Abstract
Controlling protein activity and localization is a key tool in modern biology. Mammalian steroid receptor ligand-binding domain (LBD) fusions have been used in a range of organisms and cell types to inactivate proteins of interest until the cognate steroid ligand is applied. Here, we demonstrate that the glucocorticoid receptor LBD confers ligand-gated control of a heterologous gene expression system (Q system) and the DAF-16 transcription factor in Caenorhabditis elegans. These experiments provide a powerful tool for temporal control of protein activity, and will bolster existing tools used to modulate gene expression and protein activity in this animal.
Keywords: Glucocorticoid receptor, ligand-binding domain, drug inducible, gene expression, Caenorhabditis elegans
THE ability to temporally and spatially control gene expression and protein activity/localization are essential tools in modern genetics. Heterologous systems, such as Gal4-upstream activation sequence (UAS) have been widely used in Drosophila melanogaster, allowing researchers exquisite control over the time and tissue in which the transgene is expressed (Brand and Perrimon 1993). The Q system—which adapts the Neurospora crassa transcriptional activator, QF, and its repressor, QS, to gene activation via derepression of QS by quinic acid—offers an additional system for transgene expression analysis, lineage tracing, and mosaic analysis in mammalian cells and in fly models (Potter et al. 2010; Riabinina et al. 2015). Other methods to control gene expression include tissue-specific expression of the Cre recombinase fused to the estrogen receptor ligand-binding domain (LBD) (Feil et al. 1996), CRISPR interference and CRISPR activation (Gilbert et al. 2013; 2014; Qi et al. 2013), tetracycline-inducible gene regulation systems (TET on/off) (Gossen and Bujard 1992; Gossen et al. 1995), and tags to stabilize or destabilize proteins, in some cases in an inducible manner (Dohmen et al. 1994; Banaszynski et al. 2006; Nishimura et al. 2009; Bonger et al. 2011; Holland et al. 2012; Morawska and Ulrich 2013; Zhang et al. 2015).
In Caenorhabditis elegans, the most common method to induce gene expression is to fuse heat-shock promoters upstream of a gene of interest; following acute heat shock, gene expression is robustly induced (Stringham et al. 1992). Modifications to this approach include the FLP-out system (Voutev and Hubbard 2008) and the HSF-1 system, which allows for some tissue specificity of gene induction (Bacaj and Shaham 2007). These approaches have been powerful in C. elegans, but have some caveats: (1) in addition to activating the transgene of interest, heat shock also provokes regulation of heat shock-responsive genes and can affect cellular transcription and translation in ways that can cause unknown and undesired physiological effects; (2) tissue-restricted expression can be achieved, but requires specific genetic backgrounds; and (3) the system results in a pulse of target gene transcription rather than sustained transgene expression, although modifications such as the FLP-out system may bypass this limitation. Bipartite gene expression systems (such as Gal4-UAS) have been historically lacking in C. elegans, likely because C. elegans is cultured at lower temperatures than Saccharomyces cerevisiae, and thus the imported Gal4-UAS system proteins are less active at these lower temperatures. The Q system, which was imported from a fungus with a more similar growth temperature range to C. elegans, was adapted to C. elegans to control gene expression (Wei et al. 2012). Additionally, a temperature-robust Gal4-UAS system was also developed for C. elegans (cGal) by optimizing the three main components of the system to function between 15 and 25° (Wang et al. 2017).
Previously, the N. crassa QF/QUAS system was used in C. elegans to mark subclasses of neurons (Wei et al. 2012). In this system, the QF transcription factor binds to a response element (Q upstream activating sequence, QUAS) to drive transcription of a downstream transgene. Constitutive expression of a transgene can be repressed through coexpression of a repressor (QS), similar to how Gal80 is used in the Gal4-UAS system. Adding quinic acid relieves QS-mediated repression and permits QF to drive expression of the transgene. Tissue specificity can be obtained using split QF constructs and/or by expressing QF and QS using tissue-specific promoters, and the system functioned effectively using single-copy transgenes (Wei et al. 2012). Some limitations of the system are that QS and QF have to be expressed constitutively and require a minimum of 6 hr for quinic acid to alleviate QS repression and allow transgene induction, with complete restoration of transgene expression taking 24 hr; this delay is not ideal for tightly controlling transgene induction in developmental contexts or during dynamic and rapid cellular events.
Using the LBD from mammalian steroid receptors permits drug-inducible control of the Cre and Flp recombinases and the Gal4-UAS system (Picard et al. 1988; Logie and Stewart 1995; Metzger et al. 1995; Kozlova and Thummel 2002). We were interested in using steroid receptor LBDs to allow for both robust repression of fused factors and rapid alleviation of this repression upon ligand addition. We therefore wished to explore whether the human glucocorticoid receptor LBD could be used to regulate two proteins in C. elegans: the QF transcriptional activator and the DAF-16/FOXO transcription factor.
Materials and Methods
Strains and maintenance
All C. elegans strains were cultivated on NGM plates using standard methods. Animals were maintained at 20°, unless otherwise denoted. The following mutant and transgenic strains were used in this study: N2 (wild type, Bristol) (Brenner 1974), EG5003 [unc-119(ed3) III; cxTi10882 IV], CB6193 [bus-8(e2885) X], PS5970 {him-5(e1490) syIs197[hsp16-41::LIN-3C complementary DNA (cDNA) + myo-2p::dsRed + pha-1(+)] V; outcrossed 4×}, and CF4087 [daf-2(e1370); outcrossed 12×]. Strains generated for this study are listed in Supplemental Material, File S1 and Table S1.
Cloning of dexamethasone-inducible constructs
Plasmids containing the full-length coding sequences for the QF activator, the 4X repeat of the quas response element, the QS repressor (pXW83, pXW82, and pXW09, respectively), and their associated selection markers, were described previously (Wei et al. 2012). The nucleotides 2967–3737 from cDNAs encoding for the human glucocorticoid receptor α (GR), which correspond to its LBD, were amplified from pFastBacGRα-6XHis (Yamamoto Laboratory, University of California, San Francisco) and fused in frame to the C-terminal end of the full-length coding sequence for QF using In-Fusion ligation (catalog #638909; Clontech). Gateway cassettes were engineered upstream of the QF-GR coding region (pGM32DEST) or QS coding region (pGM48DEST), or downstream of the QUAS response element and minimal Δpes-10 promoter (pGM34DEST) using In-Fusion ligation. Using directional primers, DNA ∼0.5- to 2-kb upstream of the start codon of atf-8, eef-1A.1, egl-17, and pro-1 genes was PCR amplified from N2 genomic DNA. Similarly, the cDNAs from peel-1 were PCR amplified from the pMA122 plasmid (plasmid #34873; Addgene), and the lin-3c gene was cloned from the pBlueScript_lin-3c plasmid (a generous gift from C. Van Buskirk, California State University, Northridge). Blunt PCR fragments were then TOPO cloned into the pENTR/D-TOPO Gateway Entry vector (catalog #K240020; Invitrogen, Carlsbad, CA) and the resultant clones were recombined with the appropriate destination vectors. All recombinant plasmids were subsequently injected into unc-119 mutant animals, with a combination of 40 ng/µl of each recombinant plasmid and pBlueScript to give a total of 100 ng/µl DNA, with the exception of quas::peel-1, which was injected at 15 ng/µl. Transgenic progeny that were both phenotypically wild type and expressed fluorescent mCherry were subsequently isolated and tested for dexamethasone (dex)-induced gene expression. In-Fusion ligation (Clontech) was used to fuse the GR LBD from pGM32DEST in frame and upstream of both eGFP and DAF-16A.
Preparation of ligand stocks
Stocks of 100 mM (1000×) dex (catalog #D1756; Sigma, St. Louis, MO) were prepared by solubilization in DMSO, followed by 0.2-µm filter sterilization. Stocks were stored at −80° until use. The 1000× dex was then added to liquid NGM (cooled to 55°) along with salts and cholesterol to a final concentration of 100 μM dex. Alternatively, dex was spread on top of existing NGM plates if the volume was known, or it was added to a worm/M9 slurry to a final concentration of 100 μM. Stock solutions of 300 mg/ml of quinic acid (pH 6–7) were similarly added to plates or worm/M9 solutions, as previously described (Wei et al. 2012). Freshly prepared 10 mM fluorescein-dex (F-dexa) stocks (catalog #D1383; ThermoFisher) were solubilized in ethanol, sterilized through a 0.2-μm filter, and stored at 4° in the dark until use.
Molecular biology
For the extraction of total RNA, selected animals were placed in TRIzol (catalog #15596026; Invitrogen) and subsequently lysed by rapid freeze-cracking. The RNeasy extraction kit (catalog #74104; QIAGEN, Valencia, CA) was used to remove contaminating genomic DNA and isolate total RNA. The iScript cDNA Synthesis Kit (catalog #170-8891; Bio-Rad, Hercules, CA) was then used to synthesize cDNAs. To measure the levels of transcripts, master mixes using cDNA templates, SsoAdvanced Universal SYBR Green Mix (catalog #1725270; Bio-Rad), and gene-specific primers were used. All reactions were performed on the Bio-Rad CFX Connect Real-Time PCR Detection System instrument (catalog #1855200). The primers used in this study are listed in File S1 and Table S2.
Fluorescence microscopy
All images were prepared for publication using ImageJ, Adobe Photoshop, and Adobe Illustrator. To score GFP expression, we transferred 10–20 mixed staged animals to NGM plates treated with vehicle or 100 μM dex. Populations were cultivated at 25° and fluorescence was assessed repeatedly every hour at ×5 to ×10 magnification using the FSM25 Kramer Fluorescence Microscope connected to an X-Cite 120Q fluorescent light source. GFP and mCherry fluorescence were performed blind to the experimental conditions and at least three independent experiments were performed.
The remaining microscopy was performed on a Carl Zeiss (Thornwood, NY) Axioplan 2 fluorescent microscope attached to Xenon excitation lamps and green/red fluorescence filter sets. Images were captured with the attached Hamamatsu ORCA-ER camera controlled by Micro-Manager, an open-source microscopy software. For these experiments, animals were paralyzed with 10 mM levamisole and mounted onto 2% agar pads. For the F-dexa experiments, mixed staged animals were washed three times for 15 min with M9 solution, soaked in an M9 + 0.05% gelatin solution containing 1–100 μM F-dexa, and rotated on the benchtop for 2 hr. Live populations were then washed an additional three times with the M9 solution before imaging. In populations expressing the hsp-16.48 promoter, mixed staged animals were gently washed off of NGM plates and resuspended in M9 with vehicle or 100 μM dex. A 30 min, 33° heat shock was performed in a thermocycler and animals were incubated at room temperature for an additional 3.5 hr on a benchtop rotator before imaging. To assess vulval morphology, populations were synchronized by extracting eggs from gravid adults by alkaline lysis, followed by hatching in M9 + 0.05% gelatin for 24–48 hr at 25°. Next, L1 hatchlings were released from the starvation-induced L1 diapause by feeding and subsequently grown to the mid-L4 larval stage. L4 larvae were then washed off of plates and treated with vehicle or 100 μM dex for 3 hr, rotating on a benchtop before imaging. Only late L4 larvae (with cuticular cap) or young adult animals were assessed for vulval phenotypes. Under a ×16 magnification of a fluorescent microscope, mCherry positive cells were located and differential interference contrast microscopy was used to assess vulval morphology. Abnormal vulval phenotypes were scored using at least one of the following criteria: holes or gaps among the P5.p, P6.p, and P7.p cells; failure of the vulval cells to evert normally, which usually resulted in large gaps; and/or protruding vulvas. Given the developmental stage at which animals were scored, it was not possible to also assess Egl phenotypes simultaneously in the same experiment. Animals were scored blind to the experimental conditions.
Behavioral assays
For measurements of behavioral adult quiescence, active young adults were transferred into either vehicle or 100 μM dex solutions in M9 buffer in a total volume of 100 μl, and then incubated on a rotator for 2 hr at room temperature. Animals were transferred to an NGM plate seeded with OP50 Escherichia coli for 30 min at room temperature and then individual adults were assessed for behavior. Behavioral quiescence was defined by two criteria: (1) the absence of pharyngeal pumping for 60 sec, and (2) the cessation of body movements for at least 30 sec, as previously described (Van Buskirk and Sternberg 2007; Monsalve et al. 2011; Hill et al. 2014; Nelson et al. 2014). Behavioral assessments were done with at least three independent trials on separate days, in parallel to the hsp::lin-3c positive control strain (PS5970), and blind to the experimental conditions.
Statistical analysis
Relative gene expression was calculated by determining the fold-change variation over control (vehicle) samples using the comparative CT method (Schmittgen and Livak 2008). Regression analysis was used to determine CT values and the mean CT value from reactions preformed in triplicate was used to determine the average fold change from the ama-1 internal control. Error bars were calculated using the error propagation of SD to the logarithmic scale. The comparisons between vehicle- and dex-treated animals (and thus limited to two conditions) were performed using an unpaired, two-tailed Student’s t-test, or a chi-square test. All P-values were calculated using the GraphPad Prism 6.0 software.
Data availability
Strains generated in this study will be available via the Caenorhabditis Genetics Center at the University of Minnesota, Twin Cities and/or through direct request to J.D.W. The destination vectors described in this study (pGM32DEST, pGM34DEST, and pGM48DEST) will be available via Addgene. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7390391.
Results
A drug-inducible expression tool for C. elegans
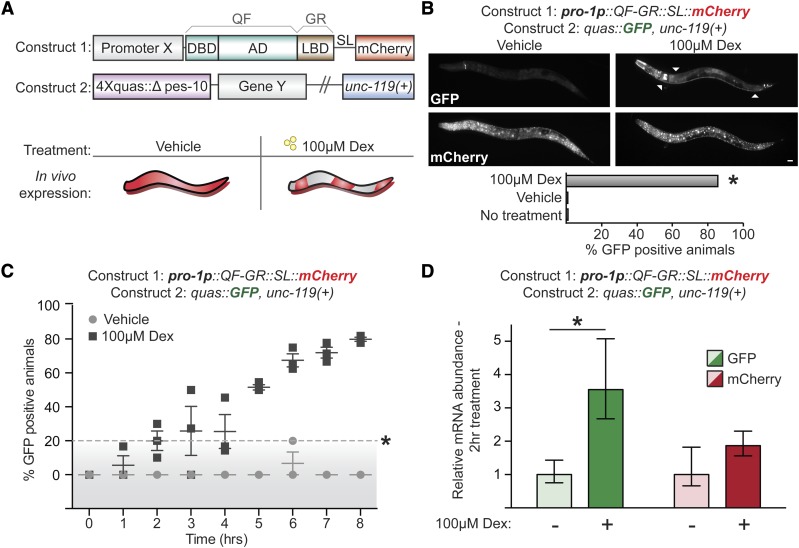
To engineer a heterologous, drug-inducible gene expression system for C. elegans, we modified the QF transcriptional activator by fusing the LBD from the human glucocorticoid receptor (GRα; National Center for Biotechnology Information gene ID: 2908) at the C terminus (QF-GR; Figure 1A). We chose the glucocorticoid receptor as C. elegans does not have any clear orthologs (Antebi 2015) and ligand binding by the GR LBD is very specific, whereas estrogen receptor LBD exhibits promiscuity (Eick et al. 2012). QF-GR should be inactive until the synthetic, nonmetabolized GR ligand dex is added (Scherrer et al. 1993). QF-GR was cloned into a plasmid with a contiguous gpd-2 splice leader (SL) and an mCherry reporter, which marks cells in which QF-GR is expressed. Although designed for Gateway cloning (Figure S1A), our system can be easily converted for any ligation-dependent or -independent cloning pipeline. Transgenes are inserted downstream of a regulatory element containing a Δpes-10 minimal promoter and a 4× repeat of the QF response element (QUAS) (Seydoux and Fire 1994; Wei et al. 2012).
Figure 1.
A drug-inducible gene expression system for C. elegans. (A) Schematic of a drug-inducible system for C. elegans, which consists of QF-GR transcriptional activator and the QUAS reporter plasmids. QF-GR consists of the full-length QF DNA-binding domain (DBD) and activation domains (AD), fused to the LBD from the human glucocorticoid receptor α (GR). The splice leader (SL) links mCherry expression to the activity of promoter X and marks for the expression of the array. The QUAS reporter consists of four tandem repeats of the QUAS response element upstream of the minimal Δpes-10 promoter and gene Y. (B) GFP and mCherry expression in animals containing QF-GR under the control of the ubiquitous pro-1 promoter and a QUAS::GFP reporter. Fluorescent micrographs depict representative animals with the indicated transgenes cultivated on either vehicle or 100 μM dex plates for 24 hr. ▵’s denote simultaneous expression of GFP in neuronal and intestinal tissues upon dex treatment. Ubiquitous mCherry expression is observed in a dex-independent manner. The percentage of GFP-positive animals observed is denoted. N ≥ 54 animals for each condition; * P < 0.001, chi-square test. Bar, 20 μm. (C) A time course scoring GFP expression in animals carrying the indicated transgenes. Mixed staged animals were cultivated on vehicle or 100 μM dex plates and GFP expression was scored hourly. Each time point denotes the mean percentage of GFP-positive animals (± SEM) from at least three independent experiments with >21 animals for each time point. Points above the dashed line denote a significant difference from vehicle-treated animals. * P < 0.01, t-test. (D) Average fold change of expression of dex-treated vs. vehicle-treated populations with the indicated transgenes. qRT-PCR measurement of GFP and mCherry transcript level after 2 hr of vehicle (−) or 100 μM dex-treatment (+) (± SEM from three biological replicates performed in triplicate). * P < 0.05, t-test.
Glucocorticoids are absorbed by C. elegans
We first tested if the GR ligand, dex, could be effectively absorbed into live animals. We used an F-dexa conjugate previously used to monitor the uptake of dex in other systems (Maier et al. 2005). As uptake of molecules in C. elegans can occur via oral ingestion and/or cuticle penetration (Valdes et al. 2012; Lee et al. 2015), we assessed uptake of F-dexa in wild-type (N2) animals and in bus-8 mutants, which have a compromised cuticular barrier (Partridge et al. 2008). Mixed-stage populations were cultured with increasing amounts of F-dexa for 2 hr and localization of F-dexa was visualized using fluorescence microscopy. In both wild-type animals and bus-8 mutants, we observed F-dexa exclusively in the pharyngeal and intestinal lumen in 97–100% (n = 1644) of animals; notably, fluorescence was not detected in the epithelia of treated animals (Figure S1B) and no fluorescence was observed in vehicle-treated populations (n = 419, * P < 0.00001, chi-square test). With increasing concentrations of F-dexa, fluorescence was detected strongly and predominately within the intestinal lumen. F-dexa labeling of intestinal tissues was only observed at the highest tested concentration (100 μM). These results indicate that 100 μM of F-dexa is sufficient for its uptake into C. elegans intestinal cells and further suggest that the intestine may be the tissue through which dex is absorbed.
The GR LBD makes QF ligand-gated
We next asked whether dex and the GR LBD could gate the activity of the heterologous QF-GR protein. We expressed QF-GR ubiquitously using the strong pro-1 promoter (Hunt-Newbury et al. 2007) and a GFP reporter was included under control of the 4xQUAS element; a cistronic mCherry cassette was used to mark cells in which the promoter was expressed. We observed induction of GFP 24 hr after cultivation with 100 μM dex, but not after vehicle treatment (Figure 1B). GFP expression was detected in 86% (n = 125) of animals expressing the mCherry reporter, as compared to 0.9% (n = 115) of vehicle-treated animals and 0% (n = 54) of animals with no drug treatment (* P < 0.001, chi-square test). GFP was detected robustly and predominately in neuronal, pharyngeal, and intestinal tissues (Figure 1B) and we also observed additional, modest GFP expression in the hypodermis and muscles of other animals (unpublished results); however, we did not observe GFP or mCherry expression in gonadal tissues. We did notice muted, yet detectable expression of GFP in a pair of unknown head neurons in both vehicle- and dex-treated populations; this background pattern of expression was consistent to that observed in animals expressing only the reporter gene (Figure 1B). Following at least 8 hr of dex treatment, the GFP and mCherry were expressed in the same cells and tissues across larval development and in adults. While the images do not overlay, this may be due to the tendency of mCherry to aggregate (Costantini and Snapp 2013); we note that the construct containing the mCherry reporter was injected at a high dose (40 ng/µl). Alternatively, temporal differences between when the mCherry and GFP messenger RNAs (mRNAs) were transcribed and translated could explain the lack of overlay. However, we noted one distinct difference in embryos: while mCherry expression was detected in eggs, GFP expression was never observed in vehicle or dex conditions (unpublished results). Finally, to determine if sustained cultivation with dex was toxic to animals, we measured the brood sizes of N2 and transgenic animals cultured on NGM plates supplemented with vehicle or 100 μM dex. We observed no significant differences in the brood sizes between vehicle- and dex-cultured populations: vehicle-treated N2 animals had an average brood size of 225 ± 7, as compared to 222 ± 13 of dex-cultured, wild-type animals (mean number of progeny per animal ± SEM; n = 10 in each condition; P = 0.85, t-test). We similarly detected no significant differences in the brood sizes of pro-1p::QF-GR transgenic animals: vehicle-treated animals had an average brood size of 116 ± 8, as compared to 98 ± 13 of dex-cultured transgenics (mean number of progeny per animal ± SEM; n = 10 in each condition; P = 0.29, t-test). The unc-119(ed3) background mutation of the pro-1p::QF-GR, quas::GFP; unc-119(+) transgenics likely contributed to the approximately twofold decrease in brood sizes observed between the rescued transgenics and wild-type animals (* P < 0.05, ANOVA test). Finally, we did not observe any abnormalities in morphology, foraging behavior, or developmental timing of the animals cultured in dex (unpublished results).
Next, we performed a time course to assess how quickly GFP was detectable by fluorescence microscopy following dex exposure. Mixed-stage animals were cultivated on plates containing either vehicle or 100 μM dex and they were scored hourly for GFP and mCherry expression. After 2 hr of drug treatment, 20% (n = 25) of animals exhibited GFP fluorescence, as compared to 0% of vehicle-treated animals (n = 21; * P < 0.03, t-test) (Figure 1C). Using quantitative PCR to measure GFP and mCherry transcript levels, we observed a 3.5-fold increase in GFP levels in dex-treated animals, as compared to vehicle-control animals (Figure 1D); mCherry mRNA levels did not change significantly within the same dex- and vehicle-treated populations. In contrast, after 30 min of ligand exposure, we did not observe any significant changes in GFP transcript levels (Figure S1C). GFP was detected initially in the nervous system and most predominately in unidentified tail neurons and the ventral cord. Notably, both the percentage of GFP-positive animals and the tissue-distribution of GFP expression increased over time, with 80% of animals (n = 38) expressing GFP in multiple tissues after 8 hr of cultivation on dex-treated plates, as compared to 0% (n = 27) of vehicle-treated populations (* P < 0.001, t-test). We similarly expressed QF-GR from an alternative ubiquitous driver, eef-1A.1, and observed dex-specific GFP induction in transgenic animals (Figure S1C). QF-GR expression from the eef-1A.1 promoter drove significant GFP reporter expression predominately in hypodermal, intestinal, and vulval tissues after 2 hr of dex exposure in 27% (n = 26) of animals, as opposed to 0% (n = 22) of vehicle-treated animals (* P < 0.04, t-test) (Figure S1D). In contrast to transgenics expressing QF-GR from the pro-1 promoter, we did not observe GFP in the nervous system in eef-1A.1::QF-GR animals. Moreover, expression of GFP in the intestinal, hypodermal, and vulval tissues appeared synchronously, and the intensity of GFP fluorescence increased with longer exposures of dex (unpublished results). Together these results indicate that QF-GR permits ligand-gated transgene expression in most major C. elegans tissues within 2 hr of dex exposure and that promoter choice is an important experimental consideration.
QF-GR allows tissue-specific transgene expression
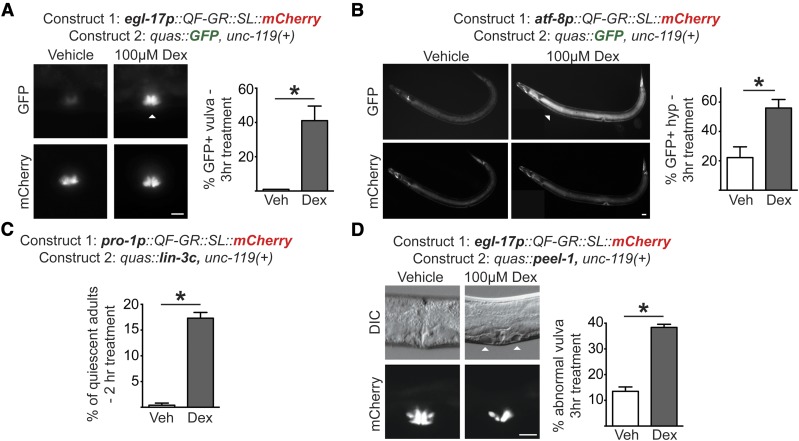
Having established that the GR LBD inactivated QF and that dex exposure allowed for QF activation, we next asked if QF-GR could drive tissue-specific transgene expression. We expressed QF-GR in vulval and hypodermal tissues using the tissue-specific egl-17 and atf-8 promoters, respectively (Hunt-Newbury et al. 2007; Ward et al. 2013). Transgenic animals were cultured in vehicle or 100 μM dex for 3 hr and the ratio of GFP-positive animals to mCherry-expressing animals was scored. We observed that 41% (n = 26) of dex-treated egl-17p::QF-GR transgenics expressed GFP in the vulva, as compared to 0% (n = 25) of vehicle-treated animals (Figure 2A). Similarly, hypodermal expression of QF-GR from the atf-8 promoter resulted in 56% (n = 41) of animals expressing GFP with 100 μM dex, as compared to 22% of vehicle-treated animals (n = 44) (Figure 2B). In both the atf-8p::QF-GR and eef-1A.1p::QF-GR transgenics, we observed a higher proportion of vehicle-treated animals expressing GFP after 3 hr of treatment compared to the pro-1p::QF-GR and egl-17p::QF-GR animals (Figure 2B and Figure S1D). These results demonstrate that dex can regulate the tissue-specific activity of QF-GR and its downstream reporter expression. Moreover, these experiments reveal that particular tissue expression of QF-GR may affect its basal activity, as previously reported for ligand-activated GAL4 in Drosophila (Osterwalder et al. 2001; Duffy 2002).
Figure 2.
The QF-GR activator drives expression of target genes in various tissues. (A and B) Tissue-specific expression from the QF-GR activator in the (A) vulva or (B) hypodermis using the egl-17 or atf-8 promoters, respectively. The QUAS reporter contains the GFP gene. Representative fluorescent micrographs of GFP and mCherry expression from animals carrying the indicated transgenes and treated with vehicle or 100 μM dex for 3 hr. ▵’s denote the tissue-specific expression of GFP. To the right of each set of images is a bar graph denoting the average percentage of GFP-positive animals after an acute 3 hr vehicle or dex treatment (± SEM; n ≥ 25 animals). * P < 0.02, t-test. Bar, 20 μm. (C) Induction of lin3c/EGF induces behavioral quiescence in adult animals. QF-GR::SL::mCherry was expressed using the ubiquitous pro-1 promoter and the QUAS reporter contains the lin-3c gene. The graph depicts the percentage of quiescent adult nematodes after 2 hr of dex treatment. Behavioral quiescence of each animal was scored by two criteria: (1) the cessation of body movements for at least 30 sec, and (2) the absence of pharyngeal pumping for 60 sec. n ≥ 180 adults; * P < 0.005, t-test. (D) QF-GR::SL::mCherry was expressed using the egl-17 promoter, which is expressed primarily in vulval cells, and the QUAS reporter contains the peel-1 toxin gene. (Left) Induction of the peel-1 gene by dex in vulval cells. Representative Nomarski micrographs of vulvae in late L4 larvae treated with vehicle or 100 μM dex for 3 hr. ▵’s mark holes in the vulva of a dex-treated animal due to presumed cell death; fluorescent images depict the vulva-specific expression of mCherry. (Right) The graph depicts the average percentage of L4s and young adults with abnormal vulval morphology after an acute 3 hr vehicle or dex treatment (± SEM; n ≥ 279 animals for each condition). * P < 0.0001, t-test. Bar, 20 μm. DIC, differential interference contrast; Veh, vehicle.
We next wished to move beyond fluorescent reporters and test whether QF-GR could be used to express transgenes to modify cell and animal physiology. Behavioral quiescence in C. elegans is characterized by cessation of feeding and movement and delayed responses to noxious odors and touch (Trojanowski et al. 2015). Lethargy in animals can be triggered, in part, by the EGF ligand, encoded by the lin-3c gene (Van Buskirk and Sternberg 2007; Hill et al. 2014). We placed lin-3c cDNA under the control of the QUAS element and expressed QF-GR ubiquitously using the pro-1 promoter (Figure 2C). We scored transgenic animals exposed for 2 hr to either vehicle or 100 μM dex, and then assessed the behavioral quiescence of adult animals as described in Materials and Methods. We observed an ∼30-fold difference in behavioral quiescence of dex-treated adults, as compared to vehicle-exposed adults (* P < 0.005, t-test). Specifically, 17% (n = 181) of adults exhibited behaviors consistent with lin-3c-induced quiescence after a 2-hr treatment of dex, as compared to 0.4% (n = 180) of vehicle-treated animals (Figure 2C). These observations indicate that the forced expression of lin-3c/EGF can be achieved with dex-inducible QF-GR.
A powerful application of Gal4-UAS in Drosophila is for the conditional cell ablation by expressing death genes, such as Reaper (White et al. 1994). In C. elegans, the peel-1 gene encodes a sperm-specific toxin which destroys both germ and somatic cells in a cell-autonomous manner. Accordingly, ubiquitous overexpression of the peel-1 gene results in animal lethality (Seidel et al. 2011). To test if our system could be used to conditionally induce animal lethality, we attempted to induce multi-tissue expression of peel-1 using pro-1p::QF-GR; however, we were unable to propagate the transgenic lines due to toxicity of the arrays, even after multiple trials with various gene dosages (unpublished results). Therefore, we asked if, alternatively, our system could be used to drive cell death in a tissue-specific fashion, as was done in other organisms (White et al. 1994; Davison et al. 2007; Obata et al. 2015). To test this, we expressed QF-GR using the egl-17 promoter, which drives expression of QF-GR primarily in 1 and 2° vulval cells, although weak hypodermal expression in L1 larvae and expression in M4 pharyngeal neurons was also reported (Burdine et al. 1998). Synchronized mid-L4-stage animals were treated for 3 hr with 100 μM dex or vehicle and scored morphologically at the late L4 and young adult stages. Abnormal vulvas were observed in 39% (n = 282) of dex-treated L4s, as compared to 14% (n = 279) of vehicle-treated L4s (Figure 2D). Continued treatment with dex resulted in lower brood sizes (unpublished results), suggesting that damage to the vulva from the peel-1 induction affected the overall fecundity of the population. Taken together, these results demonstrate that our system permits drug-inducible transgene expression of complex behavioral and morphological processes, and underscores that promoter choice affects basal expression of QF-GR.
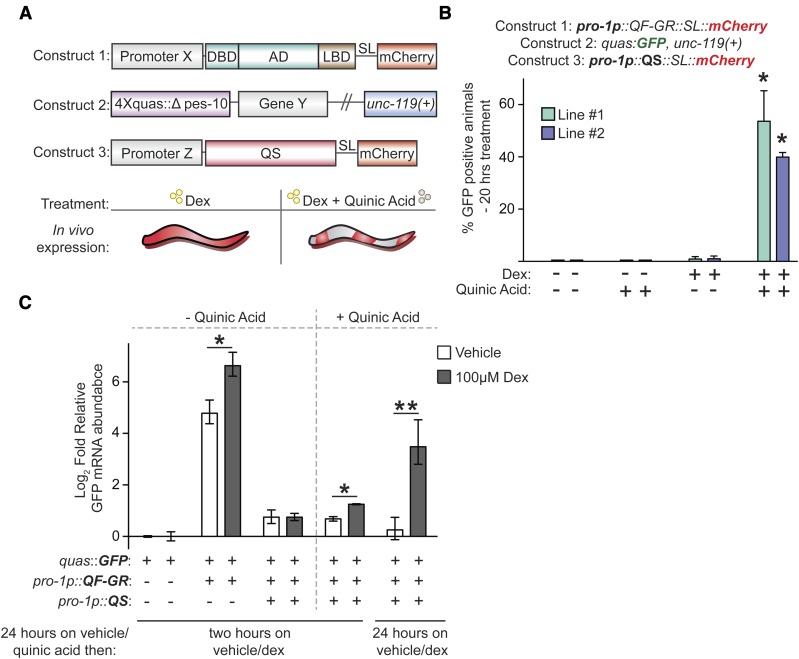
The QS repressor and quinic acid together restrain the activity of QF-GR
Neurospora QS protein inhibition of QF activity is relieved by addition of quinic acid (Wei et al. 2012). To test whether QS could inhibit the activity of QF-GR, we coexpressed QF-GR and QS, and monitored QUAS::GFP reporter activity. As expected, we failed to detect GFP fluorescence in animals cultured in either vehicle only or vehicle plus quinic acid (n ≥ 57) (Figure 3B). Following dex treatment in the absence of quinic acid, we observed GFP expression in only 0.9 and 1.0% (n ≥ 85) of animals. However, when transgenic animals were cotreated with both dex and quinic acid, we observed that 40 and 54% (n ≥ 80) of animals expressed GFP (Figure 3B). Using quantitative RT-PCR (qRT-PCR) to assess induction of transgene transcripts, after a 2 hr vehicle treatment the QF-GR activator increased GFP levels ∼28-fold above basal expression; addition of 100 μM dex increased GFP expression to 98-fold above basal expression (Figure 3). As before, QS suppressed the expression of GFP mRNAs, as transcript levels were only 1.7-fold higher in both vehicle- and dex-treated populations relative to basal expression (Figure 3C). After pretreatment with quinic acid for ∼24 hr, we measured a small but significant 2.3-fold increase in GFP transcript expression after an acute, 2 hr dex treatment. This dex-specific amplification was further intensified after a 49-hr treatment with both dex and quinic acid. We identified an 11-fold increase in GFP mRNAs in dex-treated animals, and only a 1.2-fold increase in GFP transcripts in vehicle-treated animals, as compared to the basal activity from the quas::GFP reporter. These data suggest that QS can repress transcriptional activity even in the presence of dex and that QS derepression and dex activation provide dual regulation, analogous to numerous bacterial operons that are under both negative and positive control.
Figure 3.
The QS repressor blocks activity of QF-GR. (A) Construct and experimental schematic of the addition of QS to the dex-inducible gene expression system. The composition of constructs 1 and 2 are described in Figure 1. Construct 3 encodes for the QS repressor and a splice leader that links the mCherry open reading frame to mark expression of the array. The addition of the QS repressor restricts the activity of the QF-GR activator. Derepression of QS is achieved by the addition of quinic acid; therefore, only in the presence of both dex and quinic acid will QF-GR drive target gene expression. AD, activation domains; DBD, DNA-binding domain. (B) The mean percentage of GFP-positive animals in animals carrying the indicated transgenes. QF-GR::SL::mCherry is expressed using a ubiquitously expressed pro-1 promoter and QUAS drives a GFP reporter. pro-1p also drives the expression of the QS repressor, which restricts the activity of QF (Wei et al. 2012); derepression is achieved by the addition of quinic acid. The splice leader links mCherry expression to QS expression to mark sites in which the transgene is expressed. Error bars represent the SEM from two independent experiments. n ≥ 57 animals for each condition; * P < 0.05, t-test, as compared to both vehicle-treated (−) populations. (C) qRT-PCR measurement of relative GFP transcript levels in animals carrying the indicated transgenes. The graph presents mean, log2-fold change of expression of the indicated transgene/drug combinations relative to basal expression of vehicle-treated animals carrying the quas::GFP reporter (no QF-GR or QS transgenes). Animals exposed to 7.5 mg/ml quinic acid were pretreated for 24 hr prior to dex treatment. Error bars indicate the SEM from three biological replicates performed in triplicate. * reflects the comparison by t-test of animals treated with quinic acid for 24 hr and then vehicle or dex for 2 hr; ** represents the comparison by t-test of animals treated with quinic acid for 24 hr and then vehicle or dex for an additional 25 hr (for both * and **, P < 0.05).
The GR-LBD adduct modulates DAF-16 and GFP activity in vivo
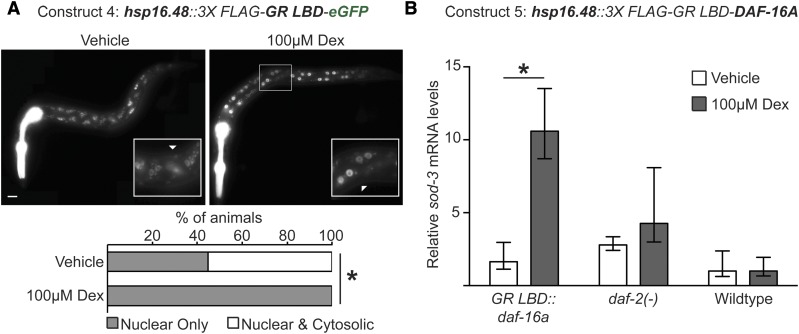
Finally, we wished to determine whether the GR LBD could be more broadly used to control the activity and localization of other C. elegans proteins. We fused the GR LBD adduct to eGFP under the control of a heat shock-inducible promoter (hsp-16.48), which drives expression predominately in the muscle and hypodermis (Stringham et al. 1992). After a 4-hr treatment with 100 μM dex, GFP expression was detected only in the hypodermal nuclei of 100% (n = 78) of animals; in contrast, GFP was visible in both the cytoplasm and nucleus in 55% (n = 94) of vehicle-treated animals (Figure 4A). This result suggests that the GR-LBD adduct may function to restrict localization of the linked protein to the cytoplasm in the absence of ligand and, upon dex-binding, translocation to the nucleus occurs more readily (Picard and Yamamoto 1987).
Figure 4.
The GR-LBD adduct modulates protein activity in vivo. (A) Representative fluorescent micrographs of L1 transgenic animals expressing the GR LBD fused in frame to the N terminus of GFP and under control of the heat shock-inducible hsp-16.48 promoter. Animals were heat shocked followed by treatment with vehicle or 100 μM dex for a total of 4 hr. Inset boxes highlight zoomed-in regions near the midbody of the animals; ▵’s denote cytoplasmic and nuclear expression of GFP. Pharyngeal fluorescence is from the co-injection marker myo-2p::tdTomato. Bottom bars represent the percentage of mixed staged, tdTomato-expressing animals with either nuclear only (shaded) or cytoplasmic and nuclear expression of GFP after 4 hr of vehicle or dex treatment (open). * P < 0.00001, chi-square test. Bar, 10 μm. (B) The relative transcript levels of the DAF-16 target gene, sod-3, after heat shock and ligand treatment for 4 hr in daf-2(e1370) mutants and wild-type animals carrying a GR LBD::DAF-16A transgene. The graph depicts the mean fold change of expression, relative to vehicle-treated N2 animals lacking any transgenes. Error bars indicate the SEM from at least four biological replicates performed in triplicate. * P < 0.03, t-test.
We next tested whether the GR LBD could similarly modulate the activity of an endogenous C. elegans protein. Transcriptionally inactive DAF-16 is localized to the cytosol and, upon translocation to the nucleus, regulates batteries of genes involved in development, aging, and metabolism (Lin et al. 1997; Ogg et al. 1997; Henderson and Johnson 2001; Kwon et al. 2010; Murphy and Hu 2013). We generated hsp16.48::3XFLAG::GR LBD::DAF-16A transgenic animals, expressed the array through acute heat shock, and assessed the levels of the DAF-16 target gene, sod-3, by qRT-PCR. We observed a robust 10.6-fold increase of sod-3 transcripts after dex treatment, and only a 1.6-fold increase in transcript levels in vehicle-treated transgenics, as compared to wild-type populations (Figure 4B). To compare the levels of sod-3 upon DAF-16 activation, we also measured sod-3 levels in daf-2(e1370) loss-of-function mutants, which genetically mimic the constitutive nuclear localization and transcriptional activation of DAF-16 (Lee et al. 2001; Lin et al. 2001). Vehicle- and dex-treated populations had 2.8- and 4.3-fold increases in sod-3 levels, respectively, as compared to wild-type animals. Notably, we only observed significant enrichment of sod-3 levels upon dex treatment in hsp16.48::3XFLAG::GR LBD::DAF-16A transgenics and not in daf-2(−) mutants or wild-type animals (* P < 0.03, t-test). There was no significant difference in sod-3 expression between vehicle-treated wild-type worms and GR-DAF-16-overexpressing worms (P > 0.05, t-test). Together, these results suggest that addition of the GR-LBD adduct allows ligand-gated control of DAF-16 activity.
Discussion
We found that the QF-GR system was sufficient to drive dex-inducible, tissue-specific expression of a GFP reporter, the toxin gene peel-1, and ubiquitous expression of lin-3c. We further demonstrate that addition of a GR LBD onto DAF-16 was sufficient to confer ligand-inducible activity of this protein, a potentially generalizable method of regulating the activity and/or localization of nuclear and cytoplasmic proteins in C. elegans.
The ability to control gene activation (Logie and Stewart 1995; Metzger et al. 1995; Kozlova and Thummel 2002; Banaszynski et al. 2006; Cho et al. 2013) and protein depletion by small molecule addition (Nishimura et al. 2009; Kanke et al. 2011; Holland et al. 2012; Zhang et al. 2015) is a powerful feature in a modern genetic toolbox. Our results demonstrate that adding the GR LBD to the QF transcriptional activator confers ligand inducibility to the Q system. While the original Q system could be induced by including the QS repressor and adding quinic acid to relieve repression, it took 6 hr to detect transgene activity and 24 hr to fully derepress the system (Wei et al. 2012); in contrast, QF-GR allows transgene induction within 2 hr (Figure 1). Although we were able to observe mRNA induction following dex treatment up to 96-fold over basal expression (Figure 3), we did frequently observe background activity of QF-GR in vehicle-treated animals, as discussed below. The QF-GR system should be particularly useful for experiments where heat-shock induction of transgenes is not possible or desirable (i.e., temperature-sensitive mutations, physiological effects of heat shock, etc.), or when sustained transgene expression is required. For example, using QF-GR rather than a heat-shock promoter to drive lin-3c would uncouple behavioral lethargy from lin-3c overexpression and lethargy as a result from heat shock (Van Buskirk and Sternberg 2007; Nelson et al. 2014).
As a tool, the GR-LBD adduct should be improved and refined going forward. As discussed above, we did frequently observe background activity of QF-GR in vehicle-treated animals. Additionally, the GR LBD did not completely restrict the GR LBD::GFP fusion to the cytoplasm, as would occur in mammalian cells (Picard et al. 1988). Possible solutions include optimizing position of the GR LBD relative to a tagged protein, or including two LBD fusions on a construct. Notably, fusing Cre to two endoplasmic reticulum LBDs in mice conferred tighter regulation and lower background with respect to recombination at floxed alleles (Zhang et al. 1996; Casanova et al. 2002). We also did not detect transgene expression in the germline. All of our experiments were performed using extrachromosomal arrays, which are frequently silenced in the C. elegans germline. However, it is also possible that our failure to observe GFP expression in the germline reflected a failure of dex to enter this tissue. New approaches to license germline expression, such as introns with periodic A/T-rich clusters (Frøkjær-Jensen et al. 2016) or removal of Piwi-interacting RNA binding sites (Zhang et al. 2018), combined with single-copy knock-ins into loci that permit germline expression would distinguish these possibilities. Empirical testing of promoters could also improve the efficacy of GR-LBD fusions.
We observed higher basal activity of QF-GR when expressed using the atf-8 and eef-1A.1 promoters, as compared to the egl-17 and pro-1 promoters (Figure 2B and Figure S1C). In Drosophila, progesterone receptor-gated Gal4 was shown to exhibit promoter-specific leakiness in the absence of the RU-486 ligand (Poirier et al. 2008), and there is leakiness dependent on the identity of the regulated transgene (Scialo et al. 2016). It is unclear what mechanisms drive leaky expression, but some possible explanations are cell-specific differences in proteins that regulate steroid receptors in the presence or absence of ligand, cell- or organism-specific differences in nuclear import/export, or that dex and/or vehicle have unknown QF-GR-independent effects on worm physiology (Picard et al. 1990; Freedman and Yamamoto 2004). That being said, all of our experiments used extrachromosomal arrays for expression of all transgenes; therefore, we cannot eliminate the possibility that high basal expression from some of the tested constructs was due to high gene dosage from these arrays.
We demonstrated that the GR LBD can be used to confer ligand gating to three proteins (QF, GFP, and DAF-16), highlighting the potential broad utility of this tool in C. elegans to regulate protein activity and/or localization. Many intensively studied transcription factors in C. elegans (DAF-16, SKN-1, PQM-1, etc.) have their activity regulated by nuclear import/export (Henderson and Johnson 2001; Lin et al. 2001; Inoue et al. 2005; Tepper et al. 2013). Fusing the GR LBD to these factors in transgenes, or knocking the GR LBD into the endogenous locus, could confer precise control over protein localization through addition/omission of ligand. The GR LBD would also be useful to add to the recently described cGal toolkit for C. elegans, adding an additional level of inducible control (Wang et al. 2017). As the cGal system currently lacks the Gal80 repressor, a negative regulator used to repress Gal4 as a “NOT” gate, use of the GR LBD could be a useful alternative NOT gate alleviated by ligand addition (Lee and Luo 1999; Wang et al. 2017). The GR LBD could also inactivate cytoplasmic proteins by redirecting them to the nucleus following ligand addition, similar to how the “anchor-away” system uses rapamycin-induced dimerization to conditionally inactivate nuclear proteins by shuttling to the cytoplasm (Haruki et al. 2008). In theory, with appropriate ligand choice to minimize cross-activation, one could use multiple steroid receptor LBDs in one experimental setting to temporally and spatially gate the activity/localization of multiple proteins.
Addition of the GR LBD to regulate protein/activity localization will be a powerful addition to the array of tools to modulate gene expression and protein function in C. elegans, including but not limited to CRISPR/Cas9 somatic genome editing (Shen et al. 2014), FLP/FRT-mediated gene (in)activation (Voutev and Hubbard 2008), and heterologous gene expression systems (Wei et al. 2012; Wang et al. 2017). Refinement of the system has the potential to permit inducible, cell-specific ablation through expression of peel-1 or other toxins.
Acknowledgments
We thank A. Scacchetti, M. Knuesel, M. Ashina, K. Ehmsen, the members of the Yamamoto, L’Etoile and Ashrafi labs (University of California, San Francisco), and the Frand lab (University of California, Los Angeles) for helpful discussions. We thank C. Van Buskirk (California State University, Northridge), N. Riehs from the Kenyon and Ashrafi laboratories (University of California, San Francisco), and X. Wei from the Shen Laboratory (Stanford) for gifts of plasmids and worm strains. G.C.M. was a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-2189-14) and a University of California Presidential Postdoctoral Fellow (PPFP 2013–2015). This work was supported by grants from the National Institutes of Health R01 (CA-020535) and R21 (ES-026068) to K.R.Y. J.D.W. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award K99 GM-107345 and R00 GM-107345. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7390391.
Communicating editor: D. Greenstein
Literature Cited
- Antebi A., 2015. Nuclear receptor signal transduction in C. elegans (June 9, 2015), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.64.2, http://www.wormbook.org. 10.1895/wormbook.1.64.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T., Shaham S., 2007. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics 176: 2651–2655. 10.1534/genetics.107.074369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L. A., Chen L.-C., Maynard-Smith L. A., Ooi A. G. L., Wandless T. J., 2006. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126: 995–1004. 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonger K. M., Chen L.-C., Liu C. W., Wandless T. J., 2011. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol. 7: 531–537. 10.1038/nchembio.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine R. D., Branda C. S., Stern M. J., 1998. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Casanova E., Fehsenfeld S., Lemberger T., Shimshek D. R., Sprengel R., et al. , 2002. ER-based double iCre fusion protein allows partial recombination in forebrain. Genesis 34: 208–214. 10.1002/gene.10153 [DOI] [PubMed] [Google Scholar]
- Cho U., Zimmerman S. M., Chen L.-C., Owen E., Kim J. V., et al. , 2013. Rapid and tunable control of protein stability in Caenorhabditis elegans using a small molecule. PLoS One 8: e72393 10.1371/journal.pone.0072393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini L. M., Snapp E. L., 2013. Fluorescent proteins in cellular organelles: serious pitfalls and some solutions. DNA Cell Biol. 32: 622–627. 10.1089/dna.2013.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. M., Akitake C. M., Goll M M. G., Rhee J. M., Gosse N., 2007. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J., Wu P., Varshavsky A., 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263: 1273–1276. 10.1126/science.8122109 [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- Eick G. N., Colucci J. K., Harms M. J., Ortlund E. A., Thornton J. W., 2012. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 8: e1003072 10.1371/journal.pgen.1003072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D., et al. , 1996. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. USA 93: 10887–10890. 10.1073/pnas.93.20.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman N. D., Yamamoto K. R., 2004. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15: 2276–2286. 10.1091/mbc.e03-11-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Jain N., Hansen L., Davis M. W., Li Y., et al. , 2016. An abundant class of non-coding DNA can prevent stochastic gene silencing in the C. elegans germline. Cell 166: 343–357. 10.1016/j.cell.2016.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., et al. , 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H., 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89: 5547–5551. 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., et al. , 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769. 10.1126/science.7792603 [DOI] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U. K., 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Johnson T. E., 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11: 1975–1980. 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M. N. G., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. 10.1016/j.cub.2014.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. J., Fachinetti D., Han J. S., Cleveland D. W., 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 109: E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., et al. , 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5: e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hisamoto N., An J. H., Oliveira R. P., Nishida E., et al. , 2005. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19: 2278–2283. 10.1101/gad.1324805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M., Nishimura K., Kanemaki M., Kakimoto T., Takahashi T. S., et al. , 2011. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 12: 8 10.1186/1471-2121-12-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T., Thummel C. S., 2002. Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development 129: 1739–1750. [DOI] [PubMed] [Google Scholar]
- Kwon E.-S., Narasimhan S. D., Yen K., Tissenbaum H. A., 2010. A new DAF-16 isoform regulates longevity. Nature 466: 498–502. 10.1038/nature09184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Zhang W., Zhang D., Yang Y., Liu B., et al. , 2015. Assessing cholesterol storage in live cells and C. elegans by stimulated Raman scattering imaging of phenyl-diyne cholesterol. Sci. Rep. 5: 7930 10.1038/srep07930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y., Hench J., Ruvkun G., 2001. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 11: 1950–1957. 10.1016/S0960-9822(01)00595-4 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., Kenyon C., 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- Lin K., Hsin H., Libina N., Kenyon C., 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28: 139–145. 10.1038/88850 [DOI] [PubMed] [Google Scholar]
- Logie C., Stewart A. F., 1995. Ligand-regulated site-specific recombination. Proc. Natl. Acad. Sci. USA 92: 5940–5944. 10.1073/pnas.92.13.5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier C., Rünzler D., Schindelar J., Grabner G., Waldhäusl W., et al. , 2005. G-protein-coupled glucocorticoid receptors on the pituitary cell membrane. J. Cell Sci. 118: 3353–3361. 10.1242/jcs.02462 [DOI] [PubMed] [Google Scholar]
- Metzger D., Clifford J., Chiba H., Chambon P., 1995. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA 92: 6991–6995. 10.1073/pnas.92.15.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Van Buskirk C., Frand A. R., 2011. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21: 2033–2045. 10.1016/j.cub.2011.10.054 [DOI] [PubMed] [Google Scholar]
- Morawska M., Ulrich H. D., 2013. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30: 341–351. 10.1002/yea.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T., and P. J. Hu, 2013 Insulin/insulin-like growth factor signaling in C. elegans (December 26, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.164.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Nelson M. D., Lee K. H., Churgin M. A., Hill A. J., Van Buskirk C., et al. , 2014. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol. 24: 2406–2410. 10.1016/j.cub.2014.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Obata F., Tanaka S., Kashio S., Tsujimura H., Sato R., 2015. Induction of rapid and selective cell necrosis in Drosophila using Bacillus thuringiensis Cry toxin and its silkworm receptor. BMC Biology 13: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., et al. , 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H., Keshishian H., 2001. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98: 12596–12601. 10.1073/pnas.221303298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge F. A., Tearle A. W., Gravato-Nobre M. J., Schafer W. R., Hodgkin J., 2008. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 317: 549–559. 10.1016/j.ydbio.2008.02.060 [DOI] [PubMed] [Google Scholar]
- Picard D., Yamamoto K. R., 1987. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6: 3333–3340. 10.1002/j.1460-2075.1987.tb02654.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Salser S. J., Yamamoto K. R., 1988. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell 54: 1073–1080. 10.1016/0092-8674(88)90122-5 [DOI] [PubMed] [Google Scholar]
- Picard D., Lindquist S., Yamamoto K. R., 1990. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348: 166–168. 10.1038/348166a0 [DOI] [PubMed] [Google Scholar]
- Poirier L., Shane A., Zheng J., Seroude L., 2008. Characterization of the Drosophila Gene-Switch system in aging studies: a cautionary tale. Aging Cell 7: 758–770. 10.1111/j.1474-9726.2008.00421.x [DOI] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L., Luo L., 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. 10.1016/j.cell.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M. N., et al. , 2015. Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12: 219–222. 10.1038/nmeth.3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer L. C., Picard D., Massa E., Harmon J. M., Simons S. S., et al. , 1993. Evidence that the hormone binding domain of steroid receptors confers hormonal control on chimeric proteins by determining their hormone-regulated binding to heat-shock protein 90. Biochemistry 32: 5381–5386. 10.1021/bi00071a013 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Scialo F., Sriram A., Stefanatos R., Sanz A., 2016. Practical recommendations for the use of the GeneSwitch Gal4 system to knock-down genes in Drosophila melanogaster. PLoS One 11: e0161817 10.1371/journal.pone.0161817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., Van Oudenaarden A., Rockman M. V., et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115 10.1371/journal.pbio.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Fire A., 1994. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120: 2823–2834. [DOI] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., et al. , 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30: 625–636. 10.1016/j.devcel.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Stringham E. G., Dixon D. K., Jones D., Candido E. P., 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell 3: 221–233. 10.1091/mbc.3.2.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T., et al. , 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154: 676–690. 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Nelson M. D., Flavell S. W., Fang-Yen C., Raizen D. M., 2015. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J. Neurosci. 35: 14571–14584. 10.1523/JNEUROSCI.1369-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes V. J., Athie A., Salinas L. S., Navarro R. E., Vaca L., 2012. CUP-1 is a novel protein involved in dietary cholesterol uptake in Caenorhabditis elegans. PLoS One 7: e33962 10.1371/journal.pone.0033962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2007. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 10: 1300–1307. 10.1038/nn1981 [DOI] [PubMed] [Google Scholar]
- Voutev R., Hubbard E. J. A., 2008. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180: 103–119. 10.1534/genetics.108.090274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu J., Gharib S., Chai C. M., Schwarz E. M., et al. , 2017. cGAL, a temperature-robust GAL4-UAS system for Caenorhabditis elegans. Nat. Methods 14: 145–148. 10.1038/nmeth.4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., Bojanala N., Bernal T., Ashrafi K., Asahina M., et al. , 2013. Sumoylated NHR-25/NR5A regulates cell fate during C. elegans vulval development. PLoS Genet. 9: e1003992 10.1371/journal.pgen.1003992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Potter C. J., Luo L., Shen K., 2012. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat. Methods 9: 391–395. 10.1038/nmeth.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Grether M. E., Abrams J. M., Young L., Farrell K., et al. , 1994. Genetic control of programmed cell death in Drosophila. Science 264: 677–683. 10.1126/science.8171319 [DOI] [PubMed] [Google Scholar]
- Zhang D., Tu S., Stubna M., Wu W.-S., Huang W.-C., et al. , 2018. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359: 587–592. 10.1126/science.aao2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z., Dernburg A. F., 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Riesterer C., Ayrall A. M., Sablitzky F., Littlewood T. D., et al. , 1996. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 24: 543–548. 10.1093/nar/24.4.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains generated in this study will be available via the Caenorhabditis Genetics Center at the University of Minnesota, Twin Cities and/or through direct request to J.D.W. The destination vectors described in this study (pGM32DEST, pGM34DEST, and pGM48DEST) will be available via Addgene. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7390391.