Abstract
The SMAD4 tumor suppressor gene product inhibits transforming growth factor-β-mediated signaling and is mutated in ~10% of colorectal carcinomas. The prognostic significance of SMAD4 mutations has been controversial. We studied the pathological and clinical characteristics of SMAD4-mutated intestinal adenocarcinomas using a retrospective case-control study design. Cases and controls were identified among 443 primary adenocarcinomas that had undergone next generation DNA sequencing (NGS) with the Ion AmpliSeq Cancer Hotspot Panel v2, which evaluates 50 cancer-related genes. Twenty-eight SMAD4-mutated (SMAD4m) patients were matched 1:2 with 56 consecutive SMAD4 wild-type (SMAD4wt) control patients from the same analysis stream. Compared with the SMAD4wt controls, the SMAD4m tumors were of higher stage (P = 0.026) and were more likely to feature mucinous differentiation (P = 0.0000), to occur in the setting of Crohn’s disease (P = 0.0041), and to harbor concurrent RAS mutations (P = 0.0178). Tumor mucin content was significantly correlated with mutations involving the MH2 domain of the SMAD4 protein (P = 0.0338). Correspondence between mutation sites and morphology was demonstrated directly in a mixed adenocarcinoma and neuroendocrine tumor where SMAD4 mutations involving different protein domains were found in histologically disparate tumor regions despite both containing identical KRAS and TP53 mutations.
Introduction
The transforming growth factor (TGF)-β signaling pathway is an important regulator of cellular and molecular processes in development and disease [1]. Among its downstream effectors, the SMAD4 tumor suppressor gene product is important in intestinal carcinogenesis. Germline mutations in SMAD4 cause juvenile polyposis syndrome (JPS) with an autosomal dominantly inherited predisposition to multiple gastrointestinal polyps and cancer [2]. SMAD4 mutations have recently been reported in 5–20% sporadic colorectal carcinomas (CRC) where they were associated with distant metastases and/or poor prognosis in some studies but not others [3–7]. Missense mutations in the MH2 domain were the most common alterations. SMAD4 mutations have also been observed in cancers with mucinous differentiation, especially those of high grade [8–11]. We carried out a retrospective case-control study aimed at characterizing the distinctive clinicopathological features of SMAD4-mutated intestinal adenocarcinomas (ACAs).
Materials and methods
Study population
We identified all primary ACAs of the large and small intestine (excluding the appendix) that underwent surgical resection and next generation sequencing (NGS) at our institution between 2013 and 2017. Information regarding the patients’ age, sex, family history, and any prior diagnosis of IBD were obtained from the electronic medical records. Patients that underwent neoadjuvant therapy before genetic analysis were excluded. For each SMAD4m tumor, the subsequent two SMAD4wt specimens in the analysis stream which contained other mutations were selected as controls.
Participant consent for this study was waived by the Institutional Review Board (IRB) of the Icahn School of Medicine at Mount Sinai.
Histology and immunohistochemistry
Tumor grading and classification were assigned according to the WHO 2010 criteria [12]. Immunohistochemical stains were performed on a Dako Omnis or Ventana Ultra instrument. All antibodies were purchased as prediluted or optimized reagents, including Chromogranin (1:200, Dako, Santa Clara, CA), and SMAD4 (1:400, Abcam, Cambridge, MA). Mismatch repair status was determined by immunohistochemical staining for expression of MLH1, PMS2, MSH2 and MSH6 (pre-diluted, Dako).
Next generation sequencing
Genomic DNA extraction was performed on paraffin-embedded tissue sections using the H&E-stained section as a guide and a cutoff of 60% tumor cellularity. DNA was amplified by multiplex PCR of targeted sequences in 50 genes using the Ion AmpliSeq Cancer Hotspot Panel v2 to generate an amplicon library. The genes included in this panel were ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11,GNAS,GNAQ, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1,MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL. The library was then clonally amplified by emulsion PCR, enriched and sequenced using the Ion AmpliSeq Cancer Hotspot Panel (v2, Thermo Fisher). The detection limit of this assay is 5% mutant alleles in a background of wild-type alleles. Reported variants from early cases were re-confirmed not to represent germline variants.
Statistical analysis
Chi-square or Fisher’s exact test was applied with statistical significance defined as P<0.05. All analyses were performed using SAS version 9.2 (SAS Institute, Cary, NC, USA).
Results
Of 443 primary intestinal ACAs (6 small bowel and 437 colorectal) that underwent sequencing, 28 (6.3%) harbored SMAD4 mutations (SMAD4m). Based on the entire cohort, SMAD4 mutations were significantly more prevalent among patients with Crohn’s disease than others (4/7 [57%] vs. 24/436 [5.5%], P = 0.0041; 3/5 [60%] vs. 24/436 [5.5%], P<0.0001 for CRCs only). The SMAD4m ACAs were then matched to ACAs with no mutations in SMAD4 (SMAD4wt) from 56 patients, serving as controls. There were no significant differences between case and control groups with respect to patient’s age, gender or tumor location (Table 1). The proportion of ACAs with nodal metastases were significantly higher among cases compared to the controls (74% vs. 46%, P = 0.036, respectively). In addition, SMAD4m cases were significantly more likely to present at a higher overall TNM stage compared to controls (P = 0.026). Further review showed a higher proportion of tumor deposits in adipose tissue (9/19 [47%] vs. 12/56 [21%], P = 0.0296), and a higher percentage of lymph node metastasis (97/389 [25%] vs. 119/1167 [10%], P<0.0001) in cases than controls.
Table 1. Clinicopathological characteristics of SMAD4m cases and SMAD4wt controls.
| SMAD4m | SMAD4wt | P_value | ||
|---|---|---|---|---|
| Patients (N) | 28 | 56 | ||
| Median age (range) | 63 (38–83) | 64 (34–85) | NS | |
| Sex | Male | 17 (61%) | 26 (46%) | NS |
| Female | 11 (39%) | 30 (54%) | ||
| Cases (N) | 28 | 57# | ||
| Tumor site |
Terminal ileum/ICV | 2 (7%) | 2 (4%) | NS |
| Cecum/Ascending colon | 10 (36%) | 21 (37%) | NS | |
| Transverse colon | 2 (7%) | 10 (18%) | NS | |
| Descending colon | 1 (4%) | 1 (2%) | NS | |
| Rectosigmoid | 8 (29%) | 23 (40%) | NS | |
| NOS | 5 (18%)* | 0 | NS | |
| Cases (N) | 22* | 56 | ||
| Nodal metastasis | None | 6 (27%) | 30 (54%) | 0.036 |
| Present | 16 (73%) | 26 (46%) | ||
| TNM stage | Stage I | 2 (9%) | 7 (13%) | 0.026 |
| Stage II | 4 (18%) | 23 (41%) | ||
| Stage III | 10 (45%) | 23 (41%) | ||
| Stage IV | 6 (27%) | 3 (5%) | ||
*Six SMAD4m tumors were metastatic with a diagnosis of colorectal cancer based on a combination of histopathology findings, clinical and imaging data.
#One control patient had two synchronous tumors, one from cecum, and one from transverse colon, which were staged according to the highest.
Compared to SMAD4wt controls, SMAD4m ACAs were significantly more likely to be classified as mucinous (>50% mucin content, 17/28 [68%] vs. 9/58 [14%], P<0.00001; Table 2) or as having mucinous features (>5% mucin content, 9/28 [32%] vs. 4/54 [7%], P = 0.0022). Importantly, this association correlated with the protein domain harboring the mutation, where 10 of 12 (83%) SMAD4m ACAs that carried mutations in the MH2 domain had mucinous features (>5% mucin content), compared with 7 of 16 (44%) SMAD4m ACAs having mucinous features when the mutation involved other SMAD4 domains (P = .0338).
Table 2. SMAD4 mutations and mucinous differentiation.
| SMAD4m (N = 28) | SMAD4wt (N = 57) | P_value | |||
|---|---|---|---|---|---|
| Mucinous | Yes | 17 (68%) | 9 (16%) | <0.00001 | |
| differentiation | No/unknown | 11 (32%) | 48 (84%) | ||
| MH2 domain (N = 12) | Other domains (N = 16) | ||||
| Mucinous | Yes | 10 (83%) | 7 (44%) | 0.0338 | |
| differentiation | No/unknown | 2 (17%) | 9 (56%) | ||
In all cases, SMAD4 mutations were accompanied by mutations in other genes (Table 3, S1 Table). The most frequent were RAS mutations, i.e., KRAS (n = 20) and NRAS (n = 2). Cumulatively, RAS mutations occurred at a higher rate in SMADm cases than in the SMAD4wt control group (79% vs 52%, P = 0.0178). Nevertheless, mucinous differentiation in SMAD4m cases occurred independently of KRAS mutation status, i.e. SAMD4m/RAS wild-type tumors and SMAD4m/RAS mutated tumors have similar frequency of mucinous features (2/5 [40%] vs. 15/23 [65%], P = 0.583). Other recurrent mutations involving TP53, APC, PIK3CA, and BRAF were less common and occurred at similar rates between the two groups. Rare mutations in FBXW7, PTEN, ATM, and CTNNB1 were also detected, but were too few for statistical comparison. A slightly higher proportion of SMAD4m than SMAD4wt tumors were MMR proficient (20/22 [91%] vs. 40/54 [74%]); however, the difference did not reach statistical significance (P = 0.103).
Table 3. Molecular characteristics of SMAD4m tumors.
| SMAD4m | SMAD4wt | P_value | ||
|---|---|---|---|---|
| Cases (N) | 28 | 56* | ||
| Genetic mutations | KRAS/NRAS | 22 (79%) | 29 (52%) | 0.0178 |
| TP53 | 13(46%) | 26 (46%) | NS | |
| APC | 11(39%) | 18 (32%) | NS | |
| PIK3CA | 3 (11%) | 14 (25%) | NS | |
| BRAF | 2 (7%) | 11 (20%) | NS | |
| FBXW7 | 2 (7%) | 5 (9%) | NS | |
| PTEN | 1 (4%) | 4 (7%) | NS | |
| ATM | 1 (4%) | 3 (5%) | NS | |
| CTNNB1 | 0 | 3 (5%) | NS | |
| Cases tested (N) | 22 | 54 | ||
| MMR by IHC | MSS | 20 (91%) | 40 (74%) | 0.103 |
| MSI-H | 2 (9%) | 14 (26%) |
* The control patient with two synchronous tumors had only transverse colon tumor sequenced.
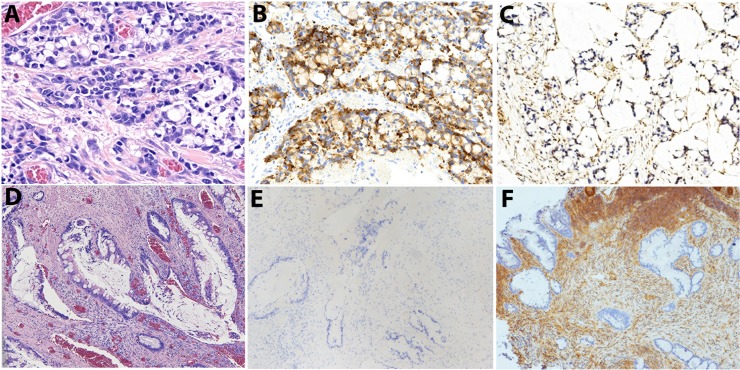
Correspondence between the site of SMAD4 mutation and tumor morphology was demonstrated directly in a case of mixed adenocarcinoma and neuroendocrine tumor (Case #20, S1 Table). In this particular case, contiguous but histologically disparate regions of the tumor comprising crypt cell neuroendocrine carcinoma (a.k.a. goblet cell carcinoid) and classical mucinous adenocarcinoma (Fig 1) harbored distinct SMAD4 mutations, MH2 domain (c.1082G>A) mutation and c.379T>A in the latter, respectively, despite harboring identical mutations of KRAS (c.35G>T) and TP53 (c.742C>T). The results suggest divergent differentiation from a single clone.
Fig 1.
A case of mucinous ACA of the ascending colon with two distinct but contiguous phenotypes: crypt cell/neuroendocrine carcinoma (A-C) and classical mucinous ACA (D-F), Immunohistochemical stains confirmed expression of Chromogranin in only the crypt cell/neuroendocrine component (B, E) and loss of SMAD4 expression in both regions of the tumor (C, F). Magnification: 200x.
Discussion
The protein products of the SMAD genes are essential mediators of the TGF-β signaling pathway, playing critical roles in growth inhibition of normal epithelial cells. Dysregulation of this pathway leads to carcinogenesis, and SMAD4 dysfunction is the most frequent cause. Earlier studies exploring the relationship between SMAD4 protein and carcinogenesis assayed loss of SMAD4 protein expression by immunohistochemical staining, which however may or may not be due to SMAD4 genetic mutations [5]. Table 4 summarizes the 10 studies that investigated the implications of SMAD4 genetic mutations in intestinal ACAs. As shown, the prognostic significance of SMAD4 mutations in intestinal ACAs was first reported in 1999 using the PCR method, yet the association between SMAD4 gene mutations and mucinous morphology was not described until 2013 [8], particularly in high-grade vs. low-grade mucinous ACAs [9,10]. A retrospective study of 90 SMAD4-mutated ACAs reported poorer survival rates in patients with SMAD4-mutated tumors, but did not include mucinous morphology as a potential risk factor [3]. Similarly, Mizuno et al reported worse survival, but did not report tumor morphology of the SMAD4-mutated cancers [6], while Khan et al reported association of mucinous morphology with SMAD4 mutation and worse prognosis [11]. Recognizing these knowledge gaps, our comprehensive study using a stringent retrospective case-control design confirms that SMAD4 mutations are associated with higher tumor stage, nodal metastasis, tumor deposits in adipose tissue, mucinous morphology, and RAS mutations.
Table 4. Summary of published studies investigating SMAD4 mutations in intestinal ACAs.
| Author, Year of publication | # of SMAD4-mutated ACAs | % of ACAs tested | Testing method | Prognostic significance | Correlation with other genes | Morphological correlation |
|---|---|---|---|---|---|---|
| Miyaki, 1999 [4] | 21 | 11.9% | PCR- SSCP | Distant metastasis | Not done | Not done |
| Alazzouzi, 2005 [5] | 5 | 6.25% | PCR | Not associated with survival | allelic imbalance in chromosome 18q21 | Not done |
| Fleming, 2012 [8] | 64 | 8.6% | Single-nucleotide polymorphism microarray analysis | No relationship to AJCC stage, T stage, N stage, or lymphovascular invasion | Not done | Mucinous morphology |
| Yoshioka, 2015 [9] | 7 | 20% | Ion AmpliSeq Cancer Hotspot Panel | Not done | Not done | High-grade mucinous morphology |
| Goswami, 2015 [7] | Not known | Not known | Next-generation sequencing hotspot mutation panel | Distant metastasis | Not done | Not done |
| Chang, 2016 [10] | 9 | 8.3% | MassARRAY-based mutation detection methods | Not done | Not done | Mucinous morphology |
| Mehrvarz Sarshekeh, 2017 [3] | 90 | 12.2% | HiSeq sequencing system hotspot testing | Associated with shorter overall survival; but not age, stage at presentation, colonic location, distant metastasis, or tumor grade | Not done | Not done |
| Mizuno, 2018 [6] | 37 | 13% | Next-generation 50-gene sequencing platform | Worse survival | RAS | Not done |
| Khan 2018 [11] | 226 | 12.3% | Ion Torrent AmpliSeq Cancer Panel Primers |
Not done | Not done | Mucinous morphology |
| Liao, 2018 | 28 | 5.6% | Next-generation 50-gene sequencing platform | Higher tumor stage, nodal and distant metastasis | RAS | Mucinous morphology |
Among all SMAD4 hotspot mutations, the MH2 domain is the most important, frequently containing missense mutations including Asp351 (D351), Pro356 (P356) and Arg361 (R361) which result in loss of function, and Ala406 (A406), Lys428 (K428), and Arg515 within the L3 loop which compromise SMAD4 binding to SMAD2/3 [8,13,14]. We found that SMAD4 mutations, particularly those involving the MH2 domain and abrogating protein function, are highly correlated with mucinous morphology. Further support for a correlation between tumor morphology and SMAD4 mutational status was obtained from a rare mixed adenocarcinoma and neuroendocrine carcinoma of the colon, in which histologically divergent tumor regions manifested distinct SMAD4 mutations despite conservation of identical KRAS and TP53 mutations.
In agreement with previous studies [4,7], we found that SMAD4m tumors were significantly more likely to have tumor deposits, nodal metastases and higher stage than corresponding SMAD4wt tumors. The mechanism is unknown but a study of in vitro CRC cell lines has implicated the effects of SMAD4 expression on tumor microenvironment [15]. Correlation between SMAD4 status and tumor stage has been described in other organs. For example, SMAD4 mutations are not typical of pancreatic intraductal papillary mucinous neoplasms but occur in up to 16% of invasive carcinomas that are associated with IPMN [16]. Likewise, low grade appendiceal mucinous neoplasms (LAMN) do not usually harbor SMAD4 mutations until there is intraperitoneal spread [17].
In this study, we also investigated the relationship between SMAD4 and other gene mutations, especially RAS genes since these two are closely associated. KRAS mutations have been reported to correlate with mucinous differentiation in CRCs [11,18], yet the mucinous differentiation in SMAD4m tumors is independent of KRAS mutation status. Tumors with RAS mutations are known to be intrinsically resistant to anti-EGFR therapy [19,20]; however, it has been shown that SMAD4 mutation is an independent factor of resistance to anti-EGFR therapy [3]. Indeed, SMAD4 inactivation also predicted worse survival in patient receiving fluorouracil-based therapy [21]. We did not find associations between SMAD4 and PTEN mutations, although a recent study showed that concurrent loss of SMAD4 and PTEN protein expression may lead to worse outcomes in patients with CRC [22]. In addition, a trend for SMAD4 mutation to associate with MMR proficiency is noted but not proved in this study, likely due to small case numbers.
Prior studies of ACA complicating Crohn’s disease either did not report or did not observe increased proportions of SMAD4 mutations compared to sporadic ACAs [23,24]. Nevertheless, the potential role of SMAD4 function in Crohn’s disease was demonstrated in a recent study showing downregulation of the SMAD4 protein in ileal epithelial cells of patients with Crohn’s disease [25]. We found a significantly high percentage of SMAD4 mutations in ACAs from patients with Crohn’s disease, with or without inclusion of small intestine ACAs, warranting future larger studies to validate and further explore this association.
In conclusion, we present a comprehensive clinicopathological and molecular characterization of SMAD4-mutated intestinal ACAs, using case-control methodology. We identified an association of SMAD4 mutations with mucinous morphology, advanced tumor stage, concomitant RAS mutations and divergent differentiation in a rare mixed adenocarcinoma and neuroendocrine carcinoma.
Supporting information
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Weiss A, Attisano L (2013) The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol 2: 47–63. 10.1002/wdev.86 [DOI] [PubMed] [Google Scholar]
- 2.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, et al. (1998) Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet. pp. 1907–1912. [DOI] [PubMed] [Google Scholar]
- 3.Mehrvarz Sarshekeh A, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR, et al. (2017) Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One 12: e0173345 10.1371/journal.pone.0173345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, et al. (1999) Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene 18: 3098–3103. 10.1038/sj.onc.1202642 [DOI] [PubMed] [Google Scholar]
- 5.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, et al. (2005) SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res 11: 2606–2611. 10.1158/1078-0432.CCR-04-1458 [DOI] [PubMed] [Google Scholar]
- 6.Mizuno T, Cloyd JM, Vicente D, Omichi K, Chun YS, Kopetz SE, et al. (2018) SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol 44: 684–692. 10.1016/j.ejso.2018.02.247 [DOI] [PubMed] [Google Scholar]
- 7.Goswami RS, Patel KP, Singh RR, Meric-Bernstam F, Kopetz ES, Subbiah V, et al. (2015) Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin Cancer Res 21: 2644–2651. 10.1158/1078-0432.CCR-14-2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. (2013) SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res 73: 725–735. 10.1158/0008-5472.CAN-12-2706 [DOI] [PubMed] [Google Scholar]
- 9.Yoshioka Y, Togashi Y, Chikugo T, Kogita A, Taguri M, Terashima M, et al. (2015) Clinicopathological and genetic differences between low-grade and high-grade colorectal mucinous adenocarcinomas. Cancer 121: 4359–4368. 10.1002/cncr.29676 [DOI] [PubMed] [Google Scholar]
- 10.Chang SC, Lin PC, Lin JK, Lin CH, Yang SH, Liang WY, et al. (2016) Mutation Spectra of Common Cancer-Associated Genes in Different Phenotypes of Colorectal Carcinoma Without Distant Metastasis. Ann Surg Oncol 23: 849–855. 10.1245/s10434-015-4899-z [DOI] [PubMed] [Google Scholar]
- 11.Khan M, Loree JM, Advani SM, Ning J, Li W, Pereira AAL, et al. (2018) Prognostic Implications of Mucinous Differentiation in Metastatic Colorectal Carcinoma Can Be Explained by Distinct Molecular and Clinicopathologic Characteristics. Clin Colorectal Cancer 17: e699–e709. 10.1016/j.clcc.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aaltonen LA, Hamilton SR, World Health Organization., International Agency for Research on Cancer. (2000) Pathology and genetics of tumours of the digestive system. Lyon, Oxford: IARC Press; Oxford University Press; (distributor). 314 p. p. [Google Scholar]
- 13.Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, et al. (2004) Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell 15: 813–823. 10.1016/j.molcel.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 14.Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. (2001) The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol 8: 248–253. 10.1038/84995 [DOI] [PubMed] [Google Scholar]
- 15.Itatani Y, Kawada K, Fujishita T, Kakizaki F, Hirai H, Matsumoto T, et al. (2013) Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology 145: 1064–1075 e1011. 10.1053/j.gastro.2013.07.033 [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein M, Noe M, Masica DL, Hosoda W, Chianchiano P, Fischer CG, et al. (2018) IPMNs with co-occurring invasive cancers: neighbours but not always relatives. Gut 67: 1652–1662. 10.1136/gutjnl-2017-315062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison JM, Hartman DA, Singhi AD, Choudry HA, Ahrendt SA, Zureikat AH, et al. (2014) Loss of SMAD4 protein expression is associated with high tumor grade and poor prognosis in disseminated appendiceal mucinous neoplasms. Am J Surg Pathol 38: 583–592. 10.1097/PAS.0000000000000194 [DOI] [PubMed] [Google Scholar]
- 18.Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y, et al. (2015) Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer 15: 340 10.1186/s12885-015-1345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765. 10.1056/NEJMoa0804385 [DOI] [PubMed] [Google Scholar]
- 20.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66: 3992–3995. 10.1158/0008-5472.CAN-06-0191 [DOI] [PubMed] [Google Scholar]
- 21.Kozak MM, von Eyben R, Pai J, Vossler SR, Limaye M, Jayachandran P, et al. (2015) Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J Clin Pathol 68: 341–345. 10.1136/jclinpath-2014-202660 [DOI] [PubMed] [Google Scholar]
- 22.Chung Y, Wi YC, Kim Y, Bang SS, Yang JH, Jang K, et al. (2018) The Smad4/PTEN Expression Pattern Predicts Clinical Outcomes in Colorectal Adenocarcinoma. J Pathol Transl Med 52: 37–44. 10.4132/jptm.2017.10.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrock AB, Devoe CE, McWilliams R, Sun J, Aparicio T, Stephens PJ, et al. (2017) Genomic Profiling of Small-Bowel Adenocarcinoma. JAMA Oncol 3: 1546–1553. 10.1001/jamaoncol.2017.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaeger R, Shah MA, Miller VA, Kelsen JR, Wang K, Heins ZJ, et al. (2016) Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology 151: 278–287 e276. 10.1053/j.gastro.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klausen P, Karstensen JG, Coskun M, Săftoiu A, Vilmann P, Cowland JB, et al. (2018) SMAD4 Protein Expression Is Downregulated in Ileal Epithelial Cells from Patients with Crohn’s Disease with Significant Inverse Correlation to Disease Activity. Gastroenterology Research and Practice 2018: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.