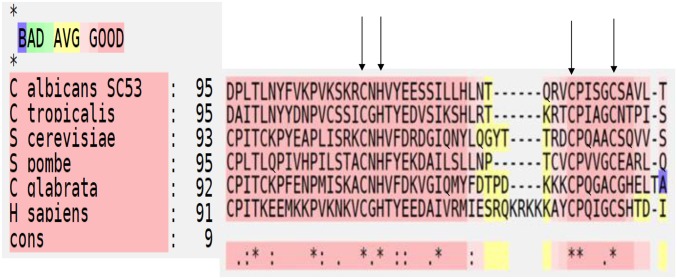
Figure 3.
T-COFFEE (tree based consistency objective function For alignment evaluation) structural protein alignment (Expresso) of Mms21/Nse2 SP [Siz/Pias (protein inhibitor of activated signal transducer and activator of transcription)] domain among different organisms. The arrows indicate residue CXH/CX4C required for Zn2+ binding. The main score (91–95) is the total consistency value. Red regions represent conserved residues while asterisks are used to label residues that are identical in the consensus sequence. Color scheme of “BAD AVG GOOD” represents level of consistency between final alignment and the library used by T-COFFEE. A score of nine (dark red regions) represents consistency score of strong alignment.