Abstract
Purpose
The prevalence and features of treatment-emergent small-cell neuroendocrine prostate cancer (t-SCNC) are not well characterized in the era of modern androgen receptor (AR)–targeting therapy. We sought to characterize the clinical and genomic features of t-SCNC in a multi-institutional prospective study.
Methods
Patients with progressive, metastatic castration-resistant prostate cancer (mCRPC) underwent metastatic tumor biopsy and were followed for survival. Metastatic biopsy specimens underwent independent, blinded pathology review along with RNA/DNA sequencing.
Results
A total of 202 consecutive patients were enrolled. One hundred forty-eight (73%) had prior disease progression on abiraterone and/or enzalutamide. The biopsy evaluable rate was 79%. The overall incidence of t-SCNC detection was 17%. AR amplification and protein expression were present in 67% and 75%, respectively, of t-SCNC biopsy specimens. t-SCNC was detected at similar proportions in bone, node, and visceral organ biopsy specimens. Genomic alterations in the DNA repair pathway were nearly mutually exclusive with t-SCNC differentiation (P = .035). Detection of t-SCNC was associated with shortened overall survival among patients with prior AR-targeting therapy for mCRPC (hazard ratio, 2.02; 95% CI, 1.07 to 3.82). Unsupervised hierarchical clustering of the transcriptome identified a small-cell–like cluster that further enriched for adverse survival outcomes (hazard ratio, 3.00; 95% CI, 1.25 to 7.19). A t-SCNC transcriptional signature was developed and validated in multiple external data sets with > 90% accuracy. Multiple transcriptional regulators of t-SCNC were identified, including the pancreatic neuroendocrine marker PDX1.
Conclusion
t-SCNC is present in nearly one fifth of patients with mCRPC and is associated with shortened survival. The near-mutual exclusivity with DNA repair alterations suggests t-SCNC may be a distinct subset of mCRPC. Transcriptional profiling facilitates the identification of t-SCNC and novel therapeutic targets.
INTRODUCTION
Prostate cancer is the most common incident cancer in men in developed countries and the eighth leading cause of cancer death globally.1 The introduction of highly potent androgen receptor (AR)–targeting therapies such as abiraterone and enzalutamide for the treatment of metastatic castration-resistant prostate cancer (mCRPC) has provided significant clinical benefit.2,3
In a subset of patients, therapeutic resistance to AR-targeting therapy is accompanied by the emergence of a histologic subtype that morphologically resembles de novo small-cell prostate cancer, a highly aggressive histologic variant present in < 1% of untreated prostate cancers at the time of diagnosis.4 It is not clear if the treatment-emergent variant, variously labeled neuroendocrine prostate cancer and aggressive variant,5,6 is the same disease entity as de novo small-cell prostate cancer. We have termed this histology treatment-emergent small-cell neuroendocrine prostate cancer or t-SCNC. Previous reports have sought to characterize t-SCNC but have been limited by the availability of prospectively collected tissue from a consecutive series of patients with sufficient follow-up to characterize incidence of t-SCNC and clinical outcomes.7,8
To understand the prevalence and characteristics of this treatment-emergent variant, and to provide a basis for tumor classification, clinical recommendations, and future development of therapies, we undertook a multi-institutional prospective study to perform biopsy of metastases from consecutively enrolled patients with progressive mCRPC.
METHODS
Patients and Study Design
A prospective IRB-approved trial (ClinicalTrials.gov identifier: NCT02432001) was conducted at five consortium sites. Eligibility criteria included progressive mCPRC by Prostate Cancer Clinical Trials Working Group 2 criteria,9 prior histologic evidence of adenocarcinoma of the prostate gland, at least one bone or soft tissue metastasis amenable to biopsy, and written informed consent. Patients were prospectively followed for overall survival. Treatment post biopsy specimen acquisition was per investigator discretion. Serum prostate-specific antigen (PSA), lactate dehydrogenase, alkaline phosphatase, hemoglobin, neuron-specific enolase (NSE), and chromogranin A (CGA) levels were measured at the time of biopsy. Repeat tumor biopsy at progression was optional.
Tissue Acquisition and Pathology Evaluation
Metastatic biopsy tissue was acquired via image-guided core needle biopsy as previously described.10 Separate needle core biopsies of the same metastatic lesion were snap frozen and formalin fixed/paraffin embedded (FFPE), respectively. Tumor RNA from frozen specimens was amplified and sequenced for gene expression analyses. FFPE biopsy specimens were evaluated by three experienced pathologists (G.T., J.H., L.T.) blinded to the clinical and genomic features for determination of consensus pathologic subclassification into three categories (pure small-cell morphology, mixed biopsy specimens with discrete regions of t-SCNC and adenocarcinoma, or biopsy specimens without any small-cell features), using recently described classification criteria (Data Supplement).11 Because the diagnosis of t-SCNC is based on morphologic criteria, immunohistochemical staining for chromogranin, CD56, or synaptophysin was not routinely performed.11 Next-generation targeted genomic sequencing of FFPE tissue was performed as previously described.12
Transcriptional Analysis and Clustering
For the unsupervised gene expression analysis, complete-linkage clustering was performed. The 5,000 most varying protein-coding HUGO Gene Nomenclature Committee genes13 were selected to compute sample-to-sample gene expression correlation values as distance metric for the hierarchical clustering. The resulting sample tree was cut into five clusters. Analysis of variance for the five sample clusters was performed, and 528 genes had an false discovery rate (FDR)-corrected P < .05.
Master regulator analysis was performed using the MARINa algorithm implemented via the viper R package.14,15 MARINa infers candidate master regulators (MRs) between two groups of samples on the basis of the expression of the regulators’ downstream targets. Sample-specific MR scores were computed with the VIPER function and visualized using TumorMap.16
t-SCNC Signature Development and Validation
RNA-Seq data from 18,538 protein-coding HUGO Gene Nomenclature Committee genes were used to distinguish t-SCNC versus adenocarcinoma. Samples with mixed histology were excluded from the learning set. Leave-pair-out cross-validation was performed on 100 models to determine model accuracy.17 The signature was subsequently applied to mixed histology tumors as well as three external mCRPC data sets and the primary prostate cancer data set of TCGA.7,8,18,19
Characterization of AR Expression and Signaling
AR protein expression was analyzed using immunohistochemical (IHC) analysis (Androgen Receptor [C6F11] XP Rabbit mAb; Data Supplement). To evaluate canonical AR transcriptional activity in each biopsy specimen, an AR expression signature was developed based on 53 AR-positive cell lines in the presence and absence of androgen.20 The derived classifier had > 90% concordance with a previously described AR signature.21
Statistical Considerations
Comparison of the continuous variables among groups was assessed by the two-sample t test, analysis of variance, Wilcoxon rank sum test, and Kruskal-Wallis test, when normality assumption did or did not hold, respectively.22-24 The statistical association between categorical variables was evaluated by χ2 and Fisher’s exact test.
Overall survival (OS) was measured from the date of development of mCRPC, as defined by Prostate Cancer Clinical Trials Working Group 2 criteria, with the prespecified primary analysis in patients previously treated with abiraterone and/or enzalutamide. Kaplan-Meier product limit method, log-rank, and Cox proportional hazards were used to characterize the relationship between OS, histology subtype, and gene cluster.
Analyses pertaining to the incidence and clinical characteristics of t-SCNC, DNA sequencing, and overall survival were conducted on a per-patient basis, using the first evaluable biopsy. Baseline and progression biopsy specimens, when available, were included as discrete samples for gene and protein expression analyses.
RESULTS
Incidence of t-SCNC
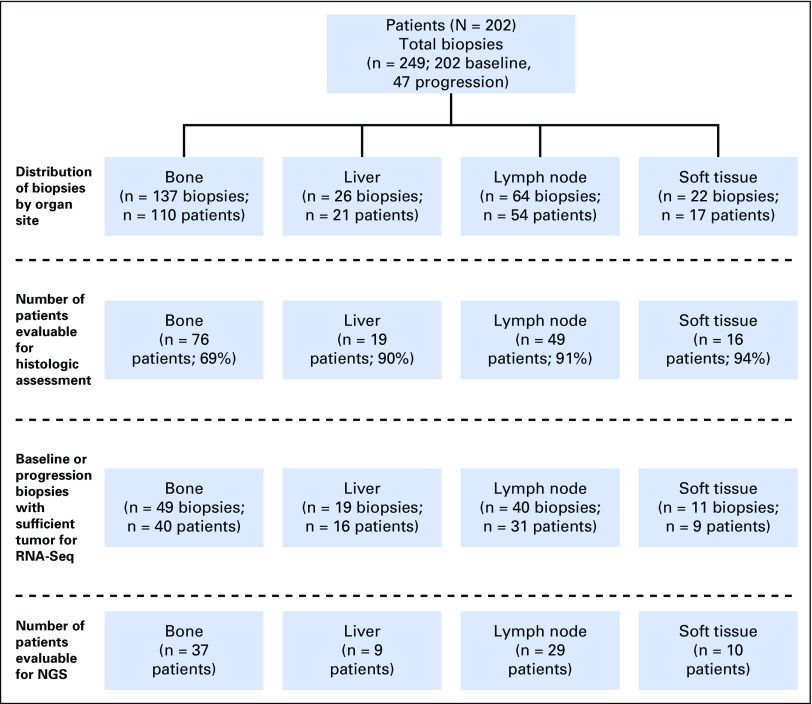
Between December 2012 and April 2016, 202 patients with mCRPC were enrolled and underwent a total of 249 metastatic tumor biopsies. The median time from mCRPC to biopsy was 17.6 months (range, 0.1 to 212.6 months). Of 202 patients enrolled, 160 (79%) had sufficient tumor present in at least one biopsy specimen to permit histologic classification. Bone metastases (n = 137) comprised 55% of all biopsy specimens, lymph node (n = 64) 26%, liver (n = 26) 10%, and other soft tissue (n = 22), 9% (Fig 1).
Fig 1.
CONSORT diagram indicating biopsy site and disposition for the various analyses. NGS, next-generation sequencing.
t-SCNC was found in 27 of 160 (17%) evaluable patients. Twenty patients harbored tumors with pure small-cell histology, and seven patients had mixed biopsy specimens with discrete regions of t-SCNC and adenocarcinoma within the same needle core (Fig 2; Data Supplement). The percentage of t-SCNC in the seven mixed cases ranged from 20% to 80%. Detection of t-SCNC was observed at similar proportions by biopsy site, including 14%, 19%, and 14% of evaluable liver, lymph node, and bone metastases, respectively (P = .76).
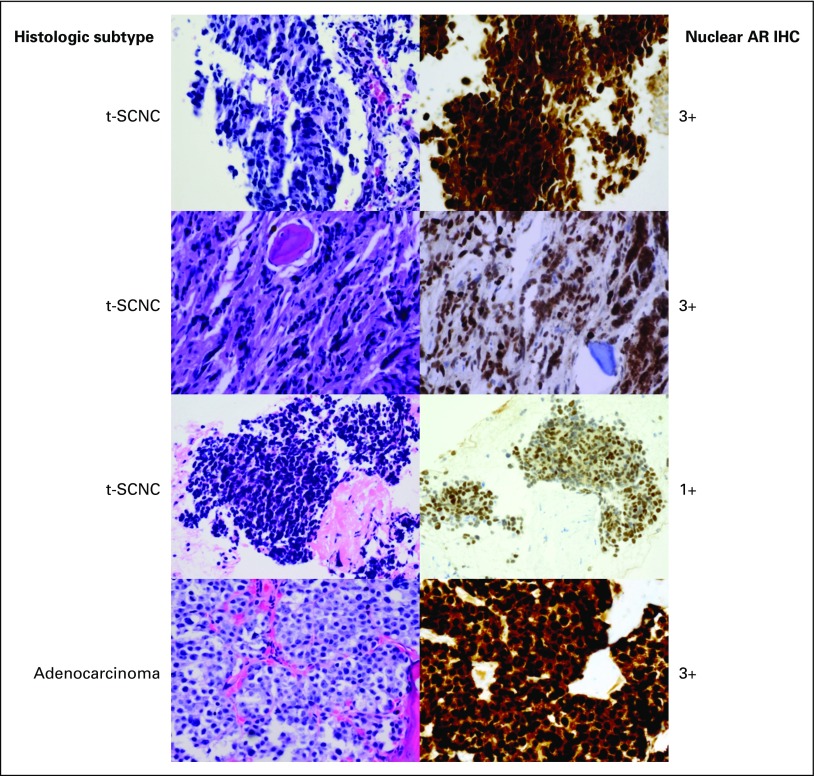
Fig 2.
Histologic appearance and immunohistochemical (IHC) staining of the androgen receptor (AR). The top three rows represent biopsy specimens with treatment-emergent small-cell neuroendocrine prostate cancer (t-SCNC) histologic classification. The top two rows have strong 3+ expression of the AR with nuclear localization. The third row demonstrates a t-SCNC biopsy specimen with low (1+) AR nuclear expression. The bottom row represents a metastatic biopsy specimen with typical adenocarcinoma morphology, with 3+ nuclear expression of the AR. Magnification, ×400.
Transcriptional Profile of t-SCNC
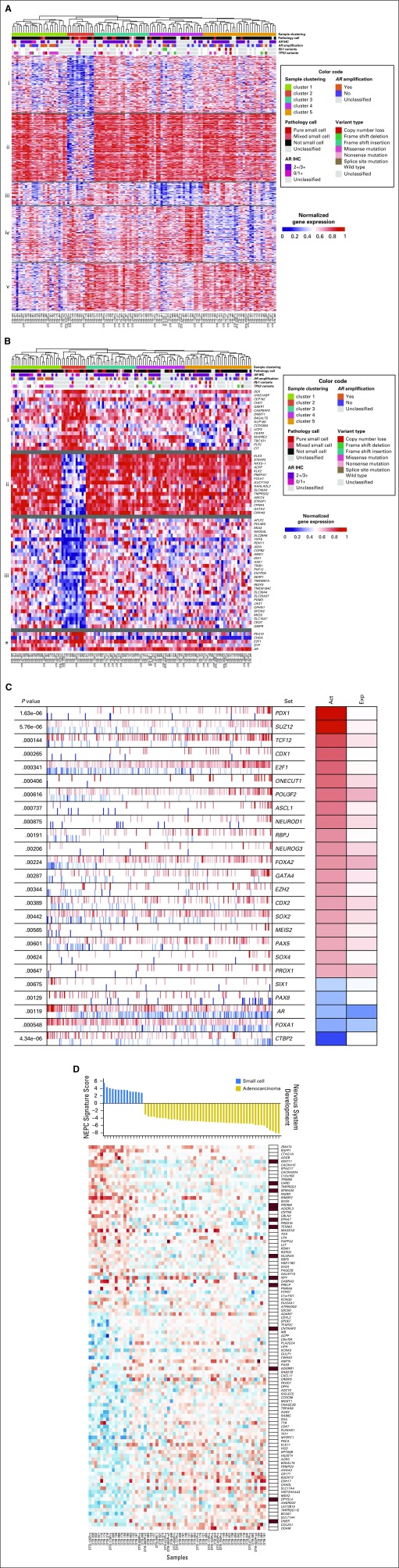
mRNA-Seq data were available from 119 baseline and progression biopsy specimens distributed across all organ sites (Fig 1), including 21 tumors with t-SCNC histologic differentiation (pure or mixed). Unsupervised hierarchical clustering of the transcriptome identified a cluster of 12 cases (cluster 2) that was enriched for the presence of t-SCNC histologic differentiation (Fig 3A). Six of 14 (43%) pure t-SCNC tumors fell within cluster 2, versus two of seven (26%) tumors with mixed histology versus four of 90 (4%) tumors with pure adenocarcinoma (P < .001).
Fig 3.
Transcriptional profile of treatment-emergent small-cell neuroendocrine prostate cancer (t-SCNC). (A) Gene expression analysis identifies t-SCNC cases in unsupervised analysis. Unsupervised hierarchical clustering of transcriptional profile of metastatic castration-resistant prostate cancer biopsy specimens (n = 119). Sample cluster 2 is enriched for presence of t-SCNC histology. The rows show the normalized gene expression of 528 genes with false discovery rate (FDR)-corrected P < .05 for analysis of variance of gene expression in the five sample clusters, k-means clustered with k = 5 (labeled i-v). Hypergeometric testing for gene sets showed the gene clusters are involved in (i) androgen response and metabolic processes; (ii) androgen response, androgen receptor (AR) activity and targets, and FOXA1 network; (iii) translation; (iv) extracellular organization; and (v) cell cycle and transport. Pathology call, AR immunohistochemistry (IHC), and variant calls of TP53 and RB1 shown in top rows. (B) Heatmap showing 61 genes with FDR-corrected P < .05 for t test of gene expression in the t-SCNC–enriched cluster 2 versus all other samples. Genes are k-means clustered, with k = 3 (i-iii), in addition showing genes of interest PEG10, CHGA, E2F1, SYP, and AR (*). Hypergeometric testing for gene set enrichment showed gene cluster i contains genes of the Hallmark E2F Targets gene set, cluster ii is dominated by genes related to androgen response and AR activity, and cluster iii contains genes of the Notch signaling pathway. (C) MARINa-inferred master regulators characterizing gene expression differences between small cell-like cluster versus other clusters. The top 25 most activated (red) and repressed (blue) transcription factors in the small-cell–like cluster samples compared with all other samples are shown, as inferred by the MARINa algorithm (FDR < 0.1). Each transcription factor’s targets are shown as tick marks projected onto the gene expression signature. Each row also shows the regulator’s P value, inferred differential activity (Act), and differential expression (Exp). (D) A newly generated t-SCNC gene expression signature distinguishes small-cell histology with 91% accuracy on leave-pair-out cross-validation. The numerical signature score is directly related to the predicted degree of t-SCNC differentiation (top). The heatmap shows median-centered gene expression of the 106 genes in the signature, and membership in neural system development pathways in column on the right. Abbreviations: NEPC, neuroendocrine prostate cancer.
A supervised analysis identified 61 genes differentially expressed between the t-SCNC–enriched cluster versus other clusters with FDR-corrected P < .05 (Fig 3B). The topmost overexpressed genes were transcriptional targets of E2F Transcription Factor 1 (E2F1; negatively regulated by Retinoblastoma 1 [RB1]). RB1 loss signature25 scores were higher in the t-SCNC–enriched cluster (Data Supplement). To further characterize the transcriptional hallmarks of cluster 2, we used the MR Inference algorithm and visualized results using TumorMap.14,16 Pancreatic-duodenal homeobox factor 1 (PDX1) was the topmost MR enriched in the t-SCNC cluster (P < .001; Fig 3C; Data Supplement). Achaete-Scute family BHLH transcription factor 1 (ASCL1), E2F1, Forkhead box A2 (FOXA2), and POU class 3 homeobox 2 (POU3F2) were also among the top MRs in the t-SCNC cluster (P < .05 for all comparisons; Fig 3C; Data Supplement).
A gene expression signature of t-SCNC was subsequently developed, with 91% internal accuracy on leave-pair-out cross-validation, and enriched for presence of neural development genes, including SEMA3, EPHA7, and TENM3 (Fig 3D). t-SCNC signature scores of biopsy specimens with mixed histology are shown in the Data Supplement. The t-SCNC signature was applied to three external mCRPC data sets and correctly categorized neuroendocrine tumors with nearly 100% accuracy (Data Supplement).7,8,18 Finally, the t-SCNC signature was applied to the TCGA database of primary tumors and correctly classified all cases as possessing adenocarcinoma histology (Data Supplement).19
Targeted Genomic Sequencing of t-SCNC
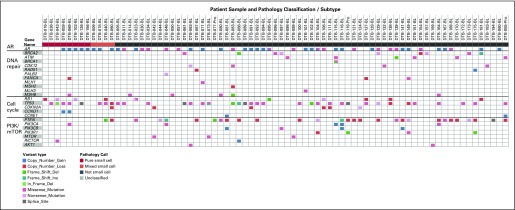
Eighty-five patients were evaluable for somatic targeted sequencing, including 12 patients (14%) with pure or mixed t-SCNC histology (Fig 4; Data Supplement). The prevalence of AR copy number gain and/or activating point mutations was similarly distributed across histologic groups (67% of t-SCNC biopsy specimens v 51% of biopsy specimens without t-SCNC; P = .304). Variants predicted to lead to loss of function in TP53 and/or RB1 were found in 10 of 12 patients with t-SCNC (83%) compared with 25 of 73 (34%) patients who did not harbor t-SCNC on biopsy (P = .0015). The presence of deleterious mutations and/or copy number loss in DNA repair pathway genes (BRCA1, BRCA2, ATM, CDK12, RAD51, PALB2, FANCA, CHEK2, MLH1, MSH2, MLH3, and MSH6) was almost entirely mutually exclusive with t-SCNC tumors (1 of 12 [8%] t-SCNC biopsy specimens v 29 of 73 [40%] biopsy specimens without t-SCNC; P = .0350; Fig 4).
Fig 4.
Variant calls (point mutations and copy number gain/loss) shown for selected subsets of genes, including androgen receptor (AR), DNA repair pathway, cell cycle, and phosphatidylinositol-3-kinase/mammalian target of rapamycin (PI3K/mTOR) pathway.
AR Expression and Activity
A total of 106 FFPE biopsy specimens were stained for AR expression (Fig 2; Data Supplement). In the overall cohort, 2+/3+ nuclear AR expression by IHC and AR amplification were both positively associated with higher AR transcriptional signature score (Data Supplement). Of 20 t-SCNC specimens, 15 (75%) demonstrated 2+/3+ nuclear AR staining, compared with 75 of 86 (87%) adenocarcinoma (P = .170). The t-SCNC biopsy specimens without strong nuclear AR staining had similar clinical features to the overall t-SCNC cohort and fell within cluster 2 of the unsupervised transcriptional analysis (Fig 3A). AR transcriptional activity was lower in tumors with t-SCNC histology (median scores of the pure t-SCNC, mixed, and not t-SCNC samples were −2.12, −1.10, and −0.24, respectively; P = .017) and was lower in cluster 2 versus other clusters (Data Supplement). Serum PSA and tissue KLK3 expression were not correlated (Pearson’s coefficient of determination, 0.0045).
Clinical Characteristics of t-SCNC
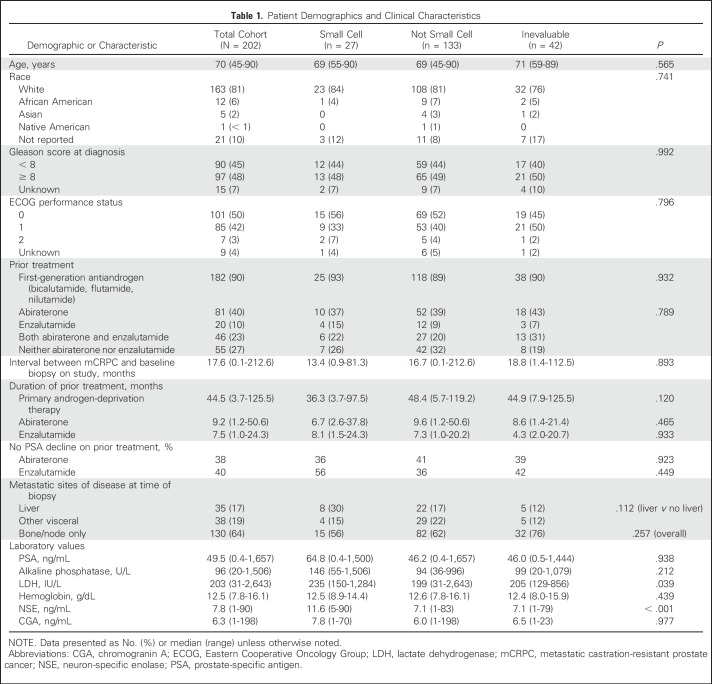
Baseline characteristics of the overall patient cohort at the time of biopsy are summarized in Table 1 and were comparable between the evaluable (n = 160) and inevaluable patients (n = 42). Nearly three fourths of the patients had developed resistance to abiraterone (n = 82; 40%), enzalutamide (n = 20; 10%), or both (n = 46; 23%) at study entry.
Table 1.
Patient Demographics and Clinical Characteristics
The majority of clinical characteristics, except for serum lactate dehydrogenase, were similar in patients harboring t-SCNC histology (whether pure or mixed) compared with those who did not (Table 1; Data Supplement). Median time from mCRPC to study entry, serum PSA, Gleason score at time of diagnosis, and sites of metastases were not significantly different in patients with t-SCNC. A comparison of the clinical features of the t-SCNC subset with two independent neuroendocrine prostate cancer cohorts5,8 is shown in the Data Supplement.
Median serum NSE was higher in patients with t-SCNC (11.6 ng/mL v 7.1; P < .001); CGA was not (median 7.8 ng/mL v 6.0; P = .977). Using receiver operating characteristic curve analysis, if serum NSE was > 6.05 ng/mL and CGA was > 3.1 ng/mL (present in 55% of patients), the sensitivity, specificity, and negative and positive predictive values for the detection of t-SCNC histology on biopsy were 95%, 50%, 98%, and 22%, respectively.
Survival Outcomes by Histologic and Genomic Subgroups
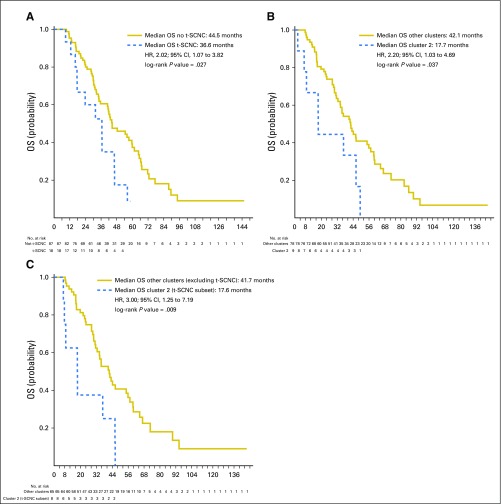
The median time from the time of development of mCRPC to death was 42.1 months, with a median follow-up of 34.1 months and 131 deaths observed. Median overall survival in the preplanned analysis of patients with prior abiraterone and/or enzalutamide treatment was significantly shorter in those with t-SCNC (median OS from date of mCRPC = 44.5 v 36.6 months; hazard ratio [HR], 2.02; 95% CI, 1.07 to 3.82; Fig 5A). A post hoc sensitivity analysis including treatment-naïve patients yielded similar results (Data Supplement). Patients with mixed tumors had similarly reduced survival as those with pure t-SCNC (median OS, 36.3 and 36.8 months, respectively v 44.5 months in patients with adenocarcinoma; log-rank P = .031). Patients whose biopsy fell within the small-cell–enriched cluster 2 on unsupervised hierarchical clustering likewise had worse survival (HR, 2.20; 95% CI, 1.03 to 4.69; Fig 5B). When histology and transcriptomic data were combined, patients with t-SCNC tumors falling within cluster 2 had significantly shorter survival than the patients without t-SCNC histology falling outside cluster 2, with a greater separation of survival curves than either histologic or genomic analysis alone (HR for overall survival, 3.00; 95% CI, 1.25 to 7.19; Fig 5C).
Fig 5.
Overall survival from date of metastatic castration-resistant prostate cancer by histologic and genomic subgroups. (A) Overall survival (OS) by histology in the preplanned evaluable cohort of patients with prior abiraterone and/or enzalutamide treatment. Blue line, treatment-emergent small-cell neuroendocrine prostate cancer cohort (t-SCNC); gold line, not t-SCNC cohort (B) Survival by unsupervised transcriptional cluster (cluster 2 v others). Blue line, cluster 2; gold line, other clusters (C) Comparison of survival between cluster 2 cases with t-SCNC histology (blue line) versus cases in other clusters without t-SCNC histology (gold line). HR, hazard ratio.
DISCUSSION
Widespread use of abiraterone and enzalutamide has had a transformative impact in the management of advanced prostate cancer, yet therapeutic resistance is a near-universal phenomenon, frequently heralded by a more aggressive clinical course. t-SCNC was identified in 17% of evaluable patients in our large, prospective series of patients with mCRPC, suggesting that this is an important mechanism in the development of treatment-resistant mCRPC. The near-mutual exclusivity between t-SCNC differentiation and presence of DNA repair mutations raises the intriguing possibility of distinct subsets of mCRPC.
The identification of mixed histologic subtypes within a single metastatic biopsy suggests that treatment-emergent small-cell neuroendocrine differentiation is a heterogeneous process. Heterogeneity may also partially account for the divergence observed between AR protein expression and inferred transcriptional activity in a subset of t-SCNC biopsy specimens. Nevertheless, although patients in this study underwent biopsy of a single metastatic site, and heterogeneity across different metastatic sites in the same patient was not evaluated, t-SCNC identified on a single biopsy, whether pure or mixed, was associated with shortened survival. Transcriptional analysis identified a subset of patients with particularly high-risk t-SCNC and had additional prognostic utility when combined with histopathologic classification. Validation of these observations in independent cohorts, as they become available, will be important.
The observed prevalence of t-SCNC is substantially higher than de novo small-cell cancer of the prostate, which occurs in < 1% of cases.4 This may reflect a transdifferentiation process after androgen-ablating therapy.26 Classic de novo SCNC is an AR-null phenotype, progressing with low serum PSA levels. In contrast, in our series of t-SCNC cases, the majority of tumors had high nuclear AR expression by IHC, and median serum PSA was > 60 ng/mL. Visceral metastases are common in de novo SCNC; in the current series, only approximately one third of patients with t-SCNC histology had liver metastases. The overlap in clinical features between patients with t-SCNC and adenocarcinoma calls into question current practice guidelines recommending metastatic biopsy to evaluate for small-cell differentiation only in cases with aggressive phenotypic features.27
In the Robinson et al18 series, the incidence of tumors with small-cell neuroendocrine differentiation was approximately 1%, compared with 17% in the current study. This may in part reflect the prospective design of the current study with inclusion of consecutively enrolled patients, in contrast to the characterization of biopsy specimens obtained within a clinical trial network described previously.18 In addition, enrollment in the current study occurred at varying time points during the course of mCRPC, with potential enrichment for patients at higher risk. Methodologic differences in pathologic evaluation of FFPE versus frozen tissue may also partially account for the difference in incidence between the two series. Application of the t-SCNC expression signature to other external data sets, as they become available, will provide additional clarity regarding the incidence of t-SCNC.
The practical limitations in obtaining metastatic tumor tissue from patients make the development of noninvasive biomarkers of t-SCNC of critical importance. In this series, the serum neuroendocrine markers CGA and NSE had a high sensitivity (95%) and negative predictive value (98%) for detecting t-SCNC but lacked specificity. If independently validated, patients with normal serum neuroendocrine markers, representing 45% of the patients in our series, may not require biopsies to detect t-SCNC. Circulating tumor cells and imaging tools may yield additional diagnostic utility in identifying t-SCNC and quantifying the degree of intra- and intertumoral heterogeneity.28,29
Persistent nuclear AR expression in the setting of lower predicted canonical AR transcriptional activity, as observed in a subset of t-SCNC biopsy specimens, suggests that AR may be under epigenetic regulation of an alternative transcriptional program. This is consistent with the observation that marked epigenetic differences exist between CRPC with and without neuroendocrine differentiation.8 The potential plasticity of the transdifferentiation process and persistent AR expression in the setting of low canonical activity raises intriguing therapeutic implications of restoring AR activity via application of epigenetic modifiers such as enhancer of zeste homolog 2 (EZH2) inhibitors.30,31 These hypotheses warrant additional investigation.
The molecular pathogenesis of t-SCNC remains incompletely defined but seems to arise in the context of TP53 and RB1 aberration from adenocarcinoma under selective pressure of AR pathway inhibition.32 We observed frequent loss of TP53 and/or RB1 at the genomic level, and upregulation of E2F, a hallmark of RB1 deficient tumors. DEK, the highest overexpressed E2F1 target gene in the t-SCNC–enriched cluster, has previously been implicated in the progression to neuroendocrine prostate cancer.33 Among tumors without small-cell differentiation, there exists a wide variability in histologic appearance, with some cases demonstrating classic adenocarcinoma features and other tumors with features suggestive of neuroendocrine differentiation. Analysis of paired longitudinal biopsies is ongoing to characterize this transitional disease state.
Despite the aggressive phenotypic features of t-SCNC, there is no standard of care for the treatment of patients who harbor this subtype. Master regulator analysis of the transcriptome identified several additional transcriptional factors implicated in the progression to neuroendocrine prostate cancer, including EZH2, POU class 3 homeobox 2 (BRN2), FOXA2, and ASCL1. FOXA2 has recently been described as a molecular marker of small-cell neuroendocrine prostate cancer.34 POU3F2, encoding the transcription factor BRN2, was recently implicated in the progression to neuroendocrine prostate cancer and inversely associated with AR expression.35 Direct enzymatic inhibitors of EZH2 are currently under clinical evaluation (ClinicalTrials.gov identifier: NCT02860286), as are drugs targeting cell-surface proteins transcriptionally regulated by these factors (eg, delta-like protein 3/ASCL1; ClinicalTrials.gov identifier: NCT02709889). The identification of PDX1, a Hox-type transcription factor that drives neuroendocrine differentiation in the pancreas,36 as the most active transcription factor in t-SCNC raises intriguing possibilities for pan–small-cell diagnostic and therapeutic strategies. With novel therapies in clinical development, there is the potential to improve disease outcomes for this high-risk and increasingly prevalent subset of mCRPC.
ACKNOWLEDGMENT
We thank Karen Knudsen and Christopher McNair for providing an RB1 loss signature25 for application to the transcriptome.
Footnotes
Supported in part by a Stand Up To Cancer Dream Team award, funded by the Prostate Cancer Foundation, Movember, and Stand Up To Cancer, Grant No. SU2C-AACR-DT0812 (E.J.S.).
Presented at the ASCO 2016 Annual Meeting, Chicago, IL, June 3-7, 2016.
Clinical trial information: NCT02432001.
AUTHOR CONTRIBUTIONS
Conception and design: Rahul Aggarwal, Jiaoti Huang, Joshi J. Alumkal, George V. Thomas, Owen N. Witte, Martin Gleave, Christopher P. Evans, Jack Youngren, Tomasz M. Beer, Matthew Rettig, Adam Foye, Charles J. Ryan, Kim N. Chi, Joshua M. Stuart, Eric J. Small
Financial support: Eric J. Small
Provision of study materials or patients: Joshi J. Alumkal, Adam Foye, Charles J. Ryan, Eric J. Small
Collection and assembly of data: Rahul Aggarwal, Jiaoti Huang, Joshi J. Alumkal, George V. Thomas, Alana S. Weinstein, Verena Friedl, Owen N. Witte, Paul Lloyd, Martin Gleave, Christopher P. Evans, Jack Youngren, Tomasz M. Beer, Matthew Rettig, Christopher K. Wong, Lawrence True, Adam Foye, Denise Playdle, Charles J. Ryan, Primo Lara, Kim N. Chi, Artem Sokolov, Joshua M. Stuart, Eric J. Small
Data analysis and interpretation: Rahul Aggarwal, Jiaoti Huang Joshi J. Alumkal, Li Zhang, Felix Y. Feng, George V. Thomas, Alana S. Weinstein, Verena Friedl, Can Zhang, Owen N. Witte, Paul Lloyd, Martin Gleave, Christopher P. Evans, Jack Youngren, Tomasz M. Beer, Matthew Rettig, Lawrence True, Charles Ryan, Primo Lara, Kim N. Chi, Vlado Uzunangelov, Artem Sokolov, Yulia Newton, Himisha Beltran, Francesca Demichelis, Mark A. Rubin, Joshua M. Stuart, Eric J. Small
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Rahul Aggarwal
Research Funding: Zenith Epigenetics, Novartis, Sanofi, Gilead Sciences
Jiaoti Huang
No relationship to disclose
Joshi J. Alumkal
Consulting or Advisory Role: Astellas Pharma, Bayer, Janssen Biotech
Research Funding: Aragon Pharmaceuticals (Inst), Astellas Pharma (Inst), Novartis (Inst), Zenith Epigenetics (Inst), Gilead Sciences (Inst)
Li Zhang
Consulting or Advisory Role: Dendreon, Fortis Therapeutics
Travel, Accommodations, Expenses: Dendreon
Felix Y. Feng
Leadership: PFS Genomics
Stock or Other Ownership: PFS Genomics
Consulting or Advisory Role: Medivation/Astellas, Dendreon, EMD Serono, Janssen Oncology, Ferring Pharmaceuticals, Sanofi, Bayer, Clovis Oncology
Patents, Royalties, Other Intellectual Property: I helped develop a molecular signature to predict radiation resistance in breast cancer, and this signature was patented by the University of Michigan, my employer. It is in the process of being licensed to PFS Genomics, a company that I helped found. (Inst)
George V. Thomas
Patents, Royalties, Other Intellectual Property: Patents at University of California Los Angeles and Oregon Health & Science University (Inst)
Alana S. Weinstein
No relationship to disclose
Verena Friedl
No relationship to disclose
Can Zhang
No relationship to disclose
Owen N. Witte
No relationship to disclose
Paul Lloyd
Stock or Other Ownership: Gilead Sciences (I), Sierra Oncology (I), Pfizer, Curis, GE Healthcare, Pacific Biosciences
Research Funding: Zenith Epigenetics (Inst)
Martin Gleave
Honoraria: Janssen, Astellas Pharma, Bayer, UBC Prostate Clinic, Sanofi
Consulting or Advisory Role: Janssen, Astellas Pharma, Bayer, AbbVie, Sanofi
Christopher P. Evans
Honoraria: Medivation, Janssen Oncology, Sanofi
Consulting or Advisory Role: Medivation, Janssen Oncology, Millennium
Speakers' Bureau: Medivation, Janssen Oncology, Sanofi
Research Funding: Medivation
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Sanofi, Astellas Pharma
Jack Youngren
Employment: GRAIL
Patents, Royalties, Other Intellectual Property: Patent for the use of tyrosine kinase receptor antagonists for the treatment of breast cancer assigned to the University of California
Tomasz M. Beer
Stock or Other Ownership: Salarius Pharmaceuticals
Consulting or Advisory Role: AstraZeneca, Dendreon, Janssen Biotech, Janssen Oncology, Janssen Research & Development, Johnson & Johnson, Janssen Japan, Clovis Oncology, AbbVie, Astellas Pharma, Pfizer, Bayer, Boehringer Ingelheim
Research Funding: Astellas Pharma (Inst), Bristol-Myers Squibb (Inst), Janssen Research & Development (Inst), Medivation (Inst), OncoGenex Pharmaceuticals (Inst), Sotio (Inst), Theraclone Sciences (Inst), Boehringer Ingelheim (Inst)
Matthew Rettig
Consulting or Advisory Role: Johnson & Johnson
Speakers' Bureau: Johnson & Johnson
Research Funding: Novartis
Patents, Royalties, Other Intellectual Property: Patent pending for small-molecule inhibitor of androgen receptor
Expert Testimony: Johnson & Johnson
Travel, Accommodations, Expenses: Johnson & Johnson
Christopher K. Wong
No relationship to disclose
Lawrence True
Research Funding: Ventana Medical Systems
Adam Foye
Consulting or Advisory Role: Sanverbos
Denise Playdle
No relationship to disclose
Charles J. Ryan
Honoraria: Janssen Oncology, Astellas Pharma, Bayer
Consulting or Advisory Role: Bayer, Ferring Pharmaceuticals
Research Funding: BIND Biosciences, Karyopharm Therapeutics, Novartis
Primo Lara
Honoraria: Pfizer
Consulting or Advisory Role: Exelixis, Pfizer, Novartis, AstraZeneca, Bayer, Genentech, Celgene, Janssen, Bristol-Myers Squibb, AbbVie, Turnstone Biologics
Research Funding: Millennium (Inst), Polaris (Inst), GlaxoSmithKline (Inst), Genentech (Inst), Aragon Pharmaceuticals (Inst), Janssen Biotech (Inst), Heat Biologics (Inst), TRACON Pharmaceuticals (Inst), Merck (Inst), Pharmacyclics (Inst), Incyte (Inst)
Kim N. Chi
Honoraria: Sanofi, Janssen, Astellas Pharma, Amgen
Consulting or Advisory Role: ESSA, Astellas Pharma, Janssen, Sanofi, Eli Lilly/ImClone, Amgen, Bayer
Research Funding: Janssen (Inst), Astellas Pharma (Inst), Amgen (Inst), Bayer (Inst), Novartis (Inst), Sanofi (Inst), Tokai Pharmaceuticals (Inst), Oncogenex Pharmaceuticals (Inst), Teva Pharmaceutical (Inst), Eli Lilly/ImClone (Inst)
Vlado Uzunangelov
No relationship to disclose
Artem Sokolov
Patents, Royalties, Other Intellectual Property: WO2017173451, targeting innate immune signaling in neuroinflammation and neurodegeneration
Yulia Newton
No relationship to disclose
Himisha Beltran
Consulting or Advisory Role: Janssen Oncology, Genzyme
Research Funding: Janssen (Inst), Stemcentryx AbbVie, Millenium (Inst)
Francesca Demichelis
Patents, Royalties, Other Intellectual Property: Co-inventor on a patent filed by The University of Michigan and The Brigham and Women’s Hospital covering the diagnostic and therapeutic fields for ETS fusions in prostate cancer. The diagnostic field has been licensed to Gen-Probe.
Mark A. Rubin
Patents, Royalties, Other Intellectual Property: NMYC in Prostate Cancer (Inst), SPOP in prostate Cancer (Inst), EZH2 in prostate cancer (Inst), ETS gene fusions in prostate cancer (Inst)
Joshua M. Stuart
Stock or Other Ownership: Nantomics
Eric J. Small
Honoraria: Janssen-Cilag
Consulting or Advisory Role: Fortis Therapeutics, Harpoon Therapeutics
Travel, Accommodations, Expenses: Janssen
REFERENCES
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Dicker D, et al. : The global burden of cancer in 2013. JAMA Oncol 1:505-527, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan CJ, Smith MR, de Bono JS, et al. : Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 368:138-148, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, et al. : Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 371:424-433, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran H, Tagawa ST, Park K, et al. : Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol 30:e386-e389, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Aparicio AM, Harzstark AL, Corn PG, et al. : Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 19:3621-3630, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aparicio AM, Shen L, Tapia EL, et al. : Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res 22:1520-1530, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran H, Rickman DS, Park K, et al. : Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 1:487-495, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltran H, Prandi D, Mosquera JM, et al. : Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22:298-305, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher HI, Halabi S, Tannock I, et al. : Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 26:1148-1159, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes MG, Foss E, Joseph G, et al. : CT-guided bone biopsies in metastatic castration-resistant prostate cancer: Factors predictive of maximum tumor yield. J Vasc Interv Radiol 28:1073-1081.e1, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Amin MB, Beltran H, et al. : Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol 38:756-767, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso C, Butler T, Rhodes K, et al. : Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn 17:53-63, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray KA, Yates B, Seal RL, et al. : Genenames.org: The HGNC resources in 2015. Nucleic Acids Res 43:D1079-D1085, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez MJ, Shen Y, Giorgi FM, et al. : Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 48:838-847, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre C, Rajbhandari P, Alvarez MJ, et al. : A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol 6:377, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton Y, Novak AM, Swatloski T, et al. : TumorMap: Exploring the molecular similarities of cancer samples in an interactive portal. Cancer Res 77:e111-e114, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Airola A, Pahikkala T, Waegeman W, et al: A comparison of AUC estimators in small-sample studies. Presented at the Third International Workshop on Machine Learning in Systems Biology, Ljubljana, Slovenia, 2009. Proceedings of Machine Learning Research, 8:3-13. [Google Scholar]
- 18. doi: 10.1016/j.cell.2015.05.001. Robinson D, Van Allen EM, Wu YM, et al: Integrative clinical genomics of advanced prostate cancer. Cell 161:1215-1228, 2015 [Erratum: Cell 162:454, 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network : The molecular taxonomy of primary prostate cancer. Cell 163:1011-1025, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazmin D, Prytkova T, Cook CE, et al. : Linking ligand-induced alterations in androgen receptor structure to differential gene expression: A first step in the rational design of selective androgen receptor modulators. Mol Endocrinol 20:1201-1217, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Mendiratta P, Mostaghel E, Guinney J, et al. : Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol 27:2022-2029, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Hastie TJ, Pregibon D: Generalized linear models, in Chambers JM, Hastie TJ (eds): Statistical Models in S. Pacific Grove, CA, Wadsworth & Brooks/Cole, 1992. [Google Scholar]
- 23.Hollander M, Wolfe DA: Nonparametric Statistical Methods. New York, NY, John Wiley & Sons, 1973 [Google Scholar]
- 24.Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457-481, 1958 [Google Scholar]
- 25.McNair C, Xu K, Mandigo AC, et al. : Differential impact of RB status on E2F1 reprogramming in human cancer. J Clin Invest 128:341-358, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin D, Wyatt AW, Xue H, et al. : High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 74:1272-1283, 2014 [DOI] [PubMed] [Google Scholar]
- 27. National Comprehensive Cancer Network Guidelines: Prostate Cancer. Version 1.2018.
- 28.Kumar A, Coleman I, Morrissey C, et al. : Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22:369-378, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltran H, Jendrisak A, Landers M, et al. : The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res 22:1510-1519, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku SY, Rosario S, Wang Y, et al. : Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355:78-83, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dardenne E, Beltran H, Benelli M, et al. : N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30:563-577, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akamatsu S, Wyatt AW, Lin D, et al. : The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Reports 12:922-936, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Lin D, Dong X, Wang K, et al. : Identification of DEK as a potential therapeutic target for neuroendocrine prostate cancer. Oncotarget 6:1806-1820, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Jin H, Zhao JC, et al. : FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 36:4072-4080, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop JL, Thaper D, Vahid S, et al. : The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 7:54-71, 2017 [DOI] [PubMed] [Google Scholar]
- 36.Oliver-Krasinski JM, Kasner MT, Yang J, et al. : The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest 119:1888-1898, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]