, FGF21, are cell-autonomously hyperactivated in GLUT8-deficient liver and in isolated primary murine hepatocytes during nutrient depletion. In vivo, PPARα knockdown in GLUT8-deficient mice normalized the exaggerated ketogenic and FGF21 secretory responses, whereas the hyperthermic response to fasting in GLUT8-deficient mice remained intact. Furthermore, hepatic PPARα knockdown in wild-type (WT) hepatocytes mirrored the perturbed mitochondrial respiration and morphology observed in GLUT8 deficiency. Together, these results define an augmented hepatic adaptive fasting response in GLUT8-deficient mice mediated in part by PPARα. Moreover, we provide evidence that the ketotic and thermic responses to fasting are differentially encoded within this GLUT8-PPARα axis.
Materials and Methods
Mouse models and treatment
All animal procedures were approved by the Washington University School of Medicine Animal Studies Committee. WT and G8KO mice harboring transgenic GFP-LC3 were obtained by crossing GLUT8-deficient mice with GFP-LC3 mice as previously described (7, 27). All mice were male and were 5 to 7 weeks old. For fasting studies, mice were housed on aspen bedding. Mice in the “fed” group were given ad libitum access to a standard chow diet. All procedures were performed in accordance with the approved guidelines by the Animal Studies Committee at Washington University School of Medicine.
Serum analyses
Fasting blood glucose was measured via glucometer using tail vein blood. For all other serum analyses, submandibular blood collection was performed immediately prior to euthanization, and serum was separated. Free triiodothyronine enzyme-linked immunosorbent assay (ELISA) (catalog no. 1650; Alpha Diagnostic International), FGF21 ELISA (catalog no. EZRMFGF21-26K; Millipore), Insulin ELISA (catalog no. EZRMI-13K; Millipore), β-hydroxybutyrate (catalog no. 700190; Cayman Chemical), triglycerides (TGs) (catalog no. TR22421; Thermo Fisher Scientific), cholesterol (catalog no. TR13421; Thermo Fisher Scientific), and free fatty acids (FFAs) (catalog nos. 999-34691, 995-34791, 991-34891, 993-35191; Wako Diagnostics) quantification were performed using commercially available reagents per manufacturer’s directions. Alanine aminotransferase and albumin levels were quantified using an LIASYS Chemistry Analyzer (AMS Diagnostics).
Hepatic lipids
Lipids were extracted from ∼100 mg hepatic tissue homogenized in 2:1 chloroform/methanol, and 0.25% to 0.5% of each extract was evaporated for at least 1 hour prior to biochemical quantification of TGs, cholesterol, and FFAs using the previously described reagents according to manufacturers’ directions.
Oil Red O staining
Methanol-fixed frozen sections from WT and G8KO mice were stained with Oil Red O according to described protocols (6, 7).
Body composition analysis
Body composition analysis was carried out in unanesthetized mice as described (5, 6, 28) using an EchoMRI 3-1 device (Echo Medical Systems) via the Washington University Diabetic Mouse Models Phenotyping Core Facility.
Indirect calorimetry
Oxygen consumption, CO2 production, respiratory exchange ratio, and heat production were measured using the Phenomaster system (TSE Systems) via the Washington University Diabetic Mouse Models Phenotyping Core Facility as described (5, 28). Metabolic parameters were documented every 13 minutes.
Antisense oligonucleotides
WT and G8KO male mice were given 25 mg/kg antisense oligonucleotides (ASOs) twice weekly for 3 weeks via intraperitoneal injection. All mice were ∼4 to 5 weeks old at the start of the injections. ASOs targeting Ppara (catalog no. 141923-83), Fgf21 (catalog no. 256216-2), or scrambled control (catalog no. 233438-7) were provided by Ionis Pharmaceuticals and used for both the in vivo and in vitro studies as indicated.
ASOs targeting Slc2a8 were designed as “gapmers” with phosphorothioate bonds and flanking 2′-O-Methyl regions at the 3′ and 5′ ends. Oligos targeting Slc2a8 (ASO1GLUT8 and ASO2GLUT8) or a scrambled control ASOs (ASOScr) were ordered from Integrated DNA Technologies. Sequences are as follows, where an asterisk (*) indicates a phosphorothioate bond and “m” indicates a 2′-O-methyl modification:
ASO1GLUT8: mC*mA*mU*mC*mC*A*C*A*G*G*C*T*C*C*G*mC*mC*mG*mC*mG
ASO2GLUT8: mG*mU*mU*mA*mC*A*G*G*C*C*A*C*T*C*C*mA*mC*mU*mC*mC
ASOScr: mU*mG*mU*mA*mC*G*A*C*C*T*A*G*C*T*A*mC*mC*mC* mC*mC
Adenoviruses
Adenovirus encoding GLUT8 with a GFP reporter was purchased from Vector Biolabs (Ad-GFP-m-SLC2A8; catalog no. ADV-272300). Two days prior to a 48-hour fast, 1.8e9 PFU adenovirus was delivered to each mouse via tail vein injection.
Cell cultures and treatment
Primary murine hepatocytes obtained from WT and G8KO mice were isolated as described (6, 8) and cultured in regular Dulbecco’s modified Eagle medium (catalog no. D5796; Sigma) containing 10% fetal bovine serum. α mouse liver 12 (AML12) cells were purchased from American Type Culture Collection [CRL-2254; Research Resource Identifier (RRID): CVCL_0140] and maintained per American Type Culture Collection guidelines. In vitro “starved” media contained 1 g/L glucose and 0.5% fetal bovine serum. Genetic knockdown was achieved via ASO transfection using Lipofectamine 3000 (catalog no. L3000015; Invitrogen).
Transmission electron microscopy
Ultrastructural analysis was performed as previously (7) with modifications. Fresh 1-mm2 liver sections were rapidly dissected and fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Polysciences Inc.) in 100 mM sodium cacodylate buffer (pH 7.2) for 1 hour at room temperature. Samples were washed in cacodylate buffer and postfixed in 1% osmium tetroxide (Polysciences Inc.) for 1 hour. Samples then were rinsed extensively in ultrapure water prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc.) for 1 hour. After several rinses in ultrapure water, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections (95 nm) were cut with an ultramicrotome (Ultracut UCT7; Leica Microsystems Inc.), stained with uranyl acetate and lead citrate, and viewed on a transmission electron microscope (1200 EX; JEOL USA Inc.) equipped with an AMT 8-megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques).
ImageJ 1.47v software (RRID: SCR_003070; National Institutes of Health) was used to trace and quantify area and roundness of mitochondrial structures in three high-powered fields (HPFs) per mouse (n = 2 mice per group). Mitochondrial spheroids were counted by a treatment-blinded observer from ∼40 high-power fields (n = 2 mice per group). “Spheroids” were defined as elongated mitochondrial structures forming an ∼270- to 360-degree curvature. Partial spheroids were defined as elongated mitochondrial structures forming an 180- to 270-degree curvature or an ∼90- to 180-degree curvature if enwrapping a cytosolic structure.
High-resolution respirometry
High-resolution respirometry was performed on skeletal muscle (soleus) and whole-liver homogenates using an Oxygraph-O2K respirometer (Oroboros Instruments). Soleus measurements were performed precisely as described (29). Liver homogenates were processed by grinding fresh liver tissue in BIOPS buffer using a dounce homogenizer, pelleting heavy cellular debris via low-speed centrifugation, and then diluting paired samples to equivalent protein concentration per Bradford assay readouts. Downstream analysis of liver homogenates was performed in Mir05 buffer precisely as described (29).
Extracellular flux analysis
In vitro respiration measurements were performed using the Seahorse XF96 Analyzer (Agilent) with the AML12 immortalized hepatocyte cell line. Cells were seeded to near confluency, treated with ASOs delivered via Lipofectamine 3000 for 24 hours, and subjected to starvation media for an additional 24 hours prior to analysis. The Seahorse Mito Stress Test kit (Agilent) was used according to manufacturer instructions. For groups treated with etomoxir, the drug was added at a final concentration of 40 μM 15 minutes prior to the start of the assay.
Ex vivo lipolysis assay
Lipolytic rate was measured as described with some modification (30). Briefly, white adipose tissue was harvested from WT and G8KO mice and diced, and 50 mg of tissue was placed in each well of a 24-well plate containing M199 media plus insulin (50 μU/mL) and dexamethasone (2.5 nM). After incubating overnight at 37°C, tissue was transferred to Krebs Ringer buffer with or without isoproterenol (0.1 μM). Media aliquots were taken hourly for up to 3 hours for glycerol quantification.
Quantitative reverse transcription polymerase chain reaction
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed as previously reported (6, 7) with some modifications. Snap-frozen livers or cultured hepatocytes were homogenized in Trizol reagent (catalog no. 15596026; Invitrogen). RNA isolated according to the manufacturer’s protocol was reverse-transcribed using the Quantitect reverse transcription kit (catalog no. 205310; Qiagen). cDNA was subjected to quantitative polymerase chain reaction using the SYBR Green master mix reagent (catalog no. 4309155; Applied Biosystems). Primers used are listed in Table 1.
Table 1.
Mouse qRT-PCR Primer Sequences
| Gene Symbol | Protein Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| Slc2a8 | GLUT8 | TTCATGGCCTTTCTAGTGACC | GAGTCCTGCCTTTAGTCTCAG |
| Slc2a2 | GLUT2 | TGTGCTGCTGGATAAATTCGCCTG | AACCATGAACCAAGGGATTGGACC |
| mt-Cytb | Cytochrome b | CTGTTCGCAGTCATAGCC | AAGAATCGGGTCAAGGTG |
| mt-Co1 | Cytochrome c oxidase I | ATGTTCTATCAATGGGAGC | TCTGAGTAGCGTCGTGGT |
| mt-Atp8 | ATP synthase 8 | ATGCCACAACTAGATACAT | TAGTGATTTTGGTGAAGG |
| mitochondrial d-loop | N/A | AATCAATGGTTCAGGTCA | ACGGAGGATGGTAGATTA |
| Actb | β-Actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| Hmgcs2 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | ATACCACCAACGCCTGTTATG | CAATGTCACCACAGACCACCA |
| Acox1 | Acyl-Coenzyme A oxidase 1 | CCTGATTCAGCAAGGTAGGG | TCGCAGACCCTGAAGAAATC |
| Cpt1a | Carnitine palmitoyltransferase 1a | AGTGGCCTCACAGACTCCAG | GCCCATGTTGTACAGCTTCC |
| Fgf21 | Fibroblast growth factor 21 (FGF21) | TACACAGATGACGACCAAGA | GGCTTCAGACTGGTACACAT |
| Atf4 | Activating transcription factor 4 | AGCAAAACAAGACAGCAGCC | ACTCTCTTCTTCCCCCTTGC |
| Tfam | Transcription factor A, mitochondrial | GCCTGAGTTTGTGTTTGCTG | CTGCTCTTTATACTTGCTCACAG |
| Nrf1 | Nuclear respiratory factor 1 | AATGTCCGCAGTGATGTCC | GCCTGAGTTTGTGTTTGCTG |
| Cox5b | Cytochrome c oxidase subunit Vb | ACCCTAATCTAGTCCCGTCC | CAGCCAAAACCAGATGACAG |
| Ndufs8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 | GTTCATAGGGTCAGAGGTCAAG | TCCATTAAGATGTCCTGTGCG |
| Uqcrc1 | Ubiquinol-cytochrome c reductase core protein I | ATCAAGGCACTGTCCAAGG | TCATTTTCCTGCATCTCCCG |
| rplp0 | 36B4/ ribosomal protein, large, P0 | TAAAGACTGGAGACAAGGTG | GTGTACTCAGTCTCCACAGA |
| Actin | Actin | GATTACTGCTCTGGCTCCTAG | GACTCATCGTACTCCTGCTTG |
Abbreviations: ATP, adenosine triphosphate; N/A, not available; NADH, nicotinamide adenine dinucleotide.
Statistics
Data were analyzed using GraphPad Prism version 7.0 (RRID: SCR_015807; GraphPad, Inc.). A P value <0.05 was defined as statistically significant. Data shown are mean ± standard error of the mean. Two-tailed unpaired homoscedastic t tests with Bonferroni post hoc correction for multiple comparisons were used for all analyses unless otherwise noted in the figure legends. Two-way analysis of variance was also used for analyses with two independent variables.
Results
Selective GLUT8 messenger RNA induction in the fasting state
We previously demonstrated that the glucose transporter inhibitor trehalose induces a hepatic pseudo-fasting response via the hexose transporter GLUT8 (8). GLUT8 deficiency blunted the starvation-like signal transduction response of hepatocytes to trehalose, leading us to investigate whether GLUT8 participates in the physiological fasting response. To examine whether physiological fasting regulates GLUT8, we quantified hepatic Slc2a8 messenger RNA (mRNA) abundance via qRT-PCR in WT mice fed ad libitum or fasted for 12, 24, or 48 hours. Slc2a8 mRNA transcript levels increased linearly over the fasting period (R2 = 0.605, P < 0.05). In contrast, Slc2a2 mRNA encoding the GLUT2 protein was not induced in response to fasting (Supplemental Fig. 1), indicating selective GLUT8 regulation at the gene expression level.
Resistance to thermic depression in fasting GLUT8-deficient mice
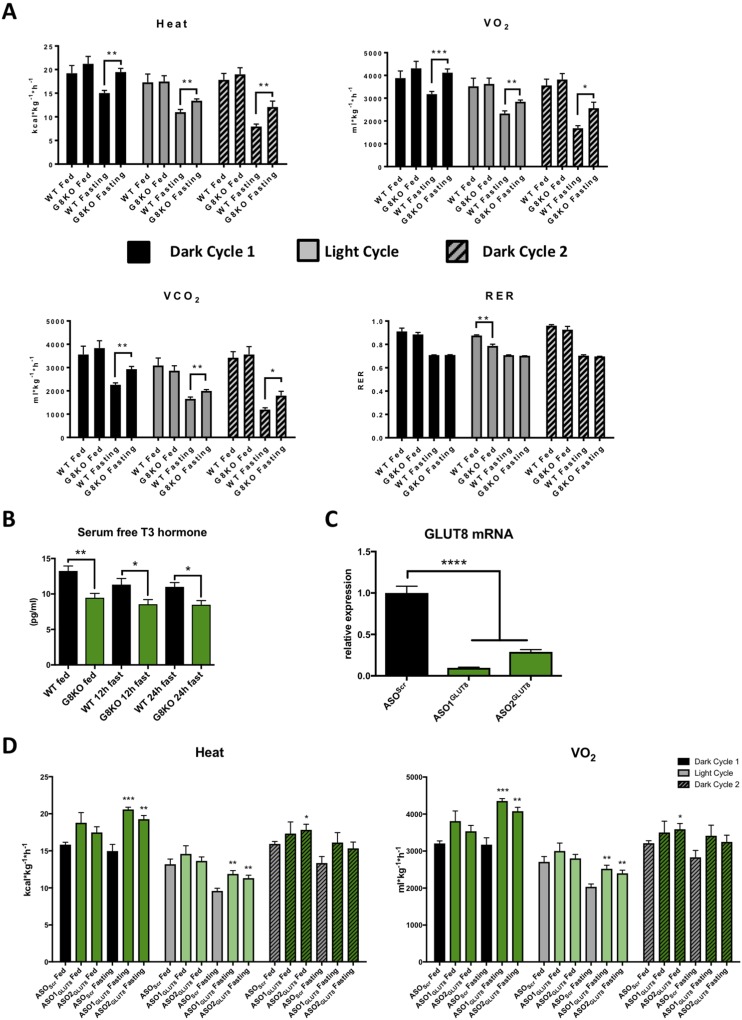
We next investigated the role of GLUT8 in whole body respiration during the fasted state. We used indirect calorimetry to quantify energy expenditure of ad libitum chow–fed or fasted WT and G8KO mice over a 48-hour period. In the absence of changes in fed-state heat production, fasting G8KO mice displayed significantly greater heat production (dark cycle 1, P = 0.001; light cycle, P = 0.007; dark cycle 2, P = 0.0098), VO2 (dark cycle 1, P = 0.001; light cycle, P = 0.0067; dark cycle 2, P = 0.0096), and VCO2 (dark cycle 1, P = 0.0011; light cycle, P = 0.0089; dark cycle 2, P = 0.0110) as compared with WT controls (Fig. 1A). No significant changes were observed in fasting respiratory exchange ratio (RER) (dark cycle 1, P = 0.7017; light cycle, P = 0.0959; dark cycle 2, P = 0.3801) or movement (dark cycle 1, P = 0.2577; light cycle, P = 0.0968; dark cycle 2, P = 0.7163) (Fig. 1A and Supplemental Fig. 2A), and circulating levels of free triiodothyronine, a prothermogenic hormone, were significantly lower in G8KO mice in both the fed and fasted states (fed, P = 0.0065; 12-hour fast, P = 0.0426; 24-hour fast, P = 0.0233) (Fig. 1B). Ad libitum–fed G8KO mice displayed significantly lower RER values during the inactive light cycle period as compared with WT mice (P = 0.0013), suggesting that G8KO mice obtain more energy from lipids than from carbohydrates during these conditions (Fig. 1A).
Figure 1.
Resistance to thermic depression in fasting GLUT8-deficient mice. (A) Indirect calorimetry measurements of heat production, VO2, VCO2, and RER in WT and G8KO mice over a 48-hour period of ad libitum chow feeding or fasting (n = 4 to 6 mice per group). (B) ELISA quantification of serum free triiodothyronine (T3) hormone levels in WT and G8KO mice that were chow fed or fasted for 12 or 24 hours (n = 4 mice per group). (C) qRT-PCR quantification of GLUT8 mRNA in primary murine hepatocytes transfected with either scrambled ASOs (ASOScr) or sequence 1 (ASO1GLUT8) or 2 (ASO2GLUT8) of ASOs targeting GLUT8 (n = 3 replicates per group). (D) Indirect calorimetry measurements of heat production and VO2 in WT mice treated with ASOScr, ASO1GLUT8, or ASO2GLUT8 and either chow fed or fasted for 48 hours (n = 6 mice per group). Data are presented as mean ± standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To more reductively examine the effects of hepatocyte-selective GLUT8 knockdown on fasting thermogenesis, we treated WT mice with scrambled or GLUT8-targeted ASOs via intraperitoneal injection, a method known to selectively target hepatic gene expression (31–33). We confirmed >70% GLUT8 knockdown in isolated hepatocytes (P < 0.0001) (Fig. 1C). Mice injected with either scrambled or GLUT8-directed oligonucleotide were subjected to indirect calorimetry measurements in the fed and fasted state. In concordance with the germline GLUT8-deficient model, fasting WT mice treated with GLUT8 ASOs displayed significantly elevated VO2 (ASO1GLUT8, P = 0.0001; ASO2GLUT8, P = 0.0018) and heat (ASO1GLUT8, P = 0.0002; ASO2GLUT8, P = 0.0018), suggesting the hyperthermic phenotype after GLUT8 disruption is at least in part liver dependent (Fig. 1D).
Enhanced lipid mobilization and ketogenesis during fasting in GLUT8-deficient mice
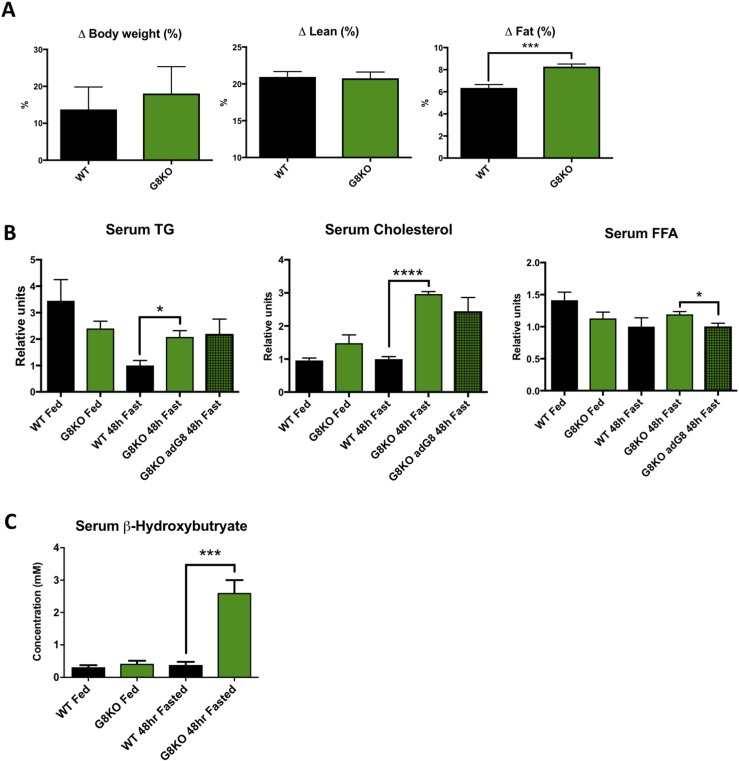
The physiological fasting response involves peripheral lipid mobilization, which shunts substrate to the liver for fat oxidation and ketogenesis. Therefore, we interrogated the role of GLUT8 in peripheral fat mobilization during fasting. GLUT8-deficient mice exhibited a greater percentage of body fat loss during the 48-hour fasting period (P = 0.0009) in the absence of differences in percentage of lean mass (P = 0.8678) or total body weight changes (P = 0.6577) (Fig. 2A). There were no differences in initial body weight or percentage of lean mass between WT and G8KO fed mice, but G8KO mice had a greater initial percent body fat as compared with WT (P = 0.0013; Supplemental Fig. 2B). The observed increase in body fat loss in G8KO mice was accompanied by elevated fasting serum TGs (P = 0.013) and cholesterol levels (P < 0.0001) in G8KO mice relative to WT, with no significant increase in fatty acids (P = 0.2921) (Fig. 2B).
Figure 2.
Enhanced lipid mobilization and ketogenesis during fasting in GLUT8-deficient mice. (A) Echo magnetic resonance image quantification of total, lean, and fat mass lost after fasting as a percentage of initial body weight (WT, n = 6; G8KO, n = 5). (B) Serum measurements of TGs, FFAs, and cholesterol after 48 hours of fasting (WT fed, n = 4; G8KO fed, n = 6; WT fasted, n = 8 to 10; G8KO fasted, n = 10 to 11; G8KO adG8 fasted, n = 4). Significant two-way analysis of variance (ANOVA) diet-genotype interactions were detected for cholesterol (P < 0.0001) and TGs (P = 0.0052). (C) Serum β-hydroxybutyrate levels in ad libitum chow-fed or 48-hour–fasted mice (WT fed, n = 6; G8KO fed, n = 4, WT fasted, n = 7; G8KO fasted, n = 9). A significant two-way ANOVA diet-genotype interaction was detected (P < 0.05). Data are presented as mean ± standard error of the mean. *P < 0.05; ***P < 0.001; ****P < 0.0001.
To gain insight into the role of the liver in the GLUT8-dependent regulation of lipid homeostasis, we overexpressed GLUT8 in a subset of G8KO via tail vein injection of an adenoviral construct. This resulted in upregulated levels of Glut8 mRNA in liver but not skeletal muscle (Supplemental Fig. 3). Reconstitution of GLUT8 in G8KO livers resulted in a trend toward decreased serum cholesterol, although this did not reach statistical significance, and a significant decrease in serum FFAs (P = 0.0329) (Fig. 2B). To investigate a potential adipose-intrinsic role of GLUT8 in lipolysis regulation, we quantified basal and β-adrenergic–stimulated lipolysis rates in adipose tissue explants and observed no differences between WT and G8KO mice (Supplemental Fig. 2C). No significant differences were observed in serum insulin levels between WT and G8KO mice in either the fed or fasted states (P = 0.1772; Supplemental Fig. 2D).
After 48 hours of fasting, G8KO mice displayed significantly elevated fasting serum ketones (P = 0.0003) (Fig. 2C). Given the relative abundance of GLUT8 as a hepatic hexose transporter and its demonstrated importance in hepatic metabolic homeostasis in response to fructose (5, 6), we assessed blood glucose levels throughout a 48-hour fasting time course. Although glucose levels were comparable between WT and G8KO mice during the first 12 hours of fasting, G8KO mice exhibited lower circulating blood glucose at 24 and 48 hours of fasting (24 hours, P = 0.0002; 48 hours, P = 0.0296) (Supplemental Fig. 2E).
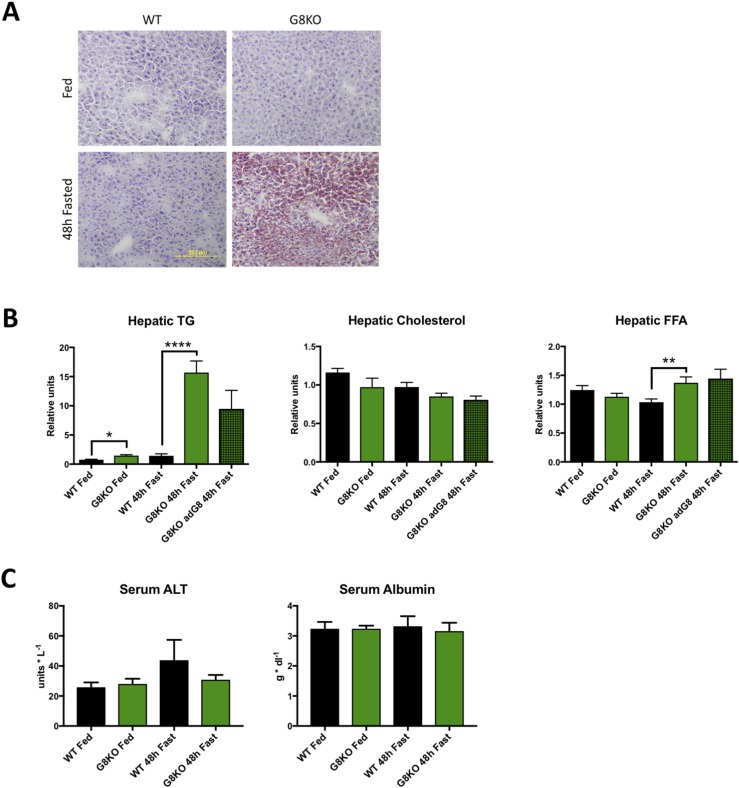
Elevated fasting hepatic lipids in G8KO mice
Enhanced peripheral lipid mobilization during fasting in G8KO mice prompted us to investigate the role of GLUT8 in fasting hepatic lipid homeostasis. Histological analysis by Oil Red O staining for neutral fat accumulation was performed in liver sections obtained from fed and fasting WT and G8KO mice. This revealed increased neutral lipid accumulation in GLUT8-deficient fasted mice as compared with WT controls (Fig. 3A). Biochemical analysis of hepatic lipids revealed significant increases in fasting FFAs (P = 0.0072) and TGs (P < 0.0001) in G8KO mice as compared with WT, whereas cholesterol levels were not statistically different (P = 0.1113). A small but statistically significant increase in hepatic TG levels was observed in G8KO mice as compared with WT during the fasted state (P = 0.0108). Overexpression of hepatic GLUT8 in G8KO mice resulted in a trend toward decreased hepatic TG levels that did not reach statistical significance (P = 0.1578) and did not significantly alter hepatic FFAs or cholesterol (Fig. 3B).
Figure 3.
Elevated fasting hepatic lipids in G8KO mice. (A) Oil Red O staining of liver sections from fed or 48-hour–fasted mice. (B) Hepatic TGs, FFAs, and cholesterol from WT and G8KO mice after 48 hours of fasting (WT fed, n = 4; G8KO fed, n = 6; WT fasted, n = 14 to 15; G8KO fasted, n = 13 to 15; G8KO adG8 fasted, n = 4). A significant two-way analysis of variance diet-genotype interaction was detected for TGs (P = 0.0047). (C) Serum measurements of alanine aminotransferase (ALT) and albumin in fed or 48-hour–fasted mice (n = 5 mice per group). Data are presented as mean ± standard error of the mean. *P < 0.05; **P =< 0.01; ****P < 0.0001.
No evidence of hepatocellular insult or synthetic dysfunction was observed in the fasted state, as quantified by circulating alanine aminotransferase (P = 0.3794) and albumin (P = 0.7217) (Fig. 3C). Together, decreased RER, decreased peripheral adiposity, and increased circulating and hepatic lipids suggest enhanced fasting-induced mobilization of peripheral lipid stores to the liver.
Mitochondrial dysmorphology and reduced hepatic oxidative function in fasting GLUT8-deficient mice
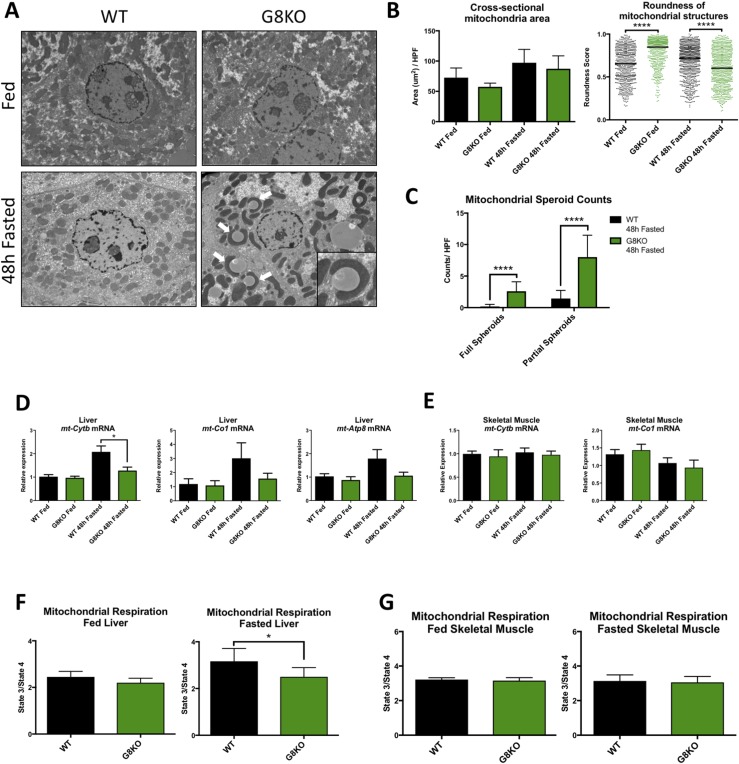
In light of the perturbed lipid homeostasis observed in GLUT8-deficient mice and the crucial role of mitochondria in fasting lipid oxidation, we evaluated mitochondrial morphology in WT and G8KO livers from fed and 48-hour–fasted mice by transmission electron microscopy. Ultrastructural analysis of hepatocyte mitochondria revealed grossly dysmorphic mitochondria in fasting G8KO mouse liver sections as well as an abundance of lipid droplets (Fig. 4A). Treatment-blinded morphometric quantification demonstrated similar mitochondrial cross-sectional area per HPF between genotypes (Fig. 4B). Mitochondrial cross-sectional analysis revealed a significant decrease in mitochondrial roundness scores in fasting G8KO mouse liver (P < 0.0001), consistent with the notable elongation of mitochondrial structures, whereas roundness was significantly increased in the fed state when compared with WT controls (P < 0.0001) (Fig. 4B). Because elongated mitochondria that form a cuplike structure around cytosol, organelles, or lipid droplets (e.g., mitochondrial “spheroids”) are a hallmark of mitochondrial damage or dysfunction (34, 35), we enumerated mitochondrial spheroid structures in fasted WT and G8KO mouse livers. Fasted G8KO livers exhibited massive mitochondrial spheroid accumulation when compared with WT mice (P < 0.0001) (Fig. 4A, white arrows; Fig. 4C).
Figure 4.
Mitochondrial dysmorphology and reduced hepatic oxidative function in fasting GLUT8-deficient mice. (A) Electron micrographs of livers of WT and G8KO fed or fasted mice with identification of mitochondrial spheroids (inset, white arrows). (B) Quantification of mitochondrial cross-sectional area and roundness from electron micrographs. (C) Enumeration of mitochondrial spheroids in electron micrographs from fasted mice. Komogorov-Smirnov test was used to analyze distribution of roundness scores. (D) qRT-PCR analysis of hepatic mitochondrial DNA-encoded mRNA for ETC proteins (n = 6 mice per group). A significant two-way analysis of variance diet-genotype interaction was detected for mt-Cytb (P < 0.05). (E) qRT-PCR analysis of skeletal muscle mitochondrial DNA-encoded mRNA for ETC proteins (WT fed, n = 4; G8KO fed, n = 4; WT fasted, n = 8; G8KO fasted, n = 8). (F) Quantification of oxygen consumption in liver homogenates from fed or 48-hour–fasted WT and G8KO mice using the Oroboros Oxygraph 2K. Respiration is reported as the state 3/state 4 respiratory control ratio and subjected to paired t test analysis (n = 4 to 6 mice per group). (G) Quantification of oxygen consumption in permeabilized soleus muscle fibers from fed or 48-hour–fasted WT and G8KO mice (n = 4 to 6 mice per group). Data are presented as mean ± standard error of the mean. *P < 0.05; ****P < 0.0001.
To further assess potential functional defects that may accompany the observed morphological changes, we quantified gene expression markers of mitochondrial mass, biogenesis, and respiratory function in livers of fed or 48-hour–fasted mice by qRT-PCR. Fasting GLUT8-deficient livers had significantly attenuated or decreased trends in expression of mitochondrial DNA-encoded mRNA transcripts for electron transport chain (ETC) subunits as compared with WT mice (mt-Cytb, P = 0.0201; mt-Co1, P = 0.2416; mt-Atp8, P = 0.1041) (Fig. 4D). However, no significant differences were observed in the genomic DNA–encoded mRNA transcripts for the ETC components Cox5b, Ndufs8, or Uqcrc1 between fed or fasted WT and G8KO mice (Supplemental Fig. 4A). Total mitochondrial biomass was similar in each group, as determined by the ratio of the mitochondrial d-loop and nuclear Actb DNA (Supplemental Fig. 4B). Similarly, mitochondrial biogenesis transcription factors Tfam and Nrf1 were similar between fed and fasted WT and G8KO mice (Supplemental Fig. 4C).
To assess organ-level mitochondrial function, we subjected liver homogenates from chow-fed or 48-hour–fasted WT and G8KO mice to high-resolution respirometry and quantified relative oxygen consumption rates. Although no significant differences in oxygen consumption were observed between fed WT and G8KO mouse liver homogenates, G8KO livers exhibited significantly impaired state 3/state 4 respiratory control ratios when compared with fasting WT controls (P = 0.0108) (Fig. 4F). These observed mitochondrial defects were selective for hepatic tissue, as demonstrated by similar oxygen consumption rates and mitochondrial DNA–encoded ETC mRNA transcripts in WT and G8KO skeletal muscle (Fig. 4E and 4G). Taken together, the data suggest that GLUT8 selectively mediates hepatic mitochondrial morphology and respiratory function during fasting, without effects on total mitochondrial biomass or biogenesis.
Cell autonomously enhanced fasting-induced PPARα signaling in GLUT8-deficient livers
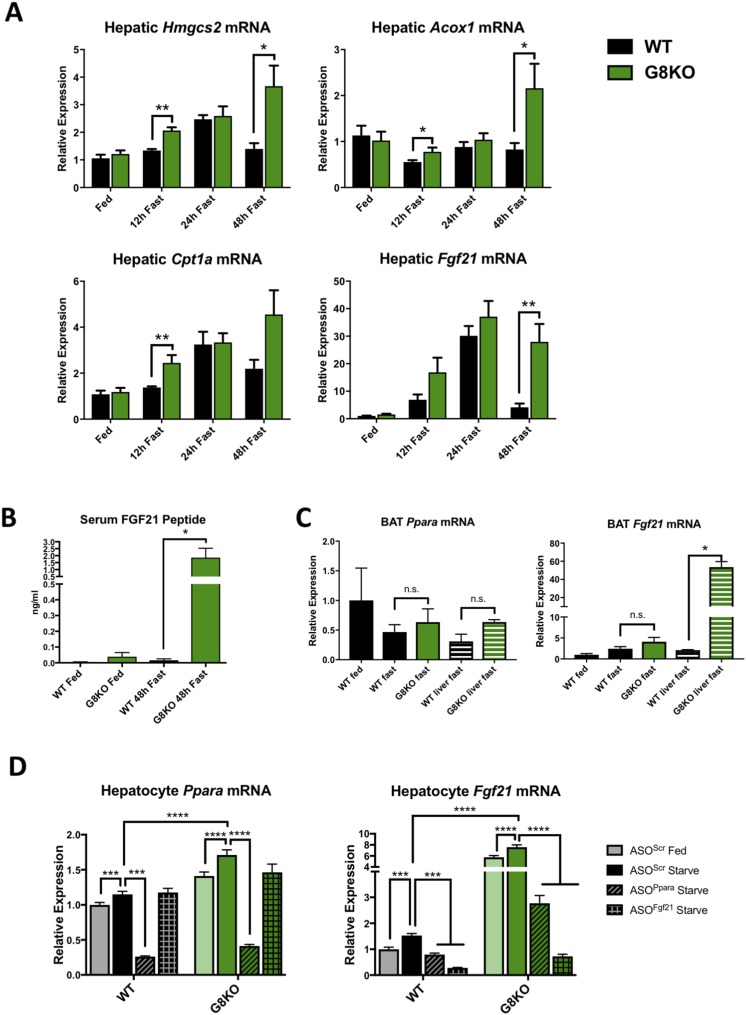
To investigate the interplay between hepatic and extrahepatic metabolic alterations observed in fasting G8KO mice, we assessed the activity of PPARα, a fasting-induced nuclear receptor that mediates hepatic fat oxidation in response to peripheral lipolysis during fasting (12, 18, 36). To that end, we quantified mRNA abundance of representative PPARα target genes (Hmgcs2, Acox1, Cpt1a, Fgf21) in livers from WT and G8KO mice fed a chow diet or fasted for 12, 24, or 48 hours. GLUT8-deficient mice exhibited significantly higher, or trends toward higher, hepatic transcript abundance for all PPARα targets at 12 hours (Hmgcs2, P = 0.0014; Acox1, P = 0.0423; Cpt1a, P = 0.0074; Fgf21, P = 0.0908) and 48 hours (Hmgcs2, P = 0.0243; Acox1, P = 0.0361; Cpt1a, P = 0.0687; Fgf21, P = 0.0011) of fasting (Fig. 5A). Given the marked increase in GLUT8-deficient expression of Fgf21 as compared with WT and given its role as a circulating hepatokine activated by both fasting and mitochondrial stress (37), we quantified circulating peptide levels of FGF21 via ELISA. After 48 hours of fasting, G8KO mice displayed higher serum FGF21 peptides levels when compared with WT (P = 0.0103) (Fig. 5B). Although circulating FGF21 is primarily liver derived, brown adipose tissue (BAT) can become a source of FGF21 secretion particularly in response to thermogenic activation (38). Thus, we quantified Fgf21 and Ppara mRNA transcripts in WT fed or 48-hour–fasted WT and G8KO BAT alongside hepatic tissue and determined that neither gene is significantly upregulated in BAT of fasting G8KO (Fig. 5C).
Figure 5.
Prolonged, PPARα-dependent activation of FGF21 and ketogenesis in G8KO mice. (A) qRT-PCR analysis of hepatic mRNA transcript levels of the PPARα targets Hmgcs2, Acox1, Cpt1a, and Fgf21 in WT and G8KO mice fed an ad libitum chow diet or fasted for 12, 24, and 48 hours (fed, n = 9 mice per genotype; 12-hour fast, n = 4 to 6 mice per genotype; 24-hour fast, n = 2 to 7 mice per genotype; 48-hour fast, n = 5 to 10 mice per genotype). (B) Serum FGF21 protein measurements in mice fed an ad libitum chow diet or fasted for 48 hours (n = 7 to 10 mice per group). A significant two-way analysis of variance diet-genotype interaction was detected (P < 0.05). (C) mRNA quantification of Ppara and Fgf21 in BAT from WT and G8KO fed or fasted mice, alongside WT and G8KO fasted liver tissue for comparison (WT fed, n = 2 mice per group; WT fast, n = 4 mice per group; G8KO fast, n = 5 mice per group; WT liver fast, n = 2 mice per group; G8KO liver fast, n = 2 mice per group). (D) qRT-PCR quantification of Ppara and Fgf21 in primary hepatocytes isolated from fed WT and G8KO mice. Cells were transfected with scrambled ASOs (ASOScr) or with ASOs targeting Ppara (ASOPpara) or Fgf21 (ASOFgf21) and treated with regular (fed) or starved media for 24 hours (n = 4 to 6 mice per group). Data are presented as mean ± standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. n.s., not significant.
Given that PPARα is activated by lipid ligands, at least in part derived from peripheral lipolysis, we next determined whether the elevated and prolonged activation of PPARα in GLUT8-deficient livers is a systemic or hepatocyte-autonomous phenotype. Primary cultured hepatocytes were obtained from either WT or G8KO mice and treated with either control or starvation media, along with transfection of ASOs targeting PPARα (ASOPpara), FGF21 (ASOFgf21), or a scrambled sequence (ASOScr). Both Ppara and Fgf21 were significantly elevated in starved G8KO hepatocytes as compared with WT, suggesting a cell-autonomous regulation of these two genes by GLUT8 (Ppara, P < 0.0001; Fgf21, P < 0.0001). Knockdown of PPARα resulted in a reduction of FGF21 but not vice versa, supporting the canonical PPARα-to-FGF21 pathway as the mechanism by which GLUT8 regulates FGF21 (Fig. 5D).
Reduced mitochondrial oxidative function in GLUT8-deficient hepatocytes is cell autonomous
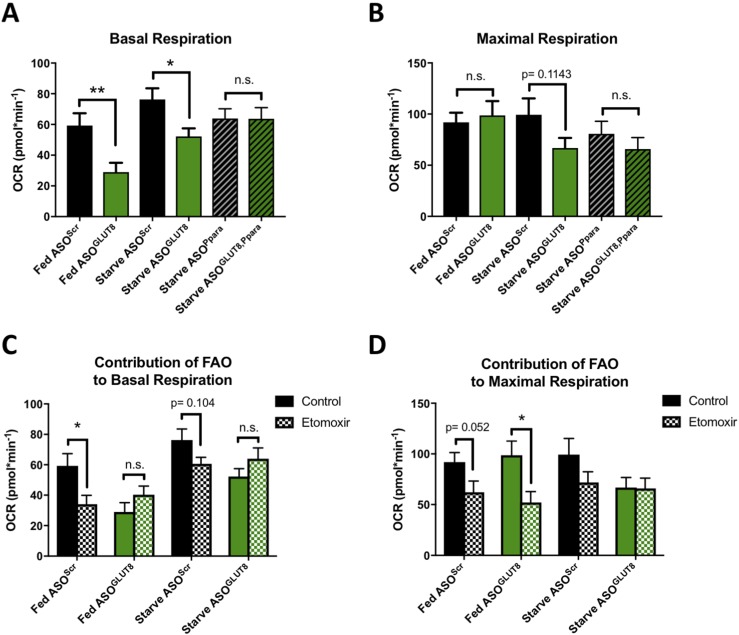
Given the observed hepatic mitochondrial impairments in vivo and the cell-autonomous role of GLUT8 in regulating the pro-oxidative transcription factor PPARα under fasted conditions, we investigated the cell-autonomous role of GLUT8 in mitochondrial oxidative function using the Seahorse XF Analyzer. AML12 immortalized mouse hepatocytes were used in lieu of primary hepatocytes due to their increased plate adherence and confluency, which is necessary for this assay. We achieved at least 60% to 75% knockdown of GLUT8 or PPARα with ASOs (Supplemental Fig. 5A), treated cells with fed or starvation media, and then subjected cells to the Mito Stress Test to assess relative respiration rates. Basal respiration was significantly lower in GLUT8 knockdown cells as compared with controls in both fed (P = 0.007) and starved (P = 0.0127) conditions (Fig. 6A). There was no significant difference in maximal respiration between scrambled control and GLUT8 knockdown under fed conditions; however, GLUT8 knockdown cells under starved conditions displayed a trend toward reduced maximal respiration as compared with scrambled controls (P = 0.1143) (Fig. 6B). GLUT8 knockdown had no effect on basal or maximal respiration in the context of PPARα deficiency (Fig. 6A and 6B). The basal extracellular acidification rate, a value that generally reflects cellular glycolytic rates, was also significantly decreased in fed GLUT8-deficient hepatocytes (P = 0.0111) and trended toward decreased in starved GLUT8-deficient hepatocytes (P = 0.0527) (Supplemental Fig. 5B).
Figure 6.
Cell-autonomous respiratory impairment in GLUT8-deficient hepatocytes. (A) Basal oxygen consumption ratio (OCR) of AML12 mouse hepatocytes with or without treatment with ASOs targeting GLUT8 (ASOGLUT8) or PPARα (ASOPpara) and cultured in fed or starved media. (B) Maximal OCR of cells as in (A). (C) Basal OCR of control [scrambled ASOs (ASOScr)] or GLUT8 knockdown (ASOGLUT8) AML12 mouse hepatocytes cultured in fed or starved media in the presence or absence of etomoxir to assess the contribution of FAO to each measurement. (D) Maximal OCR of cells as in (C) (fed ASOScr, n = 29 to 31; fed ASOGLUT8, n = 20 to 26; starved ASOScr, n = 30; starved ASOGLUT8, n = 25 to 27; starved ASOPpara, n = 33; starved ASOGLUT8, Ppara, n = 34; fed ASOScr + etomoxir, n = 21; fed ASOGLUT8 + etomoxir, n = 20; starved ASOScr + etomoxir, n = 21; starved ASOGLUT8 + etomoxir, n = 23). Data are presented as mean ± standard error of the mean. *P < 0.05; **P < 0.01. FAO, fatty-acid oxidation; n.s., not significant.
To assess the contribution of fatty-acid oxidation to each respiration measurement, fed and starved WT and GLUT8-deficient hepatocytes were treated with etomoxir, a Cpt-1 inhibitor that blocks fatty-acid oxidation (39, 40). As compared with untreated cells, etomoxir significantly reduced basal respiration in fed ASOScr cells (P = 0.0239) and trended toward decreasing basal respiration in starved ASOScr cells (P = 0.104) (Fig. 6C). In contrast, etomoxir did not reduce basal respiration under either the fed or starved conditions in ASOGLUT8 hepatocytes (Fig. 6C). Although ASOGLUT8 hepatocytes displayed significantly reduced maximal respiration upon treatment with etomoxir in the fed condition, the already low maximal respiration observed in the starved condition was not further decreased by etomoxir (Fig. 6D).
Given the observed hepatocyte-autonomous reduction in GLUT8-deficient mitochondrial respiration and the in vivo morphological changes observed in hepatic mitochondria, we assessed mitochondrial morphology of in vitro control or GLUT8-deficient AML12 hepatocytes cultured in fed or starved media. Despite a functional respiration difference, no obvious morphological differences were observed between groups (Supplemental Fig. 5C).
PPARα deficiency restores enhanced ketogenesis but does not suppress thermogenesis in G8KO mice
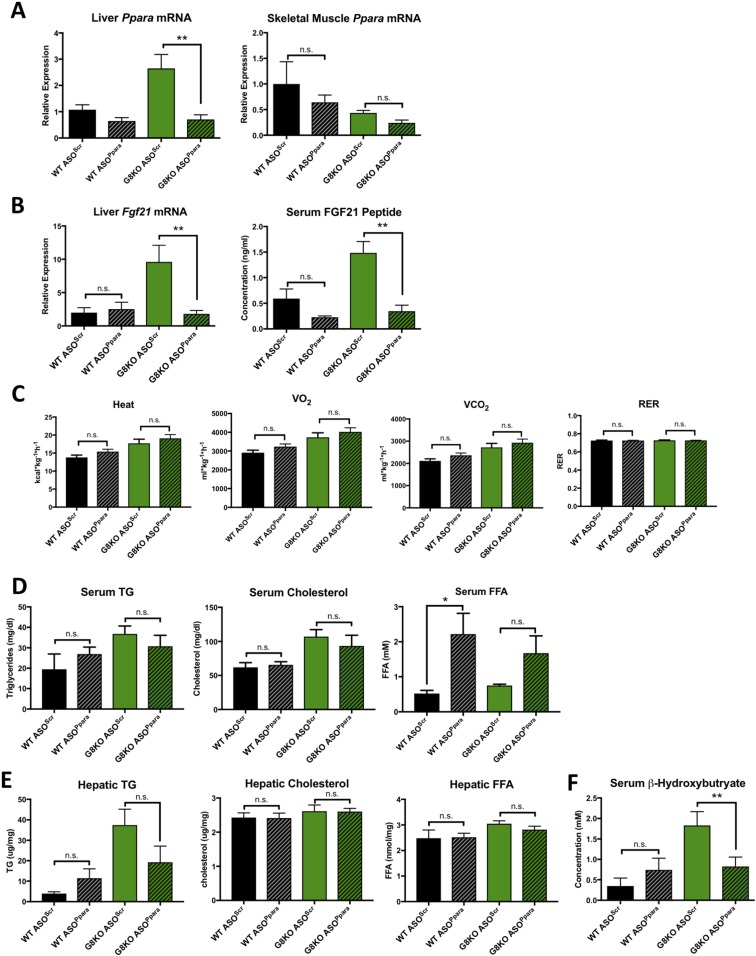
To directly test the role of PPARα in GLUT8-mediated regulation of the hepatic fasting response and FGF21 in vivo, we knocked down hepatic PPARα in WT and G8KO mice via ASOs and subjected mice to 48 hours of fasting. qRT-PCR quantification demonstrated that treatment with ASOPpara significantly reduced hepatic Ppara mRNA in fasted G8KO mice as compared with mice treated with ASOScr control (P = 0.0054) (Fig. 7A). Skeletal muscle Ppara mRNA levels were not significantly affected by ASOPpara administration (Fig. 7A). Ppara knockdown also blocked the elevation of hepatic Fgf21 mRNA (P = 0.0071) and circulating FGF21 peptide (P = 0.0023) in G8KO mice (Fig. 7B).
Figure 7.
PPARα mediaes elevated FGF21, serum ketone, and TG levels but not thermogenesis in G8KO mice. (A) qRT-PCR quantification of liver and skeletal muscle Ppara mRNA levels in 48-hour–fasted WT and G8KO mice treated with scrambled ASOs (ASOScr) or ASOs targeting Ppara (ASOPpara) (n = 3 to 6 mice per group). A significant two-way analysis of variance (ANOVA) diet-genotype interaction was detected in liver (P < 0.01). (B) Quantification of hepatic mRNA levels (left) and circulating peptide levels (right) of FGF21 in mice as in (A) (n = 3 to 6 mice per group). A significant two-way ANOVA diet-genotype interaction was detected for mRNA (P < 0.01) and peptide (P < 0.05). (C) Indirect calorimetry measurements of mice as in (A) (n = 4 to 7 mice per group). (D) Serum quantification of TGs, cholesterol, and FFAs in mice as in (A) (n = 5 to 8 mice per group). (E) Quantification of hepatic TGs, cholesterol, and FFAs in mice as in (A) (n = 5 to 8 mice per group). A significant two-way ANOVA diet-genotype interaction was detected (P < 0.05). (F) Serum β-hydroxybutyrate levels in mice as in (A) (n = 3 to 5 mice per group). A significant two-way ANOVA diet-genotype interaction was detected (P < 0.05). Data are presented as mean ± standard error of the mean. *P < 0.05; **P < 0.01. n.s., not significant.
Given the previously demonstrated role for PPARα and FGF21 in the regulation of thermogenesis (12, 41), we measured heat production and respiration in WT and G8KO fasted mice administered ASOPpara or ASOScr. Hepatic PPARα knockdown did not reverse the hyperthermic phenotype observed in fasted G8KO mice (Fig. 7C). We next assessed serum lipids and found that ASOPpara treatment did not significantly alter serum TG, cholesterol, or FFA levels in either genotype, with the exception of increased serum FFAs in ASOPpara-treated WT mice as compared with controls (P = 0.0199) (Fig. 7D). PPARα knockdown did not affect hepatic TG, cholesterol, or FFAs (Fig. 7E). Knockdown of PPARα blocked the elevation of circulating β-hydroxybutyrate in fasting G8KO mice (P = 0.0427) (Fig. 7F). We also assessed hepatic mitochondrial morphology, a known regulator of mitochondrial function and biogenesis, via electron microscopy. PPARα knockdown did not alter total mitochondrial cross-sectional area (Supplemental Fig. 6B). GLUT8-deficient and PPARα-deficient livers exhibited elongated mitochondrial structures with significantly decreased roundness scores (Supplemental Fig. 6A and 6B). Together, these data demonstrate that GLUT8 modulates the hepatic fasting response by coordinately regulating ketogenesis, lipid homeostasis, and FGF21 production via PPARα.
Discussion
The adaptive hepatic fasting response has become a major focus of investigation for its broad potential therapeutic applications (35, 42). PPARα is a central signaling molecule that acts as part of a coordinated fasting response network. Here, we demonstrate that genetically ablating the GLUT8 transporter augments the magnitude and duration of hepatocyte PPARα signaling. In comparison with GLUT2, which has received attention as a component of hepatic glucose sensing and glucose homeostasis (9, 43, 44), we demonstrate selective GLUT8 transcriptional regulation during adaptive fasting.
Studies using genetic and pharmacological manipulation of PPARα have established its role in promoting hepatic lipid oxidation and ketogenesis, at least in part through an FGF21-dependent mechanism (45). Consistent with this pathway, we observed PPARα-dependent upregulation of ketogenesis and FGF21 production in G8KO mice. In contrast, previous work by others also highlights the prothermogenic role of PPARα-FGF21 signaling axis. This was supported by data that genetic ablation of either PPARα or FGF21 reduced thermogenesis in rodent models (12, 41). Although G8KO mice exhibited both elevated fasting-induced FGF21 and heat production, hepatic PPARα knockdown blocked FGF21 upregulation without altering thermogenesis. This suggests a distinct, broadly networked mechanism by which GLUT8 regulates hepatokine production, ketogenesis, and thermogenic activity in parallel with and downstream of PPARα.
The present data demonstrating increased thermogenesis and VO2 consumption in G8KO mice is consistent with results from our previous studies of GLUT8-deficient mice both at baseline and after being fed a high-fructose diet. Increased thermogenesis was associated with elevated PPARγ levels and resistance to fructose-induced hepatic steatosis (5). Consistent with increased feeding, fasting, and fructose-fed thermogenesis in G8KO mice, the hepatic GLUT inhibitor trehalose increased whole body thermogenesis through canonical hepatic fasting response pathways, including the transcription factor EB and peroxisome proliferator–activated receptor γ coactivator 1-α (PGC1α) (26). In light of the profound evidence that PGC1α can act as a cofactor for both PPARα and PPARγ (46), we hypothesize that hepatic PGC1α is a master regulatory node downstream of pseudostarvation after GLUT blockade or deletion. Therefore, both genetic and pharmacological models of hepatic GLUT inhibition recapitulate prothermogenic effects, which suggests that hepatic glucose transport may be an attractive therapeutic target to treat diseases of energetic excess (e.g., diabetes, obesity, nonalcoholic fatty liver disease).
Our current findings therefore mandate a detailed mechanistic characterization of GLUT8-PPARα regulation. Here, we observe dysregulated mitochondrial homeostasis and lipid mobilization/oxidation in association with upregulated PPARα. Indeed, we observed markers of mitochondrial dysfunction and stress in GLUT8-deficient fasted livers and in AML12 mouse hepatocytes. GLUT8-deficient livers exhibited an accumulation of lipid droplets and mitochondrial spheroids upon fasting, a hallmark of hepatic injury or autophagic deficiency in other models (34, 35). Despite similar total mitochondrial mass between genotypes, GLUT8-deficient mice exhibited hepatic mitochondrial respiration deficiencies and decreased mitochondrial DNA–encoded mRNA transcripts for ETC proteins. Respiratory abnormalities were absent in skeletal muscle, supporting a tissue-selective role for GLUT8 in the hepatic fasting response.
Work from other groups has shown that impairments in hepatic fatty-acid oxidation result in a strikingly similar phenotype of elevated and sustained PPARα activity. PPARα is a fasting-induced nuclear receptor activated by lipid ligands derived from peripheral lipolysis (18, 36, 47). Mice deficient in the rate-limiting enzyme for fatty-acid oxidation, Acox1, display a similar phenotype of sustained PPARα activity due to the accumulation of unmetabolized lipid ligands (48). Furthermore, mice deficient in hepatic CPT2, an essential enzyme for mitochondrial long-chain fatty-acid oxidation, exhibit fasting-induced hepatic steatosis, serum dyslipidemia, and compensatory upregulation of PPARα target genes (49). Given the robust accumulation of hepatic and serum lipids we observe in fasting G8KO mice, an intriguing mechanism by which GLUT8 deficiency results in prolonged fasting-induced PPARα activity is via indirectly elevating hepatic lipid pools in the presence of oxidation-impaired mitochondria.
Numerous studies demonstrate the roles of hepatic PPARα and FGF21 in promoting mitochondrial biogenesis and catabolic processes such as oxidation (14, 18, 50). The hepatocyte-autonomous impairment in mitochondrial respiration and lipid accumulation in GLUT8-deficient models, despite elevated FGF21 and PPARα activity, supports the hypothesis that GLUT8 may serve a fundamental role in mitochondrial homeostasis that is not superseded by compensatory metabolic signals.
Several pieces of evidence indicate an essential role for hepatic GLUT8 in mitochondrial metabolism and lipid oxidation. In addition to the hepatic respiratory impairment observed in hepatic tissue from fasted G8KO mice, we demonstrated a consistent reduction in both basal and maximal respiration in an in vitro hepatocyte model of GLUT8 deficiency. Additionally, the contribution of fatty-acid oxidation to total cellular respiration was lower in GLUT8-deficient cells as compared with controls. These findings are consistent with hepatic lipid oxidative impairments contributing at least in part to the fasting steatosis and hyperlipidemia phenotypes. In vitro knockdown of PPARα resulted in trends toward reduced respiration and prevented further reduction in respiration with the additional knockdown of GLUT8. In vivo, hepatic PPARα knockdown also resulted in morphological changes in mitochondria that were similar to changes observed in GLUT8 deficiency. Although determining the precise interaction between GLUT8 and PPARα signaling will require additional investigation, these results suggest that PPARα is a principal effector downstream of GLUT8-mediated fasting responses to promote oxidative metabolism and mitochondrial function. Additionally, given the evidence that many of the downstream effects of PPARα on lipid utilization and ketogenesis are mediated in part via FGF21 (45), it will be important to clarify the role of FGF21 in GLUT8-mediated effects of fasting.
Despite evidence that GLUT8 cell autonomously regulates mitochondrial oxidative metabolism, we cannot rule out important roles for extrahepatic GLUT8 in fasting metabolism. Restoration of hepatic GLUT8 expression in G8KO mice only partially rescued fasting serum and hepatic lipid levels. This suggests that the altered lipid homeostasis in the absence of GLUT8 results from both a deficiency in hepatic lipid oxidation and altered peripheral lipid metabolism. Although we did not observe alterations in adipose-intrinsic lipolysis rates in GLUT8-deficient tissue, GLUT8 may affect lipolysis via indirect hormonal signaling. Future studies investigating extrahepatic GLUT8 will thus be important in fully understanding the physiological fasting response.
GLUT8 blockade thus modulates the adaptive hepatic fasting response in part by hyperactivating PPARα. Fasting and fasting-regulated metabolic pathways have been implicated as therapeutic targets for a number of disease states, but ongoing work is needed to fully understand the molecular underpinnings of these adaptive responses as well as critical nodes that can be leveraged for pharmacological purposes. Although inhibition of other GLUT homologs [e.g., GLUT1 and GLUT4 in particular (51, 52)] produces detrimental effects on whole-body glucose homeostasis and insulin sensitivity, we anticipate that hepatic GLUT8 is a candidate target transporter for its role as a regulator of the hepatic fasting response and may be leveraged to mitigate metabolic disease.
Supplementary Material
Acknowledgments
We thank Kelle Moley (Washington University School of Medicine) for helpful feedback, Mark Graham (Ionis Pharmaceuticals, Carlsbad, CA) for providing antisense oligonucleotides, and Laura Luecking (Washington University School of Medicine) for help with Seahorse experiments.
Financial Support: This work was supported by Washington University Child Health Research Center Grant K12HD076224 (B.J.D), Children's Discovery Institute Grant MI-FR-2014-426 (B.J.D.), the Robert Wood Johnson Foundation (B.J.D.), an AGA-Gilead Sciences Research Scholar Award in Liver Disease (B.J.D.), Washington University Adiopocyte Biology and Molecular Nutrition Core and Nutrition Obesity Research Center Grant P30DK056341 (B.J.D.), Washington University Digestive Disease Research Core Center Grant P30DK52574 (B.J.D.), and Washington University Diabetes Research Center Grant P30 DK020579 (B.J.D.). A.L.M. was supported by a Washington University Spencer T. Olin Fellowship, by Washington University National Institute of General Medical Sciences Institutional Training Grant in Cell and Molecular Biosciences Grant T32GM007067, and by National Science Foundation Graduate Research Fellowship Program Grant DGE-1143954.
Author Contributions: A.L.M. and B.J.D. designed the experiments and wrote the manuscript. A.L.M., Y.Z., E.H.F., C.B.H., O.A., T.A.P., W.L.B., and B.J.D. performed the experiments. A.L.M., Y.Z., E.H.F., T.A.P., and B.J.D. analyzed the data.
Disclosure Summary:
The authors have nothing to disclose.
Glossary
Abbreviations:
- AML12
α mouse liver 12
- ASO
antisense oligonucleotide
- BAT
brown adipose tissue
- ELISA
enzyme-linked immunosorbent assay
- ETC
electron transport chain
- FFA
free fatty acid
- FGF
fibroblast growth factor
- GLUT8
glucose transporter 8
- HPF
high-powered field
- mRNA
messenger RNA
- PGC1α
peroxisome proliferator–activated receptor γ coactivator 1-α
- PPARα
peroxisome proliferator–activated receptor α
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RER
respiratory exchange ratio
- RRID
Research Resource Identifier
- TG
triglyceride
- WT
wild-type
References
- 1. Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19(2):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crandall JP, Barzilai N. Exploring the promise of resveratrol: where do we go from here? Diabetes. 2013;62(4):1022–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5(6):493–506. [DOI] [PubMed] [Google Scholar]
- 4. Mardones P, Rubinsztein DC, Hetz C. Mystery solved: trehalose kickstarts autophagy by blocking glucose transport. Sci Signal. 2016;9(416):1–4. [DOI] [PubMed] [Google Scholar]
- 5. DeBosch BJ, Chen Z, Finck BN, Chi M, Moley KH. Glucose transporter-8 (GLUT8) mediates glucose intolerance and dyslipidemia in high-fructose diet-fed male mice. Mol Endocrinol. 2013;27(11):1887–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Debosch BJ, Chen Z, Saben JL, Finck BN, Moley KH. Glucose transporter 8 (GLUT8) mediates fructose-induced de novo lipogenesis and macrosteatosis. J Biol Chem. 2014;289(16):10989–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer AL, Higgins CB, Heitmeier MR, Kraft TE, Qian X, Crowley JR, Hyrc KL, Beatty WL, Yarasheski KE, Hruz PW, DeBosch BJ. SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy. Sci Rep. 2016;6(December):38586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DeBosch BJ, Heitmeier MR, Mayer AL, Higgins CB, Crowley JR, Kraft TE, Chi M, Newberry EP, Chen Z, Finck BN, Davidson NO, Yarasheski KE, Hruz PW, Moley KH. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016;9(416):ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seyer P, Vallois D, Poitry-Yamate C, Schütz F, Metref S, Tarussio D, Maechler P, Staels B, Lanz B, Grueter R, Decaris J, Turner S, da Costa A, Preitner F, Minehira K, Foretz M, Thorens B. Hepatic glucose sensing is required to preserve β cell glucose competence. J Clin Invest. 2013;123(4):1662–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell RC, Yuan H-X, Guan K-L. Autophagy regulation by nutrient signaling. Cell Res. 2014;24(1):42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator–activated receptor α mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, Burgess SC. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci USA. 2009;106(26):10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5(6):415–425. [DOI] [PubMed] [Google Scholar]
- 15. Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, Shanafelt AB. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115(6):1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu J, Lloyd DJ, Hale C, Stanislaus S, Chen M, Sivits G, Vonderfecht S, Hecht R, Li Y, Lindberg RA, Chen J, Jung DY, Zhang Z, Ko HJ, Kim JK, Veniant MM. FGF21 reverses hepatic steatosis, increases energy expenditure and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58(January):250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coskun T, Bina HA, Schneider MA, Dunbar JD, Hu CC, Chen Y, Moller DE, Kharitonenkov A. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149(12):6018–6027. [DOI] [PubMed] [Google Scholar]
- 18. Lundåsen T, Hunt MC, Nilsson L-M, Sanyal S, Angelin B, Alexson SE, Rudling M. PPARalpha is a key regulator of hepatic FGF21. Biochem Biophys Res Commun. 2007;360(2):437–440. [DOI] [PubMed] [Google Scholar]
- 19. Liang Q, Zhong L, Zhang J, Wang Y, Bornstein SR, Triggle CR, Ding H, Lam KS, Xu A. FGF21 maintains glucose homeostasis by mediating the cross talk between liver and brain during prolonged fasting. Diabetes. 2014;63(12):4064–4075. [DOI] [PubMed] [Google Scholar]
- 20. Markan KR, Naber MC, Ameka MK, Anderegg MD, Mangelsdorf DJ, Kliewer SA, Mohammadi M, Potthoff MJ. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes. 2014;63(12):4057–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hotta Y, Nakamura H, Konishi M, Murata Y, Takagi H, Matsumura S, Inoue K, Fushiki T, Itoh N. Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150(10):4625–4633. [DOI] [PubMed] [Google Scholar]
- 22. Hondares E, Rosell M, Gonzalez FJ, Giralt M, Iglesias R, Villarroya F. Hepatic FGF21 expression is induced at birth via PPARalpha in response to milk intake and contributes to thermogenic activation of neonatal brown fat. Cell Metab. 2010;11(3):206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37(5):907–925. [PubMed] [Google Scholar]
- 24. Kliewer SA, Mangelsdorf DJ. Fibroblast growth factor 21: from pharmacology to physiology. Am J Clin Nutr. 2010;91(1, Suppl):254S–257S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Füllgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015–1034. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Higgins CB, Mayer AL, Mysorekar IU, Razani BB, Graham MJ, Hruz PW, DeBosch BJ. Transcription factor EB (TFEB)-dependent induction of thermogenesis by the hepatocyte solute carrier 2A (SLC2A) inhibitor, Trehalose. Autophagy. 2018. [DOI] [PMC free article] [PubMed]
- 27. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeBosch BJ, Kluth O, Fujiwara H, Schürmann A, Moley K. Early-onset metabolic syndrome in mice lacking the intestinal uric acid transporter SLC2A9. Nat Commun. 2014;5:4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris L-ALS, Skinner JR, Shew TM, Pietka TA, Abumrad NA, Wolins NE. Perilipin 5-driven lipid droplet accumulation in skeletal muscle stimulates the expression of fibroblast growth factor 21. Diabetes. 2015;64(8):2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carswell KA, Lee MJ, Fried SK. Culture of isolated human adipocytes and isolated adipose tissue. Methods Mol Biol. 2012;806:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soufi N, Hall AM, Chen Z, Yoshino J, Collier SL, Mathews JC, Brunt EM, Albert CJ, Graham MJ, Ford DA, Finck BN. Inhibiting monoacylglycerol acyltransferase 1 ameliorates hepatic metabolic abnormalities but not inflammation and injury in mice. J Biol Chem. 2014;289(43):30177–30188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hall AM, Soufi N, Chambers KT, Chen Z, Schweitzer GG, McCommis KS, Erion DM, Graham MJ, Su X, Finck BN. Abrogating monoacylglycerol acyltransferase activity in liver improves glucose tolerance and hepatic insulin signaling in obese mice. Diabetes. 2014;63(7):2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wierzbicki AS, Viljoen A. Anti-sense oligonucleotide therapies for the treatment of hyperlipidaemia. Expert Opin Biol Ther. 2016;16(9):1125–1134. [DOI] [PubMed] [Google Scholar]
- 34. Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem. 2012;287(50):42379–42388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2014;11(3):187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montagner A, Polizzi A, Fouché E, Ducheix S, Lippi Y, Lasserre F, Barquissau V, Régnier M, Lukowicz C, Benhamed F, Iroz A, Bertrand-Michel J, Al Saati T, Cano P, Mselli-Lakhal L, Mithieux G, Rajas F, Lagarrigue S, Pineau T, Loiseau N, Postic C, Langin D, Wahli W, Guillou H. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016:65(7):1202–1214. [DOI] [PMC free article] [PubMed]
- 37. Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19(1):83–92. [DOI] [PubMed] [Google Scholar]
- 38. Hondares E, Iglesias R, Giralt A, Gonzalez FJ, Giralt M, Mampel T, Villarroya F. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. J Biol Chem. 2011;286(15):12983–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beeson CC, Beeson GC, Buff H, Eldridge J, Zhang A, Seth A, Demcheva M, Vournakis JN, Muise-Helmericks RC. Integrin-dependent Akt1 activation regulates PGC-1 expression and fatty acid oxidation. J Vasc Res. 2012;49(2):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clara R, Langhans W, Mansouri A. Oleic acid stimulates glucagon-like peptide-1 release from enteroendocrine cells by modulating cell respiration and glycolysis. Metabolism. 2016;65(3):8–17. [DOI] [PubMed] [Google Scholar]
- 41. Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, Spiegelman BM. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26(3):271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jaishy B, Abel ED. Lipids, lysosomes, and autophagy. J Lipid Res. 2016;57(9):1619–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leturque A, Brot-Laroche E, Le Gall M. GLUT2 mutations, translocation, and receptor function in diet sugar managing. Am J Physiol Endocrinol Metab. 2009;296(5):E985–E992. [DOI] [PubMed] [Google Scholar]
- 44. Guillemain G, Loizeau M, Pinçon-Raymond M, Girard J, Leturque A. The large intracytoplasmic loop of the glucose transporter GLUT2 is involved in glucose signaling in hepatic cells. J Cell Sci. 2000;113(Pt 5):841–847. [DOI] [PubMed] [Google Scholar]
- 45. Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS. Hepatic fibroblast growth factor 21 is regulated by PPAR α and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007:5(6):426–437. [DOI] [PubMed] [Google Scholar]
- 46. Wu H, Deng X, Shi Y, Su Y, Wei J, Duan H. PGC-1α, glucose metabolism and type 2 diabetes mellitus. J Endocrinol. 2016;229(3):R99–R115. [DOI] [PubMed] [Google Scholar]
- 47. Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138(3):476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan CY, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase: implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem. 1998;273(25):15639–15645. [DOI] [PubMed] [Google Scholar]
- 49. Lee J, Choi J, Alpergin ESS, Zhao L, Hartung T, Scafidi S, Riddle RC, Wolfgang MJ.et al. . Loss of hepatic mitochondrial long chain fatty acid oxidation confers resistance to diet-induced obesity and glucose intolerance. Cell Rep. 2017;20(3):655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Yu RT, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148(3):556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thorens B, Mueckler M. Glucose transporters in the 21st Century. Am J Physiol Endocrinol Metab. 2010;298(2):E141–E145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34(2-3):121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.