Abstract
Tumors of nearly every origin activate the expression of genes which is otherwise restricted to gametogenic cells. These genes encode proteins termed cancer/testis (CT) antigens, since expression outside of their naturally immune-privileged site can evoke an immune response. Despite extensive efforts to exploit CT antigens as immunotherapeutic targets, investigation of whether these proteins participate in tumorigenic processes has lagged. Here, we discuss emerging evidence that demonstrates that CT antigens can confer a selective advantage to tumor cells by promoting oncogenic processes or permitting evasion of tumor-suppressive mechanisms. These advances indicate the inherent flexibility of tumor cell regulatory networks to engage aberrantly expressed proteins to promote neoplastic behaviors, which could ultimately present novel therapeutic entry points.
Keywords: cancer/testis antigens, CT genes, CT antigens, cancer germline genes, tumor antigens, CT antigen function
A Brief History of Tumor Antigen Discovery
By the 1970s it was well established that the immune system was capable of recognizing tumor cells as foreign, but the identity of the tumor-specific antigens recognized by T cells was largely unknown. The turning point came in the second half of the decade when Lloyd Old’s group developed autologous typing, which permitted clonal selection of antigenic (and non-antigenic) tumor cells using sera obtained from the same patient [1]. Subsequently, Thierry Boon and colleagues used expression cloning with DNA from antigenic melanoma clones to identify gene products that could confer a cellular immune response in non-antigenic counterparts [2]. Remarkably, restoration of an antigen-coding gene, MAGEA1 (the founding member of the “melanoma antigen” family), sensitized the resistant tumor cell clones to destruction by autologous cytotoxic T lymphocytes (CTLs) [2, 3]. Subsequent analysis revealed that MAGEA1 expression was normally restricted to the testes and placenta, yet detectable in tumors of various origins [4]. In 1995, Michael Pfreundschuh’s group improved the identification of tumor-specific antigens by developing the serological analysis of recombinant cDNA expression libraries (SEREX). Here, a phage display library derived from tumor cDNA is screened with autologous patient sera [5]. This technique facilitated the discovery of numerous tumor antigens, including NY-ESO-1, the most successful target to date for cancer immunotherapy [6–8]. Through these efforts, it soon became clear that MAGEA1 represented the archetype of a family of genes with a unique testis/tumor-restricted expression pattern. Furthermore, seminal parallel discoveries suggested that their encoded proteins are indeed substrates for antigen processing machinery that leads to the presentation of cleaved peptides on the cell surface by major histocompatibility complex (MHC) molecules (Figure 1A) [9]. As the testes are an immune privileged site, CTLs most likely recognize these peptides as foreign, thereby triggering an anti-tumor immune response [10].
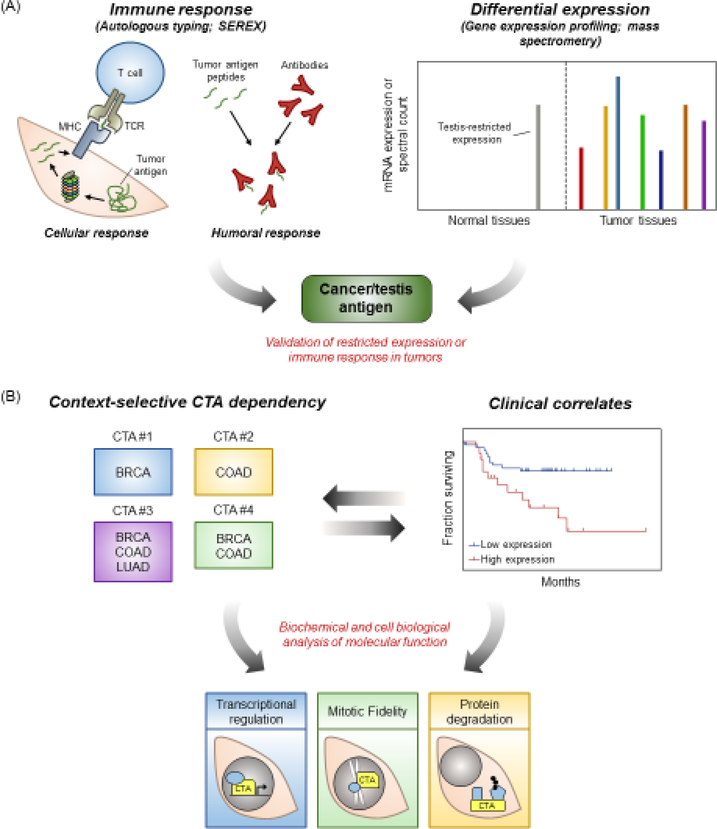
Figure 1. Workflow for the identification and functional characterization of CT antigens.
(A) CT antigens are identified either by their propensity to evoke an immune response (left) or their differential, testis-enriched expression pattern (right). (B) Differential expression analysis and patient survival data can be used to identify appropriate contexts for downstream functional analysis. Abbreviations: MHC, major histocompatibility complex; TCR, T cell receptor; BRCA, breast cancer; COAD, colon adenocarcinoma; LUAD, lung adenocarcinoma.
Birth and Growth of the CT Antigen Family
It was at the time of the NY-ESO-1 discovery that Lloyd Old coined the term cancer/testis (CT) antigen to refer to this new family of proteins [7, 11]. This name captured both the unique expression pattern of these genes (termed CT genes) and the antigenic potential of their encoded proteins (Figure 1A). It is important to note that many genes now classified as CT antigens have not been demonstrated to evoke an immune response. This was due in large part to a shift in the discovery platform for CT genes. The availability of expression profiling techniques, beginning in the early 2000s, permitted the in silico identification of genes that exhibit the classic testis/tumor-restricted mRNA expression pattern [12–17]. These unbiased analyses also revealed a subset of CT antigens whose mRNA was detectable in normal tissues, typically the brain. Thus, CT antigen classification was further refined to 3 categories: 1) testis-restricted (detectable only in testis and tumor tissues), 2) testis/brain-restricted (detectable in testis, brain, and tumor tissues), and 3) testis-selective (detectable in testis, tumor and 1–2 additional normal tissues) [13]. CT genes are also grouped into subfamilies based on sequence similarity and proximity of their chromosomal location. Interestingly, about half of the CT genes are located on the × chromosome (termed CT-X genes) and are members of multigene families (MAGE, GAGE, PAGE, SSX, CAGE, CSAG1, CTAG, and SPANX families in particular) [18]. Detailed records for most CT antigens can be found in the CT database, an online repository of information regarding the expression and immunogenic properties of CT genes and their encoded antigens, respectively (http://www.cta.lncc.br/) [19]. While the compendium of CT genes and candidate CT antigens has expanded rapidly, the laborious task of demonstrating antigenicity still remains. As such, the terms cancer/germline gene or oncofetal protein have emerged in place of CT antigen. However, it is our opinion that the terms CT antigen (often abbreviated as CTA) and CT gene be preserved as they best capture the unique expression pattern, potential antigenicity, and history of this family. Moving forward, advances in immunopeptidomics could streamline characterization of CT antigens capable of evoking an immune response and ultimately provide additional immunotherapeutic targets [20–22].
CT Antigens Have Mostly Unknown Cellular Functions
Despite long-standing efforts to exploit the antigenic properties of CT antigens for use in immunotherapy, scant information exists concerning their biological relevance to either spermatogenesis or malignancy. Provocatively, a number of studies have tied individual or sets of CT antigens to poor survival in tumors, suggesting that their expression could promote aggressive tumor growth and/or chemorefractory disease [23–26]. However, ascribing malignant functions to individual CT antigens has proven difficult for several reasons. Many of these proteins lack known motifs or domains from which their activities could be inferred. Additionally, a number of CT antigens have been characterized as intrinsically disordered, making functional predictions based on amino acid sequence alone unreliable [27]. Perhaps the most formidable challenge is that many CT genes are not well conserved, thus precluding traditional in vivo genetic studies. Indeed, male reproductive genes have undergone rapid and selective evolution in primates in comparison to other mammals, and many of those encoding CT antigens are no exception [18, 28, 29]. In particular, many CT-X antigens do not have murine counterparts. They are found in normal spermatogonia and are often classified as testisrestricted, but their expression can be readily activated by DNA demethylation in tumors, potentially making them optimal immunotherapeutic targets. However, their contribution to gametogenesis has been difficult to investigate due to a lack of model organisms for genetic studies. On the other hand, non-X CT antigens are typically expressed in spermatocytes and many have murine orthologs. The functions of these non-X CT antigens have been better characterized due to the availability of knockout mouse models. These studies have revealed functional roles in genomic maintenance, meiosis, transcriptional regulation, motility, and energy regulation during spermatogenesis [30]. Notably, CT antigen knockouts often leave animals infertile but otherwise healthy, reinforcing the notion that their inhibition in tumors would have negligible deleterious impacts on normal tissues.
Whether these anomalously expressed proteins could indeed take on functions in a non-native cellular environment and participate in tumorigenesis has been a lingering question since their initial discovery. The functional investigation of CT antigens found to be essential for tumor cell fitness, or those whose expression can predict patient outcome, has the potential to unveil previously unknown mechanisms that promote unbridled growth of cancers (Figure 1B). Indeed, the notion that activation of germline gene expression programs can drive malignancy is a long-held hypothesis first developed by Lloyd Old and colleagues [31]. In the following sections, we highlight recent findings from Drosophila melanogaster that appear to support this notion, and discuss the accumulating body of evidence that CT antigens are functional in human cancers and often essential for neoplastic processes.
CT Genes Contribute to Tumorigenesis in Drosophila melanogaster
Using a malignant brain tumor model driven by a mutation in the lethal (3) malignant brain tumor (l(3)mbt) gene, Janic and colleagues found that the expression of Drosophila CT genes drives tumor growth and that their suppression is sufficient to restrict tumor growth [32]. Notably, these Drosophila CT genes have human orthologs, including piwi (PIWIL1) and nanos (NANOS1), many of which are upregulated in human cancers [33]. An RNA interference screen performed using the l(3)mbt brain tumor model also recently identified the Drosophila gene meiotic W68 (mei-W68), an ortholog of the human CT gene SPO11, as essential for tumor growth [34]. Given the role of SPO11 in the formation of DNA double-strand breaks during meiosis, it is possible that mei-W68/SPO11 contributes to tumorigenesis via modulation of genome stability [30]. However, future work will be required to elucidate the exact mechanism through which this protein supports tumor growth in Drosophila. Studies in human cancers demonstrating the relevance of SPO11 to tumorigenesis are also lacking, despite its reported expression in tumor cells and classification as a CT antigen [19]. Nonetheless, it is becoming clear that using model organisms such as Drosophila is an attractive approach to examine the extent to which activation of germline gene expression programs contributes to tumorigenesis. However, caution must be used in presuming that genetically engineered organisms will activate the same germline genes as humans. The molecular etiology of genetically engineered tumors may bypass the selective pressures encountered during human tumor evolution. Thus, the requirement of specific CT antigens to overcome barriers to transformation may be quite different among species.
CT Antigens Participate in Diverse Cellular Processes in Human Cancer
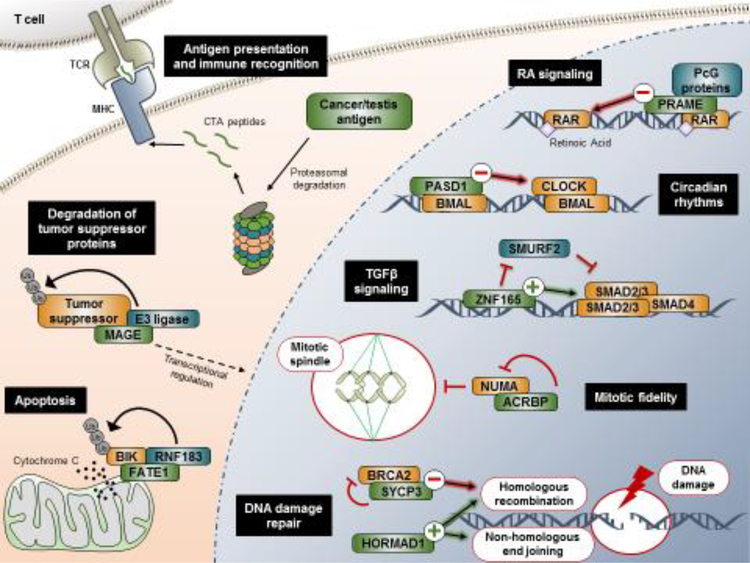
A number of studies have described individual CT antigens that contribute to neoplastic behaviors of human tumor cells (Table 1) [24, 26, 35–76]. Collectively, these findings have begun to broadly demonstrate the diversity of CT antigen function in human cancer. In many cases, these discoveries are based on large-scale functional genomic screens, proteomic analyses, or gene expression profiling [24, 35, 39, 43, 46, 51–53, 59, 67]. The utility of such approaches in identifying functional CT antigens speaks to the importance of unbiased, hypothesis-generating tools in revealing unanticipated players on the tumorigenic landscape. Interestingly, current evidence indicates that CT antigen function can be organized into three broad categories: transcriptional regulation, mitotic fidelity, and protein degradation (Table 1, Figure 1). A summary of some of these reported functions and their associated cellular processes is represented in Figure 2 (Key Figure).
Table 1.
CT antigens reported to promote neoplastic behaviors in human tumor cells
| Name | Description | Refs |
|---|---|---|
| ACRBP | Interacts with NUMA to support mitotic spindle dynamics | [35, 36] |
| AKAP4 | Supports enhanced PKA signaling and oncogenic growth | [37] |
| ATAD2 | Integral to DNA replication in tumor cells and acts as a cofactor for oncogenic MYC | [38, 39] |
| CASC5 | Essential for kinetochore assembly and chromosome segregation | [40] |
| CEP55 | Required for cell abscission during cytokinesis and interacts with PI3K to enhance its activity | [41, 42] |
| CTAG2 | Regulates invasion of breast cancer cells via interaction with pericentrin at the centrosome | [43] |
| CTCFL | Can activate expression of other CT antigens in cancer and promote alternative splicing to facilitate oncogenic growth | [44, 45] |
| CTNNA2 | Expression of the mutated (truncated) protein promotes invasion and migration | [46] |
| CCNA1 | Enhances VEGF expression and promotes metastasis | [47, 48] |
| DDX43 | Promotes unwinding and translation of SOCS1 mRNA in a manner dependent on its helicase activity, leading to PML suppression and increased growth capacity | [49] |
| DPPA2 | Capable of oncogenic transformation through transcriptional regulation | [50] |
| FATE1 | Facilitates the degradation of the pro-apoptotic protein BIK through interaction with the E3 ubiquitin ligase RNF183 | [24] |
| FMR1NB | Required for successful chromosome segregation and mitosis | [51] |
| HORMAD1 | Regulates DNA damage repair mechanisms | [26, 52] |
| IGF2BP3 | Promotes oncogenic growth through regulation of mRNA stability and mRNA-microRNA interactions | [53, 54] |
| LUZP4 | Modulates mRNA export in melanoma cells | [55] |
| LY6K | Promotes multiple neoplastic behaviors in breast cancer through regulation of TGFβ signaling | [56] |
| MAEL | Enhances the lysosome-dependent degradation of the protein phosphatase ILKAP | [57] |
| MAGEs | Large family of related proteins that form complexes with RING E3 ubiquitin ligases to facilitate oncogenic phenotypes through protein degradation or transcriptional regulation | [58, 85] |
| MPHOSPH1 | Supports cytokinesis via interaction with PRC1 | [59] |
| NUF2 | Stabilizes kinetochore-microtubule attachments to support chromosome segregation through interaction with CENP-E | [60–62] |
| NXF2 | Required for successful chromosome segregation and mitosis | [51] |
| PASD1 | Interferes with the circadian clock via interaction with BMAL | [63] |
| PBK | Enhances JNK1 activity to facilitate oncogenic transformation by H-Ras | [64] |
| PIWIL2 | Interferes with the circadian clock by modulating CLOCK:BMAL stability | [65] |
| PRAME | Represses retinoic acid (RA) signaling through interaction with RAR | [66] |
| SPAG9 | Supports mitosis through interaction with PLK1 and regulation of FOXK1 transcriptional activity | [67] |
| SPANXA/C/D | Promotes breast cancer invasion via interaction with lamin A/C | [43] |
| SSX1/2 | Forms a fusion oncoprotein with SS18 and drives synovial sarcoma via re-wiring gene regulation programs through interactions with PRC1.1 and the BAF complex | [68, 69] |
| SYCP3 | Inhibits DNA damage repair mechanisms through interaction with BRCA2 | [70] |
| TEX14 | Supports mitosis through regulation of kinetochore-microtubule attachments and the spindle assembly checkpoint | [71] |
| TFDP3 | Regulates cell cycle progression by disrupting the transcriptional activity of E2F proteins | [72] |
| TSP50 | Promotes oncogenic growth through suppression of activin signaling | [73] |
| TTK | Essential for the spindle assembly checkpoint and mitosis | [74–76] |
| ZNF165 | Regulates the TGFβ transcriptional network to promote oncogenic signaling in triple negative breast cancer | [24] |
Figure 2. A snapshot of CT antigen behavior in tumors cells.
In addition to their capacity to evoke an immune response through antigen presentation, CT antigens have been demonstrated to contribute to diverse neoplastic behaviors. CT antigens depicted here (in green) serve as key examples of how aberrant expression of these proteins can influence tumor cell regulatory networks. Abbreviations: MHC, major histocompatibility complex; TCR, T cell receptor; Ub, ubiquitin; RA, retinoic acid; PcG, polycomb group; TGFβ, transforming growth factor beta.
CT Antigens Can Regulate Tumor Cell Transcriptional Networks
One of the earliest discoveries of a molecular function for a CT antigen in human cancer was reported just over a decade ago by the Bernards group [66]. Analysis of the PRAME protein sequence revealed the presence of a conserved region of nuclear receptor boxes, suggesting a function in modulating nuclear receptor hormone signaling. Indeed, PRAME was characterized as a retinoic acid receptor (RAR) antagonist, preventing its ligand-mediated activation and facilitating recruitment of transcriptionally repressive Polycomb (PcG) proteins to target genomic loci. In this manner, PRAME acts to bypass the tumor suppressive differentiation programs directed by retinoic acid signaling. Hence, efforts aimed at pharmacologically targeting the PRAME-RAR interaction would likely be promising given the general druggability of nuclear receptors [77].
More recently, the C2H2 zinc finger transcription factor ZNF165 emerged from a loss-of-function screen as essential for viability and transforming growth factor beta (TGFβ) signaling in triple negative breast cancer (TNBC) [24]. Importantly, TGFβ signaling is highly context-dependent; it is often oncogenic in TNBC but more commonly tumor-suppressive in normal breast epithelia and other breast cancer subtypes (e.g. luminal A/B) [78, 79]. ZNF165 associates with a number of different TGFβ target gene loci to both activate and inhibit their transcription, likely tipping the balance to a more oncogenic signaling program. Given the function of both PRAME and ZNF165, a theme begins to emerge that CT antigens can be engaged to alter transcriptional networks that otherwise serve as gatekeepers to uncontrolled growth.
At least two CT antigens have now been implicated in modulating circadian rhythms in cancer cells via interactions with the CLOCK:BMAL heterodimer, which controls transcription of temporally regulated genes [80]. While the contributions of CLOCK and BMAL to tumorigenesis are not well established, it is becoming clear that disruption of the normal circadian clock is correlated with increased incidence of cancer, as key components of the cell cycle are temporally regulated [80–82]. The CT antigen PASD1, which is evolutionarily related to CLOCK, interacts with BMAL to disrupt its gene regulatory function through molecular mimicry [63]. PIWIL2, which is crucial for piRNA regulation in the developing germ cell, was recently shown to destabilize the CLOCK:BMAL heterodimer and simultaneously block its transcriptional activity in both testis and tumor cells [65, 83]. It is interesting to note that circadian rhythms are absent from the testes, possibly due to the action of CT antigens such as PASD1 and PIWIL2 [84]. Thus, the expression of PASD1 and PIWIL2 in somatic cells may indeed wreak havoc on the normal circadian regulatory network that controls crucial aspects of cellular physiology, such as cell cycle progression.
Of the large MAGE family that consists of more than 60 proteins, twenty-four are classified as CT antigens and display aberrant expression across multiple cancer types [85]. A number of reports suggest a function for this family of CT antigens in transcriptional regulation. Laduron and colleagues found that MAGE-A1, the founding member of the CT antigen family, interacts with the transcription factor SKIP and directs the histone deacetylase HDAC1 to SKIP target genes to facilitate transcriptional repression [86]. Several members of the MAGE family have also been shown to interact with the p53 tumor suppressor to modulate its transcriptional activity, via both HDAC1 recruitment and inhibition of p53 DNA binding [87, 88]. Interestingly, recent reports also suggest that many MAGE proteins interact with the E3 ubiquitin ligase TRIM28 to facilitate degradation of tumor suppressor proteins, including p53 [25, 58, 89]. It is worth noting that the functions of TRIM28, while not completely understood, include widespread transcriptional regulation in addition to its E3 ligase activity [90, 91]. MAGE proteins therefore likely modulate a balance of both transcription and protein degradation to achieve their potent activity as drivers of oncogenic growth.
An exciting, recent finding suggests that long non-coding RNAs are also part of the CT antigen family. Hosono and colleagues recently reported the discovery of a cancer/testis lncRNA (CT-lncRNA), THOR, that is capable of promoting oncogenic growth [92]. Using multiple tumor cell lines and zebrafish as a model organism, the authors demonstrated that THOR expression is sufficient to enhance malignant growth and is required for melanoma onset in the zebrafish model. THOR interacts with IGF2BP1 and enhances its ability to stabilize target mRNAs. Notably, THOR also interacts with IGF2BP3, a CT antigen previously shown to promote oncogenic growth and HIF signaling, suggesting that THOR may regulate IGF2BP3 activity in a manner similar to IGF2BP1 [24, 53, 54]. Future studies will be able to determine if THOR represents a broader subset of germline-restricted lncRNAs with the capacity to contribute to tumorigenesis via post-transcriptional regulation.
From Meiotic Recombination to Mitotic Fidelity
Numerous CT antigens appear to buttress formation of the mitotic spindle, preventing mitotic and/or genomic catastrophe (Table 1). Kinetochore assembly, which is essential for proper mitosis in rapidly dividing cancer cells, is aided by numerous CT antigens, including TEX14, CASC5, TTK, and NUF2 [40, 61, 71, 93–96]. The CT antigen ACRBP, identified using a synthetic lethal RNA interference screening approach, is essential for bipolar spindle formation in non-small cell lung cancer (NSCLC) and ovarian cancer due to its regulation of NUMA protein accumulation [35, 36]. In addition, at least two CT antigens are implicated in ensuring proper cell abscission during cytokinesis. CEP55 associates with the centralspindlin complex and is required for midbody formation and membrane fusion [41]. MPHOSPH1, on the other hand, interacts with PRC1 to support cytokinesis and drive cell growth in bladder cancer [59]. Notably, cellular or humoral immune responses to TTK, NUF2, ACRBP, CEP55, and MPHOSPH1 have been reported, indicating their potential as immunotherapeutic targets [97–102]. The development of small-molecule inhibitors targeting the functions of these CT antigens is also an attractive approach to increase the effectiveness and simultaneously decrease the toxicity of chemotherapeutic mitotic poisons [51]. For example, multiple ongoing clinical trials are in place to determine the efficacy of small-molecule inhibitors targeting TTK, which is considered to be one of the more promising anti-mitotic therapeutic targets [103]. The CT antigens CCDC110, FMR1NB, NXF2, TCC52, and OIP5 have all also been identified as positive regulators of mitosis in tumor cells, although their exact functions remain unknown [51, 104–106].
A number CT antigens, including SPO11, TEX15, SYCP1/3, and HORMAD1/2, are essential for meiosis [30]. Knowledge of their functions in meiotic recombination has led to the proposal that these CT antigens, among others, can promote oncogenic proliferation and potentially facilitate “meiomitotic” cell division in cancer [52, 107, 108]. Interestingly, at least two meiotic CT antigens have recently been demonstrated to confer an advantage to tumor cells through regulation of DNA damage repair mechanisms and therefore genomic stability. SYCP3 was recently shown to interact with BRCA2 and prevent DNA damage repair mechanisms in tumor cells, which often display severe chromosomal scarring [70]. Expression of another CT antigen, HORMAD1, was found to be highly upregulated in TNBC with high allelic imbalance. The authors implicated HORMAD1 in promoting non-homologous end joining, an error prone DNA repair pathway, and therefore driving genomic instability [52]. Recently, work from our own laboratory has shown that in NSCLC, HORMAD1 can promote homologous recombination to support DNA repair and cell division [26]. These reports suggest that HORMAD1 function in tumors is context-dependent, and that it can exert opposing effects on DNA repair mechanisms to influence malignant growth in each context. The ability of different tumors to engage the same CT antigen in a capacity suited to a specific genetic background speaks to the extraordinary adaptability of tumor regulatory networks. Importantly, the study of CT antigen function is beginning to reveal the depths and mechanisms of this adaptive capacity.
CT Antigens Regulate Protein Degradation to Antagonize Tumor Suppression Mechanisms
Although several members of the MAGE family of CT antigens are reported to regulate transcription, recent work also suggests a generalizable function for this family in coordinating protein degradation in tumor cells [58, 85]. In particular, the ~170 amino acid, highly conserved MAGE homology domain (MHD) binds RING domain E3 ubiquitin ligases. MAGEC2 and MAGE-A3/6 interact with the E3 ubiquitin ligase TRIM28 to promote proteasome-dependent degradation of the p53 and AMPK tumor suppressors, respectively [25, 58, 89]. MAGE-A4 has also been demonstrated to stabilize the E3 ubiquitin ligase RAD18, thereby promoting trans-lesion synthesis and increasing the capacity of tumor cells to cope with DNA damage [109]. Thus, a general mechanism is emerging for the function of MAGE proteins in regulating E3 ubiquitin ligase activity. Future studies to reconcile the transcriptional and post-translational functions of MAGE proteins will be important for further deducing the significance of MAGE function in tumorigenesis. Outside of the MAGE family, a similar mechanism was described for the CT antigen FATE1, which interacts with the E3 ubiquitin ligase RNF183. Studies have demonstrated that RNF183 is upregulated in tumors and targets the apoptotic proteins BIK and Bcl-XL at the mitochondria for destruction [24]. It is interesting to note that despite FATE1 having no sequence homology with members of the MAGE family, these CT antigens operate through similar mechanisms to prevent tumor-suppressor protein accumulation.
Concluding Remarks
The question as to why tumors express CT antigens has gone unanswered for many decades. Evidence presented herein indicates that their expression may be selected for during tumor evolution to bypass intrinsic proliferation and survival barriers that would otherwise lead to cell cycle arrest, senescence, or death. Continued studies in this untapped discovery space will likely expand our understanding of the remarkable plasticity in the tumor cell regulatory environment to overcome barriers to growth (see Outstanding Questions). However, with over 200 CT antigens annotated and many more candidates identified, it is a daunting task to first identify functionally relevant CT antigens and then delineate their mechanisms of action. High-throughput gain- and loss-of-function screens offer the opportunity to assess the relevance of numerous CT antigens en masse in a variety of contexts. CRISPR gene-editing techniques could serve as a powerful tool in this regard and provide increased fidelity for identifying true positives from such screening approaches. In addition, mining clinical data sets to determine correlative associations among specific CT antigens, patient outcome, and response to therapy can indicate functional relevance and reveal specific disease sites in which to study CT antigen behavior. Further in-depth examination of CT antigen function in distinct tumorigenic processes will undoubtedly provide conceptual breakthroughs regarding the mechanisms that regulate tumor cell growth and proliferation.
Highlights.
CT antigens have historically been considered promising targets for immunotherapy due to their restricted expression in tumors and immuneprivileged organs, such as the testes.
The molecular functions of CT antigens in germ cells or tumor cells are difficult to decipher and remain largely unexamined.
Accumulating evidence suggests that many CT antigens make significant contributions to tumor cell physiology and promote neoplastic behaviors.
Outstanding Questions.
What are the gene expression mechanisms that induce and control the aberrant activation of CT antigens in tumors?
Why do some tumors express very few CT antigens (e.g. brain, pancreatic), while others express many (e.g. lung, melanoma)?
Which CT antigens support neoplastic processes and what are the molecular mechanisms by which they do so?
Are CT antigens capable of initiating tumorigenesis?
Can CT antigens be directly targeted with single agent therapy or in combination with current targeted/chemotherapeutic agents?
Acknowledgments
The authors thank the members of the Whitehurst laboratory for thoughtful discussion, as well as Michael Reese and Melanie Cobb for critical reading of the manuscript. The authors regret that due to space constraints, many important studies may have been omitted here. Z.A.G., A.W.W., and the work described here from our own laboratory are supported by the NIH (R01CA196905). Z.A.G. is also supported by an NIH training grant (T32GM007062).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Old LJ (1981) Cancer Immunology: The Search for Specificity—G. H. A. Clowes Memorial Lecture. Cancer Res. 41, 361. [PubMed] [Google Scholar]
- 2.van der Bruggen P et al. (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–7. [DOI] [PubMed] [Google Scholar]
- 3.Traversari C et al. (1992) A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J. Exp. Med 176, 1453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Plaen E et al. (1994) Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics 40, 360–9. [DOI] [PubMed] [Google Scholar]
- 5.Sahin U et al. (1995) Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl. Acad. Sci. U. S. A 92, 11810–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas R et al. (2018) NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives. Front. Immunol 9, 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YT et al. (1997) A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. U. S. A 94, 1914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Angelo SP et al. (2018) Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov. 8, 944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rock KL and Goldberg AL (1999) Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol 17, 739–79. [DOI] [PubMed] [Google Scholar]
- 10.Fijak M and Meinhardt A (2006) The testis in immune privilege. Immunol. Rev 213, 66–81. [DOI] [PubMed] [Google Scholar]
- 11.Chen YT (2012) The journey from autologous typing to SEREX, NY-ESO-1, and Cancer/Testis antigens. Cancer Immun. 12. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YT et al. (2005) Identification of cancer/testis-antigen genes by massively parallel signature sequencing. Proc. Natl. Acad. Sci. U. S. A 102, 7940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann O et al. (2008) Genome-wide analysis of cancer/testis gene expression. Proc. Natl. Acad. Sci. U. S. A 105, 20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scanlan MJ et al. (2002) Identification of cancer/testis genes by database mining and mRNA expression analysis. Int. J. Cancer 98, 485–92. [DOI] [PubMed] [Google Scholar]
- 15.Wang C et al. (2016) Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat. Commun 7, 10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Silva VL et al. (2017) Genome-wide identification of cancer/testis genes and their association with prognosis in a pan-cancer analysis. Oncotarget 8, 92966–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taguchi A et al. (2014) A search for novel cancer/testis antigens in lung cancer identifies VCX/Y genes expanding the repertoire of potential immunotherapeutic targets. Cancer Res. 74, 4694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson BJ et al. (2007) Rapid evolution of cancer/testis genes on the × chromosome. BMC Genomics 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida LG et al. (2009) CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 37, D816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassani-Sternberg M et al. (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J et al. (2017) SLC45A2: A Melanoma Antigen with High Tumor Selectivity and Reduced Potential for Autoimmune Toxicity. Cancer Immunol. Res 5, 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shraibman B et al. (2018) Identification of tumor antigens among the HLA peptidomes of Glioblastoma tumors and plasma. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Rousseaux S et al. (2013) Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med 5, 186ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxfield KE et al. (2015) Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat. Commun 6, 8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pineda CT et al. (2015) Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 160, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols BA et al. (2018) HORMAD1 is a negative prognostic indicator in lung adenocarcinoma and specifies resistance to oxidative and genotoxic stress. Cancer Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajagopalan K et al. (2011) A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem 112, 3256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyckoff GJ et al. (2000) Rapid evolution of male reproductive genes in the descent of man. Nature 403, 304–9. [DOI] [PubMed] [Google Scholar]
- 29.Kouprina N et al. (2004) The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc. Natl. Acad. Sci. U. S. A 101, 3077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehurst AW (2014) Cause and consequence of cancer/testis antigen activation in cancer. Annu. Rev. Pharmacol. Toxicol 54, 251–72. [DOI] [PubMed] [Google Scholar]
- 31.Simpson AJ et al. (2005) Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5, 615–25. [DOI] [PubMed] [Google Scholar]
- 32.Janic A et al. (2010) Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330, 1824–7. [DOI] [PubMed] [Google Scholar]
- 33.Feichtinger J et al. (2014) Meta-analysis of expression of l(3)mbt tumor-associated germline genes supports the model that a soma-to-germline transition is a hallmark of human cancers. Int. J. Cancer 134, 2359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi F et al. (2017) An in vivo genetic screen in Drosophila identifies the orthologue of human cancer/testis gene SPO11 among a network of targets to inhibit lethal(3)malignant brain tumour growth. Open Biol. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitehurst AW et al. (2007) Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446, 815–9. [DOI] [PubMed] [Google Scholar]
- 36.Whitehurst AW et al. (2010) Tumor antigen acrosin binding protein normalizes mitotic spindle function to promote cancer cell proliferation. Cancer Res. 70, 7652–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V et al. (2017) Role of A-Kinase anchor protein (AKAP4) in growth and survival of ovarian cancer cells. Oncotarget 8, 53124–53136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo SJ et al. (2016) ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget 7, 70323–70335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciro M et al. (2009) ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 69, 8491–8. [DOI] [PubMed] [Google Scholar]
- 40.Cheeseman IM et al. (2008) KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell 19, 587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao WM et al. (2006) Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17, 3881–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CH et al. (2007) FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene 26, 4272–83. [DOI] [PubMed] [Google Scholar]
- 43.Maine EA et al. (2016) The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget 7, 14708–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatolin S et al. (2005) Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 65, 7751–62. [DOI] [PubMed] [Google Scholar]
- 45.Singh S et al. (2017) Intragenic DNA methylation and BORIS-mediated cancerspecific splicing contribute to the Warburg effect. Proc. Natl. Acad. Sci. U. S. A 114, 11440–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fanjul-Fernandez M et al. (2013) Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun 4, 2531. [DOI] [PubMed] [Google Scholar]
- 47.Wegiel B et al. (2005) A role for cyclin A1 in mediating the autocrine expression of vascular endothelial growth factor in prostate cancer. Oncogene 24, 6385–93. [DOI] [PubMed] [Google Scholar]
- 48.Miftakhova R et al. (2016) Cyclin A1 and P450 Aromatase Promote Metastatic Homing and Growth of Stem-like Prostate Cancer Cells in the Bone Marrow. Cancer Res. 76, 2453–64. [DOI] [PubMed] [Google Scholar]
- 49.Mathieu MG et al. (2014) The helicase HAGE prevents interferon-alpha-induced PML expression in ABCB5+ malignant melanoma-initiating cells by promoting the expression of SOCS1. Cell Death Dis. 5, e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tung PY et al. (2013) Identification of DPPA4 and DPPA2 as a novel family of pluripotency-related oncogenes. Stem Cells 31, 2330–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cappell KM et al. (2012) Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol. Cell. Biol 32, 4131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkins J et al. (2015) Genomic Complexity Profiling Reveals That HORMAD1 Overexpression Contributes to Homologous Recombination Deficiency in Triple-Negative Breast Cancers. Cancer Discov. 5, 488–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suvasini R et al. (2011) Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogenactivated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J. Biol. Chem 286, 25882–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ennajdaoui H et al. (2016) IGF2BP3 Modulates the Interaction of Invasion Associated Transcripts with RISC. Cell Rep. 15, 1876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viphakone N et al. (2015) Luzp4 defines a new mRNA export pathway in cancer cells. Nucleic Acids Res. 43, 2353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.AlHossiny M et al. (2016) Ly6E/K Signaling to TGFbeta Promotes Breast Cancer Progression, Immune Escape, and Drug Resistance. Cancer Res. 76, 3376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X et al. (2017) MAEL contributes to gastric cancer progression by promoting ILKAP degradation. Oncotarget 8, 113331–113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doyle JM et al. (2010) MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 39, 963–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanehira M et al. (2007) Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 67, 3276–85. [DOI] [PubMed] [Google Scholar]
- 60.Liu D et al. (2007) Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J. Biol. Chem 282, 21415–24. [DOI] [PubMed] [Google Scholar]
- 61.DeLuca JG et al. (2002) hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol 159, 549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayama S et al. (2006) Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 66, 10339–48. [DOI] [PubMed] [Google Scholar]
- 63.Michael AK et al. (2015) Cancer/Testis Antigen PASD1 Silences the Circadian Clock. Mol. Cell 58, 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oh SM et al. (2007) T-lymphokine-activated killer cell-originated protein kinase functions as a positive regulator of c-Jun-NH2-kinase 1 signaling and H-Ras-induced cell transformation. Cancer Res. 67, 5186–94. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y et al. (2017) Cancer/testis antigen PIWIL2 suppresses circadian rhythms by regulating the stability and activity of BMAL1 and CLOCK. Oncotarget 8, 54913–54924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Epping MT et al. (2005) The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122, 835–47. [DOI] [PubMed] [Google Scholar]
- 67.Ramkumar P et al. (2015) JNK-associated Leucine Zipper Protein Functions as a Docking Platform for Polo-like Kinase 1 and Regulation of the Associating Transcription Factor Forkhead Box Protein K1. J. Biol. Chem. 290, 29617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banito A et al. (2018) The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell 33, 527–541.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McBride MJ et al. (2018) The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 33, 1128–1141.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hosoya N et al. (2011) Synaptonemal complex protein SYCP3 impairs mitotic recombination by interfering with BRCA2. EMBO Rep. 13, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mondal G et al. (2012) Tex14, a Plk1-regulated protein, is required for kinetochoremicrotubule attachment and regulation of the spindle assembly checkpoint. Mol. Cell 45, 680–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiao H et al. (2007) Human TFDP3, a novel DP protein, inhibits DNA binding and transactivation by E2F. J. Biol. Chem 282, 454–66. [DOI] [PubMed] [Google Scholar]
- 73.Song ZB et al. (2017) Testes-specific protease 50 promotes cell proliferation via inhibiting activin signaling. Oncogene 36, 5948–5957. [DOI] [PubMed] [Google Scholar]
- 74.Abrieu A et al. (2001) Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106, 83–93. [DOI] [PubMed] [Google Scholar]
- 75.Dou Z et al. (2015) Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl. Acad. Sci. U. S. A 112, E4546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Schubert C et al. (2015) Plk1 and Mps1 Cooperatively Regulate the Spindle Assembly Checkpoint in Human Cells. Cell Rep. 12, 66–78. [DOI] [PubMed] [Google Scholar]
- 77.Burris TP et al. (2013) Nuclear Receptors and Their Selective Pharmacologic Modulators. Pharmacol. Rev 65, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Massague J (2008) TGFbeta in Cancer. Cell 134, 215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruna A et al. (2012) TGFβ induces the formation of tumour-initiating cells in claudin low breast cancer. Nat. Commun 3, 1055. [DOI] [PubMed] [Google Scholar]
- 80.Sahar S and Sassone-Corsi P (2009) Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer 9, 886–96. [DOI] [PubMed] [Google Scholar]
- 81.Filipski E and Levi F (2009) Circadian disruption in experimental cancer processes. Integr. Cancer Ther. 8, 298–302. [DOI] [PubMed] [Google Scholar]
- 82.Kelleher FC et al. (2014) Circadian molecular clocks and cancer. Cancer Lett. 342, 9–18. [DOI] [PubMed] [Google Scholar]
- 83.Roovers EF et al. (2015) Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 10, 2069–82. [DOI] [PubMed] [Google Scholar]
- 84.Morse D et al. (2003) No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol. Endocrinol 17, 141–51. [DOI] [PubMed] [Google Scholar]
- 85.Weon JL and Potts PR (2015) The MAGE protein family and cancer. Curr. Opin. Cell Biol 37, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laduron S et al. (2004) MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription. Nucleic Acids Res. 32, 4340–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marcar L et al. (2010) Mage-A Cancer/Testis Antigens Inhibit p53 Function by Blocking Its Interaction with Chromatin. Cancer Res. 70, 10362. [DOI] [PubMed] [Google Scholar]
- 88.Monte M et al. (2006) MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc. Natl. Acad. Sci. U. S. A 103, 11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang B et al. (2007) MAGE-A, mMage-b, and MAGE-C Proteins Form Complexes with KAP1 and Suppress p53-Dependent Apoptosis in MAGE-Positive Cell Lines. Cancer Res. 67, 9954. [DOI] [PubMed] [Google Scholar]
- 90.Iyengar S and Farnham PJ (2011) KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem 286, 26267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McNamara RP et al. (2016) KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol. Cell 61, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hosono Y et al. (2017) Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 171, 1559–1572.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosenberg JS et al. (2011) KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol 21, 942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jelluma N et al. (2008) Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 132, 233–46. [DOI] [PubMed] [Google Scholar]
- 95.Daniel J et al. (2011) High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl. Acad. Sci. U. S. A 108, 5384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aravamudhan P et al. (2015) The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat. Cell Biol 17, 868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mizukami Y et al. (2008) Detection of novel cancer-testis antigen-specific T-cell responses in TIL, regional lymph nodes, and PBL in patients with esophageal squamous cell carcinoma. Cancer Sci. 99, 1448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harao M et al. (2008) HLA-A2-restricted CTL epitopes of a novel lung cancer-associated cancer testis antigen, cell division cycle associated 1, can induce tumor-reactive CTL. Int. J. Cancer 123, 2616–25. [DOI] [PubMed] [Google Scholar]
- 99.Ono T et al. (2001) Identification of proacrosin binding protein sp32 precursor as a human cancer/testis antigen. Proc. Natl. Acad. Sci. U. S. A 98, 3282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tammela J et al. (2006) OY-TES-1 expression and serum immunoreactivity in epithelial ovarian cancer. Int. J. Oncol 29, 903–10. [PubMed] [Google Scholar]
- 101.Inoda S et al. (2009) Cep55/c10orf3, a tumor antigen derived from a centrosome residing protein in breast carcinoma. J. Immunother 32, 474–85. [DOI] [PubMed] [Google Scholar]
- 102.Obara W et al. (2012) Cancer peptide vaccine therapy developed from oncoantigens identified through genome-wide expression profile analysis for bladder cancer. Jpn. J. Clin. Oncol 42, 591–600. [DOI] [PubMed] [Google Scholar]
- 103.Dominguez-Brauer C et al. (2015) Targeting Mitosis in Cancer: Emerging Strategies. Mol. Cell 60, 524–536. [DOI] [PubMed] [Google Scholar]
- 104.Park HJ et al. (2007) The centrosomal localization of KM-HN-1 (MGC33607) depends on the leucine zipper motif and the C-terminal coiled-coil domain. Exp. Mol. Med 39, 828–38. [DOI] [PubMed] [Google Scholar]
- 105.Li S et al. (2008) Novel centrosome protein, TCC52, is a cancer-testis antigen. Cancer Sci. 99, 2274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Naetar N et al. (2007) LAP2alpha-binding protein LINT-25 is a novel chromatinassociated protein involved in cell cycle exit. J. Cell Sci 120, 737–47. [DOI] [PubMed] [Google Scholar]
- 107.McFarlane RJ and Wakeman JA (2017) Meiosis-like Functions in Oncogenesis: A New View of Cancer. Cancer Res. 77, 5712. [DOI] [PubMed] [Google Scholar]
- 108.Grichnik JM (2008) Melanoma, nevogenesis, and stem cell biology. J. Invest. Dermatol 128, 2365–80. [DOI] [PubMed] [Google Scholar]
- 109.Gao Y et al. (2016) A neomorphic cancer cell-specific role of MAGE-A4 in trans-lesion synthesis. Nat. Commun 7, 12105. [DOI] [PMC free article] [PubMed] [Google Scholar]