Abstract
Background
TIM-family proteins are expressed on different immune cells such as dendritic cells, macrophages, type 1 and 2 T helper (Th) cells. Therefore, they have the ability to contribute to the various intracellular signals and immune responses, importantly the regulation of Th1 and Th17 cell differentiation, which plays a remarked role in fight against inflammatory and autoimmune diseases. Association of TIM family gene polymorphisms with rheumatoid arthritis (RA) has been frequently investigated. The findings however are not entirely consistent. Therefore, we carried out the present meta-analysis to examine the association between RA and the following TIM family gene polymorphisms: rs41297579, rs1036199, rs10515746, and rs7700944.
Methods
A systematic search of Scopus, PubMed, and Web of Science databases was conducted through December 2018. Combined odds ratios (OR) with their corresponding 95% confidence intervals (CI) were calculated under different possible genetic models.
Results
A total of eight case-control studies were included in the present meta-analysis. The results demonstrated significant association of RA with TIM-3 rs1036199 polymorphism under dominant (OR, 1.93, 95% CI, 1.43–2.61) and allelic models (OR, 1.74, 95% CI, 1.31–2.30). None of the other examined polymorphisms indicated significant association with RA.
Conclusions
The present meta-analysis revealed that the TIM-3 rs1036199 polymorphism might confer susceptibility to RA. Further studies are required to reassert our findings.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by progressive inflammation of the synovial membrane of the joint capsule and tendons (synovitis). Clinical manifestations range from chronic pain, loss of joint function, and deformity, to disability and systemic complications [1, 2]. RA usually develops between 40 and 50 years of age. Like most autoimmune diseases, RA is more common in women than men (3:1 ratio) [3]. Its prevalence increases with age and varies across different regions of the world [4, 5]. Despite clear clinical manifestations of the disease, the exact etiology and pathogenesis of RA remain obscure. Generally genetic and environmental factors have been well-associated with autoimmune disorders. Particularly, the role of genetic factors in the pathogenesis of RA has been confirmed by family and twin studies. Accordingly, the heritability of RA is estimated to be about 60% [6, 7]. Furthermore, genome-wide association studies (GWAS) have identified more than 100 genetic loci related to RA [8–12]. Of note, genome-side meta-analysis by Okada and colleagues recently introduced nine new loci including B3GNT2, ANXA3, CSF2, CD83, NFKBIE, ARID5B, PDE2A-ARAP1, PLD4 and PTPN2 as genetic risk factors for RA in Japanese population [12].
The transmembrane immunoglobulin and mucin domain (TIM) family gene was first isolated in 2001 [13]. This novel gene family has been described in both mice and humans. In mice, it is located on chromosome 11B1.1 and includes eight members (TIM-1–8). While in humans, it is located on chromosome 5q33.2 and includes three members: TIM-1, TIM-3, and TIM-4. [14]. Interestingly, the TIM gene family (5q33.2) is co-located with the interleukin-4 (IL-4) gene cluster that encodes cytokines including IL-3, IL-4, IL-5 and IL-13 on the human chromosome 5. This chromosomal co-location can partly explain shared function of TIM gene family and IL-4 gene cluster in the pathogenesis of autoimmune and allergic diseases.
TIM-family proteins belong to the type I transmembrane proteins consisting the N-terminal immunoglobulin (Ig) domain of the variable type, a mucin-like domain, and a C-terminal cytoplasmic tail [15, 16]. These proteins are expressed on different immune cells and therefore have the ability to mediate various intracellular signals. TIM-4 is found on macrophages and dendritic cells (DCs) [17, 18] while TIM-1 and TIM-3 are respectively expressed by T helper 2 (Th2) cells and T helper1 cells (Th1). It is, therefore, understandable that variants of the TIM gene family might interfere with signaling pathways related to Th1 and Th2 cells whereby the delicate balance between these cells is disturbed. This deviated balance has been implicates in a variety of allergic diseases and asthma [19, 20]. To date, TIM family gene polymorphisms have been associated with allergic rhinitis [21, 22], atopy [23, 24], asthma [22, 25–27], eczema [24], and autoimmune diseases such as multiple sclerosis [28, 29].
Several studies have investigated the association between RA and TIM family gene polymorphisms. The results are, however, not conclusive. To our knowledge, no meta-analysis has evaluated the association between RA and TIM gene polymorphisms. Therefore the present meta-analysis was conducted to address the issue whether the TIM family gene polymorphisms affect susceptibility to RA.
Materials and methods
The present meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement (S1 File) and meta-analysis on genetic association studies checklist (S2 File) [30].
Searches and data sources
We searched the Scopus, PubMed, and Web of Science databases through December 2018 by combining the following search terms: (“Arthritis, Rheumatoid” OR “Rheumatoid arthritis” OR RA) AND (“Hepatitis A virus cellular receptor-1” OR HAVCR-1 OR “T-cell immunoglobulin and mucin domain containing -1” OR TIM-1 OR “Kidney Injury Molecule -1” OR KIM-1 OR “Hepatitis A virus cellular receptor-2” OR HAVCR-2 OR “T-cell immunoglobulin and mucin domain containing -3” OR TIM-3 OR “T-cell immunoglobulin and mucin domain containing-4” OR TIM-4) AND (polymorphism OR genotype OR SNP OR “single nucleotide polymorphisms”). Studies were considered eligible if they investigated the association between RA and the following TIM family gene polymorphisms: rs41297579, rs1036199, rs10515746, and rs7700944. Further, we performed hand-searching of bibliographies from relevant publications to identify potential studies that were not retrieved by the electronic search. The search strategy was not confined to English language articles and any time period.
Study selection
After removing duplicate records, two authors independently screened search results based on title and/or abstract. Then the apparently relevant papers were selected for detailed review. Finally, the authors carefully considered inclusion and exclusion criteria to include or exclude studies. Any disagreement between reviewers were resolved by consensus.
Inclusion and exclusion criteria
Eligible studies should satisfy the following inclusion criteria: a) case–control studies that investigated the association between RA and the aforementioned TIM family gene polymorphism; and b) adequate data including genotype/allele frequency in both case and control groups were provided to calculate the combined odds ratio (OR) and their corresponding 95% confidence intervals (CIs). Book chapters, review articles, duplicate reports, conference abstracts, studies lacking control group, letters to the editor, case reports, and animal studies were all excluded.
Data extraction and quality assessment
Two authors independently extracted the following data from each eligible study: the first author’s name, journal’s name, year of publication, ethnicity, country of origin, gender, study design, age of participants, methodology of genotyping, number of genotyped cases and controls, and genotype/allele frequency for in cases and controls. Genotype data were extracted for each ethnic group of people separately if data were available. Any disagreement between two reviewers were discussed and resolved by consensus. To assess the methodological quality of included studies, the Newcastle-Ottawa Scale (NOS) was used [31]. Studies with scores 0–3, 4–6 or 7–9 were of low, moderate or high-quality, respectively.
Statistical analysis
Deviation from Hardy-Weinberg equilibrium (HWE) was analyzed using the Chi-Square test in control groups [32]. For each of the examined polymorphism, pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated under different possible genetic models: dominant model, recessive model, allelic model, homozygote comparison and heterozygote comparison. To investigate the heterogeneity across included studies, we applied Cochran’s Q test and the I2 statistics [33]. There was significant heterogeneity if I2 values exceeded 50% or the Q statistic had a p value less than 0.1. If heterogeneity was detected, the random-effects model (Der Simonian–Laird approach) was applied. Otherwise, the fixed-effects model (Mantel–Haenszel approach) was used. Publication bias was assessed using the degree of funnel plot asymmetry and also by Begg’s test (a p value less than 0.05 was considered statistically significant) [34, 35]. The data analyses were carried out using STATA (version 14.0; Stata Corporation, College Station, TX) and SPSS (version 23.0; SPSS, Inc. Chicago, IL).
Results
Study characteristics
Fig 1 displays search and screening process based on the PRISMA statement [36]. A total of 137 studies were examined. Eventually eight studies meeting inclusion and exclusion criteria were considered eligible to be included in the present meta-analysis. The studies were conducted in Korea [37–39], Egypt [40], China [41–43], and Iran [44]. All eligible studies had an overall good methodological quality with NOS scores ranging from 6 to 8. The RFLP (Restriction Fragment Length Polymorphism) was applied for genotyping SNPs in most studies. Tables 1 and 2 summarizes characteristics of the included studies.
Fig 1. Flow diagram of study selection process.

Table 1. Characteristics of studies included in meta-analysis of overall RA.
| Study author | Year | Country | Ethnicity | Sex cases/controls |
Total cases/controls | Age case/control (Mean) |
Genotyping method | Quality score |
|---|---|---|---|---|---|---|---|---|
| TIM-1 rs41297579 G>A (-1454) | ||||||||
| Chae et al. | 2004 | Korea | Asian | M = 43/203 F = 252/111 |
295/314 | 38,9/50,6 | PCR–RFLP | 7 |
| Xu et al. | 2012 | China | Asian | M = NR F = NR |
118/118 | NR/NR | PCR–RFLP | 6 |
| Mosaad et al. | 2015 | Egypt | African | M = 25/NR F = 103/NR |
128/125 | 46,9/49,1 | PCR–RFLP | 6 |
| TIM-3 rs1036199 G>T(+4259) | ||||||||
| Chae et al. | 2004 | Korea | Asian | M = 43/194 F = 235/125 |
296/319 | 38,7/49,6 | PCR | 7 |
| Song et al. | 2011 | Korea | Asian | M = 41/51 F = 325/338 |
366/389 | 51/47 | RT-PCR | 8 |
| Xu et al. | 2012 | China | Asian | M = NR F = NR |
226/231 | NR/NR | PCR–RFLP | 6 |
| Xu et al. | 2012 | China | Asian | M = NR F = NR |
103/108 | NR/NR | PCR–RFLP | 6 |
| TIM-3 rs10515746 T>G (-574) | ||||||||
| Song et al. | 2011 | Korea | Asian | M = 41/51 F = 325/338 |
366/389 | 51/47 | RT-PCR | 8 |
| Xu et al. | 2012 | China | Asian | M = NR F = NR |
226/231 | NR/NR | SSP-PCR | 6 |
| Xu et al. | 2012 | China | Asian | M = NR F = NR |
103/108 | NR/NR | SSP-PCR | 6 |
| TIM-4 rs7700944 | ||||||||
| Xu et al. | 2012 | China | Asian | M = 117/102 F = 93/95 |
210/197 | NR/NR | PCR–RFLP | 7 |
| Xu et al. | 2012 | China | Asian | M = 105/112 F = 109/99 |
214/211 | NR/NR | PCR–RFLP | 7 |
| Zakeri et al. | 2013 | Iran | White | M = 16/35 F = 104/85 |
120/120 | 44,8/44,9 | T-ARMS-PCR | 6 |
| Mosaad et al. | 2015 | Egypt | African | M = 25/NR F = 103/NR |
128/125 | 46,9/49,1 | PCR–RFLP | 6 |
NR, not reported; M, male; F, female; RA, Rheumatoid Arthritis.
Table 2. Distribution of genotype and allele among RA patients and controls.
| Study author | RA cases | Healthy control | P-HWE | MAF | ||||||||
| GG | GA | AA | G | A | GG | GA | AA | G | A | |||
| TIM-1 rs41297579 G>A (-1454) | ||||||||||||
| Chae et al. | 216 | 71 | 6 | 503 | 83 | 220 | 75 | 7 | 515 | 89 | 0.839 | 0.147 |
| Xu et al. | 86 | 26 | 4 | 201 | 35 | 90 | 26 | 2 | 205 | 31 | 0.938 | 0.127 |
| Mosaad et al. | 110 | 14 | 4 | 234 | 22 | 98 | 24 | 3 | 220 | 30 | 0.309 | 0.12 |
| Study author | RA cases | Healthy control | P-HWE | MAF | ||||||||
| TT | TG | GG | T | G | TT | TG | GG | T | G | |||
| TIM-3 rs1036199 G>T(+4259) | ||||||||||||
| Chae et al. | 203 | 93 | 0 | 499 | 93 | 256 | 63 | 0 | 575 | 63 | 0/05 | 0/098 |
| Song et al. | 355 | 31 | 0 | 741 | 31 | 365 | 24 | 0 | 754 | 24 | 0/53 | 0/03 |
| Xu et al. | 198 | 28 | 0 | 424 | 21 | 244 | 7 | 0 | 455 | 7 | 0/822 | 0/013 |
| Xu et al. | 90 | 13 | 0 | 193 | 13 | 103 | 5 | 0 | 211 | 5 | 0/805 | 0/023 |
| Study author | RA cases | Healthy control | P-HWE | MAF | ||||||||
| GG | GT | TT | G | T | GG | GT | TT | G | T | |||
| TIM-3 rs10515746T>G (-574) | ||||||||||||
| Song et al. | 343 | 23 | 0 | 709 | 23 | 378 | 11 | 0 | 767 | 11 | 0/777 | 0/014 |
| Xu et al. | 203 | 23 | 0 | 429 | 23 | 212 | 19 | 0 | 443 | 19 | 0/514 | 0/041 |
| Xu et al. | 100 | 3 | 0 | 203 | 3 | 85 | 21 | 2 | 191 | 25 | 0/602 | 0/115 |
| Study author | RA cases | Healthy control | P-HWE | MAF | ||||||||
| GG | GA | AA | G | A | GG | GA | AA | G | A | |||
| TIM-4 rs7700944 G>A | ||||||||||||
| Xu et al. | 134 | 65 | 11 | 332 | 88 | 76 | 113 | 8 | 267 | 129 | 0 | 0/327 |
| Xu et al. | 77 | 135 | 2 | 289 | 139 | 135 | 70 | 6 | 341 | 85 | 0/387 | 0/194 |
| Zakeri et al. | 68 | 47 | 5 | 183 | 57 | 77 | 41 | 2 | 195 | 45 | 0/138 | 0/187 |
| Mosaad et al. | 29 | 87 | 12 | 145 | 111 | 99 | 23 | 3 | 221 | 29 | 0/25 | 0/116 |
P-HWE, p-value for Hardy–Weinberg equilibrium; MAF, minor allele frequency of control group
Quantitative synthesis
TIM-1 rs41297579 polymorphism
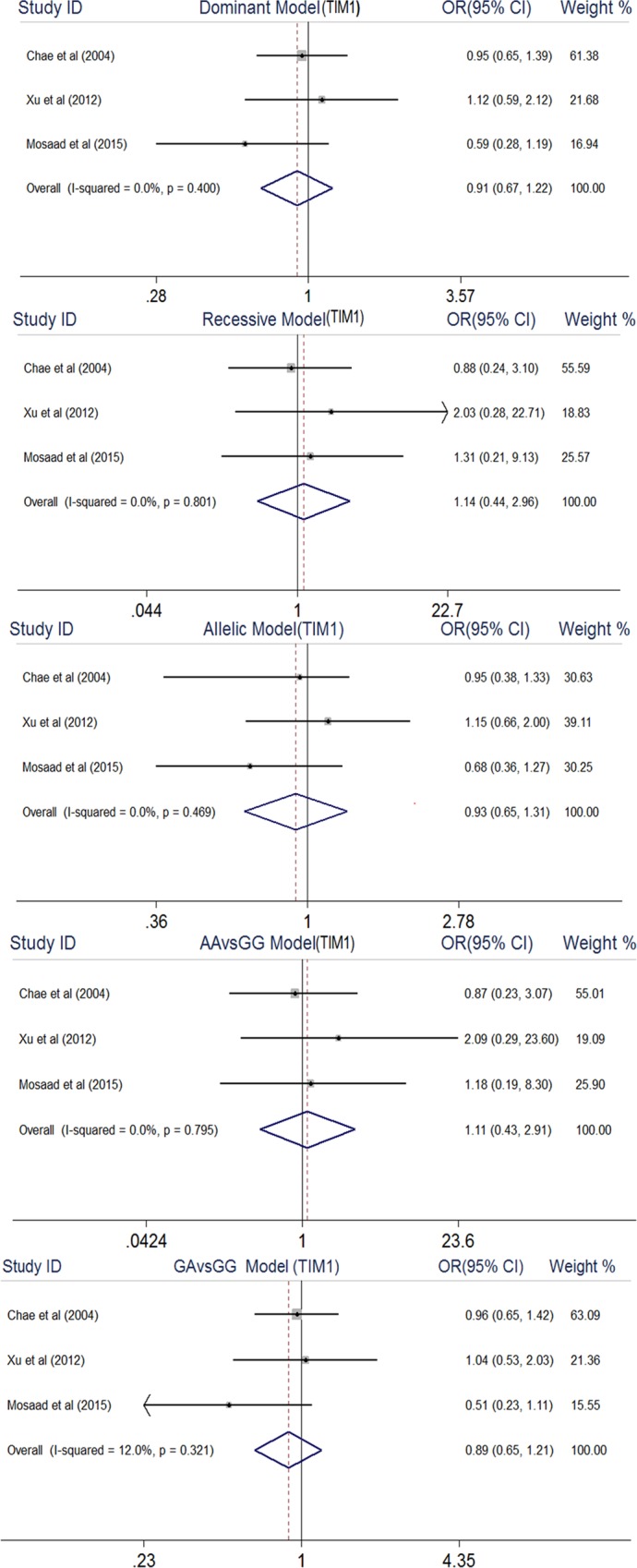
As shown in Fig 2, meta-analysis found no significant association between TIM-1 rs41297579 polymorphism and RA under all genetic models: dominant model (OR, 0.91, 95% CI, 0.67–1.22), recessive model (OR, 1.14, 95% CI, 0.44–2.96), allelic model (OR, 0.93, 95% CI, 0.65–1.31), AA vs GG (OR, 1.11, 95% CI, 0.43–2.91), and GA vs GG (OR, 0.89, 95% CI, 0.65–1.21).
Fig 2. Forest plot of association between TIM1 rs41297579 gene polymorphism and rheumatoid arthritis.
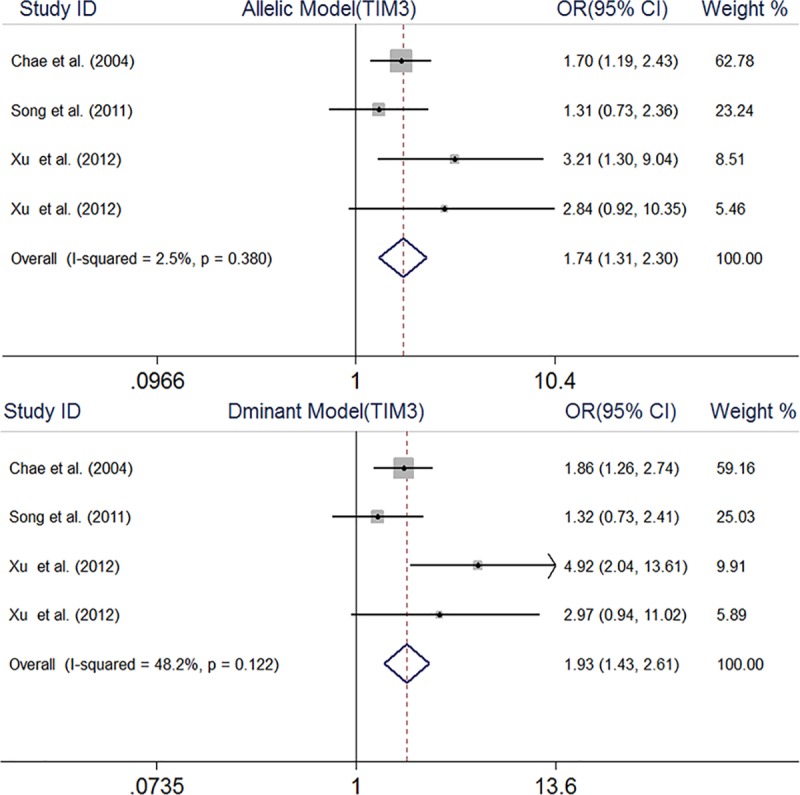
TIM-3 rs1036199 polymorphism
The TIM-3 rs1036199 polymorphism was demonstrated to associate with RA risk under dominant model (OR, 1.93, 95% CI, 1.43–2.61) and allelic model (OR, 1.74, 95% CI, 1.31–2.30) (Fig 3). Due to the TIM-3 rs1036199 GG genotype frequency of zero in both cases and controls, the recessive model and GG vs TT model were not applicable and meta-analyses of comparisons related to the dominant (GG + TG vs TT) and heterozygote models (TG vs TT) resulted in the same findings. As Thakkinstian and colleagues described in [45], the dominant model was selected.
Fig 3. Forest plot of association between TIM3 rs1036199 gene polymorphism and rheumatoid arthritis.
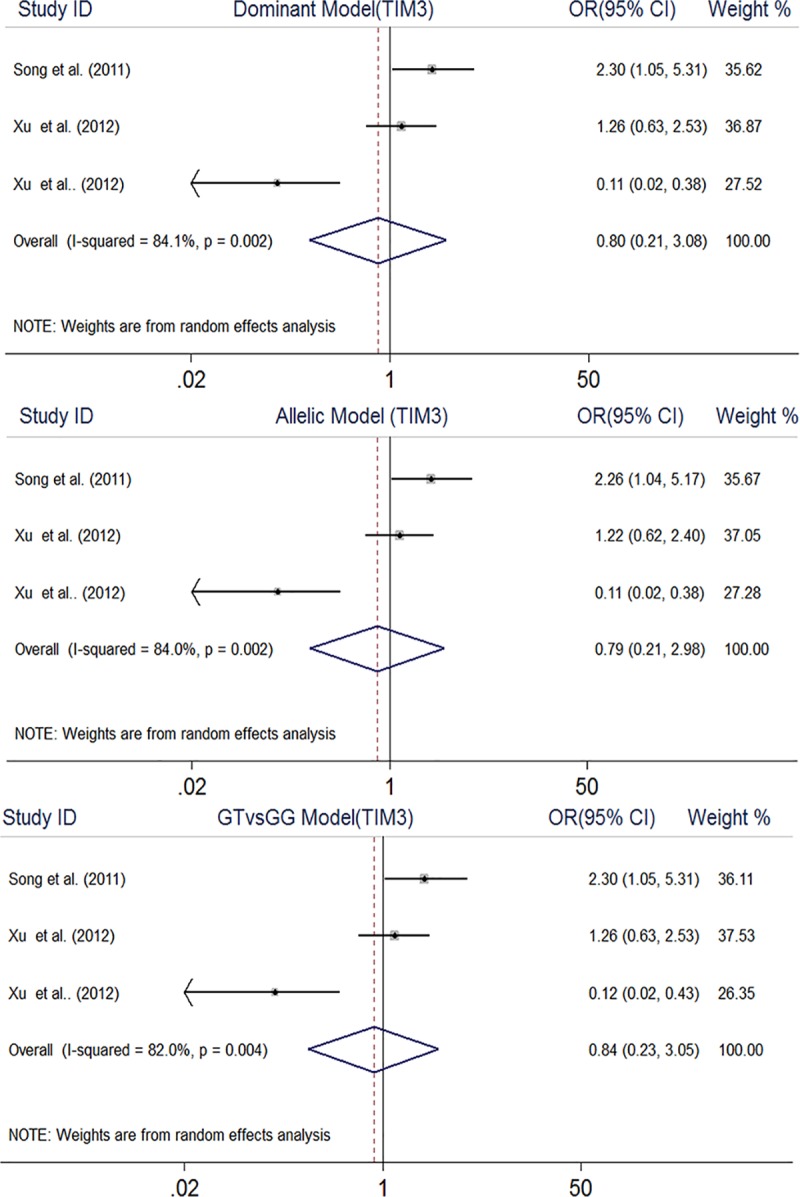
TIM-3 rs10515746 polymorphism
The pooled results rejected any association between TIM-3 rs10515746 polymorphism and RA under dominant model (OR, 0.80, 95% CI, 0.21–3.08), allelic model (OR, 0.79, 95% CI, 0.21–2.98), and GT vs. GG model (OR, 0.84, 95% CI, 0.23–3.05) (Fig 4). Similar to the TIM-3 rs1036199 polymorphism, meta-analysis under other genetic models was not performed.
Fig 4. Forest plot of association between TIM3 rs10515746 gene polymorphism and rheumatoid arthritis.
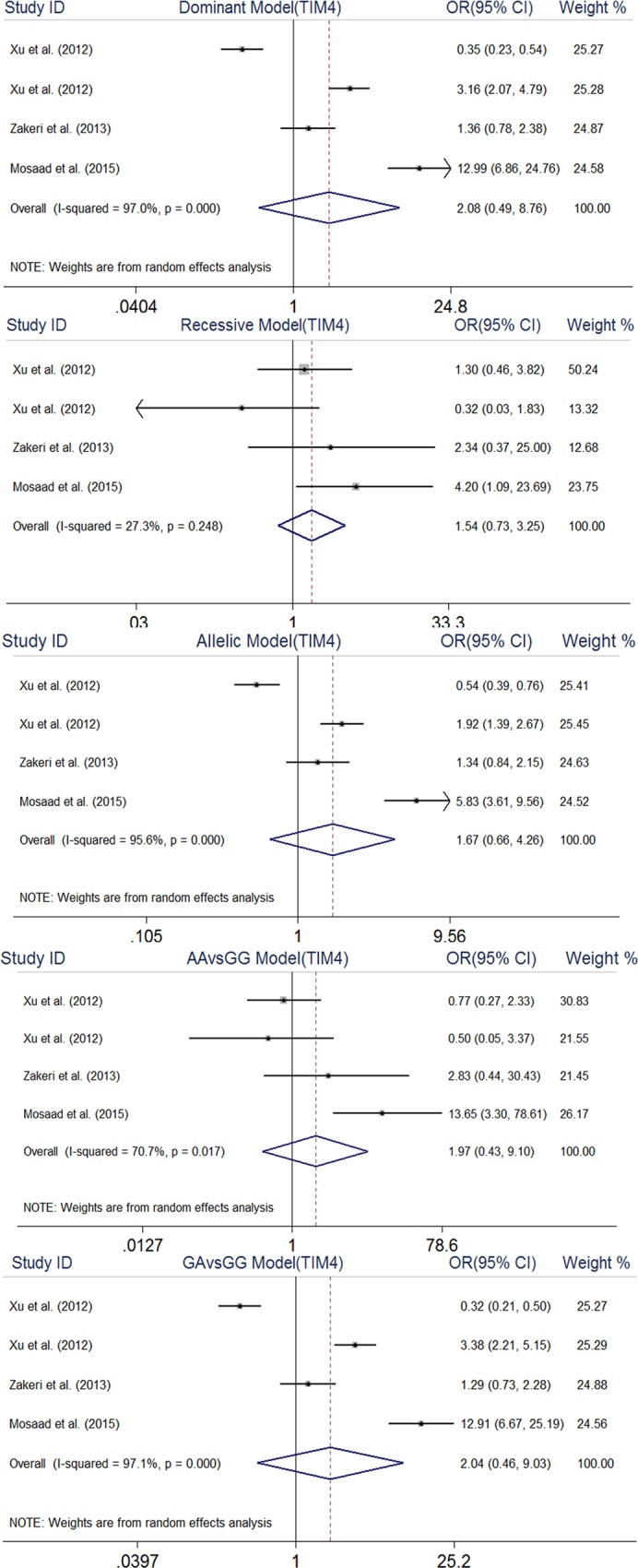
TIM-4 rs7700944 polymorphism
No significant association was found between this polymorphism and RA: dominant model (OR, 2.08, 95% CI, 0.49–8.76), recessive model (OR, 1.54, 95% CI, 0.73–3.25), allelic model (OR, 1.67, 95% CI, 0.66–4.26), AA vs. GG model (OR, 1.94, 95% CI, 0.44–8.53), and GA vs. GG model (OR, 2.04, 95% CI, 0.46–9.03) (Fig 5).
Fig 5. Forest plot of association between TIM4 rs7700944 gene polymorphism and rheumatoid arthritis.
Evaluation of heterogeneity and publication bias
For the TIM-4 rs7700944 polymorphism, there was a significant heterogeneity across included studies under all genetic models, except for recessive model: dominant model (I2 = 97%), recessive model (I2 = 27.3%, p = 0.24), allelic model (I2 = 95.6%), AA vs GG model (I2 = 91.7%), and GA vs GG model (I2 = 97.1%). Also significant heterogeneity was observed across studies included in the three genetic models applicable for TIM-3 rs10515746: dominant model (I2 = 84.1%), allelic model (I2 = 84%), and GT vs GG model (I2 = 82%). No significant heterogeneity was detected among studies included in the meta-analyses related to other investigated SNPs. As shown in Table 3, the results of Begg’s test indicated no evidence of publication bias in the present meta-analysis. Also, funnel plots are presented in Supplementary Material (S1–S4 Figs).
Table 3. Main results of pooled ORs in meta-analysis of Tim family polymorphism.
| Sample size | Test of association | Test of heterogeneity | Test of publication bias (Begg’s test) | ||||
|---|---|---|---|---|---|---|---|
| Genetic model | Case/Control | OR | 95% CI | I2 (%) | P | Z | P |
| rs41297579 | |||||||
| Dominant model | 541/557 | 0.91 | 0.67–1.22 | 0 | 0.40 | -0.52 | 0.602 |
| Recessive model | 541/557 | 1.14 | 0.44–2.96 | 0 | 0.80 | 1.57 | 0.117 |
| Allelic model | 541/557 | 0.93 | 0.65–1.31 | 0 | 0.46 | -1.57 | 0.117 |
| AA VS. GG | 541/557 | 1.11 | 0.43–2.91 | 0 | 0.79 | 1.57 | 0.117 |
| GA VS.GG | 541/557 | 0.89 | 0.65–1.21 | 12 | 0.32 | -1.57 | 0.117 |
| rs1036199 | |||||||
| Dominant model | 991/1047 | 1.93 | 1.43–2.61 | 48.2 | 0.12 | 0.68 | 0.497 |
| Allelic model | 991/1047 | 1.74 | 1.31–2.30 | 2.5 | 0.38 | 0.68 | 0.497 |
| rs10515746 | |||||||
| Dominant model | 695/728 | 0.80 | 0.21–3.08 | 84.1 | 0.002 | -0.52 | 0.602 |
| Allelic model | 695/728 | 0.79 | 0.21–2.98 | 84 | 0.002 | -0.52 | 0.602 |
| GT VS.GG | 695/728 | 0.84 | 0.23–3.05 | 82 | 0.004 | -0.52 | 0.602 |
| rs7700944 | |||||||
| Dominant model | 672/653 | 2.08 | 0.49–8.76 | 97 | ≤0.001 | 0.68 | 0.497 |
| Recessive model | 672/653 | 1.54 | 0.73–3.25 | 27.3 | 0.24 | 0.0 | 1 |
| Allelic model | 672/653 | 1.67 | 0.66–4.26 | 95.6 | ≤0.001 | 0.68 | 0.497 |
| AA VS. GG | 672/653 | 1.94 | 0.44–8.53 | 91.7 | ≤0.001 | 0.68 | 0.497 |
| GA VS.GG | 672/653 | 2.04 | 0.46–9.03 | 97.1 | ≤0.001 | 0.68 | 0.497 |
Discussion
Xie et al. 2017 [46] emphasized the association of TIM family gene polymorphisms, especially variants of the TIM1, with susceptibility to asthma through a meta-analysis study. Though some studies have investigated the association of RA with these polymorphisms, the associations remain unclear because the results are not conclusive. Therefore, we conducted this meta-analysis of data driven from eight studies that examined the relationship between RA and TIM family gene polymorphisms (rs41297579, rs1036199, rs10515746, rs7700944). Accordingly, the TIM-3 rs1036199 polymorphism was demonstrated to affect susceptibility to RA.
Analysis of the synovial fluid of RA patients revealed relatively high levels of pro-inflammatory cytokines such as IFN- γ, IL-6, IL-1, TNF-α, IL-23, and IL-12. These cytokines are able to attract leukocytes and activate joint cells such as osteoclasts and synovial fibroblasts. Activated joint cells stimulate the secretion of proteolytic enzymes, especially collagenase, which contribute to joint injury. Also, analysis of sera from patients with RA has often demonstrated the presence of rheumatoid factor (RF) or anti-citrullinated protein antibodies (ACPAs) [47]. It should however be noted that these autoantibodies are detected in various inflammatory diseases and their presence is, thus, not specific to RA. Moreover increased serum levels of C-reactive protein (CRP), an acute phase protein, are frequently found in patients with RA [48].
The TIM1−1454G>A polymorphism occurs in promoter region and therefore it is not expected to affect the function of TIM1 protein. Interestingly, Chae and colleagues reported that neither RF nor CRP were detectable in the sera of patients carrying this polymorphism [37].
The results showed significant association between TIM-3 G>T (+4259) and RA. The human TIM-3 gene coding region is consisted of seven exons. Among the various polymorphisms linked to this gene, the G>T (+4259) is the only SNP that occurs in the exon zone (exon 3). The change of allele at 4259 G>T leads to amino acid substitution from arginine to leucine. Arginine is an ionic amino acid whereas leucine is a non-polarized amino acid. Therefore the 4259 G>T polymorphism might alter the structure and function of gene. More interestingly, the TIM-3 protein is expressed on the surface of Th1 cells that serve to regulate Th1 and Th17 cell differentiation and their respective immune responses. Thus, the TIM-3 G>T (+4259) polymorphism might disturb the balance between Th1 and Th2 cell responses and thereby confer susceptibility to asthma and autoimmune diseases such as RA. Of note, Chae et al observed higher CRP values in the sera of patients harboring TIM1 +4259G>T variant [39].
The SNP rs7700944 is located in the intron region of the TIM-4 gene. The intron regions are comprised of noncoding DNA sequences which lie between coding regions in the genome. The intron regions play important role in the regulation and evolution of gene and control of alternative splicing in human genome. Evidences link polymorphisms that occur in the intron region of genes with different human diseases. For example, IL-1ß +3954 is an intronic polymorphism that has been associated with susceptibility to RA in a Chinese population [49]. After all, the present meta-analysis found no relationship between TIM-4 rs7700944 polymorphism and RA.
Although the present study was designed in a systematic manner to include all the eligible studies, there are some limitations to be mentioned. First is the concern about few number of included studies, because of which sensitivity and subgroup meta-analyses of studies stratified by sex, ethnicity, and methodology of genotyping could not be performed. Second that most of the included studies were conducted in East-Asians and so the findings are not generalizable to other race/ethnic groups. Third that our meta-analyses were confined to certain SNPs of TIM gene family due to inadequate numbers of publications.
Conclusions
In sum, the current meta-analysis provided evidence that TIM-3 G>T (+4259) gene polymorphism might increase the risk of RA. The analyses, however, failed to found significant association between RA and other polymorphisms of TIM family genes. Further studies with larger sample sizes across different race/ethnic groups are warranted to validate the findings.
Supporting information
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(DOC)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. New England Journal of Medicine. 2011;365(23):2205–19. 10.1056/NEJMra1004965 [DOI] [PubMed] [Google Scholar]
- 2.Gibofsky A. Overview of epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis. The American journal of managed care. 2012;18(13 Suppl):S295–302. [PubMed] [Google Scholar]
- 3.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356 10.1038/nature01661 [DOI] [PubMed] [Google Scholar]
- 4.Cross M, Smith E, Hoy D, Carmona L, Wolfe F, Vos T, et al. The global burden of rheumatoid arthritis: estimates from the global burden of disease 2010 study. Annals of the rheumatic diseases. 2014. [DOI] [PubMed] [Google Scholar]
- 5.Symmons D, Turner G, Webb R, Asten P, Barrett E, Lunt M, et al. The prevalence of rheumatoid arthritis in the United Kingdom: new estimates for a new century. Rheumatology. 2002;41(7):793–800. [DOI] [PubMed] [Google Scholar]
- 6.MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis & Rheumatology. 2000;43(1):30–7. [DOI] [PubMed] [Google Scholar]
- 7.Li G, Zhao J, Li B, Ma J, Zhao Q, Wang X, et al. Associations between CCL21 gene polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis. Rheumatology International. 2017:1–9. [DOI] [PubMed] [Google Scholar]
- 8.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature genetics. 2010;42(6):508–14. 10.1038/ng.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neidhart M, Karouzakis E. Genetics: a new interpretation of genetic studies in RA. Nature Reviews Rheumatology. 2014;10(4):199–200. 10.1038/nrrheum.2014.21 [DOI] [PubMed] [Google Scholar]
- 10.Orozco G, Viatte S, Bowes J, Martin P, Wilson AG, Morgan AW, et al. Novel Rheumatoid Arthritis Susceptibility Locus at 22q12 Identified in an Extended UK Genome‐Wide Association Study. Arthritis & Rheumatology. 2014;66(1):24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rooy D, Zhernakova A, Tsonaka R, Willemze A, Kurreeman B, Trynka G, et al. A genetic variant in the region of MMP-9 is associated with serum levels and progression of joint damage in rheumatoid arthritis. Annals of the rheumatic diseases. 2014;73(6):1163–9. 10.1136/annrheumdis-2013-203375 [DOI] [PubMed] [Google Scholar]
- 12.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nature genetics. 2012;44(5):511 10.1038/ng.2231 [DOI] [PubMed] [Google Scholar]
- 13.Thatayatikom A, Liu AH. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim Gene family. Pediatrics. 2003;112(Supplement 2):470–. [Google Scholar]
- 14.Kuchroo VK, Umetsu DT, DeKruyff RH, Freeman GJ. The TIM gene family: emerging roles in immunity and disease. Nature reviews Immunology. 2003;3(6):454. [DOI] [PubMed] [Google Scholar]
- 15.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunological reviews. 2010;235(1):172–89. 10.1111/j.0105-2896.2010.00903.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rennert PD. Novel roles for TIM-1 in immunity and infection. Immunology letters. 2011;141(1):28–35. 10.1016/j.imlet.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi N, Karisola P, Peña-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27(6):927–40. 10.1016/j.immuni.2007.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.宮西正憲. Identification of Tim4 as a phosphatidylserine receptor: 京都大学; 2009.
- 19.Nicholson LB, Greer JM, Sobel RA, Lees MB, Kuchroo VK. An altered peptide ligand mediates immune deviation and prevents autoimmune encephalomyelitis. Immunity. 1995;3(4):397–405. [DOI] [PubMed] [Google Scholar]
- 20.Hofstra CL, Van Ark I, Hofman G, Kool M, Nijkamp FP, Van Oosterhout AJ. Prevention of Th2-like cell responses by coadministration of IL-12 and IL-18 is associated with inhibition of antigen-induced airway hyperresponsiveness, eosinophilia, and serum IgE levels. The Journal of Immunology. 1998;161(9):5054–60. [PubMed] [Google Scholar]
- 21.Mou Z, Shi J, Tan Y, Xu R, Zhao Z, Xu G, et al. Association between TIM-1 gene polymorphisms and allergic rhinitis in a Han Chinese population. Journal of investigational allergology & clinical immunology. 2010;20(1):3–8. Epub 2010/03/18. . [PubMed] [Google Scholar]
- 22.Chae SC, Park YR, Lee YC, Lee JH, Chung HT. The association of TIM-3 gene polymorphism with atopic disease in Korean population. Human immunology. 2004;65(12):1427–31. Epub 2004/12/18. 10.1016/j.humimm.2004.07.002 . [DOI] [PubMed] [Google Scholar]
- 23.Page NS, Jones G, Stewart GJ. Genetic association studies between the T cell immunoglobulin mucin (TIM) gene locus and childhood atopic dermatitis. International archives of allergy and immunology. 2006;141(4):331–6. Epub 2006/08/31. 10.1159/000095459 . [DOI] [PubMed] [Google Scholar]
- 24.Graves PE, Siroux V, Guerra S, Klimecki WT, Martinez FD. Association of atopy and eczema with polymorphisms in T-cell immunoglobulin domain and mucin domain-IL-2-inducible T-cell kinase gene cluster in chromosome 5 q 33. The Journal of allergy and clinical immunology. 2005;116(3):650–6. Epub 2005/09/15. 10.1016/j.jaci.2005.05.004 . [DOI] [PubMed] [Google Scholar]
- 25.McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nature immunology. 2001;2(12):1109–17. 10.1038/ni739 [DOI] [PubMed] [Google Scholar]
- 26.Gao PS, Mathias RA, Plunkett B, Togias A, Barnes KC, Beaty TH, et al. Genetic variants of the T-cell immunoglobulin mucin 1 but not the T-cell immunoglobulin mucin 3 gene are associated with asthma in an African American population. The Journal of allergy and clinical immunology. 2005;115(5):982–8. Epub 2005/05/04. 10.1016/j.jaci.2005.01.035 . [DOI] [PubMed] [Google Scholar]
- 27.Chae SC, Song JH, Heo JC, Lee YC, Kim JW, Chung HT. Molecular variations in the promoter and coding regions of human Tim-1 gene and their association in Koreans with asthma. Human immunology. 2003;64(12):1177–82. Epub 2003/11/25. . [DOI] [PubMed] [Google Scholar]
- 28.Yaghoobi E, Abedian S, Babani O, Izad M. TIM-3 Rs10515746 (A/C) and Rs10053538 (C/A) Gene Polymorphisms and Risk of Multiple Sclerosis. Iranian journal of public health. 2016;45(5):644–9. Epub 2016/07/12. [PMC free article] [PubMed] [Google Scholar]
- 29.Mazrouei F, Ganjalikhani-Hakemi M, Salehi R, Alesahebfosoul F, Etemadifar M, Pouladian M, et al. Association of TIM-1 5383-5397ins/del and TIM-3 -1541C>T polymorphisms with multiple sclerosis in Isfahan population. International journal of immunogenetics. 2016;43(3):131–4. Epub 2016/04/20. 10.1111/iji.12264 . [DOI] [PubMed] [Google Scholar]
- 30.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS medicine. 2009;6(7):e1000100 10.1371/journal.pmed.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25(9):603–5. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 32.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. The American Journal of Human Genetics. 2005;76(5):887–93. 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychological methods. 2006;11(2):193 10.1037/1082-989X.11.2.193 [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994:1088–101. [PubMed] [Google Scholar]
- 36.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS medicine. 2009;6(7):e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chae S-C, Park Y-R, Song J-H, Shim S-C, Yoon K-S, Chung H-T. The polymorphisms of Tim-1 promoter region are associated with rheumatoid arthritis in a Korean population. Immunogenetics. 2005;56(10):696–701. 10.1007/s00251-004-0743-5 [DOI] [PubMed] [Google Scholar]
- 38.Song YW, Im CH, Park JH, Lee YJ, Lee EY, Lee EB, et al. T-cell immunoglobulin and mucin domain 3 genetic polymorphisms are associated with rheumatoid arthritis independent of a shared epitope status. Human immunology. 2011;72(8):652–5. 10.1016/j.humimm.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 39.Chae S-C, Park Y-R, Shim S-C, Yoon K-S, Chung H-T. The polymorphisms of Th1 cell surface gene Tim-3 are associated in a Korean population with rheumatoid arthritis. Immunology letters. 2004;95(1):91–5. 10.1016/j.imlet.2004.06.008 [DOI] [PubMed] [Google Scholar]
- 40.Mosaad Y, El‐bassiony S, El‐Ghaweet A, Elhindawy M, EL‐Deek B, Sultan W. TIM‐1 rs41297579 G> A (− 1454) and TIM‐4 rs7700944 gene polymorphisms as possible risk factor for rheumatoid arthritis: relation to activity and severity. International journal of immunogenetics. 2015;42(4):254–64. 10.1111/iji.12201 [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Yang Y, Liu X, Wang Y. Genetic variation and significant association of polymorphism rs7700944 G> A of TIM‐4 gene with rheumatoid arthritis susceptibility in Chinese Han and Hui populations. International journal of immunogenetics. 2012;39(5):409–13. 10.1111/j.1744-313X.2012.01103.x [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Yang Y, Liu X, Sun J, Wang Y. Polymorphisms of the TIM-1 gene are associated with rheumatoid arthritis in the Chinese Hui minority ethnic population. Genet Mol Res. 2012;11(1):61–9. 10.4238/2012.January.9.7 [DOI] [PubMed] [Google Scholar]
- 43.Xu J, Yang Y, Liu X, Wang Y. The− 1541 C> T and+ 4259 G> T of TIM‐3 polymorphisms are associated with rheumatoid arthritis susceptibility in a Chinese Hui population. International journal of immunogenetics. 2011;38(6):513–8. 10.1111/j.1744-313X.2011.01046.x [DOI] [PubMed] [Google Scholar]
- 44.Zakeri Z, Hashemi M, Pourhosseini SME, Eskandari-Nasab E, Bahari G, Taheri M. Association between the rs7700944 polymorphism in the TIM-4 gene and rheumatoid arthritis in Zahedan, southeast Iran. Revista Brasileira de Reumatologia (English Edition). 2013;53(4):341–5. [PubMed] [Google Scholar]
- 45.Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J. A method for meta‐analysis of molecular association studies. Statistics in medicine. 2005;24(9):1291–306. 10.1002/sim.2010 [DOI] [PubMed] [Google Scholar]
- 46.Xie X, Shi X, Chen P, Rao L. Associations of TIM-1 Genetic Polymorphisms with Asthma: A Meta-analysis. Lung. 2017;195(3):353–60. 10.1007/s00408-017-0006-5 [DOI] [PubMed] [Google Scholar]
- 47.Lawrence J, Ball J. Genetic studies on rheumatoid arthritis. Annals of the rheumatic diseases. 1958;17(2):160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amos R, Constable T, Crockson R, Crockson A, McConkey B. Rheumatoid arthritis: relation of serum C-reactive protein and erythrocyte sedimentation rates to radiographic changes. Br med J. 1977;1(6055):195–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You C-g, Li J-f, Xie X-d, Zhu Y, Li P-q, Chen Y-r. Association of interleukin-1 genetic polymorphisms with the risk of rheumatoid arthritis in Chinese population. Clinical Chemical Laboratory Medicine. 2007;45(8):968–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


