Abstract
Three new 3-amino-6-hydroxy-2-piperidone (Ahp)-containing cyclic depsipeptides named loggerpeptins A–C (1–3) along with molassamide (4) were discovered from a marine cyanobacterium, extending the structural diversity of this prevalent scaffold of cyanobacterial serine protease inhibitors. Molassamide (4), containing the 2-amino-butenoic (Abu) unit in the cyclic core, was the most potent and selective analogue against human neutrophil elastase (HNE). Given the growing evidence supporting the role of HNE in breast cancer progression and metastasis, we assessed the cellular effects of compounds 3 and 4 in the context of targeting invasive breast cancer. Both compounds inhibited the cleavage of the elastase substrate CD40 in biochemical assays; however, only 4 exhibited significant cellular activity. Since CD40 and other receptor proteolytic processing culminates in NFKB activation, we assessed the effects on the expression of target genes, including ICAM-1. ICAM-1 is also a direct target of elastase, and in our studies compound 4 attenuated both elastase-induced ICAM-1 gene expression and ICAM-1 proteolytic processing by elastase, revealing a potential dual effect on migration through modulation of gene expression and proteolytic processing. Molassamide (4) specifically inhibited the elastase-mediated migration of highly invasive triple negative breast cancer cells.
Keywords: Breast cancer, elastase, marine cyanobacteria, metastasis, protease inhibitors
The chemical exploration of a marine cyanobacterium collected from Loggerhead Key, Florida led to the discovery of three new elastase inhibitors, loggerpeptins A–C (1–3), along with molassamide (4) that may have a value in the context of cancer. Molassamide (4), the most potent analogue, was shown to inhibit the cleavage of CD40, the expression and cleavage of its downstream target ICAM-1, reverse HNE-induced morphological changes as well as the elastase-induced migration of invasive breast cancer cells.
Introduction
Cyanobacterial serine protease inhibitors, composed of 19-membered ring cyclodepsipeptides bearing the modified glutamic acid residue 3-amino-6-hydroxy-2-piperidone (Ahp), are among the most predominant secondary metabolites isolated from marine cyanobacteria.[1,2] More than 100 members of this class have been discovered, all of which are characterized by the conserved backbone and a variable side chain, exhibiting inhibitory activities against the proteases elastase, chymotrypsin, and trypsin.[1,3] Some of these secondary metabolites contain the 2-amino-2-butenoic acid (Abu) moiety incorporated within the macrocycle, which contributes to enhanced potency towards elastase inhibition.[4] Lyngbyastatins 4–10[4,5] and symplostatins 5–10[6] are examples of compounds bearing the Abu residue and considered among the most potent marine derived elastase inhibitors. The structural basis towards elastase inhibition was demonstrated in prior studies via SAR and X-ray cocrystallization studies, highlighting the importance of the Ahp unit and N-Me-Tyr in the cyclic core, as well as a polar functionality in the pendant side chain for the greatest activity.[6–8]
Human neutrophil elastase (HNE) is a serine protease with broad substrate specificity, which hydrolyzes elastin and other extracellular matrix proteins such as fibronectin, type IV collagen, and proteoglycans.[9] HNE has been shown to be implicated in several diseases such as asthma, chronic obstructive pulmonary disease (COPD), and cancer.[10,11] In the context of cancer, HNE has been shown to play a role in tumor progression leading to metastasis and is associated with poor prognosis and relapse in cancer patients.[12] The prognostic importance of HNE in cancer is attributed to its ability to degrade the extracellular matrix as well as cleave substrates involved in tumor promoting pathways such as migration, cell cycle regulation, and inflammation.[13]
Sivelestat is the only clinically approved elastase inhibitor on the market used in Japan and South Korea for the treatment of acute lung injury associated with systemic inflammatory response syndrome;[14–16] however, due to its marginal clinical effects, its approval has been stalled in Europe and United States.[17] Therefore, developing new small molecule inhibitors of elastase is of great importance for the therapeutic management of elastase-implicated pathogenesis.
We previously investigated the cellular effects of Ahp-containing cyanobacterial cyclodepsipeptides on bronchial epithelial cells for application to treat inflammatory conditions in respiratory diseases and subsequently also reported a total synthesis strategy to enable further biological studies and catalyze the development of these compounds.[6,18] Others have also developed and reported synthetic approaches of tailored Ahp-cyclodepsipeptide supporting the facts that this scaffold is a promising starting structure for the generation of customized serine protease inhibitors.[19,20] However, there are no rigorous studies describing the cellular effects of these compounds in cancer. Herein, we describe the isolation and structure determination of new members of this family of serine protease inhibitors and report the evaluation of their inhibitory effects on downstream cellular substrates of elastase that are implicated in breast cancer progression.
Results and Discussion
Isolation and structure elucidation
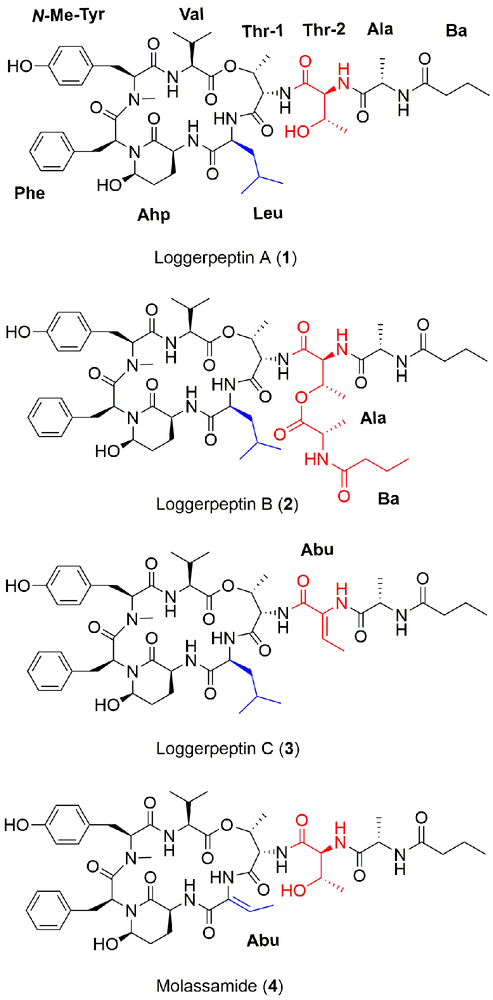
Samples of the cyanobacterium DRTO-73 were collected from Loggerhead Key, Florida on May 21, 2013. The freeze-dried sample was extracted directly with the non-polar solvents EtOAc:MeOH (1:1) three times. The extract (17.4 g) was then partitioned between hexanes and 80% MeOH in H2O. The 80% MeOH in H2O fraction was evaporated and further partitioned between n-BuOH and H2O. The n-BuOH fraction (2.1 g) was concentrated in vacuo and later subjected to silica gel chromatography using a step gradient system starting the elution with CH2Cl2 followed by increasing i-PrOH in CH2Cl2, and finally with MeOH. The purification of 50% i-PrOH/DCM silica gel fraction by reversed-phase HPLC resulted in the isolation of several related analogues (1–4), named loggerpeptins A–C (1–3), in addition to the known compound molassamide (4)[21] (Scheme) as colorless amorphous solids.
Scheme.
1–3) and molassamide (4) isolated from a marine cyanobacterium.
HRESIMS analysis of loggerpeptin A (1) in the positive mode revealed an adduct ion peak at m/z of 1015.5087 [M + Na]+ suggesting a molecular formula of C50H72N8O13. The 1H NMR spectrum suggests the presence of a tertiary amide (N-Me) singlet (δH 2.72, δC 30.69) and six secondary amide NH doublets (δH 7.03, 7.40, 7.7, 7.81, 8.01, 8.37). The 2D NMR data revealed the presence of valine, N-methyltyrosine, phenylalanine, 3-amino-6 hydroxy-2-piperidone (Ahp), two threonine units, alanine, and a butanoic acid (Ba) moiety (Table S1). The sequence of the amino acids was determined with the aid of ROESY data to be: Ba, Ala, Thr-2, Thr-1, Leu, Ahp, Phe, N-Me-Tyr, Val. The structure shows similarity in the core structure to the known compound kempopeptin A.[22] However, it differs in the side chain where Ala and Ba moieties replace the Pro and Ac units present in kempopeptin A.[22]
HRESIMS of loggerpeptin B (2) in the positive mode exhibited an adduct ion peak at m/z of 1156.5901 [M + Na]+ suggesting a molecular formula of C57H83N9O15. The combination of both 1D and 2D NMR data suggested similar core structure to compound 1 with additional Ala and Bu units extended from Thr-2 (Table S2).
The HRESIMS of loggerpeptin C (3) in the positive mode revealed an adduct ion peak at m/z of 997.4978 [M + Na]+ suggesting a molecular formula of C50H70N8O12. NMR analysis revealed the presence of a methine quartet (δH 6.48, δC 129.8) which showed a COSY correlation to a Me doublet (δH 1.63, δC 12.91). The latter showed an HMBC correlation to a quaternary sp2 carbon (δC 131.0) and a carbonyl (δC 164.7) and a ROESY correlation to a broad NH singlet (δH 9.26). Those signals are characteristic of an Abu unit (Table S3). The NH (δH 9.26) of the Abu unit showed a ROESY correlation to the alpha proton of alanine. Also, the methine quartet (δH 6.48, δC 129.8) of the Abu showed a ROESY correlation to the NH (δH 7.05) of Thr-1. Therefore, the Abu unit was found to replace Thr-2 that was present in compound 1. The sequence of the amino acids was determined with the aid of ROESY and HMBC data to be: Ba, Ala, Abu, Thr-1, Leu, Ahp, Phe, N-Me-Tyr, Val.
HRESIMS of compound 4 in the positive mode exhibited an adduct ion peak at m/z of 985.4618 [M + Na]+ suggesting a molecular formula of C48H66N8O13 consistent with that of molassamide, a cyanobacterial compound isolated from the cyanobacterium Dichothrix utahensis.[21] Analysis of 1D and 2D NMR data suggested the presence of an Abu unit, which replaced the Leu moiety present in the three other structures of loggerpeptins A–C (1–3) (Table S4; Figure 1). This was confirmed by ROESY correlations between the CH quartet (δH 6.56, δC 132.01) of the Abu and the alpha proton of Ahp (δH 3.83, δC 48.43) and also the ROESY peak between the Me (δH 1.51, δC 13.08) of Abu and the alpha proton of Thr-1 (δH 4.60, δC 55.47). This structure resembles lyngbyastatin 7 with respect to the cyclic part of the molecule but differs in the side chain.[4]
Figure 1.
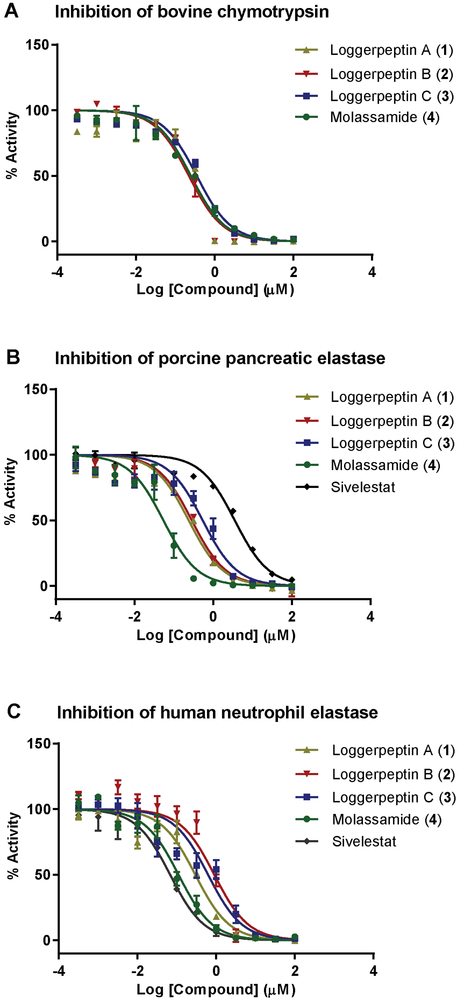
Dose response curves for loggerpeptins A–C (1–3), molassamide (4) and sivelestat against serine proteases: A) Bovine pancreatic chymotrypsin, B) Porcine pancreatic elastase, and C) Human neutrophil elastase. The dose-response is presented as % fold inhibition against solvent control (DMSO). Data is presented as mean ± SD, n=3.
The absolute configuration of the stereocenters in the four compounds (1–4) was established by enantioselective HPLC-MS analysis of the acid hydrolyzates, which showed retention times corresponding to l-Thr, l-Ala, l-Val, l-Leu, l-Phe, and N-Me-L-Tyr. In order to establish the absolute configuration of the Ahp unit, CrO3 oxidation was carried out which resulted in the liberation of glutamic acid. The reaction product was then subjected to acid hydrolysis, which was subsequently subjected to enantioselective HPLC-MS analysis liberating l-Glu, thus establishing the configuration of the Ahp unit as 3S. The relative configuration of the Ahp unit was established based on ROESY correlations within the Ahp unit and proton-proton coupling constants which were observed in kempopeptin A and other related Ahp-containing cyanobacterial secondary metabolites (3S,6R). The geometry of the Abu unit was established based on examination of the ROESY spectrum, which revealed a correlation between the CH (δH 6.48) of the Abu and NH of the Thr-1, indicating E configuration.
Compounds 1–4 represent the classical diversity of the compounds with some notable novelties. Loggerpeptin C (3) is the first compound of this class with an Abu unit in the side chain, while molassamide (4) with the Abu moiety in the cyclic core closely resembles the most potent elastase inhibitors, including lyngbyastatin 7.[4]
Inhibition of Serine Proteases In Vitro
The similarity of loggerpeptins A–C (1–3) and molassamide (4) in terms of structure, characterized by the presence of a 19-membered cyclic hexadepsipeptide core containing the modified glutamic acid residue Ahp, suggested that they would have serine protease inhibitory activities with selectivity towards chymotrypsin and elastase. The antiproteolytic activities of 1–4 were tested against bovine pancreatic chymotrypsin, porcine pancreatic elastase, and human neutrophil elastase (HNE) (Figure 1). All compounds inhibited the enzymes tested with IC50 values in the sub-micromolarrange (Table 1).
Table 1.
IC50 values of loggerpeptins A–C (1–3), molassamide (4) and sivelestat against serine proteases
| Compound | Bovine Pancreatic Chymotrypsin |
Porcine Pancreatic Elastase |
Human Neutrophil Elastase |
|---|---|---|---|
| Loggerpeptin A (1) | 0.24 ± 0.01 μM | 0.24 ± 0.01 μM | 0.29 ± 0.04 μM |
| Loggerpeptin B (2) | 0.22 ± 0.02 μM | 0.28 ± 0.01 μM | 0.89 ± 0.09 μM |
| Loggerpeptin C (3) | 0.35 ± 0.02 μM | 0.54 ± 0.13 μM | 0.62 ± 0.38 μM |
| Molassamide (4) | 0.24 ± 0.01 μM | 0.05 ± 0.01 μM | 0.11 ± 0.01 μM |
| Sivelestat | NDa | 3.55 ± 0.14 μM | 0.06 ± 0.01 μM |
Not determined; Data are presented as the mean ± SD, n=3.
Compounds 1–4 exhibited similar IC50 values against bovine chymotrypsin, which were comparable to the reported IC50 value of kempopeptin A against the same enzyme (IC50 0.3 μM).[22]
In the porcine pancreatic elastase inhibition assay, loggerpeptins A and B (1 and 2) inhibited elastase with comparable IC50 values, whereas loggerpeptin C (3) was 2-fold less potent than 1 and 2. Molassamide (4) was the most potent analogue with IC50 value of 50 nM. The differences in activity highlight the importance of the residues adjacent to the Ahp unit on its N-terminal side, which confer selectivity towards serine proteases. It has been shown through previous crystallographic and SAR data that the presence of a hydrophobic residue (Leu) between Ahp and Thr-1 confers selectivity towards elastase and chymotrypsin.[3,7] Also it has been reported that the presence of the Abu unit in that position enhances the potency towards elastase, which was the case for compound 4.[4] Interestingly compounds 1–3, and 4 were found to be ~12 fold and ~70 fold more potent against porcine elastase compared to sivelestat (IC50 3.55 μM), respectively (Table 1).
When the antiproteolytic activities of 1–4 were evaluated against the disease relevant human neutrophil elastase (HNE), loggerpeptins B (2) and C (3) were 3 and 2-fold less potent than 1, respectively. Also, molassamide (4) was the most potent analogue (IC50 110 nM). Compared to sivelestat (IC50 0.06 μM), compounds 1–4 were found to be ~2–15 fold less potent in inhibiting HNE (Table 1). As the IC50 value for a covalent inhibitor would vary depending on the pre-incubation time with the enzyme, we also obtained the progress curve of HNE treated with sivelestat without pre-incubation step (Figure S1). Regarding the mode of inhibition of the Ahp-containing depsipeptides, prior studies and X-ray cocrystallization data of another Ahp containing elastase inhibitor, lyngbyastatin 7, did not show covalent bond formation between the Abu moiety and HNE.[6,20]
In all subsequent biological and molecular studies, we focused on loggerpeptin C (3) which was isolated in higher yield, compared to loggerpeptins A (1) and B (2), to allow rigorous biological evaluation, and on molassamide (4) as it was the most potent analogue.
Molecular Docking
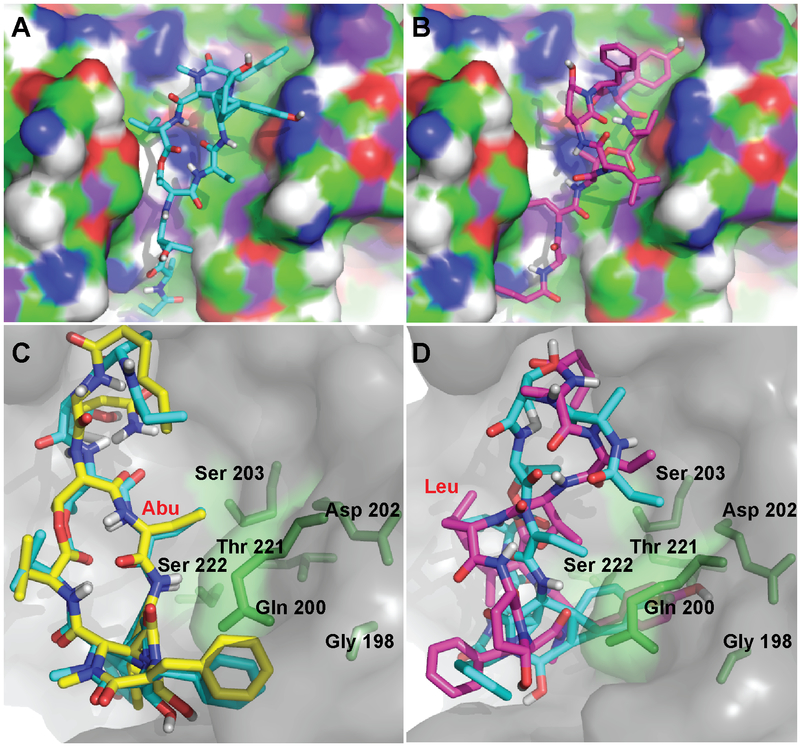
Consistent with prior studies,[7,8,23] the differences in the antiproteolytic activity between loggerpeptin C (3) and molassamide (4) against porcine and HNE was rationalized by molecular docking experiments. The docking was carried out using Autodock Vina[24] and the published crystal structures of porcine (4GVU)[23] and human (3Q76)[25] neutrophil elastases were used. Both compounds where found to exhibit comparable binding within the binding pocket of the enzyme (Figures 2A and B). In compounds 3 and 4, the Leu/Abu and the N-terminal amino acid residues (Thr-1, Thr-2/Abu, and Ala) were found to occupy the subsites S1 through S4 of elastase, respectively. At the S1 subsite of elastase, the Leu in 3 and Abu in 4 occupy the oxanion hole of the enzyme. These compounds exploit the same binding pattern previously reported in several other elastase-inhibitor complexes, such as scyptolin, FR901277, and lyngbyastatin 7.[6,8,26] Also, the orientation of the macrocycle in these compounds was comparable as demonstrated in the overlay of the docked structures of molassamide (4) and lyngbyastatin 7 (Figure 2C). In the docked structures of molassamide (4) in both receptors, the Abu moiety at the S1 subsite was found to be in close proximity to the Ser residues (Ser 203 and Ser 222), potentially resulting in previously reported CH/π interactions,[6,26] hence, contributing to the enhanced potency compared to loggerpeptin C (3) bearing Leu moiety at the same position (Figures 2C and D). Additionally, the Abu moiety was found to be in close proximity to polar residues including Ser 222, Thr 221, Asp 202, and Gln 200, suggesting potential H-bonding interactions (Figures 2C and D). This is consistent with the previously reported co-crystal structure of lyngbyastatin 7 with porcine pancreatic elastase, where the Abu moiety was found in close distance to Ser 203 contributing to a non-bonded CH/π interaction.[6] Additionally, several H-bonding interactions were also reported between the Abu of lyngbyastatin 7 and Gly 201, Ser 222, and indirectly to Thr 44.[6]
Figure 2.
Docked structures of A) loggerpeptin C (3) and B) molassamide (4) into HNE (PDB: 3Q76). Overlay of docked structures of C) molassamide (4; blue) and lyngbyastatin 7 (yellow) and D) loggerpeptin C (3; pink) and molassamide (4; blue) into porcine pancreatic elastase (PDB: 4GVU). The residues of the elastase enzyme in close proximity to the Abu unit are displayed in stick representation (green).
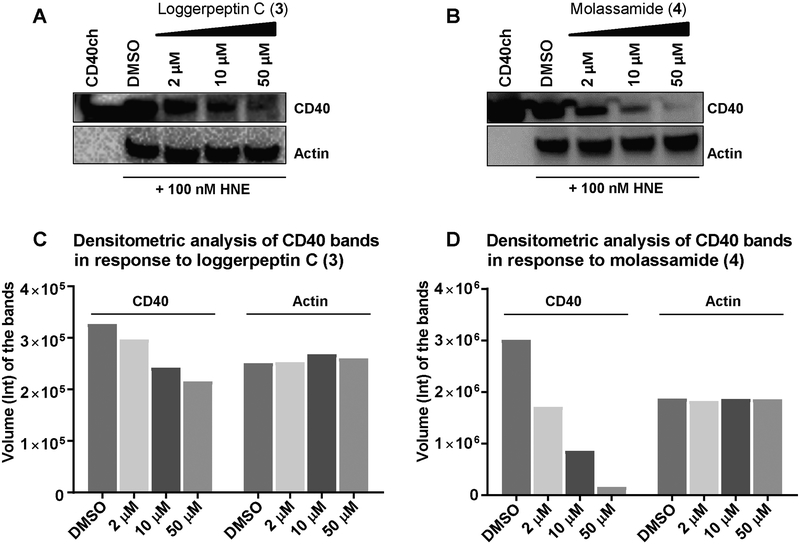
Inhibition of Cleavage of CD40 by Human Neutrophil Elastase In Vitro
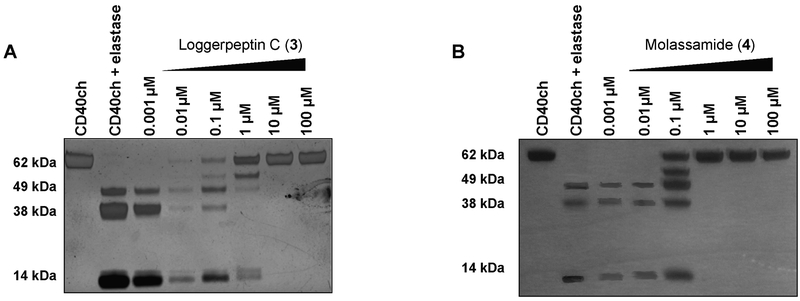
CD40 is a 44 KDa membrane protein and a member of tumor necrosis factor (TNF) receptor superfamily and known to be expressed in breast carcinomas.[27,28] CD40 was previously identified, in other studies, to be an extracellular substrate of elastase.[29] We investigated the ability of HNE to cleave CD40 using CD40-IgG chimera (CD40ch) because it contains the human CD40 N-terminal residues (21 to 193), including the predicted elastase cleavage sites, fused to human IgG1 fragment as an elastase substrate.[29] CD40ch was incubated with or without HNE (molar ratio of elastase to CD40 of 1:10, respectively) at 37 °C for 1.5 h and the resulting proteins were electrophoretically separated on SDS-PAGE under reducing conditions followed by silver staining. The incubation with HNE resulted in the cleavage of CD40ch into several fragments compared to the incubation of CD40ch alone (60 kDa) which remained unprocessed (Figure 3, lanes 1 and 2). A previous study predicted, through in silico structural analysis and modeling of elastase interactions with CD40, the potential elastase cleavage sites within CD40 to be at Val 91, Ala 115, and Val 138.[29] Also, this was further confirmed by in vitro CD40 processing assay and subsequent Edman degradation analysis of cleaved products, which revealed Val 138 to be the primary cleavage site.[29] In support of previous results, our data revealed higher molecular weight fragments, potentially corresponding to a sequence in the linker site and within the IgG sequence near the C-terminus, as well as a lower molecular weight fragment (~14 kDa) closely correlated with the previously established Val 138 at the N-terminus of the protein.
Figure 3.
The effects of the inhibitors (3 and 4) on the proteolytic processing of CD40 by HNE. Incubation of HNE with CD40ch (molar ratio 1:10) in 0.1 M Tris-NaCl buffer, pH 7.5 at 4 °C for 2 h in the presence and absence of different concentrations of the elastase inhibitors 3 (A) and 4 (B) followed by the addition of CD40ch and incubation for 1.5 h at 37 °C. Fragments were separated on SDS-PAGE under reducing conditions and silver stained. Note: Intensity differences across panels are attributed to differentially staining and exposure times.
To assess the effects of representative members of our protease inhibitors on the elastase-mediated cleavage of CD40, loggerpeptin C (3) and molassamide (4) were pre-incubated with HNE at different concentrations (0.001 – 100 μM) for 2 h before the addition of CD40ch to the reaction.
The presence of the inhibitors (3 and 4) resulted in a complete inhibition of CD40ch cleavage, where the ~60 kDa band corresponding to the full length was evident, up to 10 and 1 μM, respectively, with 4 being 10 fold more potent (Figures 3A and B). However, at 1–0.1 μM of 3 and 0.1 μM of 4, HNE resulted in partial cleavage of CD40 into several fragments; whereas, at lower concentrations, complete cleavage of CD40 was noted (Figures 3A and B). Our conclusions were based on qualitative assessments of the bands in the gel, as slight differences in the intensities between the bands due to the staining and development process might hinder accurate quantitative assessments.
The Effect of Elastase Inhibition on the Cleavage of CD40 in MDA-MB-231
In order to choose a cell line to further investigate the cellular effects of our elastase inhibitors, we assessed the level of expression of HNE and its extracellular (CD40) substrate by Western blot analysis of lysates harvested from a panel of breast cancer cell lines (Figure S2). Consistent with other studies,[13,30] we did not detect NE protein in any of the six breast cancer cell lines investigated. Also, the lack of NE mRNA in breast cancer cells (MCF7, T47D, HER18, MDA-MN-231, MDA-MB-453) was previously confirmed by RT-PCR.[31] However, immunohistochemical analysis of breast cancer tissues confirmed the presence of NE in tumor associated neutrophils (TAN) within the tumor microenvironment.[31] Hence, it has been proposed that within the tumor microenvironment, TAN are the primary source of NE in breast tumors.[31] On the other hand, our data showed that CD40 was detected in all breast cancer cell lines tested (Figure S2). As MDA-MB-231 cell line is known to exhibit the highest level of NE uptake,[30] and because its substrate CD40 was detected in MDA-MB-231 lysates, we selected this cell line to further investigate the cellular effects of loggerpeptins C (3; a representative of the newly isolated inhibitors) and molassamide (4; the most potent analogue).
We examined the ability of 3 and 4 to inhibit HNE processing of CD40 within a cellular context using MDA-MB-231 breast cancer cell line. Because we did not detect HNE protein in MDA-MB-231 lysates, we added purified recombinant HNE (100 nM) to the culture medium in the presence of different concentrations of compounds 3 and 4. Following 24 h incubation, cell lysates were harvested and analyzed by Western blot using anti-CD40 antibody that detects the N-terminal fragment of the protein (Figures 4A and B). Our data showed a dose-dependent reduction in the level of processed N-terminal fragment of the CD40 protein in the presence of 4 compared to HNE alone, with strong inhibition (~70%) at 10 μM and virtually complete inhibition achieved at 50 μM. The results were further confirmed by densitometric analysis of the bands (Figure 4D). However, loggerpeptin C (3) lacked significant activity in this cellular context at 10 μM and only possessed marginal activity at 50 μM, demonstrating the importance of the Abu moiety adjacent to the Ahp unit in the cyclic core, consistent with the enzyme assays. To rule out the potential effect of cell viability on the expression level of CD40, the effects of HNE and molassamide (4) on the viability of MDA-MB-231 cells were examined. There was no reduction in cell viability by HNE and 4 (Figure S3).
Figure 4.
The effects of compounds 3 and 4 on the cleavage of CD40 by HNE in MDA-MB-231 cells A, B) MDA-MB-231 cells were seeded in 12-well plates and following 24 h the medium was replaced with serum free medium containing different concentrations of 3 (A) and 4 (B) or solvent control in the presence of 100 nM HNE. Following 24 h incubation, lysates were harvested and analyzed by Western blot. C,D) Densitometric analysis of CD40 and actin bands in panels A and B.
The Effect of Elastase Inhibition on NFKB Target Gene Expression
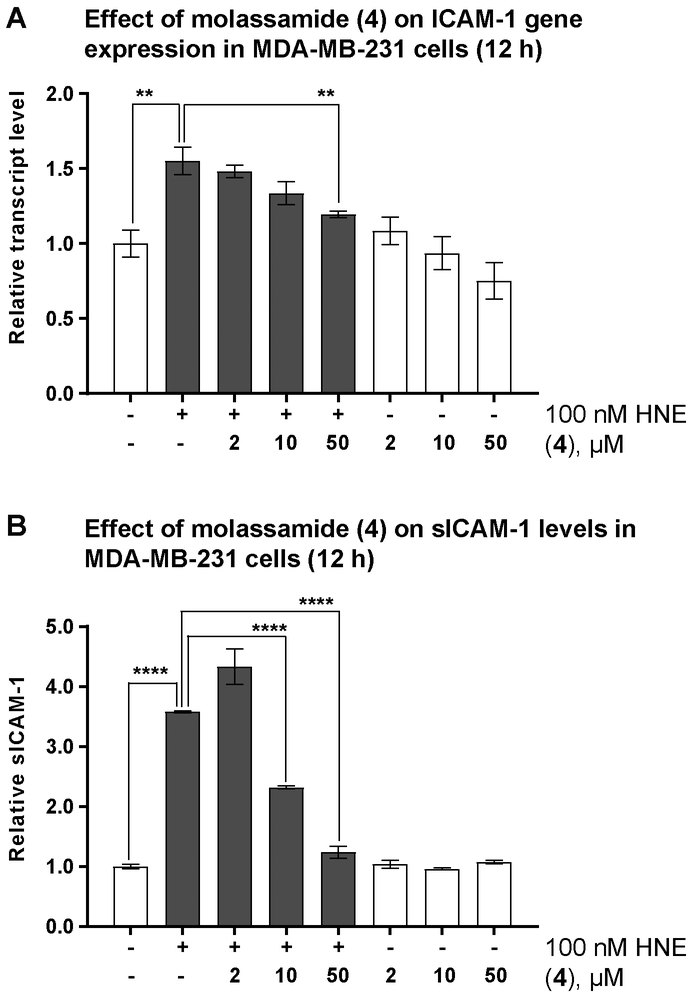
Transcription factor NFKB is known to play critical roles in the development, progression, proliferation, and survival of breast cancer cells.[32,33] Prior studies have shown that activated CD40 stimulates the recruitment of TNF receptor-associated factors (TRAF1 and TRAF2: positive regulators, TRAF3: negative regulator of NFKB) to the intracellular domain of CD40.[27,34] The elastase-cleaved CD40 signaling is mediated through the binding and interactions of TRAF1 and 2 within the intracellular domain of CD40 resulting in nuclear translocation, upregulation of NFKB transcriptional activity, and increase in the expression of NFKB target genes. We investigated the effects of HNE on the expression levels of several NFKB target genes known to be involved in promoting breast cancer cells migration (ICAM-1, CXCR4, CD44).[29,35–37] MDA-MB-231 cells were treated with 100 nM HNE for different time points and gene expression was assessed by RT-qPCR. Our data showed 2-fold increase in the expression levels of the Intracellular Adhesion Molecule-1 (ICAM-1) after 12 and 24 h in response to HNE (Figure S4) and 1.6-fold increase for CXCR4 after 6 h, while CD44 transcript levels were unaffected in our experiments. ICAM-1 is a cell surface glycoprotein known to be highly expressed in breast cancer tissues and involved in the progression of metastasis.[35] We examined, therefore, the ability of the most potent analogue, molassamide (4), to downregulate elastase-induced ICAM-1 expression in MDA-MB-231 cells (Figure 5A). The results revealed dose-dependent decrease in the expression levels of ICAM-1 in response to 4, suggesting a potential involvement of the ligand-activated full-length CD40 in the negative regulation of NFKB transcription activity and subsequently downregulation of NFKB target gene expression. In the absence of HNE, no significant change in the expression levels was noted in response to 4 (Figure 5A).
Figure 5.
The effects of elastase inhibition on ICAM-1gene expression and cleavage. A) Effects of molassamide (4) on ICAM-1 gene expression in the presence and absence of 100 nM HNE. B) Effects of molassamide (4) on the levels of sICAM-1 in the presence (grey bars) and absence (white bars) of 100 nM HNE. MDA-MB-231 cells seeded in 6-well plates, incubated overnight then starved for additional 24 h. The medium was then replaced with serum free DMEM containing different concentrations of 4 and 100 nM HNE. After 12 h, RNA was extracted and the medium was collected. The transcript levels and the levels of sICAM-1 were then assessed by RT-qPCR and ICAM-1 AlphaLisa, respectively. The asterisks denote significance of P < 0.05 relative to solvent control co-treated with HNE using two-tailed unpaired t test (** denotes P ≤ 0.01, *** denotes P ≤ 0.001, and **** denotes P ≤ 0.0001).
Although, in this study, the effects of elastase inhibition on NFKB target gene expression were evaluated using mainly compound 4, we previously assessed and reported the effects of sivelestat and other members of the Ahp-containing cyclodepsipeptides with Abu in the cyclic core (symplostatin 5 and lyngbyastatin 7), using epithelial lung airway model system.[6,18] The effects in both cellular systems appear to be similar and therefore our data support potential applications of this class of elastase inhibitors in not only inflammatory conditions but also in cancer.
The Effect of Elastase Inhibition on ICAM-1 Cleavage
Disruption in cell-cell adhesion is known to play a critical role in breast cancer metastasis. The cell-surface receptor, ICAM-1, is a regulator of cell-cell adhesion and affected by elastase, being cleaved to generate a soluble form (sICAM-1) that is secreted into the medium.[38] Prior studies have shown the important role of ICAM-1 in mediating tumor cell adhesion and metastasis.[35] To determine the effects of HNE on the shedding of ICAM-1 from the cell surface, the levels of sICAM-1 in culture medium prepared from HNE-treated MDA-MB-231 cells were quantified by AlphaLisa. A significant increase (3.6-fold) in the level of sICAM-1 was noted upon treatment with 100 nM HNE for 12 h (Figure 5B). Next, we investigated the ability of molassamide (4) to attenuate the HNE-induced shedding of ICAM-1. Co-treatment with (4) for 12 h resulted in a dose-depended reduction (65% and 35% reduction at 50 and 10 μM, respectively) in the levels of sICAM-1 up to 10 μM (Figure 5B). On the other hand, 4 had no effects on the levels of sICAM-1 in the absence of HNE (Figure 5B), further validating the direct role of HNE as a sheddase that proteolytically processes the membrane-bound ICAM-1 to generate and release sICAM-1 into the medium. The effects of sivelestat and the Ahp-containing elastase inhibitor, symplostatin 5, on sICAM-1 levels have been previously reported; we had found these compounds to attenuate the elastase-mediated proteolytic processing of the adhesion protein ICAM-1 in bronchial epithelial cells.[6] Again, our similar findings in cancer cells support the potential of this class of elastase inhibitors in the context of cancer.
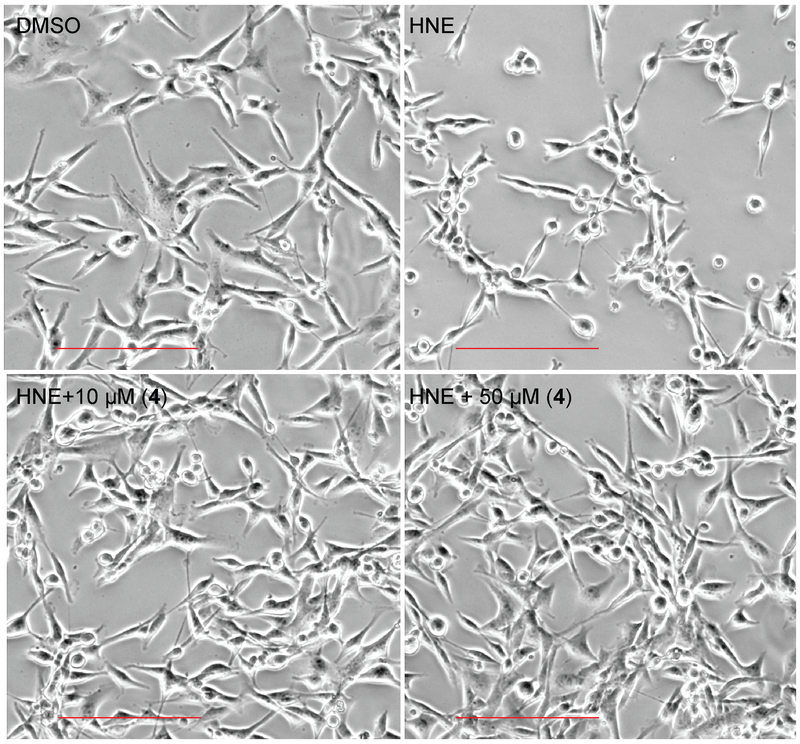
Morphological Changes of MDA-MB-231 cells Induced by HNE
Upon treatment with 100 nM HNE, a morphological change of MDA-MB-231 cells was observed after 2 h when the cells started rounding. This morphological change was not due to cell death as assessed by MTT. The HNE-induced cell rounding was prevented by co-treatment with compound 4 up to 10 μM (Figures 6, S5). Also, no morphological changes were observed upon treatment with 4 only (without HNE) up to 50 μM (Figure S5). This data further supports the previously reported effects of sivelestat and symplostatin 5,[6] suggesting a direct involvement of HNE in mediating the morphological changes, possibly by affecting adhesion molecules, components of the extracellular matrix, and potentially other unknown factors/mechanisms which remain to be investigated.
Figure 6.
The effects of molassamide (4) on HNE-induced morphological changes in MDA-MB-231 cells. MDA-MB-231 cells were treated with 4 for 3 h in the presence of 100 nM HNE. Scale bar 200 μm.
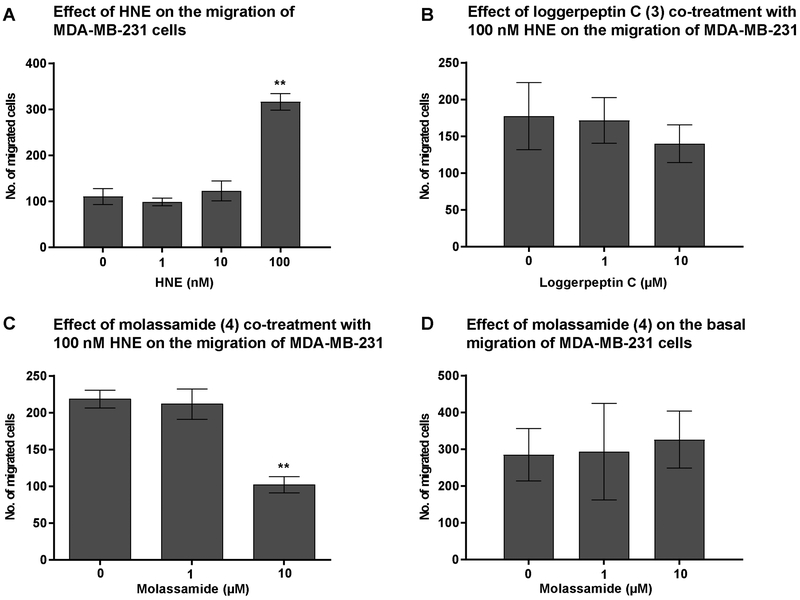
The Effect of Elastase Inhibition on the Migration of MDA-MB-231 cells
Elastase is known to promote tumor cell migration, and its inhibition in breast cancer cells was shown to limit their migratory properties.[39] We therefore assessed the effects of HNE on the migration of the highly invasive triple negative breast cancer cell line, MDA-MB-231. Our data showed a dose-dependent increase in cell migration in response to HNE, with the highest effect achieved at 100 nM (Figure 7A). The action of 4 on ICAM-1 gene expression as well as its effects as sheddase inhibitor prompted us to investigate the effects of the elastase inhibitors, loggerpeptin C (3) and molassamide (4), on the migration of MDA-MB-231 cells. The migration assay was carried out in the presence of 100 nM HNE and different concentrations of loggerpeptin C (3), molassamide (4), or solvent control. While loggerpeptin C (3) had no effect on migration (Figure 7B), consistent with the lack of potent cellular effects on CD40 cleavage, molassamide (4) at 10 μM inhibited the migration of MDA-MB-231 cells (Figure 7C). This result is in agreement with a prior study where downregulation of ICAM-1 was shown to attenuate the metastatic ability of breast cancer cells,[35] although molassamide (4) has additional elastase-related effects on shedding and gene expression (see above). Additionally, we examined the effects of 4 in the absence of HNE. Under these conditions, molassamide (4) did not exhibit any migration inhibitory activity suggesting that it specifically inhibits the elastase-induced migration but not basal migration that is mediated by other factors (Figure 7D).
Figure 7.
The effects of elastase inhibition on the migration of MDA-MB-231 cells. A) The effect of HNE on the migration of MDA-MB-231 cells. The cells were incubated with different concentrations of HNE for 48 h. B, C) The effects of loggerpeptin C (3) and molassamide (4) on the migration of MDA-MB-231 cells. MDA-MB-231 cells were incubated for 48 h with 100 nM HNE and different concentrations of 3 (B) and 4 (C). D) The effect of molassamide (4) on the migration of MDA-MB-231 cells in the absence of HNE. The effect on migration was compared to the solvent control. The asterisks denote significance of P < 0.05 relative to solvent control using two-tailed unpaired t test (** denotes P ≤ 0.01).
Although the effects of sivelestat on cancer progression were not evaluated in this study, its role in restricting cancer cells proliferation and migration were previously reported.[40–42] One study demonstrated the ability of sivelestat to inhibit the NE-induced migration of TE-13 esophageal cancer cell line.[42] Additionally, sivelestat was reported to inhibit the migration of neutrophils in HUVEC model.[43] The migration inhibitory ability of other NE inhibitors was also reported.[39,44,45] Epigallocatechin-3-gallate (EGCG) was shown to reverse HNE-induced migration in A549 cancer cells while caffeic acid phenethyl ester was reported to inhibit HNE-induced migration of PANC-1.[44,45] In breast cancer, the effects of elafin, a HNE inhibitor, on cancer growth and progression were reported, thus further supporting the role of elastase in mediating cancer progression and metastasis.[39]
Since these compounds also have the ability to inhibit bovine chymotrypsin (Table 1), we assessed the effects on the chymotrypsin-like activity of purified 20S human proteasome. Compound 4 did not inhibit the chymotrypsin-like activity of proteasomes in vitro compared to the positive control, epoxomicin (Figure S6), excluding that any of the biological effects are mediated by the proteasome, consistent with both the lack of effects on cell viability (Figure S3) and our previous results demonstrating the lack of inhibitory effects on human chymotrypsin.[6]
Conclusions
Exploring a cyanobacterial sample collected from Loggerhead Key in Florida led to the discovery of four structurally diverse cyclodepsipeptide protease inhibitors: loggerpeptins A–C (1–3) and molassamide (4), adding to the growing family of Ahp-containing secondary metabolites.[3–5,22,46] The Abu unit in the cyclic core (as in 4) appeared to be critical for the elastase selectivity and cellular activity. We have shown that 4 inhibited the migration of the highly invasive MDA-MB-231 breast cancer cells, which could be attributed to multiple on-target effects. Molassamide (4) inhibited the cleavage of the extracellular substrate of elastase, CD40. We also demonstrated the effect of elastase inhibition on NFKB target genes, one of which is ICAM-1. Our studies revealed that 4 attenuated both ICAM-1 cleavage and downregulated elastase-induced ICAM-1 gene expression. Additionally, molassamide (4) demonstrated the ability to reverse the morphological changes induced by HNE in MDA-MB-231 cells, warranting further investigation. Given the established role of HNE in cancer progression, understanding the molecular mechanisms for elastase inhibition is critical for the design and development of better inhibitors with improved potency and selectivity profiles, which might have therapeutic potential in cancers that are characterized by high levels of elastase in the tumor microenvironment.
Experimental Section
General Experimental Procedures.
The optical rotation was measured using a Perkin-Elmer 341 polarimeter. The 1H and 2D NMR spectra for compound 2 were obtained in DMSO-d6 using Agilent VNMRS-600 MHz, a 5-mm cold probe spectrometer, and Bruker 600 MHz Avance II Spectrometer (for compounds 1, 3, and 4), using a 5-mm cryogenic probe for indirect detection (Bruker Cryoprobe TXI). The spectra were referenced using the residual solvent signal [δH/C 2.5/39.5 (DMSO-d6)]. The HRESIMS data were obtained in the positive mode using Agilent LC-TOF mass spectrometer equipped with APCI/ESI multimode ion source detector. LCMS data were obtained using API 3200 (Applied Biosystems) equipped with an HPLC system (Shimadzu).
Biological Material.
Samples of the cyanobacteriumDRTO-73 were collected from Loggerhead Key, Florida on May 21, 2013. This filamentous cyanobacterium forms dense cushion-like colonies and has thin filaments with cells only 2.5-3.5 μm in width and 5-7 μm in length, which corresponds microscopically with Leptolyngbya sp. The genus Leptolyngbya is polyphyletic and in need of taxonomic revision. This cyanobacterium is pictured in the taxonomic guide to marine algae by Littler and Littler[39]. Voucher specimens are retained at the Smithsonian Marine Station at Ft. Pierce.
Extraction and Isolation.
The freeze-dried material was subjected to extraction with EtOAc–MeOH (1:1). The resulting extract was then partitioned between hexanes and 80% MeOH in H2O. The methanolic phase was evaporated to dryness and further partitioned between n-BuOH and H2O. The n-BuOH layer was concentrated and subjected to silica gel chromatography using CH2Cl2 and increasing gradients of i-PrOH (2%, 4%, 6%, 10%, 20%, 50%), 100% i-PrOH, 50% i-PrOH in MeOH, and finally with 100% MeOH. The fraction that was eluted with 50% i-PrOH–DCM was then purified by reversed-phase HPLC [column, Synergi Hydro 4u-RP, 250 × 10.0 mm; flow rate, 2.0 mL/min; PDA detection 200–800 nm] using a linear MeOH–H2O gradient (70–100% MeOH over 20 min, 100% MeOH for 10 min) to afford compound 4 (6 mg), compound 1 (4 mg), compound 3 (2.6 mg), compound 2 (2.3 mg) at tR 18.8 min, 19.4 min, 20.2 min, and 21.1 min, respectively. Those fractions were subjected to further purification using reversed-phase HPLC [column, luna 5u Phenyl-Hexyl, 250 × 10.0 mm; flow rate, 2.0 mL/min; PDA detection 200–800 nm] using a linear MeOH–H2O gradient (70–100% MeOH over 10 min, 100% MeOH for 15 min) to afford compound 4 (3.5 mg), compound 1 (0.5 mg), compound 3 (1 mg), compound 2 (0.3 mg) at tR 11.4 min, 11.5 min, 12.2 min, and 12.7 min, respectively.
Loggerpeptin A (1): Colorless amorphous solid; [α]20D –24.0 (c 0.03, MeOH); NMR data, 1H NMR, COSY, TOCSY, HSQC, HMBC, ROESY in DMSO-d6, see Table S1 and Supporting Information; HRESIMS m/z 1015.5087 [M + Na]+ (calcd for C50H72N8O13Na, 1015.5117).
Loggerpeptin B (2): Colorless amorphous solid; [α]20D –47.3 (c 0.03, MeOH); NMR data, 1H NMR, COSY, TOCSY, HSQC, HMBC, ROESY in DMSO-d6, see Table S2 and Supporting Information; HRESIMS m/z 1156.5901 [M + Na]+ (calcd for C57H83N9O15Na, 1156.5906).
Loggerpeptin C (3): Colorless amorphous solid; [α]20D –29.5 (c 0.06, MeOH); NMR data, 1H NMR, COSY, HSQC, HMBC, ROESY in DMSO-d6, see Table S3 and Supporting Information; HRESIMS m/z 997.4978 [M + Na]+ (calcd for C50H70N8O12Na, 997.5011).
Molassamide (4): Colorless amorphous solid; [α]20D –18.4 (c 0.24, MeOH); NMR data, 1H NMR, COSY, HSQC, HMBC, ROESY in DMSO-d6, see Table S4 and Supporting Information; HRESIMS m/z 985.4618 [M + Na]+ (calcd for C48H66N8O13Na, 985.4647). This data is identical to that previously reported for molassamide.[21]
Absolute Configuration.
A sample of compounds 1–4 (100 μg each) was hydrolyzed with 6 N HCl (0.5 mL) at 110 ºC for 24 hr. The hydrolyzate was concentrated to dryness, reconstituted in 100 μL of H2O, and then analyzed by chiral HPLC [column: Chirobiotic TAG (250 × 4.6 mm), Supelco; solvent: MeOH/10 mM NH4OAc (40:60, pH 5.51); flow rate 0.5 mL/min; detection by ESIMS in positive ion mode (MRM scan)]. l-Thr, l-Ala, l-Val, l-Leu, l-Phe, and N-Me-L-Tyr eluted at tR 7.2, 8.2, 8.1, 9.0, 11.7, and 17.6 min, respectively. The retention times (tR, min; MRM ion pair, parent→product) of the authentic amino acids were as follows: l-Thr (7.2; 120→74), l-allo-Thr (7.7), d-Thr (8.5), d-allo-Thr (11.3), l-Ala (8.2; 90→44), d-Ala (14.2), l-Val (8.1; 118→72), d-Val (14.4), l-Leu (9.0; 132→86), d-Leu (18.7), l-Phe (11.7; 166→103), d-Phe (17.4), N-Me-L-Tyr (17.6; 196→77), N-Me-D-Tyr (33.0). The compound-dependent MS parameters were as follows: Thr: DP 30, EP 5, CE 14, CXP 3; Ala: DP 26, EP 3, CE 19, CXP 3; Val: DP 31, EP 4, CE 14, CXP 2; Leu: DP 32, EP 7, CE 17, CXP 3; Phe: DP 31, EP 5, CE 37, CXP 2; N-Me-Tyr: DP 32, EP 3, CE 40, CXP 2. The source and gas-dependent MS parameters were as follows: CUR 50, CAD medium, IS 5500, TEM 750, GS1 65, GS2 65.
To establish the absolute configuration of the Ahp unit, a sample of compound 4 (300 μg) was dissolved in 300 μL glacial AcOH. The solution was added to 2 mg of CrO3 and kept stirring at room temperature for 5 h. The reaction mixture was then applied onto C18 SPE cartridge eluting with H2O first then MeOH. The oxidized material eluted with MeOH was hydrolyzed with 6 N HCl (0.5 mL) at 110 ºC for 24 h. The hydrolyzate was concentrated to dryness, reconstituted in 100 μL of H2O, and then analyzed by chiral HPLC [column: Chirobiotic TAG (250 × 4.6 mm), Supelco; solvent: MeOH/10 mM NH4OAc (40:60, pH 5.51); flow rate 0.5 mL/min; detection by ESIMS in negative ion mode (MRM scan)]. l-Glu was eluted at tR 6.4 min. The retention times (tR, min; MRM ion pair, parent→product) of the authentic amino acids were as follows: l-Glu (6.4; 146→102), d-Glu (7.6). The compound-dependent MS parameters were as follows: DP −20, EP −10, CE −14, CXP −3. The source and gas-dependent MS parameters were as follows: CUR 30, CAD medium, IS −4500, TEM 750, GS1 65, GS2 65. The relative configuration was assigned based on ROESY correlations.
Molecular Docking.
The docking experiments were carried out using Autodock Vina.[24] The published crystal structures used were 4GVU and 3Q76.[23,25] The 3D structures of 3 and 4 were minimized using Chem3D Pro 12.0 software (Cambridge Corporation). The receptors were prepared, using Pymol and Autodock tools, by removing H2O molecules and adding polar hydrogens. Autodock tools software was also used to generate receptor grid. The interactions of the docked structures into the receptor were analyzed using Pymol.
In Vitro Protease Inhibition Assays.
The inhibition of chymotrypsin was assessed using α-chymotrypsin from bovine pancreas (Sigma, C4129). The assay buffer was 50 mM Tris-HCl, 100 mM NaCl, and 1 mM CaCl2 (pH 7.8). The assay buffer (39 μL), enzyme solution (100 μL/ml in assay buffer, 10 μL), and various concentrations (100 μM final concentration, half log dilutions) of test compounds (1 μL, dissolved in DMSO) were pre-incubated at room temperature for 30 min before the addition of substrate solution (50 μL of 1.5 mM N-succinyl-Gly-Gly-Phe-p-nitroanilide in assay buffer). The absorbance was then measured at 405 nm every 30 s for 30 min using SpectraMax M5 plate reader (Molecular Devices). The protease inhibitor phenylmethanesulfonyl fluoride (PMSF, Sigma) was used as a positive control. The enzyme activity, expressed as a percentage, of each well was calculated based on the initial slope of each progress curve compared to that of the solvent control.
The elastase inhibition assay was carried out using porcine pancreatic elastase (Elastin Products Company, Owensville, MO) which was dissolved in assay buffer [1M Tris-HCl (pH 8.0)] to give a concentration of 75 μg/mL. In brief, the assay buffer (79 μL), elastase solution (75 μg/mL in assay buffer, 5 μL), and various concentrations (100 μM final concentration, half log dilutions) of test compounds (1 μL, dissolved in DMSO) were pre-incubated at room temperature for 15 min in a 96-well microtiter plate. Following incubation, 15 μL of substrate solution (2 mM N-succinyl-Ala-Ala-Ala-p-nitroanilide in assay buffer) was added to each well. The reaction was monitored by measuring the absorbance at 405 nm every 30 s for 30 min using SpectraMax M5 plate reader (Molecular Devices). Sivelestat was used as a positive control.
The elastase assay was also carried out using human neutrophil elastase (Elastin Products Company, Owensville, MO) which was dissolved in assay buffer [0.1 M Tris-NaCl (pH 7.5)] to give a concentration of 100 μg/mL. In brief, the assay buffer (79 μL), elastase solution (100 μg/mL in assay buffer, 5 μL), and various concentrations (100 μM final concentration, half log dilutions) of test compounds (1 μL, dissolved in DMSO) were pre-incubated at room temperature for 15 min in a 96-well microtiter plate. Following the incubation period, 15 μL of substrate solution (2 mM N-(OMe-succinyl)-Ala-Ala-Pro-Val-p-nitroanilide in assay buffer) was added to each well. The reaction was monitored by measuring the absorbance at 405 nm every 30 s for 30 min using SpectraMax M5 plate reader (Molecular Devices). Sivelestat was used as a positive control. In both assays, the enzyme activity, expressed as a percentage, of each well was calculated based on the initial slope of each progress curve compared to that of the solvent control.
In Vitro Cleavage of CD40.
Human neutrophil elastase (Elastin Products Company) (5 μL of 100 μg/mL prepared in 0.1 M Tris-NaCl buffer, pH 7.5) was pre-incubated with either 1 μL of different concentrations of compounds 3, 4 or solvent control (DMSO) in the presence of 44 μL assay buffer (0.1 M Tris-NaCl buffer, pH 7.5) at 4 °C for 2 h. Following the incubation period, recombinant human CD40-IgG chimera (R&D Systems) (50 μL of 100 μg/mL prepared in PBS) was added to the reaction to make a final molar ratio of elastase to CD40 of 1:10. The reaction mixture was then incubated at 37 °C for 1.5 h. The total volume of the reaction was 100 μL. Control reactions include elastase only and CD40 only. Following the incubation period, the reaction was stopped by freezing. 20 μL of each sample was then boiled for 5 min in 5 μL sample buffer containing 2% mercaptoethanol. CD40 fragments were then separated by SDS-PAGE (NuPAGE 4–12% Bis-Tris mini gels, Invitrogen) and stained using silver stain kit (Pierce).
Expression Levels of NE and CD40in Breast Cancer Cell Lines.
Cell lysates were harvested from a panel of breast cancer cell lines (MCF7, T47D, MDA-MB-468, BT549, MDA-MB-231, and HCC1954). Proteins (15 μg) were separated using NuPAGE 4–12% Bis-Tris mini gels (Invitrogen). The gel was transferred to polyvinylidene difluoride (PVDF) membranes, probed with CD40 (R&D), NE (Santa Cruz), and actin (Cell Signaling) antibodies, and detected with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce). The secondary anti-mouse antibody was obtained from Cell Signaling Technology (Danvers, MA).
Cleavage of CD40 by HNE in MDA-MB-231 Cells.
MDA-MB-231 cells were seeded in 12-well plates (150,000 cells/well). After 24 h, the medium (DMEM containing 10% FBS) was replaced with fresh one containing 100 nM of recombinant human neutrophil elastase (Elastin Products Company) and different concentrations of compounds 3 and 4 (prepared in DMSO) or solvent control. Following 24 h incubation, the lysates were harvested using PhosphoSafe lysis buffer (Novagen) and the BCA Protein Assay kit (Pierce) was used to measure protein concentration. Protein (17 μg) were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, probed with CD40 primary antibody (R&D) and secondary anti-mouse antibody (Cell Signaling Technology). SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) was used for the detection.
Cell Viability Assay.
MDA-MB-231 cells (12,000 cells/well) were seeded in 96-well plates. Following 24 h incubation, the cells were treated with different concentrations of compound 4, HNE, or solvent control (DMSO). After 48 h of incubation, cell viability was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide according to the manufacturer’s instructions (Promega).
RNA Isolation, Reverse Transcription, and Real Time Quantitative Polymerase Chain Reaction (qPCR).
MDA-MB-231 cells (500,000 cells/well) were seeded in 6-well plates. After 24 h, the media was replaced with serum free DMEM and the cells were incubated for additional 24 h. Following the incubation period, the media was replaced with serum free DMEM supplemented with 100 nM HNE and different concentrations of compounds 3, 4, or solvent control. After 12 h, RNA was isolated using RNeasy mini kit (Qiagen). cDNA synthesis was carried out using SuperScript II Reverse Transcriptase and Oligo(dT) (Invitrogen). qPCR was carried out after reverse transcription on a reaction solution (25 μL) prepared using 1 μL aliquot of cDNA, 12.5 μL of TaqMan gene expression assay mix, 1.25 μL of 20X TaqMan gene expression assay mix, and 10.25 μL of RNase-free water. The qPCR experiment was performed using ABI 7300 sequence detection system. The thermocycler program used was: 2 min at 50 °C, 10 min at 95 °C, and 15 s at 95 °C (40 cycles), and 1 min at 60 °C. CXCR4 (Hs00607978_s1), CD44 (Hs01075862_m1), and ICAM-1 (Hs00164932_m1) were used as a target genes and GAPDH (Hs02758991_g1) was used as endogenous control.
ICAM-1 AlphaLisa.
MDA-MB-231 cells (500,000 cells/well) were seeded in 6-well plates. After 24 h, the media was replaced with serum free DMEM and the cells were incubated for additional 24 h. Following the incubation period, the media was replaced with serum free DMEM supplemented with 100 nM HNE and different concentrations of compounds 3, 4, or solvent control. After 12 h, the media was collected in Eppendorf tubes, centrifuged to remove detached cells or debris, and used to measure sICAM-1 levels using ICAM-1 AlphaLisa Kit (PerkinElmer) according to the manufacturer’s instructions.
Morphological Changes.
MDA-MB-231 cells (500,000 cells/well) were seeded in 6-well plates. After 24 h, the media was replaced with serum free DMEM and the cells were incubated for additional 24 h. Following the incubation period, the media was replaced with serum free DMEM supplemented with 100 nM HNE and different concentrations of compounds 3, 4, or solvent control. After 3 h, brightfield photographs were taken using a Nikon Eclipse Ti-U microscope (10× magnification).
Transwell Migration Assay.
The effect on migration was examined using BD falcon cell culture inserts (353097) containing PET membrane with 8 μm pore size. Briefly, 5000 cells of MDA-MB-231 suspended in complete medium (DMEM containing 10% FBS), treated with different concentrations of compounds 3 and 4 (prepared in DMSO) or solvent control (DMSO) in the presence or absence of 100 nM of recombinant human neutrophil elastase (HNE) (Elastin Products Company). The cells were seeded (500 μL) into the inserts and 750 μL of complete medium containing the same concentrations of 3, 4, HNE, or solvent control was added into the other side of the membrane. The plate was incubated for 48 h (37 °C, 5% CO2). Following the incubation period, the non-migrated cells were removed using kimwipes and cotton swabs. Migrated cells were stained by crystal violet and the number was counted under the microscope. The assay was performed in triplicate.
Proteasome Activity Assay.
The inhibition of proteasomes was assessed using proteasome 20S assay kit measuring the chymotrypsin-like activity of proteasomes (Enzo Life Sciences) according to the manufacturer’s instructions. In brief, to ½ volume microtiter plate containing 25 μL assay buffer, 10 μL of 10 μg/mL 20S proteasome (0.1 μg/well final concentration) and 5 μL of compound 4 (50 μM final concentration) were added. The plate was then incubated at room temperature for 10 min to allow inhibitor-enzyme interactions. Following the incubation period, 10 μL of Suc-LLVY-AMC substrate was added to each well (75 μM final concentration). The fluorescence was then measured at ex 360 nm and em 460 nm using SpectraMax M5 plate reader (Molecular Devices).
Supplementary Material
Acknowledgment
The research was supported by the National Institutes of Health grant R01CA172310.
Footnotes
References
- [1].Gademann K, Portmann C, Curr. Org. Chem 2008, 12, 326–341. [Google Scholar]
- [2].Tan LT, J. Appl. Phycol 2010, 22, 659–676. [Google Scholar]
- [3].Linington RG, Edwards DJ, Shuman CF, McPhail KL, Matainaho T, Gerwick WH, J. Nat. Prod 2008, 71, 22–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H, J. Nat. Prod 2007, 70, 1593–600. [DOI] [PubMed] [Google Scholar]
- [5].Kwan JC, Taori K, Paul VJ, Luesch H, Mar. Drugs 2009, 7, 528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Salvador LA, Taori K, Biggs JS, Jakoncic J, Ostrov DA, Paul VJ, Luesch H, J. Med. Chem 2013, 56, 1276–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee AY, Smitka TA, Bonjouklian R, Clardy J, Chem. Biol 1994, 1, 113–7. [DOI] [PubMed] [Google Scholar]
- [8].Matern U, Schleberger C, Jelakovic S, Weckesser J, Schulz GE, Chem. Biol 2003, 10, 997–1001. [DOI] [PubMed] [Google Scholar]
- [9].Sato T, Takahashi S, Mizumoto T, Harao M, Akizuki M, Takasugi M, Fukutomi T, Yamashita J, Surg. Oncol 2006, 15, 217–22. [DOI] [PubMed] [Google Scholar]
- [10].Korkmaz B, Horwitz MS, Jenne DE, Gauthier F, Pharmacol. Rev 2010, 62, 726–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roghanian A, Sallenave J-M, Aerosol Med J. Pulm. Drug Deliv 2008, 21, 125–44. [DOI] [PubMed] [Google Scholar]
- [12].Akizuki M, Fukutomi T, Takasugi M, Takahashi S, Sato T, Harao M, Mizumoto T, Yamashita J, Neoplasia 2007, 9, 260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mittendorf EA, Alatrash G, Qiao N, Wu Y, Sukhumalchandra P, St John LS, V Philips A, Xiao H, Zhang M, Ruisaard K, et al. , Cancer Res 2012, 72, 3153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Imaki K, Okada T, Nakayama Y, Nagao Y, Kobayashi K, Sakai Y, Mohri T, Amino T, Nakai H, Kawamura M, Bioorg. Med. Chem 1996, 4, 2115–34. [DOI] [PubMed] [Google Scholar]
- [15].Kawabata K, Suzuki M, Sugitani M, Imaki K, Toda M, Miyamoto T, Biochem. Biophys. Res. Commun 1991, 177, 814–820. [DOI] [PubMed] [Google Scholar]
- [16].Aikawa N, Ishizaka A, Hirasawa H, Shimazaki S, Yamamoto Y, Sugimoto H, Shinozaki M, Taenaka N, Endo S, Ikeda T, et al. , Pulm. Pharmacol. Ther 2011, 24, 549–554. [DOI] [PubMed] [Google Scholar]
- [17].Zeiher BG, Artigas A, Vincent J-L, Dmitrienko A, Jackson K, Thompson BT, Bernard G, STRIVE Study Group, Crit. Care Med 2004, 32, 1695–702. [DOI] [PubMed] [Google Scholar]
- [18].Luo D, Chen Q-Y, Luesch H, J. Org. Chem 2016, 81, 532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stolze SC, Meltzer M, Ehrmann M, Kaiser M, Chem. Commun 2010, 46, 8857. [DOI] [PubMed] [Google Scholar]
- [20].Köcher S, Rey J, Bongard J, Tiaden AN, Meltzer M, Richards PJ, Ehrmann M, Kaiser M, Angew. Chemie Int. Ed 2017, 56, 8555–8558. [DOI] [PubMed] [Google Scholar]
- [21].Gunasekera SP, Miller MW, Kwan JC, Luesch H, Paul VJ, J. Nat. Prod 2010, 73, 459–462. [DOI] [PubMed] [Google Scholar]
- [22].Taori K, Paul VJ, Luesch H, J. Nat. Prod 2008, 71, 1625–9. [DOI] [PubMed] [Google Scholar]
- [23].Salvador LA, Taori K, Biggs JS, Jakoncic J, Ostrov DA, Paul VJ, Luesch H, J. Med. Chem 2013, 56, 1276–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Trott O, Olson AJ, J. Comput. Chem 2010, 31, 455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hansen G, Gielen-Haertwig H, Reinemer P, Schomburg D, Harrenga A, Niefind K, J. Mol. Biol 2011, 409, 681–691. [DOI] [PubMed] [Google Scholar]
- [26].Nakanishi I, Kinoshita T, Sato A, Tada T, 1999, 53, 434–445. [DOI] [PubMed] [Google Scholar]
- [27].Pullen SS, Miller HG, Everdeen DS, Dang TT, Crute JJ, Kehry MR, Biochemistry 1998, 37, 11836–45. [DOI] [PubMed] [Google Scholar]
- [28].Wingett DG, Vestal RE, Forcier K, Hadjokas N, Nielson CP, Breast Cancer Res. Treat 1998, 50, 27–36. [DOI] [PubMed] [Google Scholar]
- [29].Aronchik I, Bjeldanes LF, Firestone GL, Cancer Res 2010, 70, 4961–4971. [DOI] [PubMed] [Google Scholar]
- [30].Chawla A, Alatrash G, Philips AV, Qiao N, Sukhumalchandra P, Kerros C, Diaconu I, Gall V, Neal S, Peters HL, et al. , Cancer Immunol. Immunother 2016, 65, 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mittendorf EA, Alatrash G, Qiao N, Wu Y, Sukhumalchandra P, St. John LS, Philips AV, Xiao H, Zhang M, Ruisaard K, et al. , Cancer Res 2012, 72, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou Y, Eppenberger-Castori S, Marx C, Yau C, Scott GK, Eppenberger U, Benz CC, Int. J. Biochem. Cell Biol 2005, 37, 1130–44. [DOI] [PubMed] [Google Scholar]
- [33].Madhusoodhanan R, Natarajan M, Veeraraghavan J, Herman TS, Aravindan N, Cancer Biol. Ther 2009, 8, 765–73. [DOI] [PubMed] [Google Scholar]
- [34].Hostager BS, Bishop GA, J. Immunol 1999, 162, 6307–11. [PubMed] [Google Scholar]
- [35].Di D, Chen L, Wang L, Sun P, Liu Y, Xu Z, Ju J, Oncol. Rep 2016, 35, 1541–1548. [DOI] [PubMed] [Google Scholar]
- [36].Helbig G, Christopherson KW, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE, Nakshatri H, J. Biol. Chem 2003, 278, 21631–21638. [DOI] [PubMed] [Google Scholar]
- [37].Smith SM, Lyu YL, Cai L, PLoS One 2014, 9, DOI 10.1371/journal.pone.0106966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Van de Stolpe A, van der Saag PT, J. Mol. Med 1996, 74, 13–33. [DOI] [PubMed] [Google Scholar]
- [39].Hunt KK, Wingate H, Yokota T, Liu Y, Mills GB, Zhang F, Fang B, Su C-H, Zhang M, Yi M, et al. , Breast Cancer Res 2013, 15, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kumagai K, Saikawa Y, Takeuchi H, Suda K, Fukuda K, Nakamura R, Takahashi T, Kawakubo H, Wada N, Miyasho T, et al. , Anticancer Res 2013, 33, 3653–9. [PubMed] [Google Scholar]
- [41].Ho A-S, Chen C-H, Cheng C-C, Wang C-C, Lin H-C, Luo T-Y, Lien G-S, Chang J, Oncotarget 2014, 5, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wada Y, Yoshida K, Tsutani Y, Shigematsu H, Oeda M, Sanada Y, Suzuki T, Mizuiri H, Hamai Y, Tanabe K, et al. , Oncol. Rep 2007, 17, 161–7. [PubMed] [Google Scholar]
- [43].Sakai S, Tajima H, Miyashita T, Nakanuma S, Makino I, Hayashi H, Nakagawara H, Kitagawa H, Fushida S, Fujimura T, et al. , Dig. Dis. Sci 2014, 59, 787–794. [DOI] [PubMed] [Google Scholar]
- [44].Li X, Xiaokaiti Y, Wu H, Chen Y, Yang H, Duan J, Li X, Pan Y, Tie L, Zhang L, Sci. Rep 2015, 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Duan J, Xiaokaiti Y, Fan S, Pan Y, Li X, Li X, Oncol. Rep 2017, 37, 3019–3025. [DOI] [PubMed] [Google Scholar]
- [46].Al-Awadhi FH, Salvador LA, Law BK, Paul VJ, Luesch H, Mar. Drugs 2017, 15, DOI 10.3390/md15090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.