Abstract
In the life cycle of the fungal pathogen Candida albicans, the formation of filamentous cells is a differentiation process that is critically involved in host tissue invasion, and in adaptation to host cell and environmental stresses. Here, we have used the Gene Replacement And Conditional Expression library to identify genes controlling invasiveness and filamentation; conditional repression of the library revealed 69 mutants that triggered these processes. Intriguingly, the genes encoding the small ubiquitin-like modifier (SUMO) E3 ligase Mms21, and all other tested members of the sumoylation pathway, were both nonessential and capable of triggering filamentation upon repression, suggesting an important role for sumoylation in controlling filamentation in C. albicans. We have investigated Mms21 in detail. Both Mms21 nulls (mms21Δ/Δ) and SP [Siz/Pias (protein inhibitor of activated signal transducer and activator of transcription)] domain (SUMO E3 ligase domain)-deleted mutants displayed invasiveness, filamentation, and abnormal nuclear segregation; filament formation occurred even in the absence of the hyphal transcription factor Efg1. Transcriptional analysis of mms21Δ/Δ showed an increase in expression from two- to eightfold above that of the wild-type for hyphal-specific genes, including ECE1, PGA13, PGA26, HWP1, ALS1, ALS3, SOD4, SOD5, UME6, and HGC1. The Mms21-deleted mutants were unable to recover from DNA-damaging agents like methyl methane sulfonate, hydroxyurea, hydrogen peroxide, and UV radiation, suggesting that the protein is important for genotoxic stress responses. In addition, the mms21Δ/Δ mutant displayed sensitivity to cell wall and thermal stresses, and to different antifungal drugs. All these findings suggest that Mms21 plays important roles in cellular differentiation, DNA damage and cellular stress responses, and in response to antifungal drugs.
Keywords: Candida albicans, filamentation, Mms21, stress response, sumoylation
CANDIDA albicans is one of the major causes of hospital-acquired infections in the USA, and the cost in treatment is nearly 2.6 billion USD annually (Stevens et al. 2014). This opportunistic fungal pathogen has evolved to survive inside its human host, and switching among a variety of cellular forms appears important for its ability to adapt to host cell responses and different environmental stresses (Staib et al. 2001; Gasch 2007). These different phenotypic forms include white, opaque (Slutsky et al. 1987), gastrointestinally induced transition (GUT) (Pande et al. 2013), and gray (Tao et al. 2014) yeast forms, and a filamentous form that appears critical for the fungus to invade host tissues (Calderone and Fonzi 2001). The white yeast form, with small, rounded budding yeast cells, is thought to be important for pathogen dissemination via the blood stream and to thus have a role in virulence (Jacobsen et al. 2012). The invasive or filamentous form is induced in response to environmental cues such as nutrient limitation or the presence of serum. When an appropriate signal is transduced, yeast cells can initiate invasion by forming either hyphae or pseudohyphae (Csank et al. 1998; Felk et al. 2002). These filamentous cells can invade host tissues and ultimately gain entry into the blood stream to cause life-threatening systemic infections (Felk et al. 2002). Thus, in addition to the yeast form, the filamentous form is also critical for C. albicans virulence (Lo et al. 1997).
To combat this pathogen, we need a better understanding of its biology including the regulation of yeast–filamentous switching. To date, only about one quarter of the C. albicans genes with Saccharomyces cerevisiae orthology have been significantly characterized (http://www.candidagenome.org/), so the majority still need to be investigated if we are to have a good understanding about the physiology of this organism, an understanding that can ultimately help find improved treatments for C. albicans infections.
Because invasiveness, the ability of C. albicans cells to invade host tissues in vivo or agar medium in vitro, is critical for fungal pathogenicity, we have explored, using the Gene Replacement And Conditional Expression (GRACE) library (Roemer et al. 2003), the C. albicans genome to identify genes controlling invasiveness and filamentation. Through tetracycline-induced gene repression of the GRACE library, we have identified 69 negative regulators of invasiveness and filamentation. Intriguingly, the genes encoding the small ubiquitin-like modifier (SUMO) protein Smt3 and all other tested members of the sumoylation pathway [Uba2 and Aos1 (SUMO E1), Ubc9 (SUMO E2), and Mms21 (SUMO E3)] were both nonessential and capable of triggering filamentation upon repression. The SUMO protein Smt3 and the E1 enzyme Aos1 have been previously implicated as repressors of filament formation in C. albicans (O’Meara et al. 2015).
Sumoylation, the covalent attachment of SUMO (Small Ubiquitin-like Modifier) to target proteins, is an important post-translational modification that plays key roles in processes as distinct as transcription regulation, stress responses, cellular differentiation, and protein transport (Seeler and Dejean 2003; Wohlschlegel et al. 2004; Zhou et al. 2004). The steps in the sumoylation process are very similar to those of ubiquitination. Initially a precursor SUMO protein, for example Smt3 in S. cerevisiae, is cleaved at its C-terminus by specific SUMO proteases to expose diglycine, followed by SUMO activation by a SUMO E1 enzyme heterodimer (Aos1-Uba2) in an ATP-dependent step resulting in a thioester bond between Uba2 and SUMO. The E1 enzymes then transfer activated SUMO to the E2-conjugating enzyme (Ubc9) forming a thioester bond between the E2 and the SUMO diglycine. The E2 can directly transfer activated SUMO to the lysine (K) of many target proteins recognizing the consensus sequence ΨKXE (where Ψ is a hydrophobic amino acid and X any amino acid) through the formation of an isopeptide bond (Gong et al. 1997; Johnson and Blobel 1997; Johnson 2004). Sumoylation of a majority of proteins is carried out by specific SUMO E3 ligases (Meulmeester et al. 2008). Mms21 in S. cerevisiae, and its ortholog Nse2 in both humans and Schizosaccharomyces pombe, is an E3 ligase containing the so-called Siz/PIAS (SP)-RING domain that is exclusively found in SUMO E3s. Mms21 has important roles in post-translational modification, homologous recombination, DNA repair, and the maintenance of chromosomal integrity and telomeric length (McDonald et al. 2003; Pebernard et al. 2004; Andrews et al. 2005; Potts and Yu 2005, 2007; Zhao and Blobel 2005; Duan et al. 2009b).
Mms21 is also a member of the DNA repair Smc5/6 complex, and our GRACE screening also revealed Smc6 and Qri2 of this complex to be nonessential filamentation repressors. In S. cerevisiae, the Smc5/6 complex is required for nucleolar exclusion of Rad52 foci to prevent rDNA hyperrecombination (Torres-Rosell et al. 2007). In C. albicans, deletion of Rad52 causes filamentation and cell cycle-related defects with increased hyphal-specific gene (HSG) expression that is independent of Efg1 (Andaluz et al. 2006). The deletion of the SUMO protein Smt3 also results in similar cell cycle and growth defects (Leach et al. 2011). In C. albicans, DNA replication checkpoints are critical for filament formation in the presence of DNA-damaging agents (Shi et al. 2007). Defects in the cell division, DNA damage, and stress response machinery have been implicated in increased polarized growth and white–opaque switching (Bachewich and Whiteway 2005; Bachewich et al. 2005; Alby and Bennett 2009). Wos1, a newly characterized SUMO E3 ligase in C. albicans, affects white–opaque switching through sumoylation of Wor1 (Yan et al. 2015). As Mms21 is an essential part of the Smc5/6 complex in S. cerevisiae, where this complex is critical for DNA replication checkpoints and damage repair, we explored the roles of Mms21 in C. albicans physiology to shed some light on the process the yeast–filamentous switching control, and to investigate the importance of Mms21 in response to different genotoxic and cellular stresses.
Materials and Methods
Strains and culture conditions
Strains used in this study are described in Supplemental Material, Table S2. For general growth and maintenance, strains were cultured in fresh YPD medium (1% w/v yeast extract, 2% w/v Bacto peptone, 2% w/v dextrose, and 80 mg/liter uridine with the addition of 2% w/v agar for solid medium) at 30°. Cell growth, transformation, and DNA preparation were carried out using standard yeast procedures. Different types of phenotypic analyses were carried out in yeast growth conditions. The conditions used for microscopy and stress susceptibility are described below.
GRACE library and invasive assay
The GRACE library is a collection of ∼2500 conditional mutants created in the CaSS1 strain background for the purpose of finding essential genes and effective drug targets in C. albicans (Roemer et al. 2003). The parental CaSS1 strain and all the derivatives constitutively express a chimeric transactivation fusion protein consisting of the Escherichia coli tetR-binding domain and the S. cerevisiae GAL4 activation domain. Conditional repression of individual mutant strains can be achieved by supplementing the medium with tetracycline, and permanent loss of the transactivation module can be identified by growing them on 5-FOA-containing medium to detect loss of the URA3-linked module.
The GRACE library was transferred from frozen glycerol stocks into 96-well plates containing liquid YPD with 1 mg/ml nourseothricin using sterile 96-pin replicators and incubated overnight at 30°. For the invasive assay, overnight-grown copies of the library were inoculated onto YPD agar with added tetracycline at a concentration of 100 μg/ml and incubated at 30° for 4 days, followed by 10 sec washing of each colony with a stream of water. The colonies that stayed on the plates after washing were scored as invasive and those that were washed away were considered noninvasive. Images of plates and colonies were scanned at 600 dots per inch (dpi) using an Epson Perfection v500 photo scanner.
Deletion of the MMS21 and Mms21 SP domains
Oligonucleotides and plasmids used in this study are shown in Table S3. The two alleles of MMS21 were deleted in the SN148 strain (wild-type) background by a standard gene deletion method using two-step PCR disruption (Dignard et al. 2007). The first allele was replaced with HIS1 and the second allele with ARG4 from the pFA-HIS1 and pFA-ARG4 plasmids, respectively (Gola et al. 2003). Since both plasmids have a common DNA backbone, oligonucleotides Amj37Fw and Amj38Rv (Table S3) were used to prepare the cassettes for the deletion of the MMS21 gene. The HIS1 marker was transformed into the parent strain SN148 using the standard lithium acetate method. The correct insertion of HIS1 was confirmed by PCR analysis using genomic DNA from positive colonies and the resulting strain was named MMS21/mms21Δ. The second allele was deleted with ARG4 in strain MMS21/mms21Δ using same transformation method, and was confirmed by PCR to generate the mms21Δ/Δ or null mms21 mutant.
Deletion of the Mms21 SP domain in strain MMS21/mms21Δ was performed using plasmid pFa-TAP-Arg4 (Lavoie et al. 2008) using a similar strategy. The SP domain was deleted from strain MMS21/mms21Δ with a PCR-generated cassette, using oligonucleotides Amj73Fw and Amj72Rv, and subsequently confirmed by PCR. Western analysis of the mutant protein using anti-TAP antibodies was carried out to confirm the correct sizes.
Reintegration of MMS21 into the null mms21 mutant
The null mms21 mutant was complimented at the RP10 locus with a copy of MMS21 from a wild-type strain using the CIp10 plasmid (Murad et al. 2000). For reintegration, a 1465-bp MMS21 fragment from genomic DNA was amplified by PCR using oligonucleotides AmjHindIIIFw and AmjXhoIRv, and Expand high-fidelity polymerase (Roche). The amplified PCR fragment was digested with restriction enzymes HindIII and XhoI, and cloned into plasmid CIp10 in E. coli strain DH5α. Correct integration of MMS21 into the plasmid was confirmed by sequencing; this construct was designated CIp10-MMS21. CIp10-MMS21 was then linearized by digestion with StuI prior to transforming the null mms21 mutant. The URA-positive colonies were analyzed by PCR and successful MMS21 integration into the null mms21 mutant was confirmed.
Microscopic analysis
For cell morphology, overnight cultures grown in YPD at 30° were subjected to phase differential interference contrast (DIC) microscopy. For nuclear segregation analysis, DAPI (Sigma [Sigma Chemical], St. Louis, MO) staining of live cells was performed without permeabilization. Overnight cultures were resuspended at a starting optical density of 600 nm (OD600) = 0.1 in synthetic complete medium (0.67% w/v yeast nitrogen base with ammonium sulfate, 2% w/v dextrose, and 1.59% w/v complete amino acids mixture, pH 7.0) and incubated for 3.5–6 hr (until successful completion of the first cellular division of both wild-type and mutant strains) followed by the addition of 2 μg/ml DAPI into each tube. Calcoflour staining (2 μg/ml) was performed using a similar strategy. Cells were examined by DIC at 63× and fluorescent microscopy at ×100 magnification using a Leica DM 6000 microscope mounted with a Hamamatsu Orca R2 CCD camera.
Stress phenotypes
To test sensitivities of strains to different agents and to temperature, overnight cultures grown in YPD at 30° were serially diluted in 10-fold stages at room temperature to a dilution concentration of 106 and 103 cells/ml, and 5 μl of each dilution was spotted on YPD agar containing one of the following; MMS (0.01% v/v), hydroxyurea (HU, 10 mM), hydrogen peroxide (5 mM), Congo red (150 μg/ml), amphotericin B (1 μg/ml), clotrimazole (0.5 μg/ml), flucanozole (10 μg/ml), posacanozole (10 μg/ml), and NaCl (1 M). Following 90-sec UV light exposure, plates were then incubated at 30° for 4 days, except for caspofungin (0.75 μg/ml)-containing plates, which were incubated for 7 days at 30°. Heat sensitivity of strains was tested on YPD agar at 37° and 42° for a period of 4 days.
Microarray profiling
The mms21Δ/Δ and SN148 cultures were grown in YPD overnight at 30°, diluted to OD600 of 0.1 in YPD at 30°, grown to an OD600 of 0.8–1.2, then harvested and stored at −80° until RNA extraction. Total RNA isolation and cDNA preparation were performed as previously described (Tebbji et al. 2014). C. albicans whole-genome arrays were obtained from the National Research Council Canada Biotechnology Research Institute and were hybridized in a hybridization chamber at 42° for overnight incubation. Microarray slides were scanned using a GenePix4000B microarray scanner and the images were analyzed in GenePix Pro 7; the output data were processed with Microsoft Excel 2013 and MultiExperiment Viewer.
RNA-sequencing
The mms21Δ/Δ and SN148 cultures were grown in YPD overnight at 30°, diluted to OD600 of 0.1 in YPD at 30°, and then grown to an OD600 of 0.8–1.2 on a 220-rpm shaker. Total RNA was extracted using the QIAGEN (Valencia, CA) RNeasy minikit protocol, and RNA quality and quantity were determined using an Agilent bioanalyzer. Paired-end sequencing (150 bp) of extracted RNA samples was carried out at the Quebec Genome Innovation Center located at McGill University using an Illumina miSEQ sequencing platform. Raw reads were preprocessed with the sequence-grooming tool cutadapt version 0.4.1 (Martin 2011) with the following quality trimming and filtering parameters (`–phred33–length 36 -q 5–stringency 1 -e 0.1`). Each set of paired-end reads was mapped against the C. albicans SC5314 haplotype A, version A22 downloaded from the Candida Genome Database (http://www.candidagenome.org/) using HISAT2 version 2.0.4. SAMtools was then used to sort and convert SAM files. The read alignments and C. albicans SC5314 genome annotation were provided as input into StringTie v1.3.3 (Pertea et al. 2015), which returned gene abundances for each sample. Raw and processed data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (Edgar et al. 2002).
Data availability
Strains and plasmids are available upon request. Table S1 contains 69 invasive strains recovered from GRACE library screening. Strains used in this study are listed in Table S2. Plasmids and primers used with their respective sequences are listed in Table S3. Table S4 contains 373 hits from transcription profiling, while Table S5 represents RNA-seq up- and down-regulated genes need to change in figshare portal too. Figure S1 contains efg1 mms21 double-deletion filamentation. Transcription profiling data are available at GEO with the accession number: GSE116544. RNA-sequencing (RNA-seq) data are accessible through GEO Series accession number: GSE121210. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7385114.
Results
Large-scale invasive growth screening in C. albicans
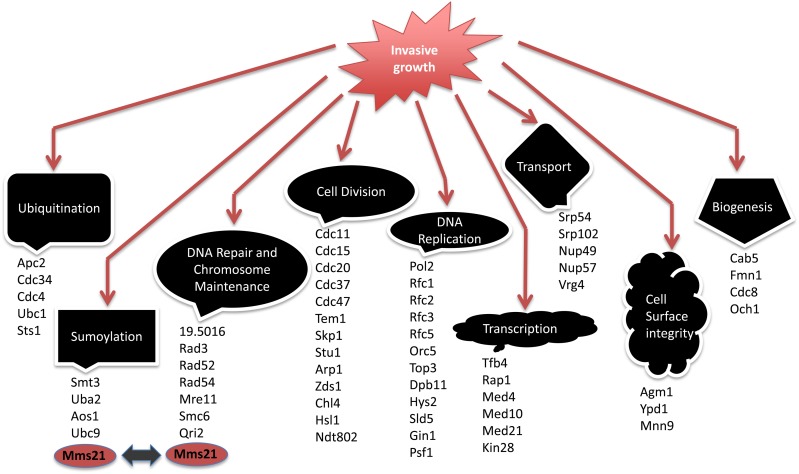
To determine a subset of genes that play roles in C. albicans invasive growth, the GRACE strains (Roemer et al. 2003) were conditionally repressed on tetracycline-containing (100 μg/ml) YPD agar at 30° for 96 hr, followed by a 10-sec wash of each colony with a stream of water. We identified those strains that were not washed off the plates and displayed wrinkled colonies during growth under the repressing conditions as potentially containing repressed genes that were involved in the regulation of invasive growth. We identified 69 strains from the 2357 single mutants in the GRACE collection that were invading the agar medium during tetracycline repression, suggesting that those genes code for functions that inherently repress invasiveness and filamentation (Table S1). The genes observed included several transcription regulators and, overall, were implicated in a variety of cellular processes including cell division, chromosome maintenance, DNA repair, transport, ubiquitination, and sumoylation (Figure 1).
Figure 1.
Invasive mutants collection from GRACE screening. A total of 69 mutants were scored invasive from GRACE library screening of which 60 mutants are shown here with their respective cellular processes. GRACE, Gene Replacement And Conditional Expression.
These data emphasized the inherent complexity involved in controlling invasiveness and filamentation in C. albicans. Many of the negatively acting components were also involved in the control of cell cycle and division, highlighting a correlation between normal cell division and filamentation. Some of the invasive growth repressors, specifically those involved in cell division, chromosome maintenance, and DNA repair, have been previously implicated in negative control of filamentation (O’Meara et al. 2015). We also found a considerable number of genes that have not been previously implicated in the control of filamentation that include transcription regulators, genes involved in cellular transport and biogenesis, and some members of the ubiquitination and sumoylation pathways (Table S1).
Members of the sumoylation pathway negatively regulate filamentation
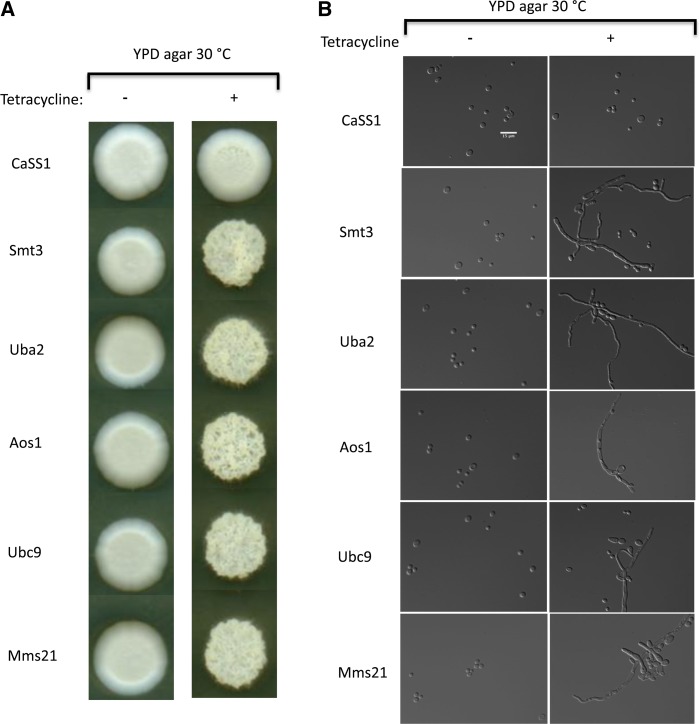
Sumoylation is involved in the processing of many proteins including elements involved in cellular differentiation (Zhou et al. 2004). We examined all members of the SUMO regulatory circuit in the GRACE collection for invasive growth and filament formation following their repression with tetracycline. All tetracycline-repressed strains containing members of the SUMO pathway were viable, and all showed invasiveness and filamentation, forming highly invasive wrinkled colonies on agar medium (Figure 2A). The cellular morphology of all the repressed mutants was highly filamentous, with cells displaying both hyphal and pseudohyphal phenotypes, while the parental strain cells remained in the yeast form (Figure 2B). The SUMO protein Smt3 and the E1 enzyme Aos1 have been previously implicated as repressors of filament formation (O’Meara et al. 2015), while Uba2 (a second E1), Ubc9 (an E2), and Mms21 (an E3) had not been previously reported; this suggests that the SUMO pathway as a whole has a critical role in the regulation of filamentous growth. Since we also found Smc6 and Qri2 of the Smc5/6 complex to be filamentation and invasiveness repressors, and Mms21 is also part of this complex, we investigated Mms21 as a key player of filamentation control.
Figure 2.
Members of the SUMO pathway negatively regulate filamentation. (A) Putative members of the sumoylation pathway in C. albicans were grown at 30° on YPD agar with and without 100 μg/ml tetracycline for conditional repression. Each mutant repression caused fuzzy colonies except for the wild-type CaSS1. (B) Representative DIC images of repressed and nonrepressed SUMO mutants at ×63 magnifications using a Leica DM 6000 microscope; mutant cells displayed filamentous (hyphal and pseudohyphal) morphology, while the wild-type CaSS1 showed normal yeast morphology. Bar, 15 μm. SUMO, small ubiquitin-like modifier.
Mms21 contains a conserved SUMO SP domain
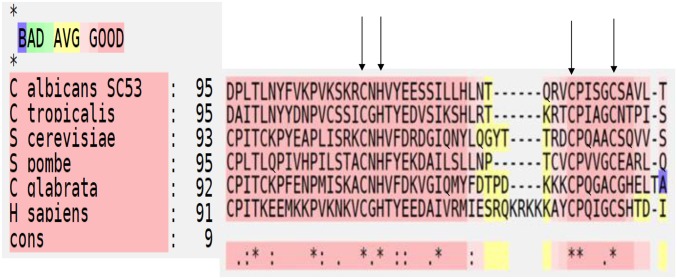
The C. albicans MMS21/C3_06200C_A gene resides on chromosome 3 (http://www.candidagenome.org/) and encodes a 272-amino acid protein that is 32% identical to its S. cerevisiae ortholog, 39% identical to S. pombe Nse2, and 32% identical to human Nse2. Mms21 contains an SP (Siz/PIAS) RING domain that is exclusively found in SUMO E3s and is involved in the final conjugation step of SUMO to the lysine of the substrate protein. The SP domain of C. albicans Mms21 is predicted to start at S192 and run until L239. As shown in Figure 3, the SP domain is highly conserved among different organisms when we performed a T-COFFEE (tree based consistency objective function For alignment evaluation) structural protein alignment (Armougom et al. 2006; Di Tommaso et al. 2011). This domain contains the motif CXH/CX4C, which is required for zinc ion (Zn2+) binding to create protein stability and is important for E3 ligase function (Yunus and Lima 2009).
Figure 3.
T-COFFEE (tree based consistency objective function For alignment evaluation) structural protein alignment (Expresso) of Mms21/Nse2 SP [Siz/Pias (protein inhibitor of activated signal transducer and activator of transcription)] domain among different organisms. The arrows indicate residue CXH/CX4C required for Zn2+ binding. The main score (91–95) is the total consistency value. Red regions represent conserved residues while asterisks are used to label residues that are identical in the consensus sequence. Color scheme of “BAD AVG GOOD” represents level of consistency between final alignment and the library used by T-COFFEE. A score of nine (dark red regions) represents consistency score of strong alignment.
Mms21-deleted cells are viable and highly invasive
Mms21 is an essential SUMO E3 ligase in both S. cerevisiae (Zhao and Blobel 2005) and S. pombe (McDonald et al. 2003).Mms21 is also part of the chromosome maintenance and the DNA repair Smc5/6 complex; some members of this complex (O’Meara et al. 2015) have been previously implicated in controlling filamentous growth in C. albicans. We have confirmed from our GRACE screening that inactivation of Mms21 derepressed invasiveness, suggesting that Mms21 may be a link between invasive growth and important cellular processes like chromosome maintenance, DNA repair, and sumoylation.
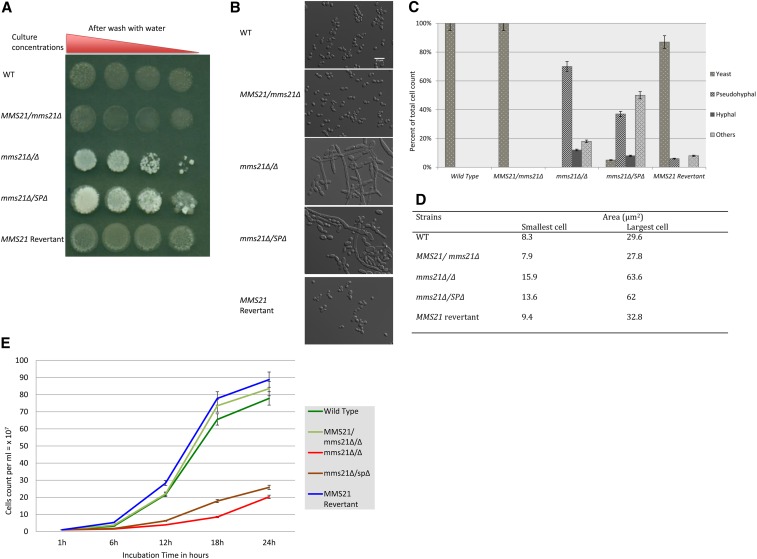
To confirm our finding of Mms21 as a nonessential protein in C. albicans required to repress invasiveness and filamentation, we generated a complete knockout of both the alleles in the strain SN148 using the standard two-step deletion approach. One copy of the gene was replaced with the selection marker HIS1 while the other was replaced by ARG4. The complete removal of both functional alleles of MMS21 and their replacement with the selection markers was confirmed by PCR. These mms21 Δ/Δ (null) mutants grew slowly in liquid YPD medium and formed small but highly invasive wrinkled colonies on YPD agar under standard yeast growing conditions (Figure 4A). A functional copy of MMS21 from the wild-type SN148 strain was reinserted into the RP10 locus of the mms21 Δ/Δ mutants; this recovered the normal smooth yeast phenotype from the mutant-specific wrinkly invasive phenotype. Thus, Mms21 is a nonessential protein in C. albicans, but a key player in morphogenesis control.
Figure 4.
Colony and cell morphologies of MMS21 mutants. (A) A 1:10 serial dilution of overnight culture of mutants was spotted on to YPD agar and then incubated at 30° for 4 days. Individual colonies were washed with a stream of water for 10 sec; mms21Δ/Δ and Mms21 SP-deleted mutants were found to be invasive. (B) DIC images of mutants grown under yeast conditions at ×63 magnification; Bar, 15 μm. The mms21Δ/Δ and mms21Δ/SPΔ mutants have displayed psuedohyphae, hyphae, and others (large cells nearly identical to gut, gray, or opaque cells), while the MMS21 revertant strain recovered the WT yeast phenotype. (C) A clustered column graph showing the percentage of different cell morphologies displayed by mutants. The mms21Δ/Δ and mms21Δ/SPΔ mutants have increased filamentous phenotypes. (D) A table of the cell-size area of the log-phase growing strains measured in the DAPI channel using ImageJ Fiji 1.0. (E) A graph showing cell counts of mutants and WT cultures in YPD at 30° for 24 hr. The initial cell count was started at 1 × 107 cells/ml. Both mms21 mutants doubled their first cell population at the 8-hr mark, while the revertant and the WT doubled every 2 hr. Error bars are based on the SD from two biological replicates of each data point reading taken every 6 hr. SP, Siz-/Pias (protein inhibitor of activated signal transducer and activator of transcription) domain; WT, wild-type.
Mms21 SP domain-deleted mutants are also invasive
Our GRACE screening suggested an important role for sumoylation in repressing invasiveness and filamentation. To test whether specific E3 ligase catalytic activity is involved in this control, we replaced the 92 amino acids of the C-terminal SP domain of the functional MMS21 allele in a deletion heterozygote strain with a C-terminal TAP tag and an ARG4 selectable marker. The mms21Δ/SPΔ mutant was confirmed by both PCR and western blot analysis. These mms21Δ/SPΔ mutants also formed highly invasive and slow-growing small wrinkled colonies on YPD agar plates under yeast growth conditions (Figure 4A), suggesting that Mms21-mediated sumoylation plays a major role in C. albicans invasive growth control.
mms21Δ/Δ and mms21Δ/SPΔ cells displayed yeast to filamentous phenotypes, increased cell size, and cell cycle-related defects
A structurally and enzymatically functional Mms21 is required for normal growth. Mms21 deletion in S. cerevisiae, S. pombe, and humans is lethal, while mutations in its C-terminal SP domain causes severe growth defects (Andrews et al. 2005; Zhao and Blobel 2005; Hoch et al. 2008). Moreover, mutations in the N-terminal region of the S. cerevisiae Mms21 affected its interaction with Smc5 (Smc5/6 complex) and also caused growth defects (Duan et al. 2009a). We have found that the mms21Δ/Δ and mms21Δ/SPΔ cells displayed abnormal phenotypes at 30° when cultured in liquid YPD. Individual mutant cells were morphologically aberrant, ranging from enlarged and elongated yeast-like cells to pseudohyphal or hyphal cells (Figure 4B). There were 12% hyphal, 70% pseudohyphal, and 18% abnormal (large irregular yeast-like) phenotypic variants in the mms21Δ/Δ mutants, with none of the cells displaying true normal yeast morphology despite growing at 30° (Figure 4C). The mms21Δ/SPΔ mutants, on the other hand, were 8% hyphal, 37% pseudohyphal, 50% abnormal, and 5% yeast-type. In the wild-type background, 100% of the cells exhibited the normal yeast form. In both the complete deletion and the SP-deleted strains, the mutant cells grew as clusters of attached cells that were difficult to dissociate. We measured and compared (see Materials and Methods) the cell size of the mutants in log-phase yeast growth conditions. The yeast-form-like mms21Δ/Δ cells ranged from 15.9 to 63.6 μm2 in size, while mms21Δ/SPΔ yeast-like cells were in the range of 13.6–62 μm2 (Figure 4D). The size of these mutants was nearly twice the size of the wild-type and the mms21 revertant strain cells, which ranged from 7.9 to 32.8 μm2. Thus, either Mms21 deletion or its SP domain’s removal has a significant impact on the cellular morphology of C. albicans.
We measured the cell density of cultures of these mutants after a 24-hr incubation period by counting the number of cells using a hemocytometer to avoid issues caused by cell filamentation during spectrophotometric growth measurements. We used a starting cell count of 1 × 107 cells/ml. The mms21Δ/SPΔ mutants reached 25 × 107 cells/ml while the mms21Δ/Δ mutants acquired cellular densities of only 20 × 107 cells/ml after 24-hr incubation at 30° in liquid YPD. The wild-type and the revertant strains reached much higher cell densities, near or above 80 × 107 cells/ml (Figure 4E). Furthermore, measuring cell densities using a spectrophotometer (OD 600 nm) revealed that the wild-type strain reached an OD of 1 from 0.1 within 6 hr of incubation under yeast growth conditions, while mms21Δ/Δ mutants took ∼14 hr to acquire an OD of 1. These data highlight the impact of a structural and enzymatically functional Mms21 on the growth of C. albicans.
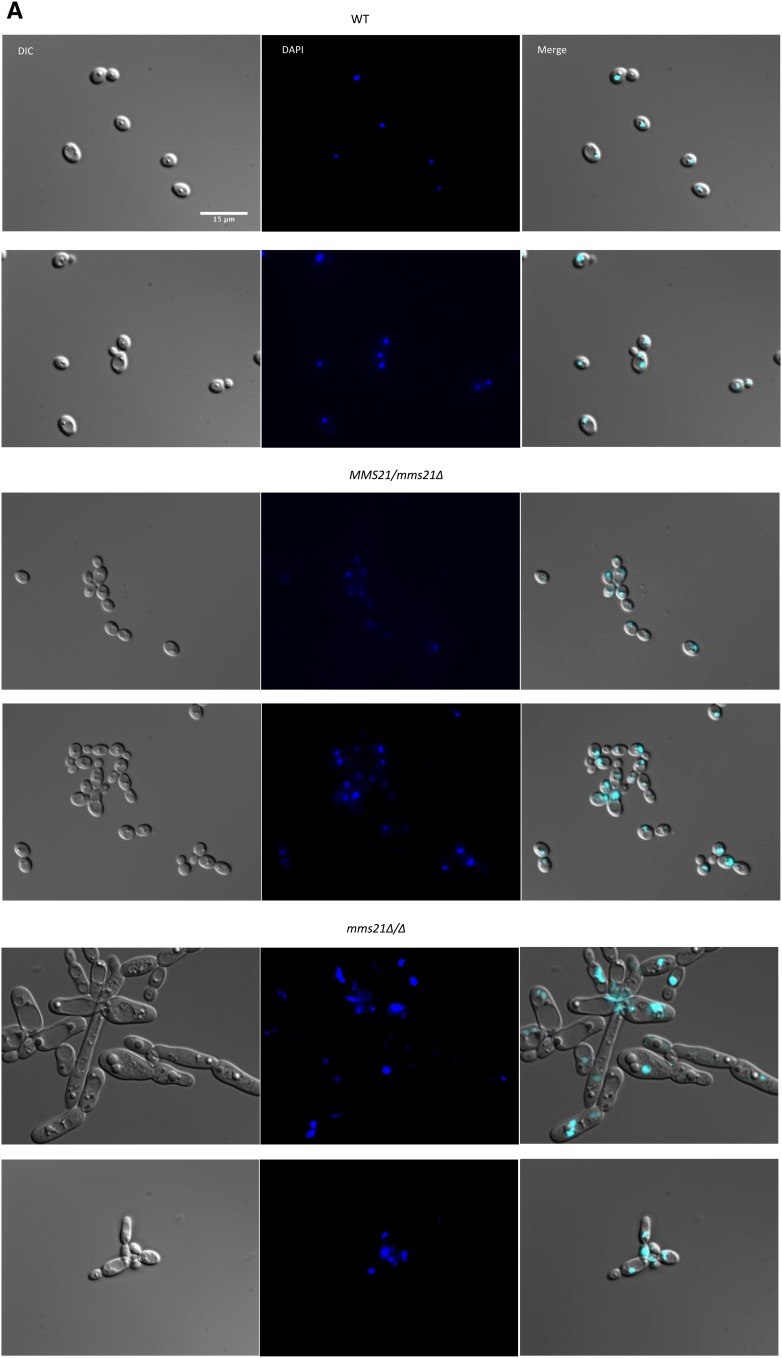
In S. cerevisiae, the SUMO E3 ligase activity encoded by MMS21 is essential for nuclear integrity and the proper organization of telomeres (Zhao and Blobel 2005). Deletion of the C. albicans SUMO protein Smt3 resulted in significant nuclear segregation and chitin distribution defects (Leach et al. 2011). To examine the effects of Mms21 null and its SP domain deletion on nuclear segregation, cells (n = 200 for each) of the mutant and wild-type strains were stained with DAPI and observed under a microscope. Interestingly, both null and SP-deleted mutants showed variable nuclei within a high proportion of cells (Figure 5A). In the case of the wild-type strain, 92% (n = 62) of large-budded cells have two nuclei, one in each bud and mother cell, while 91% (n = 62) of small-budded cells have nuclei at the junction of mother and bud cell separation. For the heterozygote strain with one functional MMS21 allele, similar numbers were observed, i.e., 94% (n = 68) for large-budded cells and 91% (n = 66) for small-budded cells, respectively. Both these strains showed a normal pattern of yeast cell morphogenesis where large-budded cells with two nuclei were in late M phase, while small-budded cells were in late G2 phase of the cell cycle (Berman 2006). Interestingly, both null and SP-deleted mutants showed variable defects in nuclear segregation, with large-budded cells containing only one nucleus either in the mother or in the bud cell, or nonbudded cells having two nuclei prior to bud formation. Unlike the wild-type cells with more prominent circular nuclei, a large number of mutant cells have irregularly shaped nuclei (Figure 5A). These abnormal nuclear variations were 68% (n = 136) in null and 47% (n = 96) in SP-deleted mutant cells, respectively, indicating that cells might be in late S/G2 arrest or have a defective M cycle, perhaps due to a failure in the DNA repair machinery causing abnormal nuclear content and a respective increase in cell size.
Figure 5.
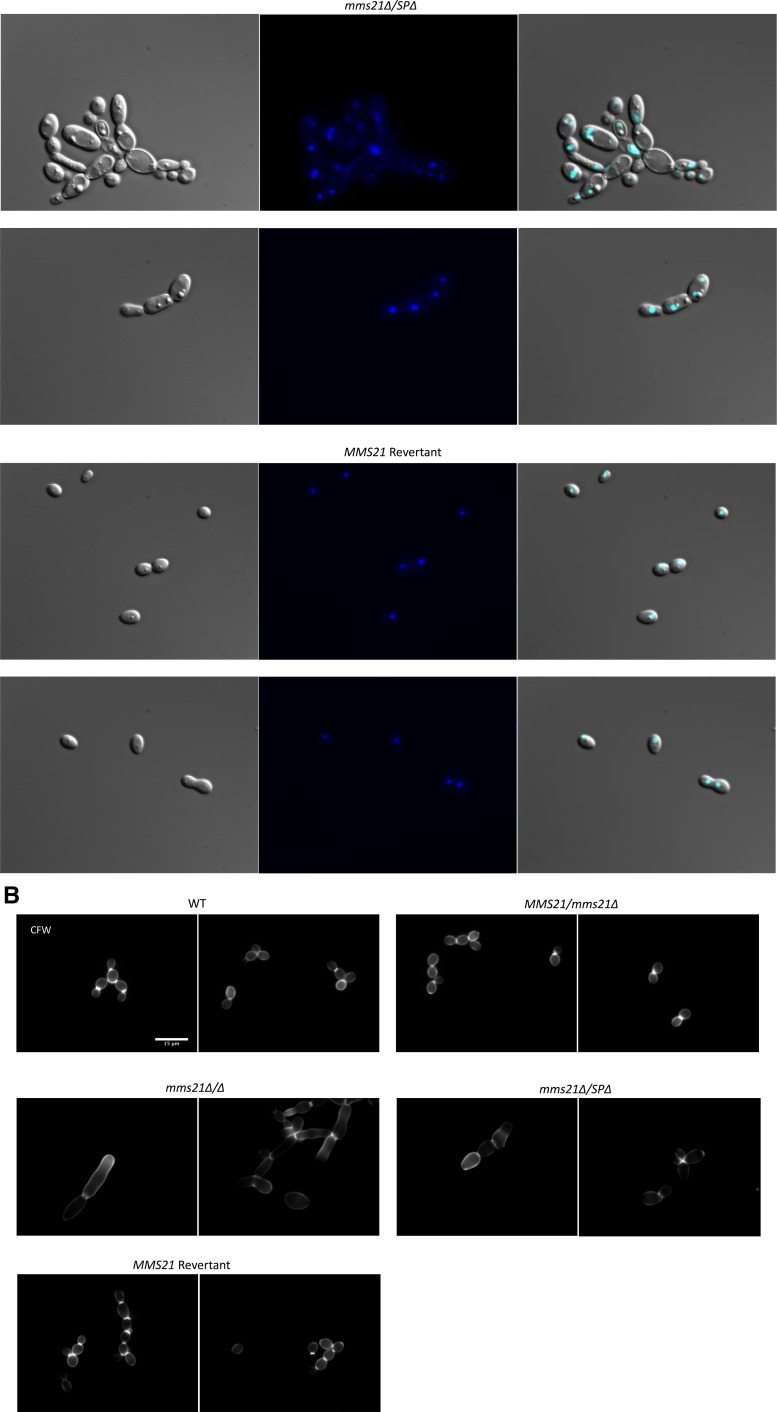
Staining of the MMS21 mutants with DAPI and calcoflour. Cultures were allowed to grow for 3.5 hr for WT and revertant strains, and 6 hr for mutants under yeast growth conditions, and then stained with 2 μg/ml DAPI or 2 μg/ml calcoflour. Individual cells were examined under ×100 magnification using a Leica DM 6000 microscope, Bar, 15 μm. Only (more or less) yeast-looking cells of mutants were considered for better comparison to the WT. (A) WT cells and strains with one WT allele of MMS21 showed normal nuclear segregation. The mms21Δ/Δ and mms21Δ/SPΔ mutants displayed considerable variation in the number of nuclei present in the individual cells. (B) Calcoflour-stained cells where WT cells display even chitin distributions and prominent septa, while both mms21 mutants have abnormal chitin composition and inconspicuous septa. Reinsertion of Mms21 into the null mutant resulted in highly noticeable septa. WT, wild-type.
Septal ring formation is central to faithful nuclear segregation between mother and daughter cells (Berman 2006). We tested all of our strains for the chitin composition of the cell wall and septa using calcofluor staining. All of the wild-type cells (n = 214) showed normal chitin distribution in their cell walls and at septal junctions. The null and SP-deleted mutant cells displayed uneven chitin distribution, while the septum separating a bud and a mother cell was not as prominent and distinct as in wild-type cells (Figure 5B). The null mutants had 46% (n = 201) while SP-deleted cells had 40% (n = 194) of the cells showing chitin and septal abnormalities, suggesting that septa and chitin deposition were perturbed in the mutants rather than occurring at the start of nuclear and cellular division. Overall, the modifications in chitin deposition and in nuclear structure are not simply representative of the enhanced level of filamentation, but are likely a direct consequence of loss of MMS21 function, as filamenting wild-type cells do not show the abnormalities seen in the mms21 mutants. These results are consistent with previous findings that the Mms21 protein is required for proper nuclear and cell segregation in budding yeast. All these defects in cellular morphology (slow growth, enlarged cell size, filamentation, and abnormal nuclear segregation) were eliminated by complementing the null mutant with a functional copy of MMS21, showing that Mms21 is required for normal cellular physiology in C. albicans.
Mms21 is required for the genotoxic and cellular stress responses
MMS21 was first identified as a gene in S. cerevisiae whose inactivation conferred sensitivity to MMS, X-rays, and UV radiation (Prakash and Prakash 1977). In yeast and other organisms, Mms21 has been found to be part of the Smc5/6 DNA repair complex, where Mms21 is required for sumoylating other complex members (such as Smc5) as well as for providing structural stability to the complex (McDonald et al. 2003; Andrews et al. 2005; Potts and Yu 2005; Zhao and Blobel 2005). Genotoxic stress compromises the genome integrity of an organism, and in C. albicans defects in response to such stresses have been implicated in increased polarized growth and white–opaque switching (Bachewich et al. 2005; Shi et al. 2007; Alby and Bennett 2009). In C. albicans, several proteins have been reported to be sumoylated during hydrogen peroxide-induced oxidative stress (Leach et al. 2011). As our phenotypic screening revealed that there is an increase in filamentation in Mms21-deleted mutants, we assessed whether these mutants were defective in response to different types of stress.
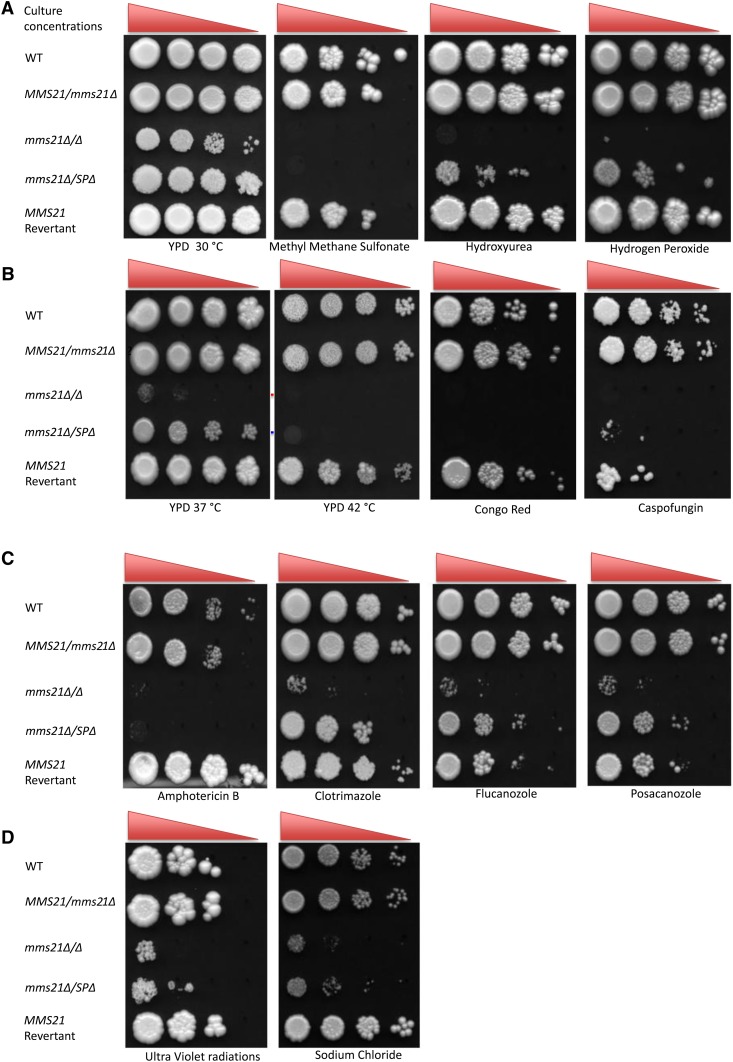
We assayed the response of mms21Δ/Δ and mms21Δ/SPΔ cells to genotoxic stress-causing agents including the alkylating agent MMS, the DNA replication-stalling chemical HU, the oxidative stress agent hydrogen peroxide, and thymine dimer-causing UV radiation (Figure 6, A and D). Both mms21 null and SP-deleted mutants were highly sensitive to MMS and UV, showing that the loss of the Mms21 protein or its putative SUMO SP domain function blocks cells from recovering from MMS- and UV-induced cellular stress. The null mutant, but not the SP-deleted mutant, never recovered from HU- and hydrogen peroxide-induced stresses, highlighting the importance of Mms21 for a potential structural role in maintaining the Smc5/6 complex as intact and fully functional for its subsequent DNA damage response. The reinsertion of the MMS21 gene into the null mutant resulted in the wild-type response to these stresses.
Figure 6.
Genotoxic and cellular stress assay. A 1:10 serial dilution of overnight cultures grown in yeast growth conditions were spotted (highest cell concentrations being 1 × 106 cells/ml and lowest being 1 × 103 cells/ml) onto YPD agar plates containing different chemicals and were incubated at 30° for 4 days except for caspofungin (7 days). (A) strains were subjected to genotoxic stress of MMS (0.01% v/v), hydroxyurea (10 mM), and hydrogen peroxide (5 mM). (B) Strains response to heat (37 and 42°) and cell wall stress caused by Congo red (150 μg/ml) and caspofungin (0.75 μg/ml). (C) Determination of resistance to different cell membrane-damaging drugs: amphotericin B (1 μg/ml), clotrimazole (0.5 μg/ml), flucanozole (10 μg/ml), and posacanozole (10 μg/ml). (D) sensitivity to UV radiation and NaCl (1 M). Experiment was repeated three times for each sample for consistent results. WT, wild-type.
Global sumoylation analysis has revealed several sumoylated proteins, including various heat shock proteins in both S. cerevisiae and C. albicans (Zhou et al. 2004; Leach et al. 2011). The null mutants and cells lacking the SP domain necessary for E3 activity were unable to grow at 42°, consistent with Smt3 being implicated in sumoylating heat shock proteins in C. albicans. As well as not growing at 42°, cells lacking both alleles of Mms21 showed extremely poor growth at 37° (Figure 6B). The fact that mms21Δ/MMS21 and the Mms21 revertant grew normally at 37° implies that at least one fully functional allele of Mms21 gene may be required for C. albicans to thrive under normal human body temperature.
We also surveyed the effects of different cellular stress-causing agents in Mms21-defective mutants. The cell wall is the first line of defense for most of unicellular organisms, including bacteria and fungi. The cell wall of C. albicans contains an estimated 2–6% chitin, which is very important for the rigidity of the cell to resist damaging external factors (Ruiz-Herrera et al. 2006). Congo red and the antifungal drug caspofungin cause cell wall stress by targeting glucan and chitin synthase (Roncero and Duran 1985; Ghannoum and Rice 1999). The mms21 null mutant was highly sensitive to both Congo red and caspofungin (Figure 6B). Mutants were also tested against cell membrane stress-causing ergosterol inhibitors (Ghannoum and Rice 1999). The null and SP-deleted mutants were highly sensitive to amphotericin B as well to ergosterol biosynthesis inhibitors including fluconazole, clotrimazole, and posaconazole (Figure 6C). Moreover, both mutants also grew slowly under salt stress (Figure 6D). Altogether, these results show that Mms21 plays important roles in C. albicans physiology by helping the pathogen to cope with genotoxic and cellular stresses.
Expression profiling of mms21Δ/Δ
Since the null mms21 mutant has a significant effect on the physiology of C. albicans through an increase in invasiveness and filament formation, and displays slower growth rates, aberrant nuclear segregation, and weak stress responses, we assayed the overall differences in gene expression between the mms21Δ/Δ mutant and the wild-type under yeast growth conditions using custom microarrays. Using a statistical significance analysis with a P-value < 0.05 and a cutoff of twofold, we have identified 373 genes whose levels of expression were significantly affected in MMS21 inactivation; 151 genes were up- and 222 were downregulated (Table S4). The majority of the upregulated genes were involved in ribosome and ribonucleoprotein biogenesis, RNA processing, and the response to reactive oxygen species, while many were associated with nucleic acid and heterocycle metabolic processes. Around 15% of the upregulated genes were associated with C. albicans filamentation and virulence. Among the downregulated genes were genes involved in oligopeptide transport, hexose transport, the stress response, nitrogen utilization, oxidation–reduction processes, and fatty acid metabolism. An increase in the expression of ribosomal protein genes, and a decrease in the expression of oligopeptide and hexose transport genes, have been implicated in C. albicans infection of epithelial tissues (Spiering et al. 2010).
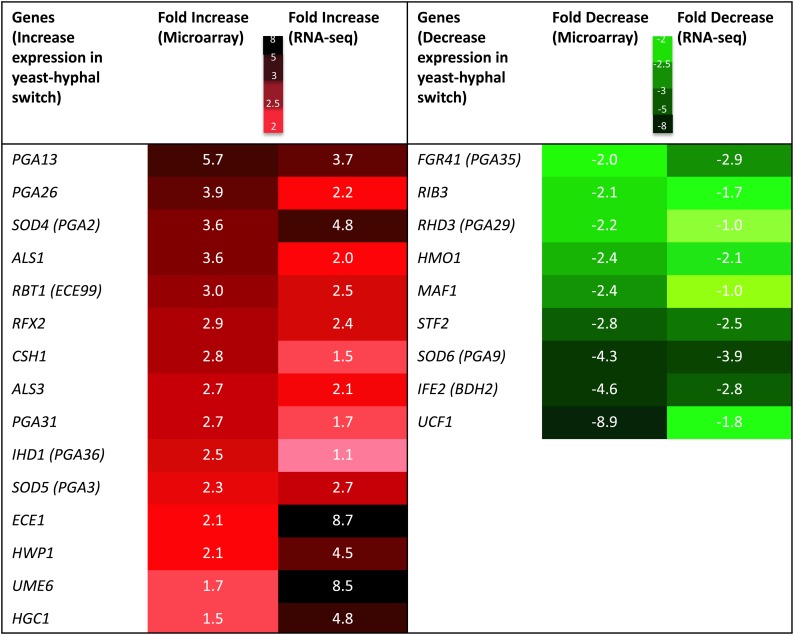
There are a number of significant genes that are up- and downregulated during C. albicans yeast–hyphal transition (Nantel et al. 2002; Kadosh and Johnson 2005), and when the cells are grown under hypoxia (Stichternoth et al. 2011). We have found many genes that increase or decrease in their expression correlated with the yeast–hyphal transition and virulence (Figure 7). Important genes in this list include PGA13, PGA26, PGA31, ALS1, ALS3, ECE1, HWP1, SOD4, and SOD5. The genes for two important glycosyl phosphatidyl inositol) mannoproteins that constitute 30–40% of the cell wall by weight (Pga13 and Pga26) were highly upregulated in our mutant (5.8- and 3.9-fold, respectively); these are key players in C. albicans morphogenesis and virulence (Laforet et al. 2011; Gelis et al. 2012). Similarly, there is upregulation of genes for Hwp1 (hyphal wall protein 1), Als1 (agglutinin like sequence 1), and Ece1 (extent of cell elongation1), all proteins that are highly important for fungal adherence to the host cells, the development of hyphae, and virulence (Staab et al. 1999; Tsuchimori et al. 2000; Sheppard et al. 2004; Moyes et al. 2016). The expression of the SOD5 (superoxide dismutase 5) gene was 2.3-fold upregulated in the mms21 null strain and this gene is also upregulated during hyphal development (Martchenko et al. 2004). This increase in expression of SOD5 could be related to the adaptation to the reactive oxygen species in the absence of Mms21.
Figure 7.
A list of genes that are highly significant for hyphal development and virulence, and their fold increase and decrease in expression in mms21 null against wild-type cells using microarray and RNA-sequencing analysis (RNA-seq). Color coding is used to highlight an increase or decrease in gene expression. Dark red regions represent highly increased gene expression and dark green regions represent highly decreased gene expression.
We followed up on our microarray profiling by performing whole-genome transcriptome sequencing (RNA-seq) analysis of mms21Δ/Δ and wild-type cells under yeast growth conditions. By using a P-value of < 0.05 and fold cutoff of 2.0, we found 188 up- and 97 downregulated genes (Table S5). Analysis using the Candida Genome Database Gene Ontology Slim Mapper revealed that, among upregulated genes, 21.3% represent RNA metabolic process and 16% represent genes involved in ribosome biogenesis. Among the downregulated genes, 17.5% were related to carbohydrate metabolic processes, 16.5% to transport, and 14% to stress responses. Our RNA-seq data were consistent with our microarray data for the aforementioned HSG expression analysis (Figure 7). Furthermore, the RNA-seq analysis revealed Ume6, a true hyphae transcription factor (Zeidler et al. 2009), to be more than eightfold upregulated, and Hgc1, a hyphal-specific cyclin (Zheng et al. 2004), to be 4.8-fold upregulated.
Transcription profiling also revealed that several genes of the stress response machinery were downregulated, including DAP1, RAD2, RAD4, BUB3, and HNT2, which supports our findings of reduced genotoxic and cellular stress responses in null mms21 mutants. Overall, our transcription data suggest that in the absence Mms21, there is a profound change in the expression levels of several important genes that contribute to C. albicans morphogenesis and virulence.
Filamentation in mms21Δ/Δ is independent of Efg1
Efg1 is required for hyphal protein expression under hyphae-inducing conditions (Sohn et al. 2003). Intriguingly, our microarray data show no change in the level of expression of this important filamentation transcription factor in the null mms21 mutant. Efg1 lies upstream of Hwp1 and its deletion eliminates the yeast–hyphal transition (Sharkey et al. 1999). To test whether the filamentation exhibited by the mms21 null mutant is independent of Efg1, we deleted MMS21 in an efg1Δ/Δ strain as previously described (Regan et al. 2017). As shown in Figure S1, the efg1Δ/Δ mutant cells were locked in the yeast form, but deleting both alleles of MMS21 (efg1Δ/Δ mms21Δ/Δ) resulted in filament formation as in the mms21 null cells. Therefore, filamentation caused by the absence of Mms21 is independent of the pathway controlled by Efg1.
Discussion
We have characterized, as a component of invasiveness, the C. albicans ortholog of Mms21, initially identified as part of the essential DNA damage repair SMC5/6 complex of yeast and also as an E3 ligase of the sumoylation pathway in humans, S. cerevisiae, and S. pombe (McDonald et al. 2003; Andrews et al. 2005; Potts and Yu 2005; Zhao and Blobel 2005). We found MMS21 to be nonessential in C. albicans, which is consistent with the findings that the unique SUMO protein-encoding locus SMT3 is nonessential as well (Leach et al. 2011). Although the SUMO pathway is dispensable in C. albicans, the loss does generate growth-related defects. Either deleting MMS21 or removing its active site domain directs C. albicans to form invasive filaments under normal yeast growing conditions. Intriguingly, microarray analysis of the mms21Δ/Δ mutant compared with wild-type cells under yeast growth conditions revealed two- to sixfold increases in the expression of several genes involved in filamentation and virulence. Previously, a large-scale homozygous mutant study in C. albicans had found that mms21-deleted mutants showed normal competitive fitness in mice (Noble et al. 2010), though they did not detect changes in cell morphology upon deletion.
The yeast–hyphal (and hyphal–yeast) transition in C. albicans can be triggered by both environmental conditions and internal stimuli. There are a number of filamentation transcription factors and repressors that are important for this transition. The hyphal G cyclin 1 (Hgc1) is a key protein involved in hyphal induction (Zheng et al. 2004); this protein is regulated by the cyclic AMP pathway, which is one of the key pathways controlling hyphal morphogenesis (Harcus et al. 2004; Lu et al. 2013). This pathway also involves transcription factor Efg1 and is repressed by Nrg1-Tup1 (Kadosh and Johnson 2005). Ume6 is another transcription factor that is required for true hyphal elongation, as it acts downstream of Efg1 and other activators like Czf1 and Cph1, while it is also repressed by Nrg1-Tup1 (Zeidler et al. 2009). We found that the null mms21 mutant formed filaments in the absence of Efg1, and our microarray and RNA-seq data also showed no significant changes in the expression of Efg1, Czf1, Cph1, and the repressors Nrg1-Tup1 in the null mutant. However, Hgc1 and Ume6 were both upregulated, indicating that these two proteins and the HSGs were transcriptionally induced independently of Efg1. The activated Ume6 in particular may have derepressed the HSGs, thereby affecting the cell wall architecture to form hyphae as previously proposed (Zeidler et al. 2009).
In S. cerevisiae, Mms21 is part of the DNA repair Smc5/6 complex and mutations in this complex result in G2/M phase cells arrest (Menolfi et al. 2015). Moreover, the Smc5/6 complex interacts and sumoylates the DNA damage repair Rad52 protein for recombination repair at rDNA (Torres-Rosell et al. 2007). In C. albicans, Rad52 deletion resulted in filamentation with cells arresting in the G2/M phase (Andaluz et al. 2006). Since an unperturbed Smc5/6 complex is required to regulate the Rad52 protein and subsequent recombination, it is possible that Mms21 deletion is affecting this regulation, thus resulting in G2/M cycle arrest. In S. cerevisiae, it has been proposed that extending the G2 phase or delaying the formation of the active cyclin-dependent kinase complex would result in pseudohyphae formation (Lew and Reed 1995). Therefore, this increased filamentation in the absence of Mms21 appears to be a result of defects in cell division and the DNA damage repair machinery, as seen for other genes (Bachewich et al. 2003; Bachewich and Whiteway 2005). In fact, often the input signals that cause filamentation impose some sort of cell stress, whether these signals are changes in temperature or pH, or use of N-acetylglucosamine or serum (Brown and Gow 1999). Mms21-deleted cells are also under stress, thus inducing filamentation response genes and eventual filamentation.
Besides the hyphal and pseudohyphal morphology displayed by the mutant strains, many of the cells were enlarged, elongated, and irregularly shaped (Figure 4, B and C), and growing at a slow rate when compared to the wild-type. We also observed that many of these mutant cells were multinucleated, suggesting a defect in proper nuclear segregation (Figure 5A). Similar defects had been previously attributed to improper SUMO ligase activity of Mms21 in other organisms (Andrews et al. 2005; Sergeant et al. 2005; Zhao and Blobel 2005; Duan et al. 2009a). Moreover there were abnormal chitin contents and inconspicuous septa in these mutant cells (Figure 5B). These defects in our mutants likely arise from the inability of these cells to carry out proper sumoylation of the septin system, as in S. cerevisiae (Johnson and Blobel 1999); specific septin-related proteins, although not the septins themselves, have been reported to interact with the SUMO Smt3 in C. albicans (Martin and Konopka 2004; Leach et al. 2011). Mms21 is also part of the DNA repair Smc5/6 complex, which is required for the timely removal of DNA-mediated sister chromatid linkages in S. cerevisiae (Bermudez-Lopez et al. 2010). All these abnormal cellular morphologies could have arisen due to mutant cell problems in late G2 or M phase of the cell cycle, because mutations in Mms21 caused defects in the Smc5/6 complex and in the sumoylation pathway.
We have observed that both mms21Δ/Δ and mms21Δ/SPΔ mutants showed impaired responses to different genotoxic stress-causing agents (Figure 6, A and D). The Smc5/6 complex in humans repairs DNA double-strand breaks by homologous recombination, by recruiting the Smc1/3 cohesin complex to the sites of double-strand breaks (Potts et al. 2006). Both mms21Δ/Δ and mms21Δ/SPΔ mutants were highly sensitive to both DNA-alkylating MMS and UV radiation, indicating that not only is the whole functional Mms21 domain required to overcome the DNA damage caused by MMS and UV, but also its active C-terminal SP domain. These findings were consistent with previous reports that the SUMO ligase function of Mms21 is required for DNA damage repair in S. cerevisiae (Zhao and Blobel 2005; Rai et al. 2011). In the case of other genotoxic stresses, the null mms21 mutant cells were more sensitive to the genotoxic stress than the mutation that was specifically defective in the sumoylation active site. This is possibly because the N-terminus of Mms21 is interacting with Smc5 in the Smc5/6 complex and this interaction (to provide structural stability to the complex) is required for the normal DNA repair function of the Smc5/6 complex. Compromise of this interaction has been found to be lethal in S. cerevisiae, while mutating the Mms21 C-terminus containing the E3 ligase domain has less severe consequences (Duan et al. 2009a). Based on our results and previous findings that the Smc5/6 complex is required to cope with genotoxic stresses, we suggest that the structural stability role of Mms21 is more important than its E3 ligase activity in cellular recovery from different types of genotoxic stresses.
The mms21Δ/Δ and mms21Δ/SPΔ mutants were found to be highly sensitive to a temperature of 42° (Figure 6B). Leach et al. (2011) showed several heat shock proteins to be sumoylation targets in C. albicans, including Hsp60, Hsp104, Ssc1, Ssb1 and Sse1. We detected no temperature sensitivity when we deleted Siz1, a second putative SUMO E3 ligase (J. Feng, A. Islam and M. Whiteway, unpublished data). There is thus a possibility that these proteins are modified via Mms21-mediated sumoylation to help C. albicans cope with increasing temperatures. Moreover, Mms21 was also found to be important for the tolerance of C. albicans to amphotericin B and ergosterol biosynthesis inhibitors, thus maintaining the cell membrane permeability and pathogen resistance to these drugs.
Overall, from our GRACE library screen, we have found Mms21 to be nonessential and a repressor of invasiveness. We have further investigated the roles of Mms21 in C. albicans by deleting both the entire gene as well its C-terminal SUMO SP domain. By creating mms21 null (mms21 Δ/Δ) and Mms21 SP domain-deleted (mms21Δ/SPΔ) mutants, we have found out that the full Mms21 protein and its C-terminal SUMO activity act as repressors of invasiveness and filamentation. Both these mutants have also shown cell cycle-related defects, with increases in division time and in defective nuclear segregation. Furthermore, we have shown that Mms21 is critical for the C. albicans response to genotoxic stresses, and thus plays a vital role in preserving genome integrity. Mms21 is also important for the normal response of cells to cell wall, cell membrane, and thermal stresses. Our data show that Mms21 plays important roles in cellular differentiation, and genotoxic and cellular stress responses, thus contributing to the pathogenicity and survival of this opportunistic fungal pathogen. It will be exciting to identify the molecular targets of Mms21-mediated sumoylation and establish how they impact on these various cellular processes.
Acknowledgments
We thank Adnane Sellam and Yuan Sun for help with data analysis, and Chloe Triplet van Oostende and Chris Law at Concordia University Centre for Microscopy and Cellular Imaging for help with microscopy. This work was supported by Canadian Institutes of Health grant MOP 42516, and a Natural Sciences and Engineering Research Council Canada Research Chair award Tier 1 950-228957 and a Discovery award RGPIN/4799 to M.W.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7385114.
Communicating editor: A. Mitchell
Literature Cited
- Alby K., Bennett R. J., 2009. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20: 3178–3191. 10.1091/mbc.e09-01-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaluz E., Ciudad T., Gomez-Raja J., Calderone R., Larriba G., 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 59: 1452–1472. 10.1111/j.1365-2958.2005.05038.x [DOI] [PubMed] [Google Scholar]
- Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., et al. , 2005. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25: 185–196. 10.1128/MCB.25.1.185-196.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom F., Moretti S., Poirot O., Audic S., Dumas P., et al. , 2006. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34: W604–W608. 10.1093/nar/gkl092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C., Whiteway M., 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4: 95–102. 10.1128/EC.4.1.95-102.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C., Thomas D. Y., Whiteway M., 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14: 2163–2180. 10.1091/mbc.02-05-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C., Nantel A., Whiteway M., 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57: 942–959. 10.1111/j.1365-2958.2005.04727.x [DOI] [PubMed] [Google Scholar]
- Berman J., 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9: 595–601. 10.1016/j.mib.2006.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Lopez M., Ceschia A., de Piccoli G., Colomina N., Pasero P., et al. , 2010. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 38: 6502–6512. 10.1093/nar/gkq546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., Gow N. A., 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7: 333–338. 10.1016/S0966-842X(99)01556-5 [DOI] [PubMed] [Google Scholar]
- Calderone R. A., Fonzi W. A., 2001. Virulence factors of Candida albicans. Trends Microbiol. 9: 327–335. 10.1016/S0966-842X(01)02094-7 [DOI] [PubMed] [Google Scholar]
- Csank C., Schroppel K., Leberer E., Harcus D., Mohamed O., et al. , 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66: 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignard D., El-Naggar A. L., Logue M. E., Butler G., Whiteway M., 2007. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot. Cell 6: 487–494. 10.1128/EC.00387-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., et al. , 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39: W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Sarangi P., Liu X., Rangi G. K., Zhao X., et al. , 2009a. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell 35: 657–668. 10.1016/j.molcel.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Yang Y., Chen Y. H., Arenz J., Rangi G. K., et al. , 2009b. Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5–6 subcomplex and the Hinge regions of Smc5 and Smc6. J. Biol. Chem. 284: 8507–8515. 10.1074/jbc.M809139200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felk A., Kretschmar M., Albrecht A., Schaller M., Beinhauer S., et al. , 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70: 3689–3700. 10.1128/IAI.70.7.3689-3700.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., 2007. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast 24: 961–976. 10.1002/yea.1512 [DOI] [PubMed] [Google Scholar]
- Gelis S., de Groot P. W., Castillo L., Moragues M. D., Sentandreu R., et al. , 2012. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 49: 322–331. 10.1016/j.fgb.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A., Rice L. B., 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12: 501–517. 10.1128/CMR.12.4.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gola S., Martin R., Walther A., Dunkler A., Wendland J., 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20: 1339–1347. 10.1002/yea.1044 [DOI] [PubMed] [Google Scholar]
- Gong L., Kamitani T., Fujise K., Caskey L. S., Yeh E. T., 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272: 28198–28201. 10.1074/jbc.272.45.28198 [DOI] [PubMed] [Google Scholar]
- Harcus D., Nantel A., Marcil A., Rigby T., Whiteway M., 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15: 4490–4499. 10.1091/mbc.e04-02-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch N. C., Santos R. S., Rosa R. M., Machado R. M., Saffi J., et al. , 2008. Allelism of Saccharomyces cerevisiae gene PSO10, involved in error-prone repair of psoralen-induced DNA damage, with SUMO ligase-encoding MMS21. Curr. Genet. 53: 361–371. 10.1007/s00294-008-0192-z [DOI] [PubMed] [Google Scholar]
- Jacobsen I. D., Wilson D., Wachtler B., Brunke S., Naglik J. R., et al. , 2012. Candida albicans dimorphism as a therapeutic target. Expert Rev. Anti Infect. Ther. 10: 85–93. 10.1586/eri.11.152 [DOI] [PubMed] [Google Scholar]
- Johnson E. S., 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73: 355–382. 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G., 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272: 26799–26802. 10.1074/jbc.272.43.26799 [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G., 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147: 981–994. 10.1083/jcb.147.5.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D., 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16: 2903–2912. 10.1091/mbc.e05-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforet L., Moreno I., Sanchez-Fresneda R., Martinez-Esparza M., Martinez J. P., et al. , 2011. Pga26 mediates filamentation and biofilm formation and is required for virulence in Candida albicans. FEMS Yeast Res. 11: 389–397. 10.1111/j.1567-1364.2011.00727.x [DOI] [PubMed] [Google Scholar]
- Lavoie H., Sellam A., Askew C., Nantel A., Whiteway M., 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9: 578 10.1186/1471-2164-9-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Stead D. A., Argo E., Brown A. J., 2011. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Mol. Biol. Cell 22: 687–702. 10.1091/mbc.e10-07-0632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I., 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129: 739–749. 10.1083/jcb.129.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., et al. , 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949. 10.1016/S0092-8674(00)80358-X [DOI] [PubMed] [Google Scholar]
- Lu Y., Su C., Solis N. V., Filler S. G., Liu H., 2013. Synergistic regulation of hyphal elongation by hypoxia, CO(2), and nutrient conditions controls the virulence of Candida albicans. Cell Host Microbe 14: 499–509. 10.1016/j.chom.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M., Alarco A. M., Harcus D., Whiteway M., 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15: 456–467. 10.1091/mbc.e03-03-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martin S. W., Konopka J. B., 2004. SUMO modification of septin-interacting proteins in Candida albicans. J. Biol. Chem. 279: 40861–40867. 10.1074/jbc.M406422200 [DOI] [PubMed] [Google Scholar]
- McDonald W. H., Pavlova Y., Yates J. R., III, Boddy M. N., 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 278: 45460–45467. 10.1074/jbc.M308828200 [DOI] [PubMed] [Google Scholar]
- Menolfi D., Delamarre A., Lengronne A., Pasero P., Branzei D., 2015. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 60: 835–846. 10.1016/j.molcel.2015.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F., 2008. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30: 610–619. 10.1016/j.molcel.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Moyes D. L., Wilson D., Richardson J. P., Mogavero S., Tang S. X., et al. , 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532: 64–68. 10.1038/nature17625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J., 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16: 325–327. [DOI] [PubMed] [Google Scholar]
- Nantel A., Dignard D., Bachewich C., Harcus D., Marcil A., et al. , 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13: 3452–3465. 10.1091/mbc.e02-05-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D., 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42: 590–598. 10.1038/ng.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara T. R., Veri A. O., Ketela T., Jiang B., Roemer T., et al. , 2015. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 6: 6741 10.1038/ncomms7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande K., Chen C., Noble S. M., 2013. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 45: 1088–1091. 10.1038/ng.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., McDonald W. H., Pavlova Y., Yates J. R., III, Boddy M. N., 2004. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell 15: 4866–4876. 10.1091/mbc.e04-05-0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H., 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25: 7021–7032. 10.1128/MCB.25.16.7021-7032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H., 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14: 581–590. 10.1038/nsmb1259 [DOI] [PubMed] [Google Scholar]
- Potts P. R., Porteus M. H., Yu H., 2006. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25: 3377–3388. 10.1038/sj.emboj.7601218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L., 1977. Increased spontaneous mitotic segregation in MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 87: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Varma S. P., Shinde N., Ghosh S., Kumaran S. P., et al. , 2011. Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J. Biol. Chem. 286: 14516–14530. 10.1074/jbc.M110.157149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan H., Scaduto C. M., Hirakawa M. P., Gunsalus K., Correia-Mesquita T. O., et al. , 2017. Negative regulation of filamentous growth in Candida albicans by Dig1p. Mol. Microbiol. 105: 810–824. 10.1111/mmi.13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Jiang B., Davison J., Ketela T., Veillette K., et al. , 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50: 167–181. 10.1046/j.1365-2958.2003.03697.x [DOI] [PubMed] [Google Scholar]
- Roncero C., Duran A., 1985. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163: 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Elorza M. V., Valentín E., Sentandreu R., 2006. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 6: 14–29. 10.1111/j.1567-1364.2005.00017.x [DOI] [PubMed] [Google Scholar]
- Seeler J. S., Dejean A., 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4: 690–699. 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., et al. , 2005. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol. Cell. Biol. 25: 172–184. 10.1128/MCB.25.1.172-184.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey L. L., McNemar M. D., Saporito-Irwin S. M., Sypherd P. S., Fonzi W. A., 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181: 5273–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. C., Yeaman M. R., Welch W. H., Phan Q. T., Fu Y., et al. , 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279: 30480–30489. 10.1074/jbc.M401929200 [DOI] [PubMed] [Google Scholar]
- Shi Q. M., Wang Y. M., Zheng X. D., Lee R. T., Wang Y., 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18: 815–826. 10.1091/mbc.e06-05-0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., et al. , 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169: 189–197. 10.1128/jb.169.1.189-197.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K., Urban C., Brunner H., Rupp S., 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47: 89–102. 10.1046/j.1365-2958.2003.03300.x [DOI] [PubMed] [Google Scholar]
- Spiering M. J., Moran G. P., Chauvel M., Maccallum D. M., Higgins J., et al. , 2010. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot. Cell 9: 251–265. 10.1128/EC.00291-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab J. F., Bradway S. D., Fidel P. L., Sundstrom P., 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283: 1535–1538. 10.1126/science.283.5407.1535 [DOI] [PubMed] [Google Scholar]
- Staib P., Wirsching S., Strauss A., Morschhauser J., 2001. Gene regulation and host adaptation mechanisms in Candida albicans. Int. J. Med. Microbiol. 291: 183–188. 10.1078/1438-4221-00114 [DOI] [PubMed] [Google Scholar]
- Stevens V., Geiger K., Concannon C., Nelson R. E., Brown J., et al. , 2014. Inpatient costs, mortality and 30-day re-admission in patients with central-line-associated bloodstream infections. Clin. Microbiol. Infect. 20: O318–O324. 10.1111/1469-0691.12407 [DOI] [PubMed] [Google Scholar]
- Stichternoth C., Fraund A., Setiadi E., Giasson L., Vecchiarelli A., et al. , 2011. Sch9 kinase integrates hypoxia and CO2 sensing to suppress hyphal morphogenesis in Candida albicans. Eukaryot. Cell 10: 502–511. 10.1128/EC.00289-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Du H., Guan G., Dai Y., Nobile C. J., et al. , 2014. Discovery of a “white-gray-opaque” tristable phenotypic switching system in candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 12: e1001830 10.1371/journal.pbio.1001830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbji F., Chen Y., Richard Albert J., Gunsalus K. T., Kumamoto C. A., et al. , 2014. A functional portrait of Med7 and the mediator complex in Candida albicans. PLoS Genet. 10: e1004770 10.1371/journal.pgen.1004770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., et al. , 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9: 923–931. 10.1038/ncb1619 [DOI] [PubMed] [Google Scholar]
- Tsuchimori N., Sharkey L. L., Fonzi W. A., French S. W., Edwards J. E., Jr., et al. , 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68: 1997–2002. 10.1128/IAI.68.4.1997-2002.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., III, 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279: 45662–45668. 10.1074/jbc.M409203200 [DOI] [PubMed] [Google Scholar]
- Yan M., Nie X., Wang H., Gao N., Liu H., et al. , 2015. SUMOylation of Wor1 by a novel SUMO E3 ligase controls cell fate in Candida albicans. Mol. Microbiol. 98: 69–89. 10.1111/mmi.13108 [DOI] [PubMed] [Google Scholar]
- Yunus A. A., Lima C. D., 2009. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35: 669–682. 10.1016/j.molcel.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler U., Lettner T., Lassnig C., Müller M., Lajko R., et al. , 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9: 126–142. 10.1111/j.1567-1364.2008.00459.x [DOI] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782 (erratum: Proc. Natl. Acad. Sci. USA 102: 9086). 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wang Y., Wang Y., 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23: 1845–1856. 10.1038/sj.emboj.7600195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ryan J. J., Zhou H., 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279: 32262–32268. 10.1074/jbc.M404173200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Table S1 contains 69 invasive strains recovered from GRACE library screening. Strains used in this study are listed in Table S2. Plasmids and primers used with their respective sequences are listed in Table S3. Table S4 contains 373 hits from transcription profiling, while Table S5 represents RNA-seq up- and down-regulated genes need to change in figshare portal too. Figure S1 contains efg1 mms21 double-deletion filamentation. Transcription profiling data are available at GEO with the accession number: GSE116544. RNA-sequencing (RNA-seq) data are accessible through GEO Series accession number: GSE121210. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7385114.