Abstract
Amyloid-β precursor protein (APP) is central to the pathogenesis of Alzheimer’s disease, yet its physiological function remains unresolved. Accumulating evidence suggests that APP has a synaptic function mediated by an unidentified receptor for the shed APP ectodomain (sAPP). Here, we showed that the sAPP extension domain directly bound the sushi 1 domain specific to the gamma-aminobutyric acid type B receptor subunit 1a (GABABR1a). sAPP-GABABR1a binding suppressed synaptic transmission and enhanced short-term facilitation in hippocampal synapses via inhibition of synaptic vesicle release. A 17 amino acid peptide corresponding to the GABABR1a binding region within APP suppressed spontaneous neuronal activity in vivo. Our findings identify GABABR1a as a synaptic receptor for sAPP and reveal a physiological role for sAPP in regulating GABABR1a function to modulate synaptic transmission.
One Sentence Summary:
Amyloid-β precursor protein suppresses vesicle release from presynaptic boutons by binding to the sushi domain of the GABAB1a receptor.
Amyloid-β Precursor Protein (APP), a type 1 transmembrane protein, was first identified more than 30 years ago (1–4) as the precursor to the amyloid-β peptide, the primary constituent of amyloid plaques found in the brains of Alzheimer’s disease (AD) patients. APP undergoes ectodomain shedding by α-, β-, or η- secretase to release soluble APP (sAPPα, sAPPβ, or sAPPη respectively) (5, 6). Evidence suggests that the synaptic function of APP (7–13) is carried out by sAPP (14, 15). sAPPα affects synaptic transmission and plasticity, including a reduction in synaptic activity and an enhancement of LTP (16–19). Moreover, sAPPα is sufficient to rescue synaptic defects in App KO mice, including defects in spine density (20), LTP (21, 22), and spatial learning (21). Together, this has led to speculation of a yet unidentified cell-surface receptor for sAPP to mediate its synaptic function (15, 23, 24).
Proteomics screen for synaptic interactors of sAPP identifies GABABR
We first confirmed, using biochemical fractionation and structured illumination imaging, that APP was abundantly expressed at presynaptic terminals (25) of excitatory and inhibitory hippocampal synapses (Fig. S1A,B). Next, to identify candidate synaptic receptors for sAPP, we performed an extensive series of affinity purification experiments using recombinant sAPP-Fc (C-terminal Fc-tag; affinity purified from transfected-HEK293T supernatants; Fig. S2A,B) to pull down interacting proteins from synaptosome extracts, followed by mass spectrometric analysis of bound proteins (AP-MS) (Fig. 1A) (26). We consistently identified, among a few intracellular proteins (Fig 1B, S3A,B, Table S1), the gamma-aminobutyric acid type B receptor subunit 1 (GABABR1) as the most abundant and reproducible cell-surface protein, using sAPPα or sAPPβ as bait, in wildtype (WT) and in App/Aplp1 knockout (KO) synaptosome extracts (Fig. 1B, S3A,B, Table S1). Supporting our observations, APP has previously been identified in a GABABR interactome analysis (27). Together, the sAPP AP-MS experiments identified GABABR as the leading candidate for a synaptic, cell-surface receptor for sAPP.
Fig. 1. sAPP selectively binds the sushi 1 domain of GABABR1a.
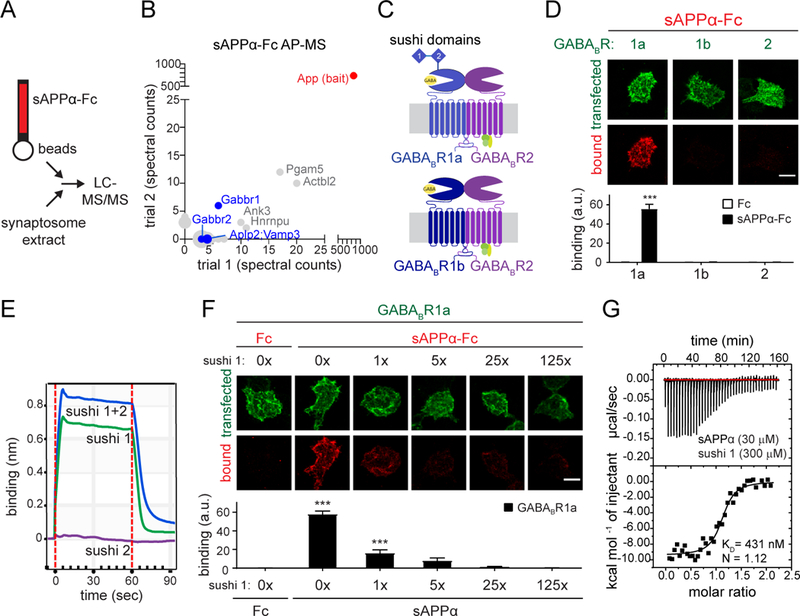
(A) Cartoon illustrating AP-MS workflow. (B) Spectral counts of proteins identified by mass spectrometry from 2 independent sAPPα-Fc pull-downs on rat synaptosome extracts. Only proteins which were absent in the Fc controls and present with > 2 spectral counts in a single trial are included. Cell-surface proteins are highlighted in blue. (C) Cartoon of GABABR subunits and isoforms. (D) Confocal images (upper) and quantifications (lower) of immunostaining for sAPPα-Fc or Fc binding to GABABR1a- , 1b- , or 2-expressing HEK293T cells (n=24). (E) Binding of sAPPα purified protein to immobilized Fc-tagged sushi 1, sushi 2, or sushi 1+2 peptides by biolayer interferometry. (F) Confocal images (upper) and quantifications (lower) of immunostaining for Fc control or sAPPα-Fc binding to GABABR1a-expressing HEK293T cells in the presence of increasing concentrations of untagged sushi 1 peptide (n=24–31). (G) Binding of purified sAPPα and sushi 1 proteins (Fc-tag enzymatically removed from both constructs) by isothermal titration calorimetry (ITC). The number of total cells from 3–4 independent experiments is defined by n. Graphs show means ± SEM. Two-way (D) or one-way (F) ANOVA with Bonferroni’s post hoc analysis. ***P < 0.001. Scale bar 10 μm.
The extension domain of APP binds the sushi 1 domain of GABABR1a
GABABR, the metabotropic receptor for the inhibitory neurotransmitter GABA, regulates presynaptic neurotransmitter release and postsynaptic membrane excitability (28). It consists of two subunits: GABABR1 which binds GABA, and GABABR2 which couples to G proteins (29). Two major isoforms, GABABR1a and GABABR1b, differ by two N-terminal sushi repeats only present in the a-variant (29) (Fig 1C). To validate the proteomics results, we performed cell-surface binding assays, applying recombinant sAPPα-Fc to HEK293T cells expressing the GABABR ectodomain on the plasma membrane using the pDisplay vector. sAPPα-Fc, but not Fc alone, bound strongly to GABABR1a-, but not to GABABR1b-, or GABABR2-, expressing cells (Fig. 1D). Biolayer interferometry experiments using recombinant sAPPα (Fc-tag enzymatically removed; Fig. S2C-F) and GABABR1a sushi domains showed that the sushi 1 peptide was sufficient for binding sAPPα (Fig. 1E). Accordingly, excess sushi 1 peptide blocked binding of sAPPα-Fc to GABABR1a-expressing cells (Fig. 1F). Isothermal titration calorimetry (ITC) determined the dissociation constant (KD) for sAPPα-sushi 1 = 431 nM (Fig. 1G). These data show that sAPPα binds directly and selectively to the sushi 1 domain of GABABR1a with sub-micromolar affinity.
The ectodomain of the APP695 isoform contains several functional domains (Fig. 2A). Surprisingly, growth factor like domain (GFLD)-Fc, copper binding domain (CuBD)-Fc, and E2-Fc each failed to bind GABABR1a-expressing cells (Fig. 2B). However, a peptide corresponding to the natively unstructured linker region between the APP695 E1 and E2 domains (Fig. 2A) strongly binds to GABABR1a-expressing cells (Fig. 2B). The linker region includes the acidic domain (AcD) and the recently defined extension domain (ExD), which is a flexible, partially structured region (30). The binding affinity of the purified ExD-AcD fragment (Fc-tag enzymatically removed) to sushi 1 in ITC experiments (Fig. 2C) was in the same range as that of full-length sAPPα binding to sushi 1 (Fig. 1G). To further narrow down the minimal domain in the APP linker region required for sushi 1 binding, we generated ExD-Fc and AcD-Fc fragments. ExD-Fc, but not AcD-Fc, bound to GABABR1a-expressing cells (Fig. 2B), identifying the ExD as the minimal domain required for sushi 1 binding. Consequently, deletion of the ExD in sAPPα (sAPPαΔExD-Fc) abolished binding to GABABR1a-expressing cells (Fig. 2B). sAPPβ-Fc and sAPPη-Fc, a product of the recently described η-secretase processing pathway (6), which both contain the ExD, also bound to GABABR1a-expressing cells (Fig 2D). APP family members APP-like protein 1 and 2 (APLP1 and APLP2) (31) on the other hand lack a conserved ExD and failed to bind GABABR1a-expressing cells (Fig. 2E). Thus, the sAPP ExD is necessary and sufficient to bind to the GABABR1a sushi 1 domain.
Fig. 2. The extension domain of sAPP binds GABABR1a.
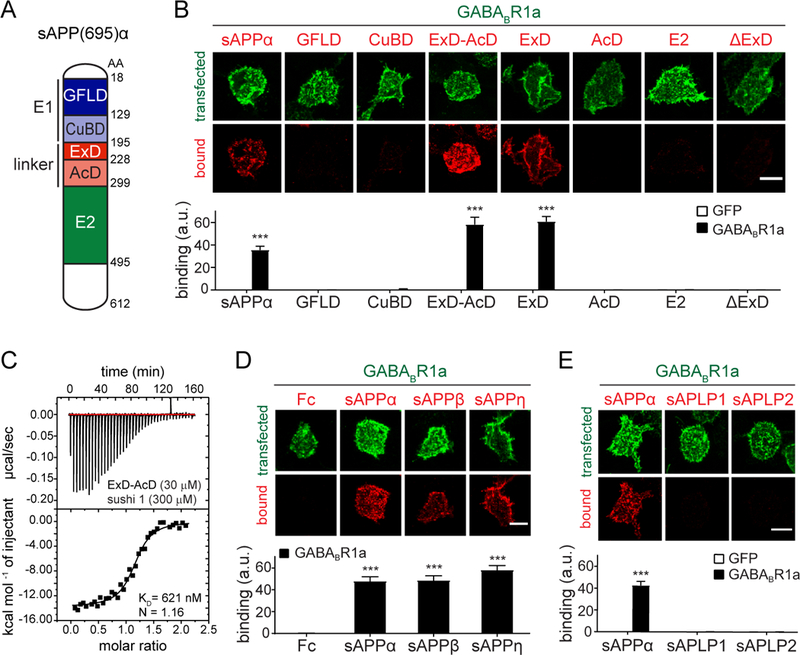
(A) Cartoon of sAPPα domains. (B) Confocal images (upper) and quantifications (lower) of immunostaining for sAPPα-Fc, GFLD-Fc, CuBD-Fc, ExD-AcD-Fc, ExD-Fc, AcD-Fc, E2-Fc or sAPPαΔExD-Fc binding to GFP- or GABABR1a-expressing HEK293T cells (n=24–32). (C) Binding of purified ExD-AcD-Fc and sushi 1 proteins by ITC. (D) Confocal images (upper) and quantifications (lower) of immunostaining for Fc control, sAPPα-Fc, sAPPβ-Fc binding to GABABR1a-expressing HEK293T cells (n=24–30). (E) Confocal images (upper) and quantifications (lower) of immunostaining for sAPPα-Fc, sAPLP1-Fc, of sAPLP2-Fc (red) binding to GFP or GABABR1a-expressing HEK293T cells (green) (n=24). The number of total cells from 3–5 independent experiments is defined by n. Graphs show means ± SEM. Two-way (B,E) or one-way (D) ANOVA with Bonferroni’s post hoc analysis. ***P < 0.001. Scale bar 10 μm.
sAPP suppresses presynaptic vesicle release probability via GABABR1a
Sushi domain-containing GABABR1a is the predominant isoform localized to presynaptic compartments at excitatory synapses (32–34), where it functions to inhibit neurotransmitter release (28). To test whether sAPPα can modulate GABABR function, we simultaneously measured miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs), which were separated on the basis of their distinct decay kinetics as described (35), in cultured mouse hippocampal neurons (12–17 days in vitro (DIV)) (Fig. 3A). Consistent with previous observations (36, 37), acute exposure of hippocampal neurons to 30 μM baclofen, a GABABR agonist, reduced the frequency of mEPSCs by 63 ± 5% (n=14 cells; P < 0.001) (Fig. S4A,B). Likewise, 250 nM sAPPα (Fc-tag removed) reduced the frequency of mEPSCs by 39 ± 5% (n=13 cells; P < 0.001) (Fig. 3B,C), an effect that was already apparent at 25nM (Fig. S4D,E), without affecting mEPSC amplitude (Fig. S4C). sAPPβ similarly reduced mEPSC frequency (Fig. S4D,E). Acute application of the APP695 ExD-AcD fragment reduced mEPSC frequency to a similar degree as sAPPα (Fig. 3D, S4F), whereas application of sAPPαΔExD had no effect (Fig. 3D, S4F), indicating that the extension domain of sAPP is necessary and sufficient for the suppression of spontaneous glutamatergic synaptic transmission by sAPPα. Accordingly, acute application of sAPLP1, which lacks a conserved ExD, did not reduce mEPSC frequency (Fig. S4G), although we observed a minor (17 ± 9 %; n=17 cells; P < 0.05) reduction in mIPSC frequency (Fig. S4H). Pretreatment with the GABABR antagonist CGP55845 (CGP, 5 μM) attenuated the sAPPα-mediated reduction of mEPSC frequency (Fig. 3E, S4I), showing that the effect is mediated by GABABR.
Fig. 3. sAPPα reduces the release probability of synaptic vesicles via presynaptic GABABR1a.
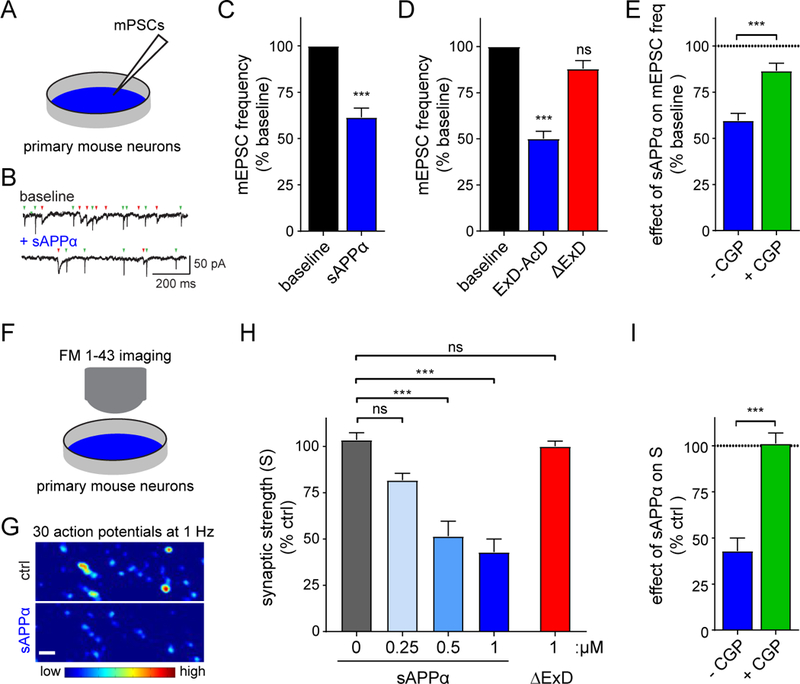
(A) Cartoon of mPSC measurements in cultured hippocampal mouse neurons reported in B-E. (B,C) Example traces of mEPSCs (green arrowheads) and mIPSCs (red arrowheads) (B) and average mEPSC frequency (C) normalized to baseline recorded from primary neurons before (baseline) and after treatment with sAPPα (250 nM, Fc-tag enzymatically removed, n=13, N = 3, paired t-test). (D) Same as C but with either ExD-AcD, or sAPPαΔExD (Fc-tag enzymatically removed, n=17–20, N=3, one way ANOVA with Dunnett’s post hoc analysis). (E) Same as C but with sAPP and either without (blue) or with (green) preincubation with CGP55845 (CGP, 5 μM), a GABABR antagonist. Dotted line denotes baseline (n=14–17, N=3 unpaired t-test). (F) Cartoon of FM1–43 measurements in cultured hippocampal mouse neurons reported in G-I. (G) High-magnification ΔF images before and after application of sAPPα (Fc-tag enzymatically removed, 1 μM) to primary neurons. (H) Summary of the dose-dependent inhibitory effect of sAPPα on presynaptic strength (S) (N= 5–8, one way ANOVA analysis with post hoc Tukey’s analysis). (I) Summary of sAPPα effect on presynaptic vesicle recycling in hippocampal neurons with or without CGP (normalized to control (ctrl)) (N =8). The number of neurons is defined as n, and the number of independent experiments or mice is defined as N. Graphs show means ± SEM. * P < 0.05, ** P < 0.1 *** P < 0.001.
GABABR1a also localizes to GABAergic boutons (34). Consistent with previous observations (37, 38), acute exposure of hippocampal neurons to 30 μM baclofen reduced the frequency of mIPSCs by 62 ± 5% (n=14 cells; P < 0.001) (Fig. S5A). Acute application of 250 nM sAPPα to hippocampal neurons reduced the frequency of mIPSCs by 44 ± 5% (n=13 cells; P < 0.001) (Fig. 3B, S5B). Application of sAPPα caused a minor (14% reduction in mIPSC amplitude (Fig. S5C), possibly due to a small post-synaptic effect of sAPPα on GABABR1a at post-synaptic GABAergic sites (39). The APP695 ExD-AcD fragment, but not sAPPαΔExD, reduced mIPSC frequency to a similar extent as sAPPα (Fig. S4F, S5D). The effect of sAPPα on mIPSC fequency was blocked by pretreatment with the GABABR antagonist CGP55845 (CGP, 5 μM) (Fig. S4I, S5E). Taken together, these data show that sAPPα acutely reduces both glutamatergic and GABAergic quantal synaptic transmission through a GABABR1a isoform-dependent mechanism.
sAPPα might exert its effect on synaptic transmission by interfering with a complex of full-length APP and GABABR1a. In neurons lacking APP however, sAPPα still reduced mEPSC and mIPSC frequency (Fig. S6A,B), excluding this possibility. Application of 30 μM baclofen similarly reduced mEPSC and mIPSC frequency in App/Aplp1 dKO cultures (Fig. S6C,D) as in WT cultures (Fig. 3C, S5B), suggesting that the absence of full-length APP does not cause major alterations in GABABR localization to presynaptic terminals. However, the possibility that full-length APP also interacts with and affects GABABR signaling separate from the effects of sAPPα reported here cannot be excluded.
The decrease in mEPSC frequency but not amplitude following acute sAPPα application suggests a change in presynaptic release properties. We therefore assessed the effect of sAPPα on presynaptic vesicle recycling using the fluorescent membrane dye FM1–43. We measured presynaptic strength by measuring the density (D) of FM+ boutons per image area and the change in fluorescence intensity (ΔF) of FM1–43 signals at individual boutons of cultured hippocampal neurons using a combined FM1–43 loading/unloading stimulation paradigm (Fig. 3F). Application of sAPPα decreased the total presynaptic strength (S = ΔF × D) across synaptic populations (Fig. 3G, S7A) in a dose-dependent manner (Fig. 3H), reaching 57 ± 7 % (n=8 experiments; P < 0.001) reduction at 1 μM sAPPα. This decrease was not observed with deletion of the ExD (sAPPαΔExD, 1 μM) (Fig. 3H, S7B) and was occluded by the GABABR antagonist CGP54626 (CGP, 10μM) (Fig 3I, S7C), indicating that GABABR1a mediates the presynaptic inhibition induced by sAPPα.
sAPP enhances short-term plasticity at Schaffer collateral synapses in a GABABR1a-dependent manner
We next assessed the effect of sAPPα on synaptic transmission in an intact circuit at CA3-CA1 Schaffer collateral (SC) synapses, which exclusively contain GABABR1a receptors (32). We measured field EPSPs (fEPSPs) evoked by low frequency stimulation (0.1 Hz) at varying intensities (30–150 μA) in CA1 stratum radiatum after 90 min pre-incubation of acute hippocampal slices with or without 1 μM sAPPα (Fig 4A). Treatment with sAPPα reduced fEPSP amplitude and decreased the slope of the input/output (i/o) curve by 23% (Fig. S8A), indicating that sAPPα suppresses basal synaptic transmission at SC synapses. To specifically assess if sAPPα affects presynaptic properties, we applied a burst of 5 stimuli at 3 different frequencies (20, 50, and 100 Hz) to induce short-term facilitation, which inversely correlates with the probability of neurotransmitter release. Facilitation was higher for each frequency tested in sAPPα-incubated compared to control slices (Fig. 4B, S8B,C). Analysis of the paired-pulse ratio (PPR) for the first 2 stimuli showed an increased PPR for each frequency following sAPPα treatment (Fig. 4C), indicating a decreased release probability. Deletion of the ExD (sAPPαΔExD 1 μM) abolished the sAPPα-mediated effect on the i/o curve (Fig. S9D), short-term facilitation (Fig. 4D, S8E,F), and PPR (Fig. 4E). In addition, preincubation of slices with the GABABR antagonist CGP54626 (CGP, 10 μM) abolished the sAPPα-mediated decrease in the slope of the i/o curve (Fig. S8G) and occluded the sAPPα-induced increase in short-term facilitation and PPR at each frequency (Fig. 4F,G, S8H,I), demonstrating GABABR-dependence of these effects. Altogether, these results indicate that sAPPα controls vesicle release at SC synapses by acting on presynaptic GABABR1a.
Fig. 4. sAPP enhances short-term plasticity at Schaffer collateral synapses in a GABABR1a-dependent manner.
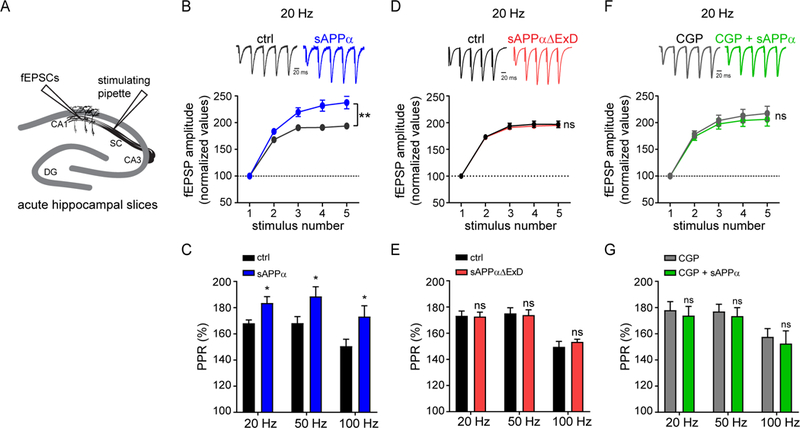
(A) Cartoon of fEPSC measurements in acute mouse hippocampal slices reported in B-G. (B) Representative traces (upper) and average fEPSP amplitude (lower) recorded at Schaffer collaterals (SC) in response to high-frequency burst stimulation at 20 Hz in mouse hippocampal slices incubated without (n = 12, N = 7) or with sAPPα (1 μM, Fc-tag enzymatically removed) (n = 10, N = 7). fEPSPs were normalized to the peak amplitude of the first response. (C) Paired-pulse ratios (PPR) for the first two pulses at each frequency (20 Hz, 50 Hz, and 100 Hz). (D) Same as B but in slices incubated without (n = 10, N = 4) or with sAPPαΔExD (1 μM, Fc-tag enzymatically removed, n = 9, N = 4). (E) Same as C. (F) Same as B but in slices incubated with CGP 54626 (CGP, 10μM) alone (n = 9, N =4) and slices incubated with CGP + sAPPα (n = 8, N = 4). (G) Same as C. The number of slices is defined as n, and the number of independent experiments or mice is defined as N. Graphs show means ± SEM. * P < 0.05, ** P < 0.1 *** P < 0.001. Two-way ANOVA analysis.
A short peptide within the APP ExD suppresses synaptic vesicle release via GABABR1a
A GABABR1a isoform-specific modulator has potential therapeutic implications for a number of neurological disorders involving GABABR signaling (29). Since we observed that purified protein corresponding to the linker region of APP (Fig. 2A) was sufficient to mimic the effects of sAPPα on mEPSC frequency (Fig. 3D), we set out to identify the minimally active region within the ExD. Alignment of the sAPP ExD (amino acids (AA) 195–227 of APP695) from seven vertebrate species revealed the strongest conservation within a 17AA stretch (204–220AA; Fig. 5A). The corresponding synthetic APP 17mer peptide bound sushi 1 of GABABR1a with a KD of 810nM (Fig. 5B), in the same range as the binding affinity of the entire linker region (Fig. 2C). Shortening the peptide to a synthetic 9mer consisting of APP695 residues 204–212 (APP 9mer) lowered the KD to 2.3 μM (Fig. 5C); whereas residues 211–220 failed to bind sushi 1 (Fig. S9A). Thus, a conserved, minimal 9AA sequence within the sAPP ExD is sufficient for direct binding to the sushi 1 domain of GABABR1a.
Fig. 5. A short peptide within the APP ExD suppresses synaptic vesicle release via GABABR1a.
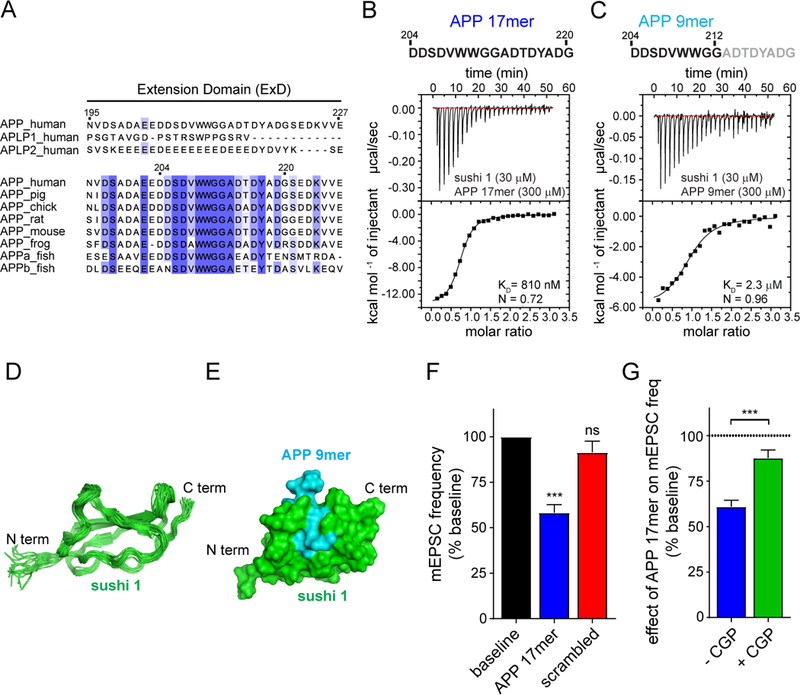
(A) Sequence alignment for the extension domain (ExD) of human APP with APLPs and with 7 vertebrate APP sequences. (B,C) ITC binding experiments of purified sushi 1 and synthetic peptides within the ExD corresponding to (B) 204–220AA or (C) 204–212AA of APP695. (D) An ensemble of 20 lowest-energy NMR structures of the sushi 1 domain of GABABR1a when bound to the APP 9mer peptide. (E) A structural model of the complex between the sushi 1 domain of GABABR1a (green) and the APP 9mer peptide (cyan) shown as the molecular surface. Protein termini are indicated by the labels. (F) Average mEPSC frequency normalized to baseline recorded from mouse primary neurons before (baseline) and after treatment with 17mer APP peptide (250 nM, APP695 204–220AA) (n= 20, N=3) or scrambled 17mer control peptide (250 nM, n= 18, N= 4) (one way ANOVA with Dunnett’s post hoc analysis). (G) Quantification of the effect of 250 nM 17mer APP peptide (APP695 204–220AA) on mEPSC frequency normalized to baseline (K) either without (n=14; N=3) or with preincubation with CGP55845 (CGP, 5 μM; n=16, N=3) (unpaired t-test). Dotted line denotes baseline. The number of neurons is defined by n. The number of independent experiments is defined by N. Graphs show means ± SEM. * P < 0.05, ** P < 0.1 *** P < 0.001.
To gain further insight in the binding of the APP 9mer to the GABABR1a sushi 1 domain, we used nuclear magnetic resonance (NMR) spectroscopy. As previously reported (40), we observed that the sushi 1 domain of GABABR1a is natively unstructured (Fig. S9B). Strikingly, APP 9mer binding stabilized the sushi 1 domain of GABABR1a, allowing determination of its solution structure (Figure 5D, S9C) and generation of a structural model of the complex (Fig. 5E). In our model, a valine and tryptophan at 208–209AA of APP695 bind within a pocket of sushi 1, formed by the loops and the short beta-strand in the N-terminal part of the protein (32–53 AA of full-length GABABR1a) (Fig. S9D). Thus, APP binding induces a conformational change in the natively unstructured sushi 1 domain of GABABR1a. This structure-function relationship strongly supports the physiological relevance of the interaction.
As the affinity for sushi 1 was better retained in the APP 17mer compared to the 9mer (Fig. 5B,C), we next tested whether the APP 17mer could functionally mimic sAPPα. Acute application of the APP 17mer peptide, but not of a scrambled 17mer control peptide, reduced mEPSC frequency in hippocampal neurons to a similar degree as sAPPα (Fig. 5F, S9E) and was already apparent at 25 nM (Fig. S9F). Pretreatment with the GABABR antagonist CGP55845 (CGP, 5μM) blocks this effect (Fig. 5G, S9G). Together, these findings show that the APP 17mer peptide mimics the effects of sAPPα on GABABR1a-dependent inhibition of synaptic vesicle release.
APP 17mer peptide suppresses neuronal activity of CA1 pyramidal cells in vivo
In the final series of experiments, we utilized the APP 17mer peptide as a tool to examine the effects of sAPP-GABABR signaling on neuronal activity in vivo. Using two-photon calcium imaging, we measured calcium transients of CA1 hippocampal neurons in anesthetized Thy1-GCaMP6s mice before (baseline) and after a 60–90 min superfusion of the exposed hippocampus with either baclofen (30 μM), APP 17mer (5μM), or scrambled 17mer control peptide (5μM) (Fig. 6A). Application of 30 μM baclofen caused a dramatic decrease in the frequency of calcium transients compared to baseline (Fig. S10A-C), indicating that activation of GABABRs strongly suppresses neuronal activity in CA1 pyramidal neurons in vivo. Consistent with our results in cultured hippocampal neurons, application of the APP 17mer significantly reduced the frequency of calcium transients compared to baseline (Fig. 6B-D, Movie S1). The frequency of calcium transients was restored back to baseline following a two-hour wash-out of the peptide (Fig. S10D-F), indicating that the suppression of CA1 neuron activity by the APP 17mer peptide is reversible. Furthermore, the scrambled 17mer control peptide did not affect the frequency of calcium transients (Fig. 6E-G; S10G-I, Movie S2). Together, these results indicate that APP inhibits neuronal activity in vivo and that the GABABR1a binding domain is sufficient for such inhibition.
Fig. 6. A 17AA peptide corresponding to the GABABR1a binding region within APP suppresses neuronal activity in vivo.
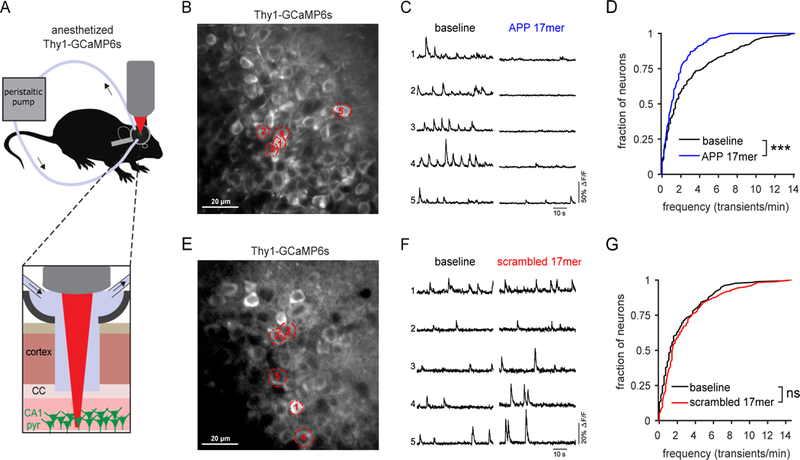
(A) Cartoon of in vivo 2-photon calcium imaging of CA1 hippocampus of anesthetized Thy1-GCaMP6s mice with superfusion of APP 17mer, or scrambled control 17mer. (B) in vivo image of CA1 hippocampal neurons of Thy1-GCaMP6s mice. Representative neurons indicated with dotted outline. (C) Calcium traces of five representative neurons, labeled in panel A, before (baseline) and during bath application of 5 μM APP 17mer peptide corresponding to the GABABR1a binding region within APP (APP 17mer). (D) Cumulative distribution of the frequency of calcium transients at baseline (black line) and during APP 17mer bath application (blue line) (n=277; N=3). (E) in vivo image of CA1 hippocampal neurons of Thy1-GCaMP6s mice. (F) Calcium traces of five representative neurons, labeled in panel D, before (baseline) and during bath application of 5μM scrambled 17mer control peptide (scrambled 17mer). (G) Cumulative distribution of the frequency of calcium transients at baseline (black line) and during scrambled 17mer bath application (red line) (n=183; N=3). Wilcoxon rank sum test. The number of neurons is defined by n. The number of mice is defined by N. *** P < 0.001; NS P>0.05
Discussion
Our studies reveal that sAPP acts as a GABABR1a-specific ligand to suppress synaptic vesicle release. Consequently, sAPP modulates hippocampal synaptic plasticity and neurotransmission in vivo. APP is among the most abundant proteins in synaptic boutons (25), and deletion of App in mice leads to synaptic deficits (7–9, 21, 22). Synaptic activity enhances proteolytic processing of APP (41, 42) and GABABR is a key regulator of homeostatic synaptic plasticity (43). Hence, our observations raise the possibility that the sAPP-GABABR1a interaction acts as an activity-dependent negative feedback mechanism to suppress synaptic release and maintain proper homeostatic control of neural circuits. While AD-causing mutations in APP all affect Aβ generation, it is not entirely clear whether other aspects of APP function contribute to AD. Network abnormalities such as hyperexcitability and hypersynchronization precede clinical onset of AD in human patients (44). Some studies indicate that sAPP levels may be altered in AD (14). Interestingly, a GABABR antagonist has been shown to improve memory in animal models and patients with mild cognitive impairment (45–47). Moreover, as most transgenic AD mouse models overexpress sAPP, the role of sAPP in synaptic phenotypes of transgenic APP mice should be considered, particularly given evidence that network hyperexcitability in these mice is independent of Aβ production (48).
GABABR signaling has been implicated in a number of neurological and psychiatric disorders including epilepsy, depression, addiction, and schizophrenia (49). Selective binding partners of the GABABR1a sushi domains are of potential therapeutic interest due to localization and functional differences of GABABR1 isoforms (32, 50) as well as the adverse effects of current non-specific agonists (29). The identification of sAPP as a functional GABABR1a-specific binding partner provides a target for the development of therapeutic strategies for modulating GABABR1a-specific signaling in neurological and psychiatric disorders. The identification of short APP peptides that confer structure in the GABABR1a sushi 1 domain and modulate neurotransmission in vivo are major steps towards development of a GABABR1a isoform-specific therapeutic.
Summary of Methods
To identify candidate synaptic interactors for sAPP, affinity purification experiments were performed using recombinant sAPP-Fc to pull down interacting proteins from synaptosome extracts, followed by mass spectrometric analysis of bound proteins. Cell surface binding assays, biolayer interferometry, and isothermal titration calorimetry were used to determine domains of interaction and apparent binding affinities between sAPP to GABABR. Nuclear magnetic resonance spectroscopy was used to generate a structural model of the APP-GABABR complex. The function of the sAPP-GABABR interaction was investigated by accessing spontaneous postsynaptic currents and FM1–43 dye labeling in mouse hippocampal cultures, short-term facilitation in acute hippocampal slices, and 2-photon in vivo calcium imaging in CA1 hippocampus of anesthetized Thy1-GCaMP6 mice. The details of each of these methods are described in the supplementary materials.
Supplementary Material
Acknowledgments:
We thank Genevieve Conway, An Snellinx, Katleen Craessaerts, Katrien Horré, Kristel Vennekens, Véronique Hendrickx, and Jonas Verwaeren for technical help. We thank Charlotte Martin, Nuno Apóstolo, Giuseppe Condomitti, Gabriele Marcassa, and Iordana Chrysidou for experimental assistance. We thank Lieven Buts for help with NMR structure calculations; Marc Aurel Busche for advice on in vivo calcium imaging experiments; Patrick Vanderheyden, Sven Zels, Isabelle Beets, Liliane Schoofs, Henry Dunn, Kirill Martemyanov for advice on and/or performing GPCR activity experiments; Ulrike Mueller for the APP/APLP1 KO mice.
Funding: This work was supported by Alzheimer’s Association Research Fellowship (AARF-16–442885, HCR), Stichting Voor Alzheimer Onderzoek Pilot Grant (16011, HCR); Agency for Innovation by Science and Technology in Flanders (IWT 141698, AS), National Science Foundation BRAIN EAGER MCB-1450895,and IOS-1755189 (DC), and Robert Wood Johnson Foundation Grant # 74260 to the Child Health Institute of New Jersey (DC); RO1AG061787 (JNS); European Research Council (ERC) (724866, IS); Vlaams Initiatief voor Netwerken voor Dementie Onderzoek (VIND, Strategic Basic Research Grant 135043) (BDS); KU Leuven Methusalem Grant (BDS and JdW); ERC Starting Grant (311083) and FWO Odysseus Grant (JdW). BDS is supported by the Arthur Bax and Anna Vanluffelen chair for Alzheimer’s disease, “Opening the Future” of the Leuven Universiteit Fonds (LUF) and by the “Geneeskundige Stichting Koningin Elisabeth”.
Footnotes
Author contributions: H.C.R, B.D.S, and J.d.W. conceived the study. All authors planned experiments. H.C.R, D.D.M, A.S., S.F. I.V.M., A.V, E.C., I.V., J.N., F.M.R, and K.D.W. performed the experiments. All authors interpreted data. H.C.R., B.D.S., and J.d.W. wrote the first version of the manuscript. All authors contributed to and approved the final version.
Competing interests: EP 16180433.1 “Therapeutic agents for neurological and psychiatric disorders”
Data and materials availability: Resonance assignments were deposited in the BMRB data bank (accession number 27581) and the 20 lowest-energy structures were deposited in the PDB bank (accession code 6HKC).
References and Notes:
- 1.Goldgaber D, Lerman M, McBride OW, Saffiotti U, Gajdusek DC, Characterization and Chromosomal Localization of a cDNA Encoding Brain Amyloid of Alzheimer’s Disease. Science. 235, 877–880 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Robakis NK, Ramakrishna N, Wolfe G, Wisniewski HM, Molecular Cloning and Characterization of a cDNA Encoding the Cerebrovascular and the Neuritic Plaque Amyloid Peptides. Proc. Natl. Acad. Sci. 84, 4190–4194 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. R. Tanzi RE, Gusella JF, Watkins PC, Bruns GA, St George-Hyslop P, Van Keuren ML, Patterson D, Pagan S, Kurnit DM, Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 235 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Kang J et al. , The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 325 (1987), pp. 733–6. [DOI] [PubMed] [Google Scholar]
- 5.Haass C, Kaether C, Thinakaran G, Sisodia S, Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2 (2012), doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willem M et al. , η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature. 526, 443–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson GR et al. , Age-related cognitive deficits, impaired long-term potentiation and reduction in synpatic marker density in mice lacking the beta-amyloid precursor protein. Neuroscience. 90, 1–13 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Seabrook GR et al. , Mechanisms contributing to the deficits in hippocampal synaptic plasticity in mice lacking amyloid precursor protein. Neuropharmacology. 38, 349–359 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Tyan S-H et al. , Amyloid precursor protein (APP) regulates synaptic structure and function. Mol. Cell. Neurosci. 51, 43–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzjohn SM et al. , Similar levels of long-term potentiation in amyloid precursor protein - null and wild-type mice in the CA1 region of picrotoxin treated slices. Neurosci. Lett. 288, 9–12 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Wang Z, Wang B, Justice NJ, Zheng H, Amyloid Precursor Protein Regulates Ca v 1 . 2 L-type Calcium Channel Levels and Function to Influence GABAergic Short-Term Plasticity. Neuroscience. 29, 15660–15668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B et al. , The Amyloid Precursor Protein Controls Adult Hippocampal Neurogenesis through GABAergic Interneurons. J Neurosci. 34, 13314–13325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Wang J, Jiang J, Zheng X, Justice NJ, APP modulates KCC2 expression and function in hippocampal GABAergic inhibition, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mockett BG, Richter M, Abraham WC, Müller UC, Therapeutic Potential of Secreted Amyloid Precursor Protein APPsα. Front. Mol. Neurosci. 10, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller UC, Deller T, Korte M, Not just amyloid : physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. (2017), doi: 10.1038/nrn.2017.29. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa K, Barger SW, Blalock EM, Mattson MP, Activation of K+ channels and suppression of neuronal activity by secreted beta-amyloid-precursor protein. Nature. 379 (1996), pp. 74–78. [DOI] [PubMed] [Google Scholar]
- 17.Ishida A, Furukawa K, Keller JN, Mattson MP, Secreted form of beta-amyloid precursor protein shifts the frequency dependency for induction of LTD, and enhances LTP in hippocampal slices. Neuroreport. 8, 2133–7 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Taylor CJ et al. , Endogenous secreted amyloid precursor protein-α regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol. Dis. 31, 250–260 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Xiong M et al. , Secreted amyloid precursor protein-alpha can restore novel object location memory and hippocampal LTP in aged rats. Neurobiol. Learn. Mem. 138, 291–299 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Weyer SW et al. , Comparative analysis of single and combined APP/APLP knockouts reveals reduced spine density in APP-KO mice that is prevented by APPsα expression. Acta Neuropathol. Commun. 2, 36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ring S et al. , The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 27, 7817–7826 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hick M et al. , Acute function of secreted amyloid precursor protein fragment APPsα in synaptic plasticity. Acta Neuropathol. 129, 21–37 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Saitoh T et al. , Secreted form of amyloid-beta protein precursor is involved in the growth regulation of fibroblasts. Cell. 58, 615–622 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Reinhard C, Borgers M, David G, De Strooper B, Soluble amyloid-β precursor protein binds its cell surface receptor in a cooperative fashion with glypican and syndecan proteoglycans. J. Cell Sci. 126, 4856–61 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Wilhelm BG et al. , Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 344, 1023–8 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Savas JN et al. , Ecto-Fc MS identifies ligand-receptor interactions through extracellular domain Fc fusion protein baits and shotgun proteomic analysis. Nat. Protoc. 9, 2061–2074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwenk J et al. , Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat. Neurosci. 19 (2015), doi: 10.1038/nn.4198. [DOI] [PubMed] [Google Scholar]
- 28.Gassmann M, Bettler B, Regulation of neuronal GABAB receptor functions by subunit composition. Nat. Rev. Neurosci. 13, 380–394 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Pin J-P, Bettler B, Organization and functions of mGlu and GABAB receptor complexes. Nature. 540, 60–68 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Coburger I et al. , Analysis of the overall structure of the multi-domain amyloid precursor protein (APP). PLoS One. 8, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shariati SAM, De Strooper B, Redundancy and divergence in the amyloid precursor protein family. FEBS Lett. 587, 2036–2045 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Vigot R et al. , Differential Compartmentalization and Distinct Functions of GABAB Receptor Variants. Neuron. 50, 589–601 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biermann B et al. , The Sushi domains of GABAB receptors function as axonal targeting signals. J. Neurosci. 30, 1385–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldmeier PC, Kaupmann K, Urwyler S, Roles of GABAB receptor subtypes in presynaptic auto- and heteroreceptor function regulating GABA and glutamate release. J. Neural Transm. 115, 1401–1411 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Wierda KDB, Sørensen JB, Innervation by a GABAergic neuron depresses spontaneous release in glutamatergic neurons and unveils the clamping phenotype of synaptotagmin-1. J. Neurosci. 34, 2100–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scanziani M, Capogna M, Gähwiler BH, Thompson SM, Presynaptic inhibition of miniature excitatory synaptic currents by baclofen and adenosine in the hippocampus. Neuron. 9, 919–927 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Iyadomi M, Iyadomi I, Kumamoto E, Tomokuni K, Yoshimura M, Presynaptic inhibition by baclofen of miniature EPSCs and IPSCs in substantia gelatinosa neurons of the adult rat spinal dorsal horn. Pain. 85, 385–393 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Rohrbacher J, Jarolimek W, a Lewen U Misgeld, GABAB receptor-mediated inhibition of spontaneous inhibitory synaptic currents in rat midbrain culture. J. Physiol. 500 ( Pt 3, 739–49 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulrich D, Bettler B, GABAB receptors: synaptic functions and mechanisms of diversity. Curr. Opin. Neurobiol. 17, 298–303 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Blein S et al. , Structural analysis of the complement control protein (CCP) modules of GABAB receptor 1a: Only one of the two CCP modules is compactly folded. J. Biol. Chem. 279, 48292–48306 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Nitsch RM, Farber S. a, Growdon JH, Wurtman RJ, Release of amyloid beta-protein precursor derivatives by electrical depolarization of rat hippocampal slices. Proc. Natl. Acad. Sci. U. S. A. 90, 5191–5193 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamenetz F et al. , APP Processing and Synaptic Function. Neuron. 37, 925–937 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Vertkin I et al. , GABAB receptor deficiency causes failure of neuronal homeostasis in hippocampal networks. Proc. Natl. Acad. Sci. U. S. A. 112, E3291–3299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palop JJ, Mucke L, Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat. Rev. Neurosci. 17, 777–792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y et al. , Implications of GABAergic Neurotransmission in Alzheimer’s Disease. Front. Aging Neurosci. 8, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Froestl W et al. , SGS742: The first GABAB receptor antagonist in clinical trials. Biochem. Pharmacol. 68, 1479–1487 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Helm KA et al. , GABAB receptor antagonist SGS742 improves spatial memory and reduces protein binding to the cAMP response element (CRE) in the hippocampus. Neuropharmacology. 48, 956–964 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Born HA et al. , Genetic Suppression of Transgenic APP Rescues Hypersynchronous Network Activity in a Mouse Model of Alzeimer’s Disease. J. Neurosci. 34, 3826–3840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bettler B, Kaupmann K, Mosbacher J, Gassmann M, Structure M, Molecular Structure and Physiological Functions of GABA B Receptors. Fisiol Rev. 84, 835–867 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Foster JD, Kitchen I, Bettler B, Chen Y, GABAB receptor subtypes differentially modulate synaptic inhibition in the dentate gyrus to enhance granule cell output. Br. J. Pharmacol. 168, 1808–1819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips GR et al. , The presynaptic particle web: Ultrastructure, composition, dissolution, and reconstitution. Neuron. 32, 63–77 (2001). [DOI] [PubMed] [Google Scholar]
- 52.De Strooper B, Umans L, Van Leuven F, Van Den Berghe H, Study of the synthesis and secretion of normal and artificial mutants of murine amyloid precursor protein (APP): Cleavage of APP occurs in a late compartment of the default secretion pathway. J. Cell Biol. 121, 295–304 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sala Frigerio C et al. , ??-Secretase cleavage is not required for generation of the intracellular C-terminal domain of the amyloid precursor family of proteins. FEBS J. 277, 1503–1518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Wit J et al. , LRRTM2 Interacts with Neurexin1 and Regulates Excitatory Synapse Formation. Neuron. 64, 799–806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savas JN et al. , The Sorting Receptor SorCS1 Regulates Trafficking of Neurexin and AMPA Receptors. Neuron. 87, 764–780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.a J. Link et al. , Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Washburn MP, Wolters D, Yates JR, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–7 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW, Extremely Long-Lived Nuclear Pore Proteins in the Rat Brain. Science (80-. ). 335, 942–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP, Evaluation of Multidimensional Chromatography Coupled with Tandem Mass Spectrometry ( LC / LC - MS / MS ) for Large-Scale Protein Analysis : The Yeast Proteome. J. Proteome Res. 2, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- 60.D. Cociorva, D. L. Tabb, J. R. Yates, in Current Protocols in Bioinformatics, Andreas, D, Baxevanis, Eds. (2007). [DOI] [PubMed]
- 61.Tabb DL, McDonald WH, Yates JR, DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics. J. Proteome Res. 1, 21–26 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.JK E, AL M, JR Y., An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 5, 976–89 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Heber S et al. , Mice with Combined Gene Knock-outs Reveal Essential and Partially Redundant Functions of Amyloid Precursor Protein Family Members. J. Neurosci. 20, 7951–7963 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abramov E et al. , Amyloid-beta as a positive endogenous regulator of release probability at hippocampal synapses. Nat. Neurosci. 12, 1567–1576 (2009). [DOI] [PubMed] [Google Scholar]
- 65.A. Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax, NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 66.E. D. Vranken WF; Boucher W; Stevens TJ; Fogh RH; Pajon A; Llinas M; Ulrich EL; Markley JL; Ionides J; Laue, The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 67.R. W. Cheung MS; Maguire ML; Stevens TJ; Broadhurst, DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202, 223–233 (2010). [DOI] [PubMed] [Google Scholar]
- 68.L. Güntert, P; Buchner, Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR. 62, 453–471 (2015). [DOI] [PubMed] [Google Scholar]
- 69.G. L. Brünger AT; Adams PD; Clore GM; DeLano WL; Gros P; Grosse-Kunsteleve RW; Jiang J-S; Kuszewski J; Nilges M; Pannu NS; Read RJ; Rice LM; Simonson T; Warren, Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Cryst. Ser. D. 54, 901–921 (1998). [DOI] [PubMed] [Google Scholar]
- 70.W. M.P., The Transferred NOE In: Webb GA (eds) Modern Magnetic Resonance. Springer, Dordrecht: (2008). [Google Scholar]
- 71.G. M. Schwieters, CD; Kuszewski J; Tjandra N; Clore, The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 66–7r (2003). [DOI] [PubMed] [Google Scholar]
- 72.Dana H et al. , Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS One. 9 (2014), doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pologruto TA, Sabatini BL, Svoboda K, ScanImage: Flexible software for operating laser scanning microscopes. Biomed. Eng. Online. 2, 1–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Özkan E et al. , An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 154 (2013), doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anton van der Merwe P, Neil Barclay A, Transient intercellular adhesion: the importance of weak protein-protein interactions. Trends Biochem. Sci. 19, 354–358 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Blein S et al. , Structural analysis of the complement control protein (CCP) modules of GABAB receptor 1a: Only one of the two CCP modules is compactly folded. J. Biol. Chem. 279, 48292–48306 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


