Abstract
This work, describes for the first time, a simple biosensing design to yield an ultrasensitive electrochemical biosensor for a cancer biomarker detection, miRNA-155, with linear response down to the attomolar range. MiRNA-155 was selected for being overexpressed in breast cancer.
The biosensor was assembled in two stages: (1) the immobilization of the anti-miRNA-155 that was thiol modified on an Au-screen printed electrode (Au-SPE), followed by (2) blocking the areas of non-specific binding with mercaptosuccinic acid. Atomic force microscopy (AFM) and electrochemical techniques including cyclic voltammetry (CV), impedance spectroscopy (EIS) and square wave voltammetry (SWV) confirmed the surface modification of these devices and their ability to hybridize successfully and stably with miRNA-155.
The final biosensor provided a sensitive detection of miRNA-155 from 10 aM to 1.0 nM with a low detection limit (LOD) of 5.7 aM in real human serum samples. Good results were obtained in terms of selectivity towards breast cancer antigen CA-15.3 and bovine serum albumin (BSA). Raw fluid extracts from cell-lines of melanoma did not affect the biosensor response (no significant change of the blank), while raw extracts from breast cancer yielded a positive signal against miRNA-155.
This simple and sensitive strategy is a promising alternative for simultaneous quantitative analysis of multiple miRNA in physiological fluids for biomedical research and point-of-care (POC) diagnosis.
Keywords: Breast cancer, Anti-miRNA-155, miRNA-155, Biosensors, Eletrochemistry
1. Introduction
Cancer is a public health concern worldwide. One in three women and one in two men will develop cancer during lifetime in developed countries (Siegel et al., 2012). Early detection is the only known approach that may improve these indicators. Traditional diagnostic tools for breast cancer detection include: i) clinical and physical examinations, ii) imaging mammography, iv) ultrasound magnetic resonance imaging and v) histopathology. Clinical and physical examination have been shown insufficient, driving patients into mammography or ultrasound studies. In turn, mammography has limited sensitivity, yielding a high rate of false positive results. This approach may also lead to an accumulated exposure to radiation, which is considered an additional and significant risk factor (Yahalom et al., 2013). Ultrasound is a non-invasive and safe tool, but it cannot replace mammograms, especially in women above 40, and is unable to screen many of the cancers. Histopathology is an invasive approach, of interest when the disease is installed.
Thus, there is urgent need to develop non-invasive, simple and low risk methods for screening/diagnosing breast cancer. Considering the high prevalence of cancer diseases and the high frequency of analysis to be performed in clinical context, there is a strong desire that such tests are made available in POC point-of-care (POC) (Riaz et al., 2013; Grieshaber et al., 2008). Currently, strong efforts are being developed to monitor serum biomarkers as early detection of cancer (Mirabelli & Incoronato, 2013; Chung et al., 2014).
In breast cancer, several serum markers may be used in clinical practice, including carcinoembryonic antigen (CEA), carbohydrate antigen 15-3 (CA 15-3), circulating cytokeratins, such as tissue polypeptide antigen (TPA), tissue polypeptide specific antigen (TPS) and cytokeratin 19 fragment (CIFRA-21-1), and the proteolytically cleaved ectodomain of the human epidermal growth factor receptor 2 (s-HER2). The most widely used serum biomarker is CA 15-3, however, due to its lack of sensitivity at an early stage of the disease, it cannot be used exclusively. In addition, this marker can lead to false results after initiation of treatment without clinical correlation. Also, other biomarkers like TPA and CEA are less sensitive than CA-15.3 (Sun et al., 2012).
But recent technological advances have led to the identification of a new class of biomarkers named miRNAs that may lead to a novel strategy in early cancer screening (He et al., 2015; Volinia et al., 2012). MiRNAs contain 18-24 nucleotides in length (Johnson & Mutharasan, 2014) and play an essential role in biological processes such as development, cellular proliferation, apoptosis and response to stress, and tumorigenesis. Aberrant expression levels of miRNAs have been observed in many solid tumours, including breast cancer (Fu et al., 2011), accounting their dysregulated expression in cancer. In addition, miRNAs are stable in the blood and have high/low expressions that can be correlated to chemo-resistance, (Kong et al., 2010; Tavallaie et al., 2015).
In breast cancer, several studies have supported an abnormal expression of miRNA-155 (MIR-155) in patients with the disease (Mattiske et al., 2012). The overexpression of MIR-155 was considered as a breast cancer risk factor (Zeng et al., 2014), being associated with clinical-pathological markers, tumour subtype, poor survival rates, metastasis events and invasive properties of breast cancer, as well as high tumour grade, advanced stage and lymph node metastasis (Mattiske et al., 2012). MIR-155 is involved in controlling several mechanisms of cell survival, cell growth, radio/chemo-resistance (Liu et al., 2015; Mattiske et al., 2012), inhibiting target genes such as FOXO3A, RhoA, and SOCS1. The use of miRNA-155 as a potential biomarker in breast cancer opens the possibility towards a simple serological test for prognosis/diagnosis and follow-up of breast cancer under therapy.
Biosensors are today a successful route towards POC testing, allowing fast results and a direct sample reading, without the need for transporting samples into the laboratory (Campuzano et al., 2014; Labib and Berezovski, 2015; Bohunicky and Mousa, 2011). Devices relying in electrochemical transduction are currently among the attractive biosensors reaching portability, accounting their simplicity, sensitivity, low cost, small size, rapidity of response, ease of use, and possibility of reading samples directly over a wide range of concentrations (Xia and Zhang, 2014). In general, these features of electrochemical biosensors depend of the recognition element immobilized on the transducer, which is responsible for interacting with the analyte in a selective mode (Choi et al., 2010; Grieshaber et al., 2008).
In electrochemical miRNA detection, the recognition elements are mostly linear oligonucleotide strands of DNA/RNA nature with which the miRNA target hybridizes (Catuogno et al., 2011; Johnson & Mutharasan, 2014; Tavallaie, De Almeida, & Gooding, 2015). The transduction event measures the changes in the electrode or interfacial properties occurring upon hybridization, by means of electrochemically-active reporter species, including small redox molecules (such as ferric species, guanine oxidation) or enzyme-substrate pairs (such as horseradish peroxidase/hydrogen peroxide) (Johnson & Mutharasan, 2014), reaching LODs typically ranging fento-picomolar levels. Concurrent methods are mostly devoted to biosensors using other transduction modes (optical or electromechanical) or molecular-based approaches, including polymerase amplification, microarray (coupled to amplification approaches), spectroscopy, sequencing, cloning, and Northern blotting (Johnson & Mutharasan, 2014). Biosensors with other modes of transduction are typically linked to higher detection levels and molecular based approaches involve complex operations that are either expensive or far from POC requirements.
Regarding the specific electrochemical determination of miRNA-155, there is a limited number of works reported in the literature. The first one was presented by Wu et al., 2013, employing a conductive composition of Nafion, thionine (Thi) and Pd nanoparticles, displaying electrocatalytic activity for H2O2, to provide an LOD of ~1900 fM. Zhu et al., 2014, combined high base-mismatch selectivity of ligase chain reaction with reporting probes labeled with two different quantum dots. This elegant and complex approach had an LOD ranging from 12 to 31 fM. Later, Wu et al., 2015, presented another biosensor based on catalyzed hairpin assembly target recycling and cascade enzymatic electrocatalysis for signal amplification, in a quite complex approach, to yield an LOD of 0.35 fM. Hu et al., 2015, presented a horseradish peroxidase and graphene quantum dots combination, this time giving rise to a 0.14 fM LOD. Very recently, Azimzadeh et al., 2015, proposed the modification of a glassy carbon electrode surface by a thiolated probe-functionalized with gold nanorods and decorated on the graphene oxide material, having as reporter label Oracet Blue. The LOD was however higher than those already reported, equal to 0.6 fM. Overall, the previous attempts to produce a sensitive device for miRNA detection have been effective but quite complex in terms of biosensor assembly and no significant improvements in terms of analytical features have been gained within time.
Thus, a simple and low cost concurrent approach would be appreciated, especially if a direct reading of serum samples is rendered possible and if the biosensor assembly is set to a minimum complexity by suitable optimization of critical variables. The biorecognition element used herein is the complementary oligonucleotide of synthetic origin (anti-miRNA-155) that was simply immobilized on Au Screen printed electrodes (Au-SPE). Non-specific binding was blocked with mercaptosuccinic acid modification, for subsequent hybridization of the anti-MIR with the target analyte, MIR-155. All studies regarding the chemical modification and optimization of the biosensor design, electrical characterization, and analytical application are presented herein.
2. Experimental section
2.1. Apparatus
The electrochemical measurements were performed with a potentiostat/galvanostat from Metrohm Autolab, PGSTAT320N, controlled by NOVA 1.11 software. The gold-screen printed electrodes (Au-SPEs) were purchased from DropSens (DS-C220AT), built with (i) a counter electrode made of gold; (ii) a reference electrode and electrical contacts made of silver; and (iii) a gold working electrode with 4 mm diameter. Also, the Au-SPEs were interfaced in a switch box from DropSens, allowing their connection to the potentiostat.
UV/Vis studies were made in the spectrophotometer Evolution 220 UV-visible from Thermo Scientific. AFM studies were made in a Nanoscope IVA from Veeco Metrology Multimode.
2.2. Reagents
The water was ultrapure Mili-Q laboratory grade (conductivity <0.1µS/cm) or autoclaved with 0.1% diethylpyrocarbonate (DEPC) in order to increase stability of the miRNA by decreasing RNase activity.
All reagents were used without further purification. Potassium hexacyanoferrate-III (K3[Fe(CN)6]), potassium hexacyanoferrate-II (K4[Fe(CN)6]) trihydrate, magnesium chloride (MgCl), and sodium phosphate (Na2HPO4), were from Riedel-deHaen; Serum cornay human was from PZ CORMAY S.A., Poland; Phosphate buffer saline (PBS) and bovine serum albumin (BSA) were from Amresco; Calcium chloride (CaCl2) and potassium chloride (KCl) from Merck; Ethylenediamine tetraacetic acid (EDTA) from BDH; Sodium chloride (NaCl) and absolute ethanol (99.5%) from Panreac; Hydroxymethyl-aminomethane (Tris) from Fisher BioReagents; Sodium citrate from Analar Normapur; Sulphuric acid (H2SO4), dithiothreitol (DTT), mercaptosuccinic acid (MSA, 97%), DEPC, and cancer antigen 15-3 (CA-15.3) from Sigma.
The oligonucleotide probes were anti-miRNA-155 (complementary strand that was thiol modified and with adenine spacers) and miRNA-155, purified by HPLC and obtained from the Metabion. The sequences were as follows: anti-miRNA-155 (anti-miRNA155), 5’-HS-AAA AAA AAC CCC UAU CAC GAU UAG CAU UAA-3’; miRNA-155 (miRNA-155), 5’-UUA AUG CUA AUC GUG AUA GGG GU-3’.
2.3. Solutions
All solutions were prepared in ultrapure water, autoclaved with 0.1% DEPC. The stock solution of oligonucleotide (1.16 µg/ml, anti-miRNA) was prepared in SSPE buffer containing 0.02 M EDTA, 2.98 M NaCl and 0.2 M phosphate buffer (20× diluted, pH 7.4). The stock solution of 0.6 µg/mL of miRNA-155 was prepared in Tris-HCl, containing 0.02 M Tris, 0.14 M NaCl, 0.001 M MgCl2, 0.005 M KCl and 0.001 M CaCl2 (pH 7.4). Less concentrated standards were obtained by accurate dilution of the previous solution in 0.01 M Tris buffer containing 0.001 M EDTA and 0.05 M NaCl (pH 7.4). The immobilization and hybridization buffer were prepared in the same buffer solution. The buffer for regeneration (SSC) contained 3.0 M NaCl and 0.3 M trisodium citrate (0.1× diluted, pH 7.4).
Solutions of interfering species (for selectivity study) were prepared in the same buffer used in the immobilization and hybridization stages. The concentration of miRNA-155 was set to 0.6 µg/ml and the interfering species were BSA (0.30µg/ml) and CA-15.3 (30U/mL). The miRNA-155 solutions were prepared in real human serum (Cornay Human Serum), 1000× diluted. The electrochemical studies were made in a solution of 5.0×10-3 M of K3[Fe(CN)6] and 5.0×10-3 M of K4[Fe(CN)6] redox probe, prepared in PBS buffer, pH 7.4.
2.4. Preparation of electrochemical biosensor on Au-SPE
Before modification, the working Au-SPE surface was cleaned by washing with absolute ethanol, followed by CV electrochemical treatment with 0.5 M H2SO4, for 10 cycles (500 mV/s; -0.1 to 1.5V). Finally, the Au surface was washed with ultrapure water to ensure the removal of unwanted chemical species.
The synthesis of the biosensor was made in two steps. Step (1) consisted in incubating anti-miRNA-155 (1µM/mL) with DTT (0.1M) on the Au-SPE working area. The probe solution was prepared in buffer of pH 7.4 that contained Tris, EDTA and NaCl, previously heated at 90ºC for 5 minutes, and incubated for 2h at room temperature. The electrode surface was then washed with ultrapure water several times. In step (2), the non-specific binding areas were blocked by incubating the sensory surface with 0.002 µg/mL MSA, for 2h, at room temperature. The hybridization of the probe with the target miRNA-155 was made with several miRNA-155 standards solutions, prepared in buffer or in blank sera. Each standard was incubated for 30 minutes at 37ºC.
2.5. UV Characterization
The hybridization stage occurring between probe and target miRNA was followed by UV-Vis spectrophotometry. It was done by measuring the spectral data of each individual solution (Anti-miRNA-155, miRNA-155; and mixture of both) in the spectral range from 220 to 300 nm.
2.6. AFM analysis
The morphological analysis of biosensor was performed by AFM in tapping mode. These studies were conducted on planar Au surfaces employed in regular surface plasmon resonance (SPR) measurements. For this, anti-miRNA-155, Anti-miRNA-155/MSA and anti-miRNA-155/MCA/miRNA155 materials were assembled on SPR planar gold chips following the same procedure as that described for the Au-SPEs. The analysis of AFM images were collected and handled by the nanoscope software, coupled to the same equipment.
2.7. Electrochemical Procedures
All electrochemical assays were conducted in triplicate. CV assays were made for scanning potentials from -0.5 to +0.5 V, at a scan-rate of 50 mV/s. EIS assays were performed at an open circuit potential, using a sinusoidal potential perturbation with an amplitude of 0.01V, and 50 data points, logarithmically distributed over 0.1 to 100000 Hz frequency range. The EIS data was fitted into a Randles equivalent circuit, using 1.11 Nova Software from Autolab. SWV assays were conducted for a potential range from -0.2 to +0.8 V, with a frequency of 25 Hz and a step height of 50mV.
The electrical properties of modified surfaces were followed-up by CV, EIS and SWV assays, carried out in a redox probe solution of 5.0×10-3 M of [Fe(CN)6]3- and 5.0×10-3 M of [Fe(CN)6]4-, prepared in PBS buffer, pH 7.4. Calibration curves used EIS and SWV assays and included standard solutions of miRNA-155, prepared in Tris buffer pH 7.4 (ranging from 1.0 fM to 10 nM), or in human serum (ranging from 0.01 aM to 10 nM). The LOD was calculated as X+3σ, where X was the average value of the EIS or SWV blank signals (obtained in the absence of miRNA-155) and σ the known standard deviation of EIS or SWV blank signal consecutive readings (Harvey, 2000).
Selectivity studies were conducted by competitive assay between miRNA-155 with two different biomolecules that can be found in biological fluids, such as BSA (0.30 µg/mL) and CA-15.3 (30 U/mL).
2.8. MiRNA-155 assay
The performance of the biosensor was analysed by the standard addition method, in order to determine the miRNA-155 concentration in spiked human blank serum. MiRNA-155 concentrations in samples were set to different levels, equal to 100 fM and 100 pM, always lying within the linear range of the device. The human serum was diluted in TRIS buffer. All the assays were conducted in duplicate and analytical data was collected in EIS.
3. Results and Discussion
3.1. Biosensor assembly
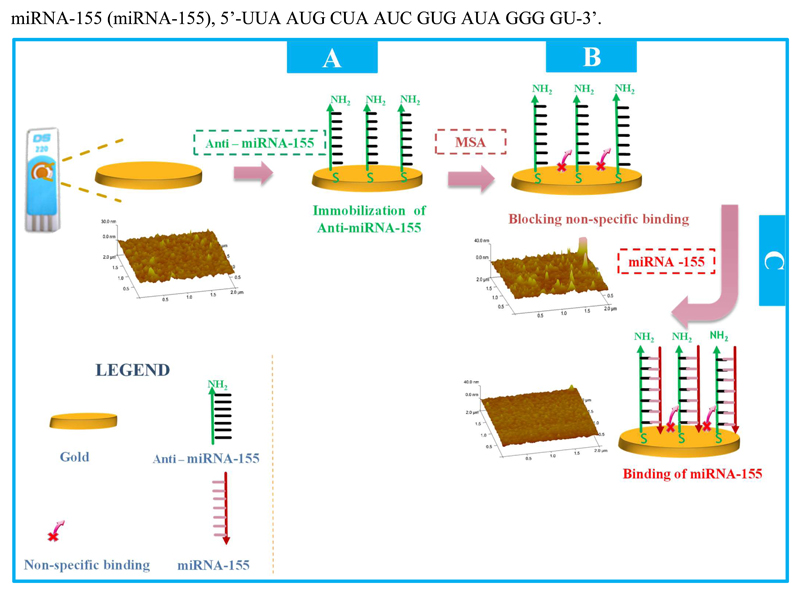
The schematic construction of the miRNA-155 biosensor is shown in Figure 1, and involved three main stages: (1) Au-SPE pre-treatment; (2) oligonucleotide immobilization and (3) non-specific binding blockage. The miRNA-155 hybridization assay was the final approach made with the biosensor, and concerned the analytical stage.
Figure 1.
Initially, Au-SPEs were cleaned first by washing with ethanol, two times, and after by electrochemical cleaning, making use of consecutive CV assays in a H2SO4 solution. This oxidative stage allowed the oxidation of any organic species allocated on the surface and improved the electrochemical properties of the bare Au electrode, by decreasing the peak-to-peak separation of the Fe redox probe and increasing peak currents.
The probe was then covalently bond to the clean SPE by incubating the Au working electrode in a solution of anti-miRNA-155 with a thiol group at its 5’end, in a hydrated chamber, at room temperature, and for a set time period. (Figure 1A). The probe had several consecutive adenine nucleotides between the –SH end and the complementary sequence to miRNA-155, acting as spacer and conferring mobility to the probe (in order to reduce steric hindrance at the hybridization stage). Before incubation, the anti-miRNA155 probe was treated with 0.10M DTT for 15 minutes, in order to disrupt disulphide bonds, and warmed at 90ºC for 5 minutes to ensure that the strand had a linear pattern. This last heating stage was found crucial, as without it the amount of immobilized probe was much smaller (less than 10%) and irreproducible.
Next, the non-specific binding was blocked with MSA (Figure 1B). This compound has an –SH bond allowing its covalent binding to free Au areas existing on the working electrode. This stage was fundamental to avoid the direct interaction between any biomolecule in a sample and the gold.
The analytical event involved the hybridization between the probe and miRNA-155. It was made by incubating the working electrode area with different concentrations of target miRNA (Figure 1C). The different standard solutions were incubated on the working Au-SPE surface at 37ºC, for a set amount of time.
The relevant data regarding the morphological characterization of the biosensor and the UV analysis (please see Figure S1, confirming the hybridization between anti-miRNA-155 and miRNA-155) is presented and discussed in the Supplementary Section (identified as S.1 and S.2).
3.2. Electrochemical follow-up of the gold surface modifications
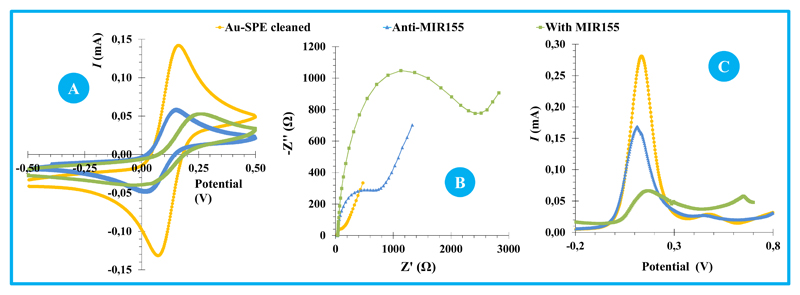
The chemical modification of the Au-SPE surface produced alterations in its electrical features, evaluated by monitoring the electron transfer properties of a standard redox system. Different redox systems were tested for this purpose (Ruthenium and Iron), prepared in different buffer conditions. A redox probe of [Fe(CN)6]4-/[Fe(CN)6]3- was selected, at pH 7.4, as the electrical changes monitored under these conditions generated more sensitive alterations at the detected signal. The electrochemical approaches used for this purpose were CV, EIS and SWV assays and the corresponding data is shown in Figure 2.
Figure 2.
CV analysis is shown in Figure 2A. When compared to the clean surface, the Au-SPE modified with anti-miRNA and MSA showed increased peak-to-peak potential separation and decreased cathodic/anodic peak currents. This accounted an increased charge-transfer resistance at the Au-SPE surface after each modification stage. After hybridization with miRNA-155 (Au-SPE/Anti-miRNA155/miRNA-155), the peak current decreased more, and a slight shift was observed in the peak potentials, thereby confirming the occurrence of chemical changes.
The corresponding EIS measurements are shown in Figure 2B and were consistent with CV data. Randle’s equivalent circuit was used to fit the physico-chemical process occurring at the gold electrode surface and EIS spectra were represented as Nyquist plots – including a semicircle, in which the diameter corresponded to the electron transfer resistance (Rct) (Panagopoulou et al., 2010) and the linear part represented a diffusion limited process (Suni, 2008). The Randle’s equivalent circuit included several elements, in which the high frequency region is dominated by the double layer capacitance (Cdl) and the magnitude of the electrolyte solution resistance (Rs) (Liu et al., 2010); the charge-transfer resistance (Rct), which is inversely proportional to the rate of electron transfer; and the Warburg diffusion element (W) at higher frequencies, accounting for the diffusion of ions. Overall, the bare gold electrode showed a straight line or very small semicircle domain, which suggested a mass diffusion limiting step of the electron transfer process. After, the immobilization of thiol-based materials on the clean Au-SPE, the diameter of the semicircle of the Nyquist plot increased considerably. This observation was consistent with an increased electron transfer resistance at the surface, enhanced by the fact that the single strand anti-miRNA155 was negatively charged through its phosphate backbone, thereby hampering the electrode transfer event of a negatively charged redox couple (Wu et al., 2015). The hybridization of the probe with miRNA-155 also increased the Rct values, thereby confirming its effect upon the electron transfer and the increased density of negative charges occurring at the surface.
SWV data is presented in Figure 2C and is also consistent with EIS and CV assays. As it is more sensitive than CV, the changes in electrical transfer properties of the several modification stages were more evident in SWV. Voltammograms indicated a decrease in peak current for assembly and hybridization events.
Overall, CV, EIS and SWV assays were consistent, and confirmed the occurrence of chemical changes at the gold surface, both at the biosensor assembly and at the hybridization event. From a practical point of view, EIS and SWV techniques seemed more sensitive to the hybridization event, being therefore selected for the subsequent analytical application.
3.3. Analytical Performance of miRNA sensor
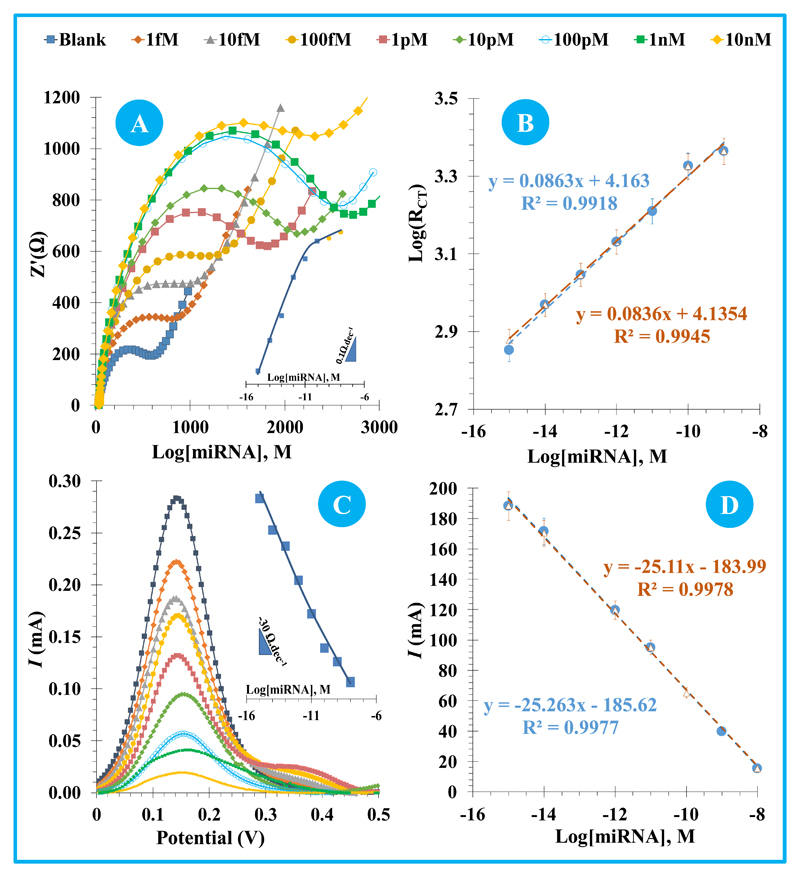
The analytical performance of the biosensor was evaluated by calibration curves in EIS and SVW measurements. For this, increasing concentrations of miRNA-155 were incubated at the surface for a specific amount of time, and the resulting Rct (in EIS) or current intensity (I, in SWV) values obtained with the iron redox probe measured against logarithm of miRNA-155 concentration. It is important to highlight that several variables were evaluated and optimized, including concentration, temperature and time given for the incubation of anti-miRNA solution on the clean gold surface, time and temperature given for the hybridization stage, buffer composition, etc.. The analytical data presented next regards only the best conditions selected.
In general, EIS (Figure 3A) measurements showed that increasing concentrations of miRNA-155 increased the electrical resistance of the surface, as the diameter of the semicircles in the Nyquist plots increased. The linear plot was made between logarithm Rct and logarithm of miRNA-155 concentration. The response of the biosensor in EIS was tested in three calibrations, made with independent sensors, in different days, and was found highly reproducible. All these showed a linear trend between 1.0 fM and 100 pM. As may be seen in Figure 3B, the second and third calibrations had an average slope of 0.086 logΩ/decade, with a standard of deviation 2.2 %. The squared correlation coefficient of all calibrations was always >0.99, and the average limit of detection was 0.54 fM.
Figure 3.
SWV assays also showed linear plots, but this time between current intensity (I, in SWV) and logarithm of miRNA-155 concentration. The linear behaviour was observed from 1.0 fM and 10 nM (Figure 3C). The increasing miRNA-155 concentrations were revealed by the decreasing peak currents of redox probe solution. The following calibrations are presented in Figure 3D, showing average slopes of 25.18 µA/decade and square correlation coefficients >0.99. The standard of deviation of the these assays was ~0,5% (Figure 3D), and the limit of detection was 2.8 aM.
Overall, the assembled biosensor showed consistent, sensitive and reproducible calibration curves, both in EIS and in SWV assays. The obtained results suggested that the biosensor displayed a highly selective and sensitive response against miRNA-155.
3.4. Regeneration of Au-SPE
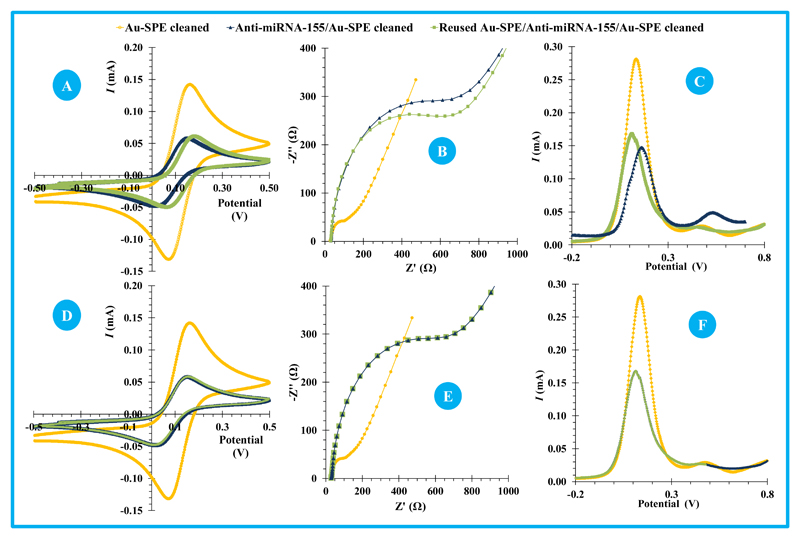
The regeneration of a biosensor after a calibration assay maybe achieved by several physical (enthalpic interaction, entropic interactions, thermal regeneration-temperature) and chemical (acid/base-mediated regeneration, detergents, glycine, urea and another buffers) approaches (Goode et al., 2014). In this work, physical (temperature) and/or chemical (buffer treatment) assays were tried out and the results presented in Figure 4.
Figure 4.
The temperature was considered as an important parameter to be tested herein since the 3D structure of oligonucleotides is typically temperature-dependent. A temperature increase leads to increased kinetic energy of the molecules, which allows binding forces to be overcome after reaching the suitable temperature. The first assay was made by heating the Au-SPE up to 90ºC, the same temperature that ensured previously that the anti-miRNA-155 was linear and free to hybridize. The 90 ºC is indeed the typical denaturation temperature used in PCR (Polymerase chain reaction) assays, for which no irreversible denaturing or decoupling of oligonucleotide base pairs would be expected (Champaign, 2013).
After 90ºC for 5 minutes, only little alterations were detected compared to the original signal of the biosensor before its hybridization: little shift in CV peaks; (Figure 4A); little decrease of the EIS resistance (Figure 4B); and a little increase of the peak current in SWV. Still, the small changes observed in the reused Au-SPE/anti-miRNA155 biosensor seemed to be related to the silver reference electrode modification/oxidation, promoted by such a high temperature (Figure 4C). This alteration was visually perceptible.
Thus, further assays were tested at lower temperature, in conjunction with a chemical buffer. For this, the hybridized biosensor was incubated in a saline-sodium citrate buffer (SSC) diluted 10× at 60ºC. After this, the signal of the regenerated surface matched exactly that of the original biosensor, in all electrochemical techniques (Figure 4, D, E and F).
After recovering the original signal, the reused biosensor was calibrated again to evaluate its behaviour under a second calibration, and further on in a third calibration. This was made to test the ability of the complementary oligonucleotides remaining on the surface to hybridize with its target miRNA (because staying there would not grant its ability to hybridize). The calibration assays were made by EIS and SWV measurements again, and showed very similar analytical features to the original biosensor, calibrated by its first time.
In EIS assays, the linear range was observed between 1.0 fM and 100 pM, plotting logarithm Rct against logarithm miRNA-155 concentration (Figure S2). The slope was 0.0793 logΩ/decade in the second calibration and changed to 0.0786 logΩ/decade in the third calibration, while the intercept was 4.1554 logΩ in the second calibration and changed to 4.1364 logΩ in the third calibration. The standard deviation of the slope and intercept of all three calibrations performed in the same device was 5.2 and 0.3 %, meaning that the biosensor is able to generate reproducible calibrations after regeneration. Indeed, it is possible that the regeneration process has a little (but not relevant) impact on the biosensor response, as the standard deviation between second and third calibrations is much smaller (equal to 0.6 and 0.3 %, respectively).
In SWV assays the calibration was also recovered in terms of concentration range, but the variability within the three calibrations was higher. The slope of the second calibration was -26.387 μA/decade and changed to -31.929 μA/decade in the following calibration, while the intercept was 201.5 μA and changed to 257.6 μA. The standard deviation of the slope and intercept of the three calibrations (first and two regenerated) was 9.5 and 16.0 %, which was clearly high, when compared with the EIS assays.
Overall, the biosensor may be regenerated after contacting with miRNA concentrations up to 100 nM, provided that the suitable approach is taken. In addition, EIS studies seem more reliable, if the Au-SPE is to be reused after its first calibration. This is particularly suitable for making the complete analytical procedure with a single chip (which may include calibration and sample analysis, all in a single device).
3.5. Selectivity
The selectivity study was conducted by evaluating the effect of chemical compounds present in biological fluids. A competitive assay was selected for this purpose. This was done by testing a solution of 10 pM miRNA-155 as single analyte and also in conjunction with other common biomolecules in serum, keeping the same concentration of miRNA-155. The incubation time was set to 30 minutes, the same period of time used in the calibration.
The interfering compounds studied were CA-15-3 (protein biomarker that in breast cancer condition co-exists with miRNA-155 in serum) and BSA (highly concentrated protein in serum of normal/diseased person). These species were tested within their physiological levels. The % average deviation produced by each interfering species in pure miRNA-155 solutions was +3.8 % when CA-15.3 was present and -5.2 % when BSA was added (Figure S3). These assays were performed in triplicate using the same Au-SPE, accounting its reliability after regeneration in EIS measurements.
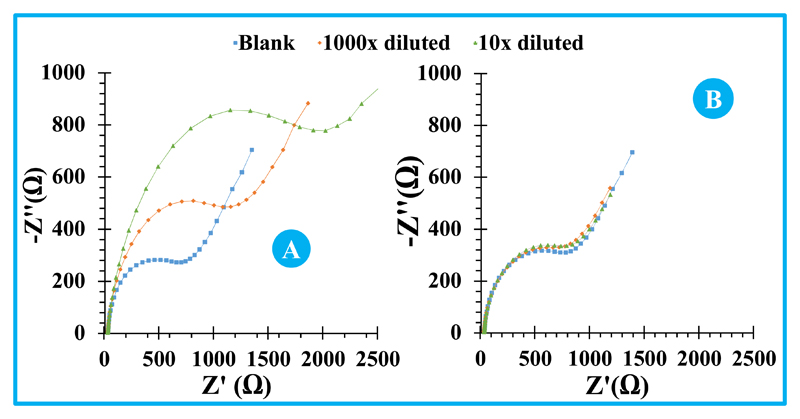
An extract from cell lines of melanoma was also tested herein. Melanoma is another cancer disease where miRNA-155 is not expected to exist, at least in significant levels. The extract was obtained by membrane cell disruption and simple filtration. The obtained signal was almost coincident to the blank (Figure 6B), therefore indicating that no significant interference would be expected by applying this biosensor to control real cell lines culture.
Figure 6.
Overall, no significant interferences were observed in these selectivity tests, neither from single analytes nor with complex real cell extracts.
3.6. miRNA-155 assay in human serum samples
The application of the biosensor to real samples was made after calibrating in standard solutions prepared in blank serum, instead of the buffer used until now. The analytical features of the resulting calibration curves in EIS and SWV (Figure 5) were also evaluated at this stage.
Figure 5.
As before, EIS calibration curves plotted logarithm Rct (in EIS) against logarithm miRNA-155 concentration. In general, the calibrations showed good analytical features in terms of lower concentration of linear range, LOD and slope (10aM, 5.7aM, 0.082 logΩ/decade) respectively (Figure 5A and 5B). Compared to the calibrations made with standard solutions prepared in buffer, this novel condition of preparing standards in serum had no significant impact in the slope (within the previous standard deviation). However, and interestingly, the use of standard solutions in buffer widened the concentration range producing a linear response. The biosensor provided linear responses down to 10 aM with lower LODs than before. The calibrations in SWV showed a similar behaviour (Figure 5C), with a decrease in the lower limit of linear range and LOD and a slope increase (1 aM, 0.18 aM, -36µA/decade, respectively).
For sample analysis, real human serum serving as blank was spiked with miRNA-155 in two different concentrations: 100 fM and 100 pM. This was performed in triplicate and only by following EIS data. The calibration data used in the analysis to calculate the concentrations in the spiked serum was the one provided by the same biosensor before regeneration. Comparing added and found amounts, the relative errors obtained were 6.9 and 7.2%, respectively. Overall, the results were found accurate in close-to-real conditions, as the device was tested in a background of control human serum of healthy individual.
Extracts from cell-lines MCF-7 from breast cancer disease were also analysed, yielding a positive signal against miRNA-155, as expected. This sample was analysed by serial dilution in buffer, diluting 1000× and 10× times the raw extract and reading without further treatment, starting by its lower concentration. The results are presented in Figure 6A, showing a proportional behaviour within the several dilutions. The obtained values confirmed the high precision of this new method, even when different dilution levels are combined. The average concentration of miRNA-155 in the cell extract was 1.02×10-11 mol/L, with a standard deviation of 8.71×10-13 mol/L that corresponded to a relative standard deviation of 8.6%.
4. Conclusions
This work described the development of a highly sensitive and inexpensive biosensor for the rapid detection of the emerging cancer biomarker miRNA155, in serum. The biosensor assembly was very simple and effective, being temperature a critical step. From the analytical perspective, the biosensor was able to detect very low concentrations of miRNA-155, down to 1-10 aM in a serum background. This was a very important feature of this biosensor, because it allows a higher degree of sample dilution and therefore surpassing the interference of any unexpected/unwanted biomolecule resulting from an abnormal physiological condition. In addition, the device could be reused along consecutive readings of new solutions, most especially in EIS readings, while showing excellent selectivity towards other proteins in biological fluids and cell extracts from another cancer disease.
This simple and sensitive strategy is considered a promising approach for the simultaneous quantitative analysis of multiple miRNA-155 in physiological fluids, in biomedical research and POC diagnosis. This device is also appropriate to integrate a multi-panel biomarker reading that includes the traditional CA15-3 biomarker, as this protein did not interfere with the analytical readings of the device.
In addition, this device opens new horizons into real time monitoring of miRNA cancer biomarker in clinical context, also taking advantage of its ability de-hybridize and allowing its reusing in an online sensor approach.
Supplementary Material
Acknowledgements
The authors acknowledge the financial support of European Research Council though the Starting Grant, ERC-StG-3P’s/2012, GA 311086 (to MGF Sales).
References
- Azimzadeh M, Rahaie M, Nasirizadeh N, Ashtari K, Naderi-Manesh H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosensors & Bioelectronics. 2015;77:99–106. doi: 10.1016/j.bios.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Bohunicky B, Mousa Sa. Biosensors: The new wave in cancer diagnosis. Nanotechnology, Science and Applications. 2011;4(1):1–10. doi: 10.2147/NSA.S13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S, Pedrero M, Pingarrón JM. Electrochemical genosensors for the detection of cancer-related miRNAs. Analytical and Bioanalytical Chemistry. 2014;406(1):27–33. doi: 10.1007/s00216-013-7459-z. [DOI] [PubMed] [Google Scholar]
- Catuogno S, Esposito CL, Quintavalle C, Cerchia L, Condorelli G, de Franciscis V. Recent advance in biosensors for microRNAs detection in cancer. Cancers. 2011;3(2):1877–1898. doi: 10.3390/cancers3021877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champaign U. Polymerase Chain Reaction. Brenner’s Encyclopedia of Genetics. (Second Edition) 2013;5(3):392–395. doi: 10.1016/B978-0-12-374984-0.01186-4. [DOI] [Google Scholar]
- Choi Y-E, Kwak J-W, Park JW. Nanotechnology for Early Cancer Detection. Sensors. 2010;10(1):428–455. doi: 10.3390/s100100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Moore K, Phillips L, Boyle FM, Marsh DJ, Baxter RC. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Research : BCR. 2014;16(3):R63. doi: 10.1186/bcr3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SW, Chen L, Man Y-G. miRNA Biomarkers in Breast Cancer Detection and Management. Journal of Cancer. 2011;2:116–122. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode JA, Rushworth JVH, Millner PA. Biosensor Regeneration : A Review of Common Techniques and Outcomes. 2014 doi: 10.1021/la503533g. [DOI] [PubMed] [Google Scholar]
- Grieshabe D, MacKenzie R, Vörös J, Reimhult E. Electrochemical Biosensors - Sensor Principles and Architectures. Sensors. 2008;8(3):1400–1458. doi: 10.3390/s8031400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey D. Modern Analytic Chemistry. 2000:797. [Google Scholar]
- He Y, Lin J, Kong D, Huang M, Xu C, Kim T-K, et al. Wang K. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clinical Chemistry. 2015;61(9):1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- Hu T, Zhang L, Wen W, Zhang X, Wang S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosensors and Bioelectronics. 2015;77:451–456. doi: 10.1016/j.bios.2015.09.068. [DOI] [PubMed] [Google Scholar]
- Johnson BN, Mutharasan R. Biosensor-based microRNA detection: techniques, design, performance, and challenges. The Analyst. 2014;139(7):1576. doi: 10.1039/c3an01677c. [DOI] [PubMed] [Google Scholar]
- Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. Journal of Biological Chemistry. 2010;285(23):17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koyun A, Ahlatcıoğlu E, İpek YK. Biosensors and Their Principles. A Roadmap of Biomedical Engineers and Milestones. 2012:117–142. doi: 10.5772/48824. [DOI] [Google Scholar]
- Labib M, Berezovski MV. Electrochemical sensing of microRNAs: Avenues and paradigms. Biosensors and Bioelectronics. 2015;68:83–94. doi: 10.1016/j.bios.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Labib M, Khan N, Berezovski MV. Protein Electrocatalysis for Direct Sensing of Circulating MicroRNAs. Analytical Chemistry. 2015;87(2):1395–1403. doi: 10.1021/ac504331c. [DOI] [PubMed] [Google Scholar]
- Liu J, Huang W, Yang H, Luo Y. Expression and function of miR- 155 in breast cancer. Biotechnology & Biotechnological Equipment. 2015;29(5):840–843. doi: 10.1080/13102818.2015.1043946. [DOI] [Google Scholar]
- Liu S, Su W, Li Z, Ding X. Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures. Biosensors and Bioelectronics. 2015;71:57–61. doi: 10.1016/j.bios.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Liu X, Duckworth Pa, Wong DKY. Square wave voltammetry versus electrochemical impedance spectroscopy as a rapid detection technique at electrochemical immunosensors. Biosensors and Bioelectronics. 2010;25(6):1467–1473. doi: 10.1016/j.bios.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiology Biomarkers and Prevention. 2012;21(8):1236–1243. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- Mirabelli P, Incoronato M. Usefulness of Traditional Serum Biomarkers for Management of Breast Cancer Patients. 2013;2013 doi: 10.1155/2013/685641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulou Ma, Stergiou DV, Roussis IG, Prodromidis MI. Impedimetric biosensor for the assessment of the clotting activity of rennet. Analytical Chemistry. 2010;82(20):8629–8636. doi: 10.1021/ac1017925. [DOI] [PubMed] [Google Scholar]
- Riaz M, van Jaarsveld MT, Hollestelle A, Prager-van der Smissen WJ, Heine AA, Boersma AW, Martens JW. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Research : BCR. 2013;15(2):R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RNA Nanotechnology. CRC Press; 2014. Retrieved from https://books.google.com/books?id=NB0yAwAAQBAJ&pgis=1. [Google Scholar]
- Siegel R, Desantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Fedewa S. Cancer Treatment and Survivorship Statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62(4):220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J. Serum MicroRNA-155 as a Potential Biomarker to Track Disease in Breast Cancer. PLoS ONE. 2012;7(10):1–8. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suni II. Impedance methods for electrochemical sensors using nanomaterials. TrAC - Trends in Analytical Chemistry. 2008;27(7):604–611. doi: 10.1016/j.trac.2008.03.012. [DOI] [Google Scholar]
- Tavallaie R, De Almeida SRM, Gooding JJ. Toward biosensors for the detection of circulating microRNA as a cancer biomarker: an overview of the challenges and successes. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2015;7(4):580–592. doi: 10.1002/wnan.1324. [DOI] [PubMed] [Google Scholar]
- Tran HV, Piro B, Reisberg S, Anquetin G, Duc HT, Pham MC. An innovative strategy for direct electrochemical detection of microRNA biomarkers. Analytical and Bioanalytical Chemistry. 2014;406(4):1241–1244. doi: 10.1007/s00216-013-7292-4. [DOI] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proceedings of the National Academy of Sciences. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Chen H, Zuo Z, Wang M, Luo R, Xu H. A Simple Electrochemical Biosensor for Rapid Detection of MicroRNA Based on Base Stacking Technology and Enzyme Amplification. 2015;10:3848–3858. [Google Scholar]
- Wu X, Chai Y, Yuan R, Su H, Han J. A novel label-free electrochemical microRNA biosensor using Pd nanoparticles as enhancer and linker. The Analyst. 2013;138(4):1060–6. doi: 10.1039/c2an36506e. [DOI] [PubMed] [Google Scholar]
- Yahalom G, Weiss D, Novikov I, Bevers TB, Radvanyi LG, Liu M, et al. Rosenberg MM. An Antibody-based Blood Test Utilizing a Panel of Biomarkers as a New Method for Improved Breast Cancer Diagnosis. Biomarkers in Cancer. 2013;5:71–80. doi: 10.4137/BIC.S13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N, Zhang L. Nanomaterials-Based Sensing Strategies for Electrochemical Detection of MicroRNAs. Materials. 2014;5366:5384. doi: 10.3390/ma7075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Fang C, Nam S, Cai Q, Long X. The Clinicopathological Significance of MicroRNA-155 in Breast Cancer : A Meta-Analysis. 2014;2014 doi: 10.1155/2014/724209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Su X, Gao X, Dai Z, Zou X. A label-free and PCR-free electrochemical assay for multiplexed microRNA profiles by ligase chain reaction coupling with quantum dots barcodes. Biosensors & Bioelectronics. 2014;53:414–9. doi: 10.1016/j.bios.2013.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.