Abstract
It has long been appreciated that tumors are diverse, varying in mutational status, composition of cellular infiltrate, and organizational architecture. For the most part, the information embedded in this diversity has gone untapped due to the limited resolution and dimensionality of assays for analyzing nucleic acid expression in cells. The advent of high-throughput, next-generation sequencing (NGS) technologies that measure nucleic acids, particularly at the single-cell level, is fueling the characterization of the many components that comprise the tumor microenvironment (TME), with a strong focus on immune composition. Understanding the immune and nonimmune components of the TME, how they interact, and how this shapes their functional properties requires the development of novel computational methods and, eventually, the application of systems-based approaches. The continued development and application of NGS technologies holds great promise for accelerating discovery in the cancer immunology field.
Keywords: transcriptomics, single-cell, next-generation sequencing
Introduction
The field of cancer immunology was born in the late 19th century, with the recognition by William B. Coley that acute bacterial infections were often associated with remissions in cancer patients. This led him to create “Coley’s toxins”, a mixture of bacteria that was administered to patients with the goal of activating the immune system to eradicate tumors [1]. Coley’s toxins constituted the first cancer immunotherapy and were variably used around the world until the mid-1900’s when nonimmune-based treatments, such as radiation and chemotherapy, became the prevailing anticancer therapies. Consequently, interest in cancer immunotherapy and, by extension, cancer immunology, diminished.
Cancer immunology has experienced a renaissance due to the advancements in therapies that realize the potential of the immune system to fight cancer. These include blockade of inhibitory or immune checkpoint receptors, adoptive cell therapies, and personalized cancer vaccines. Alongside these advancements, improvements in microfluidics and high-throughput sequencing technologies have increased the speed, efficiency, and resolution with which the nucleic acid content of cells can be read. This, coupled with advances in computational methods for data normalization and analysis, is enabling the deconvolution of the complex tumor microenvironment (TME), the discovery of tumor antigens, and the annotation of novel therapeutic targets. This article will review the state-of-the-art of next-generation sequencing (NGS) technologies, with a focus on applications of single-cell transcriptomics in cancer immunology and the challenges raised by the growing complexity of the data being generated.
Application of NGS in cancer immunology
NGS, which employs massively parallel sequencing of DNA fragments, introduced high-throughput and low-cost discrete measurement of nucleic acid profiles to the field of molecular biology, and has, for the most part, replaced microarrays. NGS technology has formed the foundation of several technologies, including whole-exome sequencing, RNA-seq, single-cell RNA-seq, and ATAC-seq. Specific examples of how some of these technologies have been applied in the cancer immunology field are discussed in Hu et al. [2]. In this review, we focus primarily on single-cell RNA-seq.
Single-cell RNA sequencing: methods overview
Single cells are captured for measurement of their transcriptional landscape using either plate-based or microfluidics-based methods. Plate-based methods involve sorting of cells into separate wells (e.g. in a 96-well plate) via fluorescence-activated cell sorting (FACS), followed by RNA-seq protocols applied to each of the wells and pooling of samples following cell barcoding (different methods pool at different steps) (Table 1). This approach enables freedom with respect to the RNA-seq protocol used and allows for index-sorting (quantification of protein expression in the cells sorted into individual wells) but is limited with respect to the number of cells that can be processed due to its time-consuming nature. Initially, microfluidics-based methods used microfluidic chips to capture single-cells into individual chambers (e.g. Fluidigm) followed by lysis, reverse transcription, and amplification for library generation. Microfluidics have been used to pair within droplets single-cells with beads carrying cell-identifying barcodes [3]. Microfluidic-based capture of single-cells and beads carrying cell-identifying barcodes into chambers has been implemented in Seq-well, which is a portable and low-cost alternative to droplet-based methods [4]. Droplet-based methods are of higher throughput than microfluidic chip-based and plate-based methods, generating thousands of single-cell transcripts at relatively low cost, but are restricted to either 3’ or 5’ end sequencing protocols.
Table 1.
Comparison of commonly used RNA sequencing protocols1
| Protocol | SMART-Seq2 | Cel-Seq2, MARS-seq, STRT |
10X Chromium, Drop-seq, Indrop |
|---|---|---|---|
| Capture method | Plate-based | Plate-based | Droplet-based |
| Transcript | Full-length | 3’ or 5’ | 3’ or 5’ |
| UMI | No | Yes | Yes |
| Throughput | Medium | Medium | High |
| TCR/BCR annotation | Yes | Possible with additional primer amplification | Specific to method |
| Pooling step | Late | Early/Late | Early |
Some methods are not cited due to space constraints.
Two methodologies (SPLiT-seq, Sci-RNA-seq) bypass the need for physical isolation of single cells by using combinatorial barcoding to perform single-cell RNA sequencing (scRNA-seq) [5,6]. Although these methods have not been applied yet in the cancer immunology field, they hold great promise for accelerating discovery, given that they can be used with fixed cells. This allows for the contemporaneous processing of samples that are collected longitudinally and mitigates batch effects stemming from serial processing of samples. This feature makes these two methods attractive for the analyses of patient samples.
In addition to the single-cell RNA-seq methods described above, single-nucleus RNA-seq methods have been introduced to overcome technical challenges associated with dissociation of single-cells from tissue. Single-nucleus methods profile the mRNA landscape within each nucleus separately and can be performed using both plate-based and droplet microfluidic-based technologies [7–9]. Single- nucleus sequencing methods have been repeatedly shown to accurately capture heterogeneity across cells and dynamic cell states, despite profiling the RNA in the nucleus only [10,11]. To date, single-nucleus RNA-seq methods have been used primarily for study of the brain but have also been used to profile tumor cells [11]. Although immune cells are difficult to profile with single-nucleus RNA-seq due to their low RNA content, future technical advances could make such methods useful given their applicability to frozen archived samples.
Single-cell RNA sequencing: deconvolving the tumor immune microenvironment
The ability to read and annotate transcriptomes at single-cell resolution, coupled with the development of computational methodologies for data analysis (see Box 1 and Fig. 1), has enabled the profiling of the different components of the TME at unprecedented depth: many cells and many transcripts. Naturally, scRNA-seq was quickly leveraged to advance our understanding of the immune component of tumors.
Box 1: Single-Cell Transcriptomics Analysis: Basic concepts.
Computational tools and packages are now available to efficiently perform a variety of analyses and produce visualizations of single-cell transcriptomics data [30]. Software packages (such as Seurat [31] (R-package), Scanpy [32] (python toolkit), R Bioconductor [33] and Biscuit [12]) are used for initial data normalization and batch correction followed by general landscape characterization of the cell population (e.g. via visualizations, clustering, and the detection of highly variable genes).
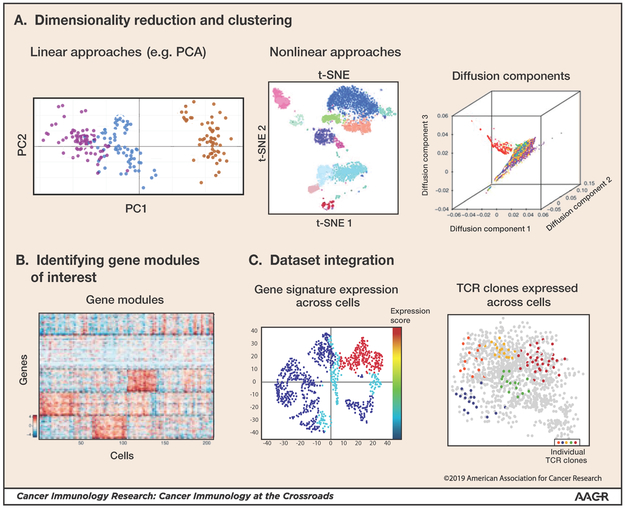
The characterization of the populations profiled by scRNA-seq includes several steps, with the goal of inferring a cell’s identity with respect to different factors of interest and gaining an understanding of the extent of diversity in the given dataset [34,35]. Due to the high dimensionality of scRNA-seq data, linear (e.g. principle components analysis (PCA)) and non-linear (e.g. diffusion components and t-distributed stochastic neighborhood embedding (tSNE)) projections are frequently used to reduce the dimensionality of the input data for subsequent analyses (Figure 1A). These techniques are useful for visualization, cell clustering, and the annotation of sets of genes that co-vary across the data. Genes that are specific to each of the identified cell clusters can be annotated using statistical models that vary in their efficiency and the extent to which they account for technical aspects of scRNA-seq [36].
Frequently, cells profiled with scRNA-seq are not naturally organized in clusters, but rather in continuous trajectories. In such cases it is advised to leverage additional methods for the extraction of informative genes and data visualization. Several packages that use diffusion maps and force directed layout or similar techniques include Destiny (implemented in R Bioconductor) [37], Monocle [38], Scanpy [32], SPRING [39], and others [40,41]. An additional approach infers the future state of a cell by leveraging the relative ratio of spliced and unspliced mRNA molecules within each cell, enabling the discovery of branching events in cell differentiation from scRNA-seq data collected at a single timepoint [42] A technique within Scanpy incorporates both clustering and trajectory inference for visualization in a unified framework [43].
A prominent component of scRNA-seq analysis involves identifying gene modules of interest: sets of genes that co-vary within the given dataset (Fig. 1B). Such gene modules can be annotated in multiple ways and are then utilized by the researcher for analyses, such as inferring the functionality of cell subsets (clusters) or identifying central candidates for perturbation. Gene modules can be identified via annotation of gene sets that are cell-cluster-specific, correlated with a dimension of interest (e.g. a specific PC or diffusion component) or co-vary across the data (as implemented in PAGODA [44]).
Following initial characterization, additional analyses are used to explore the scRNA-seq landscape. For example, gene sets identified as relevant for a cell population (cluster) or trajectory of interest can be analyzed with bioinformatics techniques to identify dominant pathways and potential regulators (via e.g. ENRICHR [45], GORILLA [46], and MSIGDB [47]). Gene sets of special interest to the researcher can be tested for their relevance to the cell populations or trajectories of interest (e.g. cell-cycle related genes [48], or gene sets associated with annotated function [49](Fig. 1C). Additionally, tailored analyses such as integration of public datasets (e.g. TCGA) or TCR/BCR information (Fig. 1C) can elucidate novel insights of the studied system.
Figure 1: Single-cell data analysis methods.
Several analysis steps are taken to generate an initial characterization of single-cell data (following or in parallel to normalization of technical noise and artifacts). (A) Various linear (e.g. PCA; principle component analysis) and non-linear (e.g. tSNE; t distributed stochastic neighborhood embedding and diffusion maps) dimensionality reduction methods can be used for identifying the main discriminants of the data of interest and for visualization. Clustering of cells by their transcriptomes can identify sets of cells that comprise units within the system. (Diffusion component illustration based on [37]) (B) Gene sets that co-vary across the data identify gene modules of interest with respect to heterogeneity and potential functionality of the cell subpopulations within a sample (figure based on [50]). (C) Integration of additional data types and sources can enable broader insights into the scRNA-seq dataset. Shown are two examples. Left: Scoring single-cells for the extent to which they express pre-defined gene signatures to infer function and characteristics of populations identified. Right: Integration of single-cell TCR information generated in parallel to the scRNA-seq data (figure based on [12]).
In breast carcinoma, a large-scale scRNA-seq study of over 45,000 cells identified increased heterogeneity of gene expression in intratumoral lymphoid and myeloid cells compared to cells in normal breast tissue, likely reflecting the responses of intratumoral immune cells to the diverse environmental signals present in tumor tissue [12]. Other scRNA-seq studies have uncovered previously unappreciated predictive properties of the immune component within the TME. In malignant glioma, scRNA-seq revealed that pre-established ways of distinguishing across glioma subtypes (IDH-A and IDH-O) are mainly accounted for by differences in the TME rather than the malignant cells themselves and that increased tumor grade was associated with differential expression of macrophage over microglia gene programs [13]. In metastatic melanoma, Nirschl et al. [14] identified a homeostatic IFNγ-dependent program that is enriched in monocytes and dendritic cells and stratifies survival. Future scRNA-seq studies of the TME will continue to advance our knowledge of the immune component of different tumors and its relationship to disease state.
Single-cell RNA sequencing: understanding T cell states in cancer
ScRNA-seq has led to important insights regarding checkpoint receptor expression in tumor-infiltrating lymphocytes (TILs) and the functional states observed in T cells in different cancers. A scRNA-seq study of human breast tumors revealed that the checkpoint receptors TIGIT and Lag-3 were present at a higher frequency on T cells than PD-1, suggesting that the former molecules may be better targets in breast tumors [15]. In a melanoma mouse model, Chihara et al. used scRNA-seq and mass spectrometry (CyTOF) to identify a coinhibitory gene module in TILs that contains novel checkpoint receptors and is cooperatively regulated by PRDM1 and c-MAF [16]. Also, in a melanoma mouse model, Singer et al. [17] showed that checkpoint receptor expression can be uncoupled from dysfunctional CD8+ T-cell phenotypes and identified distinct dysfunction and activation gene programs that separated cell populations identified with scRNA-seq. In human melanoma, scRNA-seq was used to identify a gene signature for T-cell dysfunction and inferred cell-to-cell interactions between T cells and cancer-associated fibroblasts (CAFs) [18]. Lastly, in non-small-cell lung cancer, scRNA-seq of T cells identified “exhausted” and “pre-exhausted” CD8+ T-cell populations and showed that a high ratio of pre-exhausted to exhausted cells was associated with better prognosis [19].
Analysis of TCR sequences in single cells is further shedding light on T-cell behavior in tumors. Paired scRNA-seq and TCR sequencing in breast carcinoma showed that different T-cell clones vary in their extent of activation, suggesting the presence of a continuous spectrum of T-cell activation states that is shaped by TCR usage [12]. ScRNA-seq and TCR sequence analysis of peripheral blood, tumor, and normal tissue from hepatocellular carcinoma (HCC) patients identified that exhausted CD8+ T cells and regulatory T cells (Tregs) are enriched and clonally expanded in HCC compared to normal tissues [19]. The development of TCR sequencing protocols compatible with droplet technology will further accelerate the current understanding of the relationship of T-cell clonality to functional T-cell states across different tumor types.
Epigenetics: understanding the chromatin landscape of CD8+ T cells in cancer
Coupling NGS with chromatin accessibility assays enables determination of the epigenetic and regulatory landscape of cells. Methods such as DNase-seq, Mnase-seq, and FAIRE-seq enable a genome-wide view of the epigenetic landscape but require laborious protocols and large cells counts (100K-1M cells), thus, limiting their application in cancer immunology. The introduction of ATAC-seq [20], a method that detects open chromatin by sequencing transposase-accessible regions and enables mapping of transcription factor occupancy for small cell counts and even single cells, has opened the door to the study of TILs from the epigenetic perspective.
Gaining an understanding of the epigenetic landscape of TILs that exhibit different functional states is important for understanding the underlying mechanisms that govern transition between cellular states and the reprogramming potential of TILs. Philip et al. [21] used ATAC-seq to study the epigenetic landscape of CD8+ TILs and identified two distinct CD8+ T-cell states in a murine tumor model – one that can be reversed upon in vitro activation and one that cannot. Coupling such analyses with TCR sequence data will assist in determining T-cell differentiation trajectories in the TME and how these may change upon therapeutic modulation.
NGS has been applied to analyze the spatial organization of chromatin using methods such as Hi-C. Hi-C has been used to determine the chromosomal abnormalities present in tumor cells [22,23]. This method can also be applied to study long-range DNA-DNA interactions. Chen et al. used Hi-C to identify and validate an enhancer 140kb downstream of PD-L1, which is active in tumor cells [24]. Future studies will likely apply Hi-C to study DNA organization and gene regulation in TILs.
Protein and space: The next frontiers
Two novel technologies leverage NGS to expand the dimensions of data obtained. Cellular indexing of transcriptomes and epitopes (CITE-seq) and RNA expression and protein sequencing (REAP-seq) integrate single-cell transcriptomics with limited scale proteomics [25,26]. Both of these utilize droplet microfluidics and DNA oligo-tagged antibodies to read out protein and RNA expression profiles in a single workflow. These technologies are useful for assessing transcript to protein relationships but are limited to the examination of surface protein expression only. These technologies are anticipated to be integrated into the cancer immunology field in coming years.
Technologies that allow for high-throughput transcriptomic measurements, while observing the cells’ spatial location within tissue, have also been developed. Spatial transcriptomics (ST) [27] allows for the measurement of transcriptomes within spatially resolved areas in tissue sections. This is achieved by positioning frozen tissue sections on a glass slide containing a grid with unique positional barcodes. ST can be paired with multiplex imaging and RNA-seq to map cell types and their niches in situ. Although ST does not measure transcriptomes at the single-cell level, paired scRNA-seq data from the same tissue can be used to computationally deconvolve the composition of each ST location [28]. In pancreatic cancer, ST revealed that cancer cells and stromal cells colocalize in regions disparate from pancreatic ductal and acinar cells [28]. Although this study did not map TILs, we expect ST to be used for such purposes in the future.
NICHE-seq is a different technology for adding spatial context to transcriptomic data. This technique combines two-photon microscopy, photoactivatable fluorescence, and RNA-seq to annotate single-cell transcriptomes in discrete tissue locations [29]. An attractive feature of NICHE-seq is the ability to control the timing of fluorescence photo-activation, which allows for kinetic analyses. However, a limitation of this technique is that it can only be applied to tumors that are engineered to express photoactivatable green-fluorescent protein (PA-GFP).
Perspective and concluding remarks
High-throughput data generation methods are transforming the cancer immunology field but also pose several challenges. First, they require the researcher to achieve an understanding of the data generation methods and their limitations. Second, they require the researcher to achieve a solid understanding of the analytical methods and what can be inferred from them. Third, as more hypothesis generating data is created, experimental systems suitable for validating and testing predictions made from the data will become critical. Lastly, the increasing complexity of data generated from large-scale scRNA-seq efforts, such as the human cell atlas (HCA), the immune cell atlas (ICA), and the tumor cell atlas (TCA) [38] together with the rapid increase in the dimensions that can be measured (e.g. protein and spatial variables), requires cross-disciplinary partnerships that can leverage advanced computational and systems biology approaches to discover and characterize connections between genes and cells within the TME. The continued development of NGS-based technologies and companion analytical methods are expected to rapidly propel our understanding of the immune composition of tumors through the lens of high-throughput data.
Acknowledgments
The authors would like to thank Joshua Levin and Adam Haber for helpful discussions.
Funding: Work in the author’s laboratory is supported by grants from the NIH (R01 CA187975, R01CA229400, P01 AI073748, and P01 AI039671 to A.C.A).
Footnotes
Disclosure of Potential Conflicts of Interest
A.C. Anderson is a member of the Scientific Advisory Board for Potenza Therapeutics and Tizona Therapeutics.
References
- 1.Coley WB. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. Clin Orthop Relat Res. 1893;3–11. [PubMed] [Google Scholar]
- 2.Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nature Reviews Immunology. 2018;18:168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nature Reviews Immunology. 2018;18:35–45. [DOI] [PubMed] [Google Scholar]
- 4.Gierahn TM, Ii MHW, Hughes TK, Bryson BD, Butler A, Satija R, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nature Methods. 2017;14:395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;eaam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature Methods. 2017;14:955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakken TE, Hodge RD, Miller JM, Yao Z, Nguyen TN, Aevermann B, et al. Equivalent high-resolution identification of neuronal cell types with single-nucleus and single-cell RNA-sequencing. bioRxiv. 2018;239749. [Google Scholar]
- 11.Gao R, Kim C, Sei E, Foukakis T, Crosetto N, Chan L-K, et al. Nanogrid single-nucleus RNA sequencing reveals phenotypic diversity in breast cancer. Nature Communications. 2017;8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174:1293–1308.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355:eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nirschl CJ, Suárez-Fariñas M, Izar B, Prakadan S, Dannenfelser R, Tirosh I, et al. IFNγ-Dependent Tissue-Immune Homeostasis Is Co-opted in the Tumor Microenvironment. Cell. 2017;170:127–141.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature Communications. 2017;8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018;558:454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–1511.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature Medicine. 2018;24:978–85. [DOI] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harewood L, Kishore K, Eldridge MD, Wingett S, Pearson D, Schoenfelder S, et al. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biology. 2017;18:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taberlay PC, Achinger-Kawecka J, Lun ATL, Buske FA, Sabir K, Gould CM, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016;26:719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Li C, Peng X, Zhou Z, Weinstein JN, Caesar-Johnson SJ, et al. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell. 2018;173:386–399.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology. 2017;35:936–9. [DOI] [PubMed] [Google Scholar]
- 26.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods. 2017;14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. [DOI] [PubMed] [Google Scholar]
- 28.Moncada R, Chiodin M, Devlin JC, Baron M, Hajdu CH, Simeone D, et al. Building a tumor atlas: integrating single-cell RNA-Seq data with spatial transcriptomics in pancreatic ductal adenocarcinoma. bioRxiv. 2018;254375. [Google Scholar]
- 29.Medaglia C, Giladi A, Stoler-Barak L, Giovanni MD, Salame TM, Biram A, et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. 2017;358:1622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis S List of software packages for single-cell data analysis, including RNA-seq, ATAC-seq, etc.: seandavi/awesome-single-cell [Internet]. 2018. [cited 2018 September 5]. Available from: https://github.com/seandavi/awesome-single-cell
- 31.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bioconductor - Home [Internet]. [cited 2018. September 5]. Available from: https://www.bioconductor.org/
- 34.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nature Biotechnology. 2016;34:1145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaakkola MK, Seyednasrollah F, Mehmood A, Elo LL. Comparison of methods to detect differentially expressed genes between single-cell populations. Brief Bioinform. 2017;18:735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nature Methods. 2016;13:845–8. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology. 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinreb C, Wolock S, Klein AM, Berger B. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 2018;34:1246–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 2018;555:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, et al. Reconstruction of developmental landscapes by optimal-transport analysis of single-cell gene expression sheds light on cellular reprogramming. bioRxiv. 2017;191056. [Google Scholar]
- 42.Manno GL, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf FA, Hamey F, Plass M, Solana J, Dahlin JS, Gottgens B, et al. Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. bioRxiv. 2017;208819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nature Methods. 2016;13:241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]