Variation in human complex traits is connected to variation in gene expression, and selection on complex traits can be reflected in selection on gene expression. Here, Glassberg and Gao et al. analyze polymorphic....
Keywords: Gene expression, Allele-specific expression, Stabilizing selection, Heritability
Abstract
Gene expression variation is a major contributor to phenotypic variation in human complex traits. Selection on complex traits may therefore be reflected in constraint on gene expression. Here, we explore the effects of stabilizing selection on cis-regulatory genetic variation in humans. We analyze patterns of expression variation at copy number variants and find evidence for selection against large increases in gene expression. Using allele-specific expression (ASE) data, we further show evidence of selection against smaller-effect variants. We estimate that, across all genes, singletons in a sample of 122 individuals have ∼2.2× greater effects on expression variation than the average variant across allele frequencies. Despite their increased effect size relative to common variants, we estimate that singletons in the sample studied explain, on average, only 5% of the heritability of gene expression from cis-regulatory variants. Finally, we show that genes depleted for loss-of-function variants are also depleted for cis-eQTLs and have low levels of allelic imbalance, confirming tighter constraint on the expression levels of these genes. We conclude that constraint on gene expression is present, but has relatively weak effects on most cis-regulatory variants, thus permitting high levels of gene-regulatory genetic variation.
VARIATION in human complex traits is connected to variation in gene expression (Nicolae et al. 2010; Giambartolomei et al. 2014; Gusev et al. 2014, 2016; Gamazon et al. 2015; Hormozdiari et al. 2016; Boyle et al. 2017). Selection on complex traits (e.g., complex disease) may therefore be reflected in selection on gene expression levels.
Across species, gene expression has been shown to evolve more slowly than expected under a neutral model (Chan et al. 2009; Brawand et al. 2011; Khan et al. 2013; Chen et al. 2018). An analysis of mammalian gene duplications also showed that the total expression of gene pairs in species that experienced a duplication event is similar to the expression of the corresponding single gene copy in species without duplication (Lan and Pritchard 2016). As a duplication event would be expected to dramatically increase gene expression, this suggests that stabilizing selection on gene expression may act to return the total expression of duplicate gene pairs to an optimal expression level.
Constraint on gene expression has also been shown to influence patterns of genetic variation within humans. First, some genes are unusually depleted for loss-of-function and copy number variants (CNVs) (Lek et al. 2016; Ruderfer et al. 2016). These genes are thought to be particularly constrained with respect to their expression levels.
Further, individuals with extreme expression levels for a particular gene are more likely to have rare variants in cis than individuals with average expression (Li et al. 2014, 2017; Zeng et al. 2015; Zhao et al. 2016). This suggests that large expression changes are associated with rare genetic variation. As detailed below, this is consistent with stabilizing selection acting against large-effect regulatory variants.
Despite this evidence for constraint on expression, humans exhibit substantial variation in gene expression and possess many common gene regulatory variants (Gaffney et al. 2012; Battle et al. 2014; GTEx Consortium 2017). It is therefore of interest to characterize constraint on gene expression and its effects on regulatory variation across the human genome.
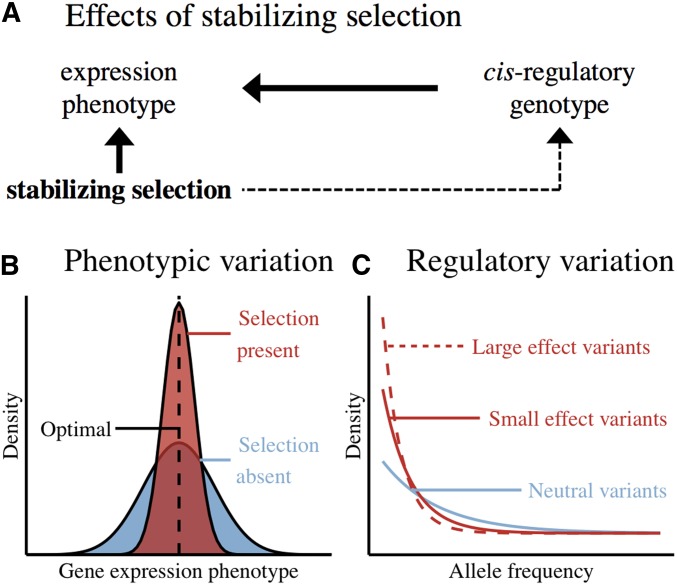
Under a model of stabilizing selection, the negative fitness effect of a cis-regulatory variant increases with its effect on expression. In other words, as large-effect variants move individuals further from an optimal gene expression value, selection acts against them, keeping them more rare than those with small effects or no effect on expression. This reduces the variance in gene expression and creates a global relationship between the allele frequencies and effect sizes of regulatory variants [Figure 1; see Simons et al. (2018) for a detailed model of stabilizing selection, genetic variation, and complex trait variance].
Figure 1.
Expected signals of stabilizing selection on gene expression. (A) Effects of stabilizing selection. (B) Phenotypic variation. (C) Regulatory variation.
This model of stabilizing selection is consistent with observed patterns of gene-regulatory variation. Past studies have noted a relationship between the allele frequencies and effect sizes of expression quantitative trait loci (eQTLs) [Battle et al. 2014, but see Tung et al. (2015) for a nonselective explanation for this relationship]. More recently, polygenic models have shown a relationship between allele frequency and the variance in gene expression explained by trait-associated variants (Hernandez et al. 2017; Zeng et al. 2018). However, the strength and breadth of this selection across genes have yet to be quantified.
Here, we test whether patterns of regulatory variation are consistent with human gene expression evolving under stabilizing selection. We analyze gene expression and genetic variation related to CNVs, eQTLs, and allele-specifically expressed (ASE) transcripts. Together, these data show that, although constraint on expression affects variation in gene expression and in regulatory genetic variation, its effects are relatively weak.
Materials and Methods
Genotypes and relative expression of CNVs
We identified loci containing CNVs in healthy individuals, as well as the number of gene copies per locus per individual, by applying LUMPY (Layer et al. 2014) and Genome STRiP (Handsaker et al. 2011) to whole blood RNA sequencing data from version 4 of the Genotype Tissue Expression Project (GTEx) (GTEx Consortium 2017). Only CNVs containing an entire protein coding sequence were retained for downstream analysis. We obtained gene expression (RPKM) measurements for each CNV in each individual across 12 tissues.
eQTL mapping
We obtained genotype and RNA sequencing data from 922 European individuals included in the Depression Genes and Networks (DGN) dataset (Battle et al. 2014; Mostafavi et al. 2014). Genotypes were imputed to 1000 Genomes as described in Kukurba et al. (2016). We then polarized genotypes relative to the human ancestral allele.
To determine the human ancestral allele, we compared the human major allele to the human-chimp ancestral allele. We obtained human major alleles from the 1000 Genomes dataset (1000 Genomes Project Consortium et al. 2015) and inferred the human-chimp ancestral allele from Ensembl multiple alignments using Ortheus (Paten et al. 2008).
At SNPs for which the human major allele and the human-chimp ancestral allele agree, we defined the human ancestral allele as the agreeing allele. At SNPs for which the human major allele and human-chimp ancestral allele disagree and the human minor allele is rare (<5% frequency), we defined the human ancestral allele as the human major allele. At SNPs for which the human major allele and human-chimp ancestral allele disagree and the human minor allele is common (>5% frequency), we defined the human ancestral allele as the human-chimp ancestral allele. At SNPs that lacked data regarding the human-chimp ancestral allele, we defined the human ancestral allele as the human major allele.
We considered SNPs with minor allele frequency >1% within a 100 kb window centered on the most upstream annotated transcription start site (TSS) to be candidate cis-eQTLs for the corresponding gene. TSSs, as annotated in Ensembl, were obtained using biomaRt (Kinsella et al. 2011).
To obtain comparable gene expression measurements across individuals, we normalized read counts at each gene by the total reads sampled per individual, and log2 transformed the resulting measurement.
To allow multiple independent eQTLs per gene, we mapped eQTLs using forward stepwise regression. For a detailed treatment of the challenges introduced by multiple regulatory variants per locus, see Zeng et al. (2017).
If any SNPs were significantly associated with gene expression ( after Bonferroni correction for the original number of candidate eQTLs for the gene), the most significantly associated SNP was added to the model. All SNPs in linkage with the newly called eQTL were excluded from the list of candidate eQTLs. This model selection procedure was repeated until no significantly associated SNPs remained.
Modeling the contribution of cis-regulatory variants to ASE
In general, ASE is measured by comparing reads expressed from each allele at a particular locus within an individual. This relies on a heterozygous site within a transcript that can be used to identify the allelic origin of each read.
We first sought to define a metric of ASE that is robust to variation in read depth across sites and agnostic to the direction of allelic bias (excess of reads from the reference or alternative allele). Here, we model allele specific expression of the gth locus in the ith individual as a squared Z-score of reads containing the alternative allele
Under a null model in which read counts follow a Binomial distribution,
| (1) |
Ai,g is the number of sampled reads containing the alternative allele, p is the underlying proportion of reads that contain the alternative allele, and Ni,g is the total number of reads sampled at the locus. The expected number of sampled reads containing the alternative allele is, then,
| (2) |
In the absence of locus- and allele-specific cis-regulation, the expected proportion of reads expressed from the alternative allele is 0.5. However, due to reference mapping bias, the observed proportion of reads from the alternative allele will be slightly lower. This ratio was estimated empirically using global reference bias.
| (3) |
where is the set of individual-gene pairs at which we measured ASE. The variance in alternative allele counts is, therefore,
| (4) |
Our ASE statistic is, therefore,
| (5) |
To explore the relationship between ASE and locus-specific regulatory variation, we allow the underlying proportion of alternative-allele reads to vary across ASE sites. Thus far, we assumed that the underlying proportion of alternative-allele reads at all loci is determined by reference bias alone. We therefore extended our model with the assumption that, within an individual, any heterozygous site in cis to an ASE site can alter expression across haplotypes and contribute to allelic imbalance.
Specifically, in each individual i at each locus j, we assumed the underlying proportion of reads containing the alternative allele to be Beta distributed. The mean proportion of alternative allele reads is determined by reference bias, as described above. However, we modeled the variance in the proportion of reads expressed from the alternative allele across individuals as a function of the number of heterozygous sites in cis to each measured ASE site.
| (6) |
is the variance in the proportion of alternative allele-reads contributed by a cis-heterozygous site, is the number of cis-heterozygous sites at the gth locus in the ith individual, and vp is the variance in the proportion of reads expressed from each allele contributed by factors other than cis-genetic variation. This could represent expression variance from trans-acting or environmental factors.
Note that is a genome-wide parameter; this assumes that, across loci, variable sites have similar effects on allelic imbalance. This model also assumes that each cis-heterozygous site contributes additively to the variance in the underlying proportion of reads expressed from the alternative allele
We then assumed that the number of alternative allele-reads sampled in individual i at locus g results from binomial sampling around the proportion of alternative allele reads at that locus.
| (7) |
In total, the observed number of reads containing the alternative allele will be Beta-Binomially distributed. The variance in alternative-allele counts is, then,
| (8) |
Under this model, we can update our squared Z-score of alleleic imbalance (Equation 5) to account for variance contributed by cis-regulatory genetic variation.
| (9) |
Note that
| (10) |
When the number of sampled reads at a locus, is large, is standard-normally distributed. Regardless of the number of sampled reads, Therefore,
| (11) |
Rearranging Equation 11,
| (12) |
As and are measurable in data, we can apply linear regression according to the above model to estimate
If stabilizing selection acts on gene expression, we would expect rare variants to contribute disproportionately to allelic imbalance. However, the above model assumes that all cis-heterozygous sites contribute equally to the variance in ASE. We therefore extended our model to allow different values of for cis-heterozygous sites with different allele frequencies. Now,
| (13) |
where b indexes allele frequency bin. is, for individual i at locus g, the number of cis-heterozygous sites with a population frequency that falls in allele frequency bin b, and is the variance in allelic imbalance explained by each variant in allele frequency bin b.
We can then estimate values of jointly using multiple regression with the counts of cis-heterozygous sites in each allele frequency bin as predictors.
Estimating the contribution of cis-regulatory variants to ASE
In this study, we analyzed ASE in 122 self-reported European individuals with RNA sequencing data and genotype calls from whole-genome sequencing from the Genotype Tissue Expression Project (GTEx Consortium 2017). We included the nine best-sampled tissues [whole blood, subcutaneous adipose, tibial artery, heart (left ventricle), lung, skin (not sun exposed), tibial nerve, skeletal muscle, and thyroid] in our analyses.
For an allelic imbalance measurement to be included in our analyses, we required the individual to have at least two reads supporting the reference and alternative alleles, respectively, within a given tissue, and at least five reads supporting each allele across the nine studied tissues. We also required the focal ASE site to be in Hardy-Weinberg equilibrium; determined using a chi-squared test with one degree of freedom and These filters help reduce false signals of ASE resulting from genotyping errors at ASE sites.
For individual-gene pairs with multiple heterozygous sites that passed these filters, the site covered by the largest number of reads was analyzed.
Human imprinted genes as listed by geneimprint (downloaded from http://www.geneimprint.com/site/genes-by-species) were excluded from downstream analyses, as were highly polymorphic human leukocyte antigens (HLA) genes (i.e., genes in the extended MHC region; bounded by SNPs rs498548 and rs2772390 plus 2 Mb extensions on both sides).
At each individual-gene pair in each tissue that met our quality control (QC) criteria, we calculated as described above. As an example, this resulted in 141,138 measurements of allelic imbalance at 18,307 unique loci in whole blood. We also calculated a combined-tissue ASE, wherein, within an individual, reads containing each allele at a focal ASE site were summed across tissues. This resulted in 343,653 measurements of combined-tissue allelic imbalance at 36,180 unique loci.
To estimate the contributions of cis-regulatory variants to ASE, we determined the number of possible gene-regulatory variants at each locus. In each individual, we considered all heterozygous sites in cis to an ASE site to have potential effects on gene regulation.
We defined sites in cis to be those that lie within 10 kb (or, when noted, 50 kb) of the most upstream TSS of a gene containing an ASE site. TSSs, as annotated in Ensembl, were obtained using biomaRt (Kinsella et al. 2011). We filtered sites not in Hardy-Weinberg equilibrium, determined using a chi-squared test with one degree of freedom and We then counted, for each individual, the number of sites in cis to each ASE site that were called as heterozygous based on whole-genome sequencing data from the GTEx Project (GTEx Consortium 2017).
For the combined-tissue data, this resulted in 1,183,405 heterozygous sites in cis to 12,159 unique genes with measured ASE, with a median of 19 cis-heterozygous sites per locus per individual.
We then estimated the average contribution of a cis-heterozygous site to allelic imbalance using linear regression, as described in Equation 12.
Due to correlation between data points (e.g., ASE was measured at the same gene in many individuals), we estimated 95% confidence intervals for the regression coefficient using a weighted jackknife as described in Busing et al. (1999), excluding measurements from all individuals for a single gene in each subsample.
To explore whether stabilizing selection acts on cis-regulatory variants in these ASE data, we tested whether there was a relationship between the allele frequency of a cis-heterozygous site and its contribution to allelic imbalance.
To do so, we binned cis-heterozygous sites by their minor allele frequency in Europeans in GTEx (GTEx Consortium 2017). We divided variants into singletons, doubletons, and spaced the remaining bins such they each contained approximately the same number of sites as the doubleton bin. The resulting bins have the following allele frequency cutoffs: (0.02, 0.04, 0.09, 0.16, 0.27, 0.4, 0.5).
We also repeated this analysis with cis-heterozygous sites binned by their minor allele frequency in Europeans in gnomAD (Lek et al. 2016). Allele frequencies in gnomAD were extracted using bcftools (Li 2011). Only variants with matching reference and alternative alleles in the GTEx and gnomAD datasets were included. Bins were spaced such that they each contained approximately the same number of sites. The resulting bins have the following allele frequency cutoffs: (0, 0.0002, 0.0013, 0.0047, 0.0125, 0.033, 0.084, 0.18, 0.33, 0.5).
For each ASE site in each individual, we counted the number of cis-heterozygous sites in each allele frequency bin. We then estimated the average contributions of cis-heterozygous sites in each bin to allelic imbalance using multiple regression, as described in Equation 13.
To explore the confidence level of these estimates, we first permuted allelic imbalance measures across all individuals and genes. Second, we permuted allelic imbalance measures across all individuals within a gene. The first tests for global artifacts of the multiple regression the second tests for variation in allele frequency spectra and allelic imbalance across genes. In all cases, the total number of variants and the number of variants in each allele frequency bin were retained as in the original data. We performed 100 permutations for each of condition and compared the resulting estimates to those obtained in the original data.
Calculating genetic variance of ASE
One way to understand the strength of selection on gene expression is to ask what proportion of the genetic variance of allelic imbalance is explained by rare variants. We therefore calculate, for each gene, the genetic variance of allelic imbalance contributed by variants in each allele frequency bin. We then calculate the total genetic variance of ASE as well as the proportion of that genetic variance attributable to each allele frequency bin.
Each variable position contributes genetic variance to allelic imbalance as follows:
| (14) |
where fi is the frequency of SNP i in allele frequency bin b and is the average variance in ASE contributed by a variant in allele frequency bin b (as estimated above).
For a given gene, the genetic variance of ASE contributed by allele frequency bin b is therefore,
| (15) |
To calculate the genetic variance of ASE captured by our model for each gene, we then sum the genetic variance contributed by each allele frequency bin.
| (16) |
The total genetic variance of ASE captured by our model can be written as
| (17) |
The proportion of the genetic variance of ASE explained by allele frequency bin b, then, is
| (18) |
Note that we do not directly estimate the heritability of allelic imbalance. However, heritability is simply the proportion of phenotypic variance that can be explained by genetic variance Therefore, the proportion of heritability explained by variants in a given allele frequency bin is equivalent to the proportion of genetic variance explained by those variants.
Data availability statement
RNA sequencing and genotype data used in eQTL calling were accessed by application through the NIMH Center for Collaborative Genomic Studies on Mental Disorders. Instructions for requesting access to data can be found at https://www.nimhgenetics.org/access_data_biomaterial.php. Inquiries should reference the “Depression Genes and Networks study (D. Levinson, PI).”
Gene expression measurements (RPKM) across 12 tissues for healthy individuals with CNVs were obtained from version 4 of the GTEx Project (GTEx Consortium 2017, dbGaP accession phs000424.v4.p1). RNA sequencing data for twins discordant for trisomy 21 were accessed from the Gene Expression Omnibus (GEO) data repository (accession GSE55426).
The 1000 Genomes phase 3 data, used in polarizing genotypes to the human ancestral allele, were obtained from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz. Details regarding the determination of human-chimp ancestral alleles are available from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/README_vcf_info_annotation.20141104.
Allele specific expression tables as well as genotype calls from whole genome sequencing, used in estimating the effects of rare regulatory variants on gene expression, were obtained from version 6 of the GTEx Project (dbGAP accession phs000424.v6.p1).
Allele frequencies from the gnomAD dataset (Lek et al. 2016) were obtained from VCFs available through the gnomAD browser http://gnomad.broadinstitute.org/downloads. Supplemental material is available at Figshare: https://doi.org/10.25386/genetics.7396673.
Results
CNVs show dosage sensitivity
Gene duplications and deletions are likely to have large effects on gene expression. If expression is under stabilizing selection, we would expect large-effect CNVs to be rare in the population. To test whether human genetic variation is affected by selection against large changes in gene expression, we analyzed the expression levels and allele frequencies of CNVs.
Using whole-genome and transcriptome sequencing of 147 individuals from version 4 of the GTEx Project (GTEx Consortium 2017), we identified 694 genes whose entire coding sequence had been duplicated in 196 polymorphic CNVs, and obtained their expression across 12 tissues.
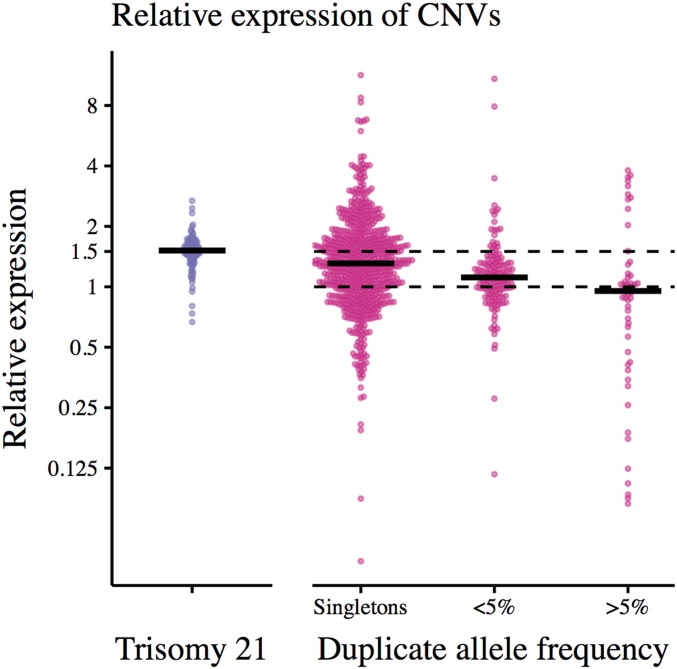
Despite high variance in expression ratios across genes, when a duplication CNV is rare (present in one individual in this sample), genes in the CNV are expressed at higher levels in heterozygous carriers (with three gene copies) than in noncarriers (with two gene copies; median expression ratio 1.31; Figure 2). By contrast, genes in common CNVs (for which >5% of sampled individuals have a duplicate gene copy) are expressed at similar levels in individuals with two and three copies (median expression ratio is 0.95; Figure 2).
Figure 2.
Expression of heterozygous carriers of a gene duplication (with three gene copies) relative to noncarriers (with two gene copies). On the left, each point corresponds to the expression level (RPKM) of one of 104 genes in an individual with trisomy 21 relative to the expression of the gene in their diploid twin. On the right, each point corresponds to the median expression of heterozygous duplication carriers relative to the median expression of noncarriers for one gene in one tissue. To reduce noise in the expression ratio, gene/tissue pairs with median noncarrier RPKM<1 were excluded (891 gene/tissue pairs retained). Duplicate frequencies obtained from GTEx. Black lines show median expression ratios across genes in each bin.
If each gene copy were expressed equally, we would expect the ratio of expression in heterozygous duplication carriers to that in noncarriers to be 1.5. The observed ratio, < 1.5, may result from a cellular buffering mechanism that reduces expression in carriers in an attempt to maintain stable expression levels.
To test for expression buffering, we obtained RNA sequencing data from a set of monozygotic twins discordant for trisomy 21 (Letourneau et al. 2014). Importantly, as trisomy 21 is a de novo expression-altering event, selection cannot affect the newly introduced expression changes.
We see that the average expression of genes on chromosome 21 is ∼50% higher in the trisomy 21 individual than in their diploid twin (Figure 2). This suggests that, before selection has time to act, when the entire gene and cis-regulatory region are duplicated, cellular buffering is negligible and expression increases proportionally with gene dosage. This is consistent with previous work showing that, for common, multi-allelic CNVs, gene expression scales linearly with copy number (Handsaker et al. 2015).
The lack of apparent expression buffering suggests that the negative relationship between the expression of duplicated genes and CNV frequency is driven by constraint. In other words, a gene duplication can become common only when expression in carriers is comparable to that in noncarriers.
Three types of regulatory variation could generate this relationship. First, during gene duplication, damage may occur to cis-regulatory elements of the duplicated gene. Damaged duplicates may lead to smaller expression increases and are therefore more likely to survive. Second, CNVs may arise on genetic backgrounds that vary in their expression. In this case, duplicates that arise on low-expression backgrounds (e.g., haplotypes with an expression-decreasing eQTL) are more likely to spread in the population. Third, haplotypes with a duplicate gene may acquire additional genetic variation; haplotypes on which compensatory, down-regulating variants arise can become common.
Regardless of the mechanism, patterns of expression variation at CNVs confirm that constraint on expression affects large-effect regulatory variants.
However, many genetic variants may have smaller effects on expression than a gene duplication. We therefore investigated whether selection on expression is sufficiently strong to affect smaller-effect variants.
Characterizing patterns of cis-regulatory variation using eQTL
First, we carried out eQTL-mapping using genotype and whole-blood RNA sequencing data from 922 European individuals from the Depression Genes and Networks cohort (Battle et al. 2014). We called cis-eQTLs using stepwise regression in 100 kb windows centered on the TSS of 12,794 autosomal, protein-coding genes (see Materials and Methods). This approach allows multiple, independent causal variants per locus. In total, we tested 3,309,888 SNPs and called 6587 significant eQTLs associated with 4734 genes (37% of genes tested).
To preserve interpretability of our effect size estimates, we did not perform any principal component or latent factor correction on our data, nor did we include any covariates in our eQTL mapping. As these additions would have improved our statistical power, the approximately twofold fewer eGenes (genes with a significant eQTL) detected here relative to previous studies is expected (Battle et al. 2014).
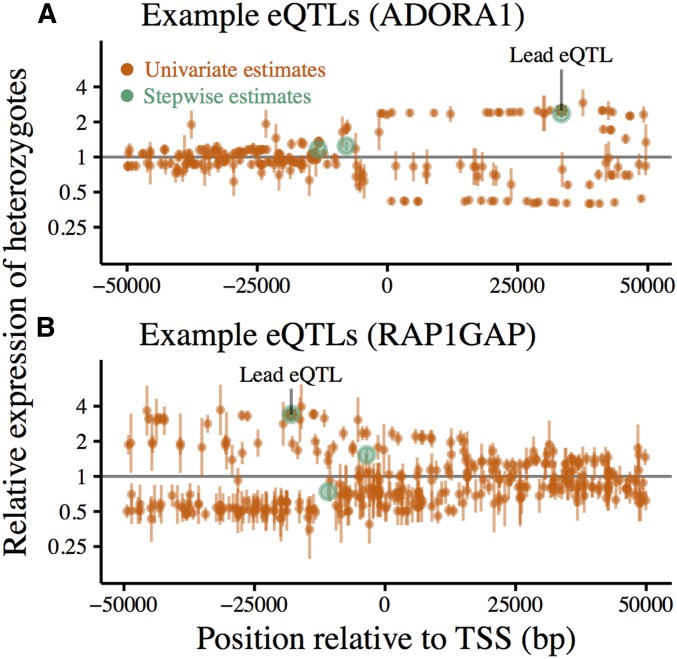
For 27.33% of eGenes, we called more than one eQTL (to a maximum of 9). This suggests that, at many loci, there may be multiple regulatory variants that would be missed when considering only the most significant eQTL (two such loci are shown in Figure 3).
Figure 3.
cis-eQTLs called using forward-stepwise mapping from whole-blood RNA sequencing in 922 European individuals. (A) eQTL mapping for ADORA1. (B) eQTL mapping for RAP1GAP. Effect sizes are polarized relative to the ancestral allele. Stepwise effect size estimates (green) are compared to those obtained by testing each SNP independently (orange) at two example loci. Vertical bars mark ± 2 SE around the estimated effect size for each SNP. Lead eQTLs (smallest P-value) from independent testing are marked with asterisks.
The median expression of individuals heterozygous for an expression-increasing eQTL was 21% higher than that of homozygotes (−18% for expression-decreasing eQTLs).
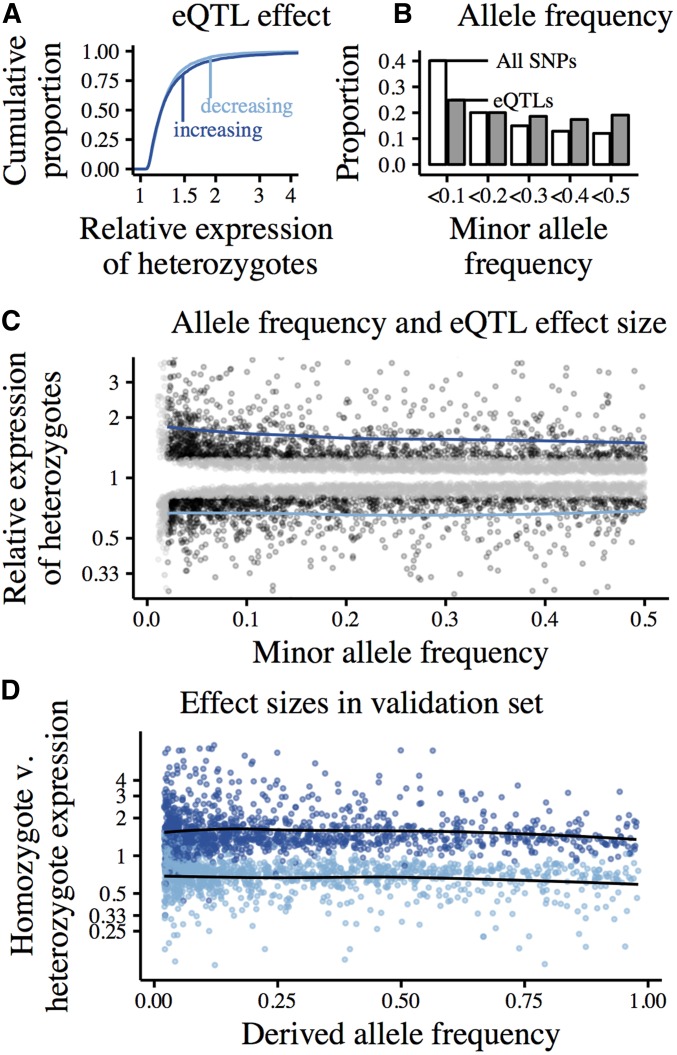
When polarizing effect sizes relative to the ancestral allele, we detected similar numbers of eQTLs predicted to increase and decrease expression (3284 expression-increasing and 3303 expression-decreasing eQTLs; not significantly different by binomial test; ). Expression-increasing and -decreasing eQTLs had comparable effect size distributions (in terms of fold-change); however, the effects of expression-increasing eQTLs tended to be slightly larger (Figure 4A; by two-sided Kolmogorov-Smirnov test). In particular, we called more large-effect, expression-increasing eQTLs; 243 eQTLs were predicted to double gene expression, 139 to cut expression in half.
Figure 4.
cis-eQTLs called using forward-stepwise mapping in 100 kb windows centered on the TSS of autosomal, protein-coding genes. Data from whole-blood RNA sequencing in 922 European individuals (DGN). Effect sizes polarized relative to the ancestral allele. (A) Cumulative distributions of estimated eQTL effect sizes, represented as the ratio of eQTL-heterozygotes to eQTL-homozygotes. Expression-increasing eQTLs (derived allele increases expression) shown in dark blue, expression-decreasing eQTLs in light blue. (B) Allele frequency spectrum of eQTLs (gray) relative to the background of all tested SNPs (white) (C) Joint distribution of eQTL MAF and estimated effect size. Black points show eQTLs with effect sizes that we are powered to detect at all frequencies (estimated effect greater than the minimum effect of eQTLs with MAF<0.02), gray points show eQTLs that are especially rare (MAF<0.02) and/or have effect sizes that we are powered to detect only at higher frequencies. Blue lines show loess-fits of MAF vs. effect size for well-powered expression-increasing and -decreasing eQTLs (black points). (D) eQTLs called in an ascertainment set of 800 individuals, effect sizes estimated in validation set of 122 individuals. Only well-powered eQTLs across allele frequencies and with MAF>0.02 are plotted. Points colored by the direction of effect in the ascertainment set; expression-increasing eQTLs in dark blue, expression-decreasing eQTLs in light blue. Loess-fits of MAF vs. effect size for expression-increasing and -decreasing eQTLs in black.
There are both technical and biological explanations for this trend. Technically, we may have less power to detect expression-decreasing eQTLs. Biologically, large decreases in gene expression may be less well-tolerated than large increases.
The joint distribution of allele frequency and effect size shows that rare eQTLs tend to have larger effects than common eQTLs (Figure 4C). The median common eQTL [minor allele frequency (MAF) > 0.1, n = 4949] was predicted to increase expression by 18% (−15% for expression-decreasing eQTLs). For rarer eQTLs (MAF < 0.1, n = 1638), the median predicted expression increase was 40% (−25% for expression-decreasing eQTLs). This is consistent with purifying selection acting against cis-regulatory variation.
However, eQTL effect sizes can be difficult to interpret. First, previous work has discussed the challenges inherent in estimating eQTL effect sizes when there are multiple causal variants at a locus (Zeng et al. 2017). Second, power to map eQTLs of the same effect varies with allele frequency. Third, a statistical phenomenon referred to as “winner’s curse” can induce a relationship between allele frequency and estimated effect size (Göring et al. 2001; Lohmueller et al. 2003; Tung et al. 2015). We explored the impacts of each of the above on our effect size estimates.
First, univariate (independent) eQTL effect size estimates were similar to those derived from our stepwise approach (Figure 3). It is therefore unlikely that the presence of multiple causal variants per locus meaningfully impacted our estimates.
Variable eQTL-detection power by allele frequency, however, did impact our calls. One might expect eQTL-based analyses to be limited to relatively common regulatory variants. Indeed, our eQTLs were, on average, more common than candidate SNPs (Figure 4B).
Further, as we lack power to call rare eQTLs of small effect, one might expect the median effect size of rare eQTLs to be inflated relative to common eQTLs. However, when considering only eQTLs with relatively large effects (that could be detected at all frequencies; details in Supplemental Material, Table S1), rare eQTLs were estimated to have larger effects than common ones (Table 1). This suggests that decreased power to detect rare eQTLs is not sufficient to explain the observed relationship between frequency and effect size.
Table 1. Summary of the effects of variable power and winner’s curse on eQTL effect size estimates and their relationship with minor allele frequency.
| Median eQTL effect | ||||
|---|---|---|---|---|
| Full Data | Ascertainment (validation) | |||
| Expression increasing | 21% | 22% (18%) | ||
| Expression decreasing | −18% | −18% (−14%) | ||
| MAF 0.02–0.1 | MAF >0.1 | MAF 0.02–0.1 | MAF >0.1 | |
| Expression increasing—filtered for power | 48% | 40% | 52% (44%) | 45% (45%) |
| Expression decreasing—filtered for power | −28% | −28% | −29% (−25%) | −29% (−29%) |
Effect sizes reported as percent change of eQTL-heterozygotes relative to the homozygous ancestral. To explore the effects of winner’s curse, individuals were split into an eQTL ascertainment set and an effect-size validation set. To explore the impact of variable power to call eQTLs of the same effect across allele frequency bins, eQTLs were filtered to remove those with estimated effects that we would lack power to call at low allele frequencies. Here, we consider eQTLs with estimated effects larger than the minimum magnitude of estimated effect for significant, rare eQTLs (MAF<0.02). To ensure conservative estimates of the relationship between effect size and allele frequency, we also remove rare eQTLs (MAF<0.02) from comparisons across allele frequency bins.
However, this relationship could also result from winner’s curse. Conditional on a variant being called as an eQTL, we expect its effect to be overestimated (Göring et al. 2001; Lohmueller et al. 2003). This is particularly true for rare variants.
To explore the effects of winner’s curse, we ascertained eQTLs and estimated their effects in separate subsamples of the DGN data. In the ascertainment set, eQTLs were estimated to have a median effect of 22% (−18% for expression-decreasing eQTLs) on expression. In the validation set, however, the median effect was 18% (−14% for expression-decreasing eQTLs; by paired t-test; Figure S1). This suggests that winner’s curse inflates eQTL effect size estimates. Comparing effect size estimates across rare (0.02 < MAF < 0.1) and common (MAF > 0.1) eQTLs revealed that winner’s curse disproportionately inflates estimates at the low end of the allele frequency spectrum (Table 1).
Indeed, effect size estimates from the validation set no longer show any relationship with allele frequency (Figure 4D, further detail in Figure S1). This is consistent with Tung et al. (2015), who also saw a significant impact of winner’s curse on effect size estimates of rare eQTLs. We conclude that eQTL effect size distributions fail to provide evidence for constraint on gene expression.
However, absence of evidence is not necessarily evidence of absence. To more carefully test for a relationship between eQTL effect size and allele frequency, we used an independent dataset of ASE.
eQTL effects on ASE
ASE, or allelic imbalance, is measured using a heterozygous site in a transcript. This site is used to identify reads from, and quantify the expression of, each haplotype within an individual.
Variation in expression across haplotypes captures the regulatory effects of all cis-heterozygous sites in an individual, regardless of their frequencies in the population. As a result, analyses of ASE include genetic variants that are too rare to be detected in eQTL studies.
On the other hand, eQTL mapping is better suited to estimate the effects of individual variants on gene regulation. ASE and eQTL mapping are, therefore, complementary tools for measuring cis-regulatory variation.
To sidestep statistical challenges in eQTL effect size estimation, we combined our eQTLs with ASE (data from GTEx version 6, GTEx Consortium 2017). We measured ASE using a squared Z-score of reads containing the alternative allele in 343,653 gene-individual-tissue trios (i.e., a gene expressed in a given tissue in a given individual; see Materials and Methods for QC filters).
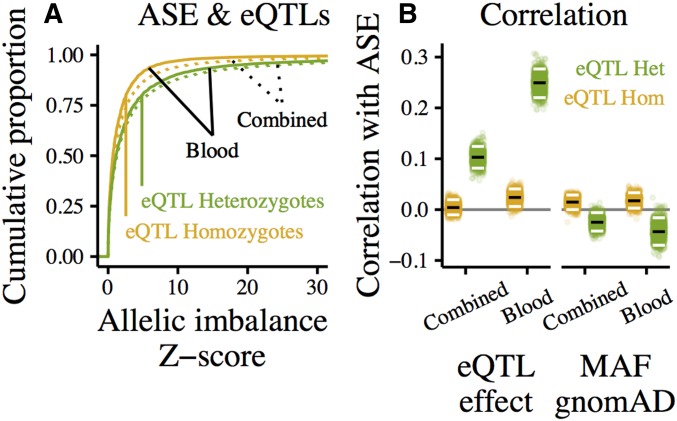
Previous work has demonstrated that ASE measurements are generally consistent with eQTLs (Pickrell et al. 2010). To confirm that here, we show that at eGenes, eQTL-heterozygotes (heterozygous for at least one eQTL) tend to have higher allelic imbalance than eQTL-homozygotes (homozygous for all called eQTLs; Figure 5A). In addition, eQTL effect sizes were correlated with allelic imbalance in eQTL-heterozygotes (Figure 5B).
Figure 5.
eQTL effects reflected in ASE at eGenes. “Blood” shows ASE in whole blood, “Combined” shows combined-tissue ASE, measured by summing reads spanning an ASE site in the same individual across tissues. (A) Cumulative distributions of ASE measured in whole blood (solid) and combined-tissue ASE (dotted), separated into eQTL-heterozygotes (green), and -homozygotes (homozygous for all called eQTLs, yellow). (B) Rank correlation between allelic imbalance and eQTL effect (left) or minor allele frequency in gnomAD (right). The effect/MAF of the most significant heterozygous eQTL (or, for eQTL-homozygotes, the most significant eQTL) was used. To determine the robustness of these correlations, each point shows the rank correlation in one of 1000 samples generated by bootstrapping over genes. Median correlations of bootstrap samples marked in black, 95% quantiles in white.
To gain additional insight into the structure of the gene regulatory landscape across tissues, we compared a single-tissue analysis of blood to a combined-tissue analysis in which reads spanning an ASE site in an individual were summed across tissues.
In both eQTL-heterozygotes and -homozygotes, combined-tissue ASE tended to be higher than ASE measured in blood (Figure 5A). This shift can be explained by differences in read depth; due to the summation of reads across tissues, average read depth per ASE site was higher in the combined-tissue data (median 130 for combined-tissue, 28 for blood; Figure S7A). Increased read depth reduces sampling variance, thus increasing our ability to detect bona fide allelic imbalance (and increasing the resulting ASE Z-score).
However, summing reads across tissues would increase ASE only if allelic imbalance were concordant across tissues. Like many other eQTL and ASE analyses (e.g., Flutre et al. 2013; Wheeler et al. 2016; GTEx Consortium 2017), our findings suggest that many gene regulatory effects are consistent (or, at least, are unlikely to be inconsistent) across tissues.
Finally, we explored whether ASE provides evidence of selection against eQTLs (Figure 5B). In both combined-tissue and blood-specific ASE, there is a subtle, but significant, negative correlation between ASE in eQTL-heterozygotes (a proxy for eQTL effect size) and the frequency of the corresponding eQTL. This suggests that selection on eQTLs is present, but weak.
Effects of cis-regulatory variants vary with allele frequency
We next examined constraint on regulatory variants not captured by eQTL mapping. We first sought to relate allelic imbalance to the amount of cis-regulatory variation at a locus within an individual. In our model, each cis-heterozygous site contributes to the variance in the ratio of reads expressed from each haplotype (detailed in Materials and Methods). In this case, we expect the number of cis-heterozygous sites to be linearly related to our ASE Z-score.
We then used linear regression to estimate the average contribution of a heterozygous site (across all genes in all individuals) to ASE. In single tissues and in a combined-tissue analysis, we observed significant, positive relationships between genetic variation and ASE (Figure 6). This suggests that variation in allelic imbalance across loci has a genetic basis, rather than being driven solely by sampling noise or environmental factors.
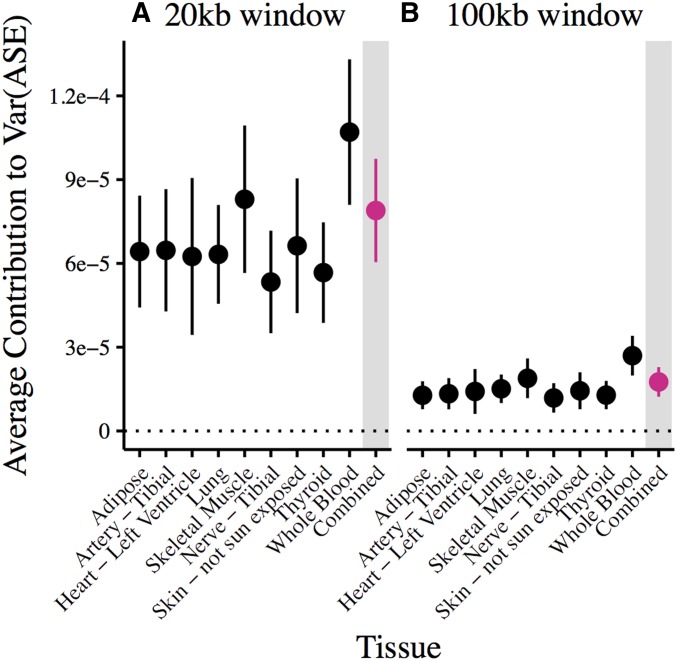
Figure 6.
Average effect of a cis-genetic variant on allelic imbalance. Estimates are based on linear regression of an individual’s number of cis-heterozygous sites within (A) 10 kb and (B) 50 kb of the TSS of a gene and measured ASE for each of the nine best sampled GTEx tissues. Vertical lines show 95% confidence intervals, estimated using a weighted jackknife as described in Busing et al. (1999). For reference, the dotted line marks zero estimated effect.
The average effect of a variant on ASE decreases as we increase the cis-regulatory window under consideration (Figure 6). This likely reflects the spatial distribution of causal regulatory variants; we and others find that eQTLs are enriched near TSSs (Figure S2; Stranger et al. 2007; Veyrieras et al. 2008). If this holds for rarer regulatory variants, widening the cis-window will decrease the proportion of causal variants included in the model and so decrease the average effect size per variant. For a given window, average per-variant effect size estimates are similar across tissues (Figure 6).
Next, we expanded our model to incorporate allele frequency information. If purifying selection keeps large-effect regulatory variants rare, we would expect a rare cis-heterozygous variant to contribute more to allelic imbalance than a common one. For each gene in each individual (individual-gene pair), we counted the number of heterozygous sites in each of nine allele frequency bins. We then used multiple regression to estimate the contribution of an average site in each frequency bin to ASE.
Correlation between the numbers of heterozygous sites across allele frequency bins would make it difficult to accurately estimate average effects for each bin. However, in these data, this correlation is very weak (maximum Pearson correlation across pairs of frequency bins is 0.07; Figure S3).
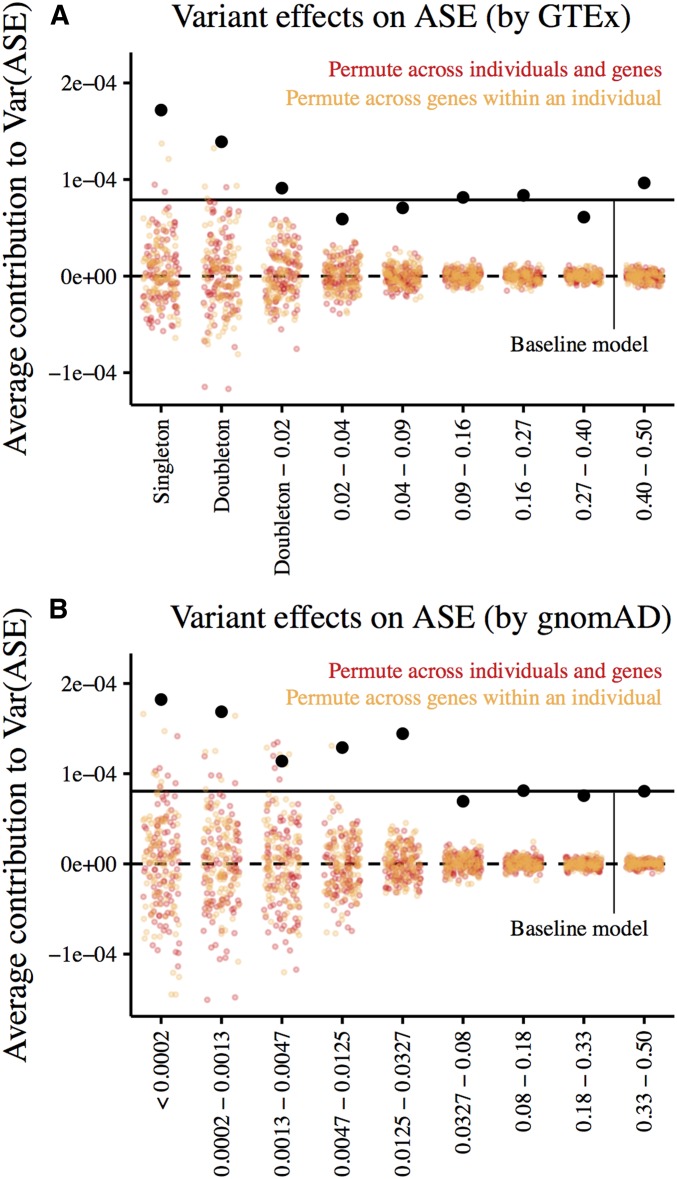
We find that the average contribution of a singleton in the GTEx dataset to the variance in ASE is ∼2.2 × greater than that of the average variant across allele frequency bins (1.8 × greater than the average common variant (MAF>0.4); Figure 7A). Permutations of ASE measurements across individuals and genes, as well as across genes within an individual, suggest that this relationship between frequency and contribution to ASE is significant.
Figure 7.
Effects of cis-genetic variants on allelic imbalance given their allele frequencies. Estimates are obtained from multiple regression on allelic imbalance using the number of cis-heterozygous sites in each allele frequency bin as predictors. Here, we include sites in a 20-kb window centered on the TSS of a gene with measured allelic imbalance. Estimates from combined-tissue ASE (reads spanning each ASE site in a single individual are summed across all tissues sampled) are shown in black. Colored points represent estimates from 100 permutations of (1) ASE measurements across genes and individuals (red) and (2) ASE measurements across genes within an individual (orange). Horizontal lines mark zero effect (dashed) and the average variant effect estimated by regressing allelic imbalance on the total number of cis-heterozygous sites (solid). (A) Estimates with variants binned by minor allele frequency in Europeans in the GTEx data (n = 122). (B) Estimates with variants binned by minor allele frequency in Europeans in gnomAD (n = 7509).
As this dataset contains only 122 individuals, allele frequency estimates may be noisy. Additionally, singletons in GTEx may not be especially rare in the population. We therefore repeated our analysis with variants binned by their minor allele frequency in the much larger gnomAD dataset (Lek et al. 2016; n = 7209 Europeans with whole genome sequencing, Figure 7B). We see similar trends with both binning strategies.
The relationship between allele frequency and contribution to allelic imbalance also persists when considering genetic variants in a much larger candidate cis-regulatory region (Figure S5). As for the average per-variant effect across allele frequencies (Figure 6), increasing the window size decreases the estimated effect for variants in each allele frequency bin. However, the average contributions of rare variants (singletons and doubletons) remain greater than those of common variants.
Importantly, this trend is so subtle that it is detectable only when reads are combined across tissues; single-tissue analyses show no clear relationship between allele frequency and estimated effect size (examples whole blood and skeletal muscle shown in Figure S6). This is likely due to the fewer genes, fewer individuals sampled per gene, and lower read depth per individual-gene pair in single tissues as compared to the combined set (for sample size comparisons, see Figure S7; for read depth comparisons, see Figure S8).
From this, we conclude that constraint on gene expression reshapes patterns of regulatory genetic variation. However, the subtlety of this signal suggests that selection on gene expression is weak.
Proportion of heritability (genetic variance) explained by rare variants
Another way to understand the strength of selection on expression is to ask what proportion of the heritability (or, what proportion of the genetic variance ) of expression can be explained by rare variants.
Two components determine how much genetic variance is explained by a variant: the variant’s squared effect size and its allele frequency (particularly its contribution to heterozygosity, ). When considering the proportion of genetic variance explained by a group of variants at a given allele frequency, the allele frequency spectrum (the proportion of variants in each allele frequency bin) is also important.
Many polygenic trait models assume that all variants, regardless of their allele frequency, contribute equally to genetic variance ( e.g., Yang et al. 2011; Bulik-Sullivan et al. 2015). As their low allele frequencies cause rare variants to contribute less to heterozygosity this type of model implicitly assumes that rare variants have larger effects than common ones.
Others (e.g., Speed et al. 2012) more explicitly account for the relationship between allele frequency (and corresponding differences in LD) and variance explained. For a more detailed comparison of heritability estimation and variance partitioning methods, see Evans et al. (2018).
While these models were not designed with singletons in mind, their varying assumptions about allele frequency, effect size, and heritability led us to consider what impact the observed 2.2 × greater effect size of singletons would have on the proportion of heritability explained by rare variants.
To explore this, we combined our average effect size estimates with allele frequency spectra at each gene to estimate the genetic variance of allelic imbalance captured by our model as well as the proportion of explained by variants in each allele frequency bin (summary of allele frequency spectra in Figure S9). Across genes, we find the median heritability explained by singletons (in a sample of 122 individuals) to be a mere 5% (Figure 8). Table 2 shows comparisons of our model to two extreme cases: (1) rare variants (singletons) and common variants (MAF>0.4) contribute equally to genetic variance (2) rare and common variants have equal effects on gene expression
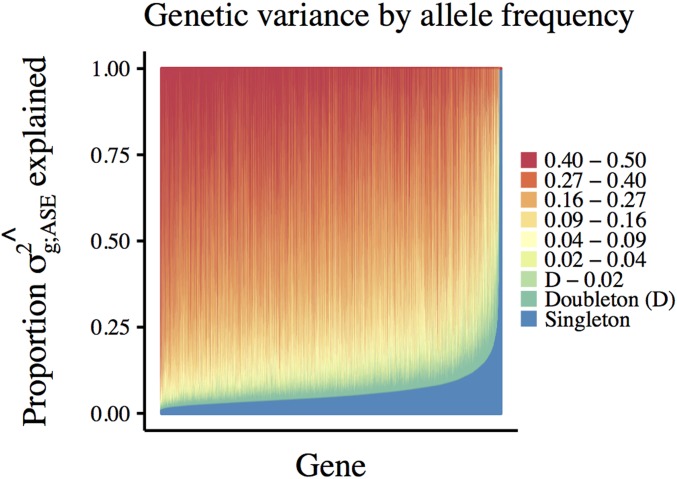
Figure 8.
Partitioning the genetic variance of allelic imbalance by allele frequency. For each gene, we combine the average heterozygosity (2 pq) and number of segregating sites in each allele frequency bin with the estimated average effect size of variants in that bin to calculate the total genetic variance of allelic imbalance captured by our ASE-model We then calculate the proportion of explained by each allele frequency bin (variants binned by minor allele frequency in GTEx). Each vertical bar represents a single gene for which we measured ASE, colors represent the proportion of genetic variance attributed to variants in each allele frequency bin.
Table 2. Singleton contributions to heritability.
| Singletons vs. common variants (MAF>0.4) | Estimates | Models | |
|---|---|---|---|
| from ASE | independent of p | ||
| Proportion explained by singletons | 4.7% | 36% | 2.4% |
| Relative squared effect size | ∼1.8 | ∼60 | 1 |
| Selection strength | Weak | Strong | None |
Estimates based on GTExV6 ASE data (cis-regulatory region +− 10kb from TSS) “ from ASE” shows estimates of the median proportion of explained by singletons based on effect sizes estimated from our ASE-model. “” shows the median expected per-gene heritability explained by singletons if singletons and common variants contributed equally to genetic variance. This is driven by the fact that 36% of all variants (median 36% of variants per gene) are singletons. The large relative effect size required for singletons to explain this proportion of heritability reflects the low variance contribution of rare variants as a consequence of their low allele frequency (2 pq). “ independent of p” shows estimates of the median proportion of heritability explained by singletons if effect size and allele frequency were independent (i.e., singletons and common variants had equal effects on gene expression).
Our model suggests that singletons explain more of the heritability of gene expression than expected under neutrality (effect size independent of allele frequency). This is due to singletons’ increased average effect size relative to common variants. However, in these data, the relative effect size difference between singletons and common variants is far less than what would be required for a singleton to contribute as much as a common variant to the heritability of gene expression.
While these findings are consistent with previous evidence of constraint on gene expression (Hernandez et al. 2017; Zeng et al. 2018), Hernandez et al. (2017) estimate a much greater contribution of singletons to expression variance, suggesting a much greater role of purifying selection on gene-regulatory variants, than we see here. At this time, we do not know the source of the discrepancy between the studies; however, the two approaches involve different analytical methods, different gene expression measurements (ASE vs. RNASeq), and samples of different sizes.
Future work will be required to understand why different approaches appear to support different conclusions with respect to the importance of rare variants on gene expression. However, in our analysis of allele-specific expression, we do not find support for rare variants being major drivers of expression variance.
Although the median heritability explained by singletons is small, we detect notable variation in singleton-heritability across genes (Figure 8). As we estimate a single, average singleton effect across all genes, this variation is due entirely to differences in allele frequency spectra.
This variation led us to wonder how much of the observed constraint (i.e., relative effect size differences between rare and common variants) is driven by a subset of particularly tightly constrained genes.
Patterns of eQTLs and ASE suggest variable constraint on expression across gene classes
To understand variation in constraint on expression across the genome, we compared patterns of regulatory variation across gene sets.
We expect genes with greater constraint on expression to have lower levels of cis-regulatory variation than genes with less-constrained expression. To test this, we compared eQTLs across gene sets predicted to have varying tolerance to large changes in gene expression. Specifically, we utilized Probability of Loss-of-Function Intolerance (pLI) (Lek et al. 2016). pLI measures, for each gene, the relative depletion of protein-truncating variants (PTVs) observed in healthy individuals compared to the expectation under a detailed mutation model.
In heterozygotes, PTVs are expected to decrease gene expression by half. Certain genes may therefore be depleted for PTVs because they are intolerant to such large expression changes. We stratified genes into two classes based on their pLI-predicted level of constraint on expression (low: pLI < 0.1, high: pLI > 0.9), and tested for differences in cis-regulatory variation.
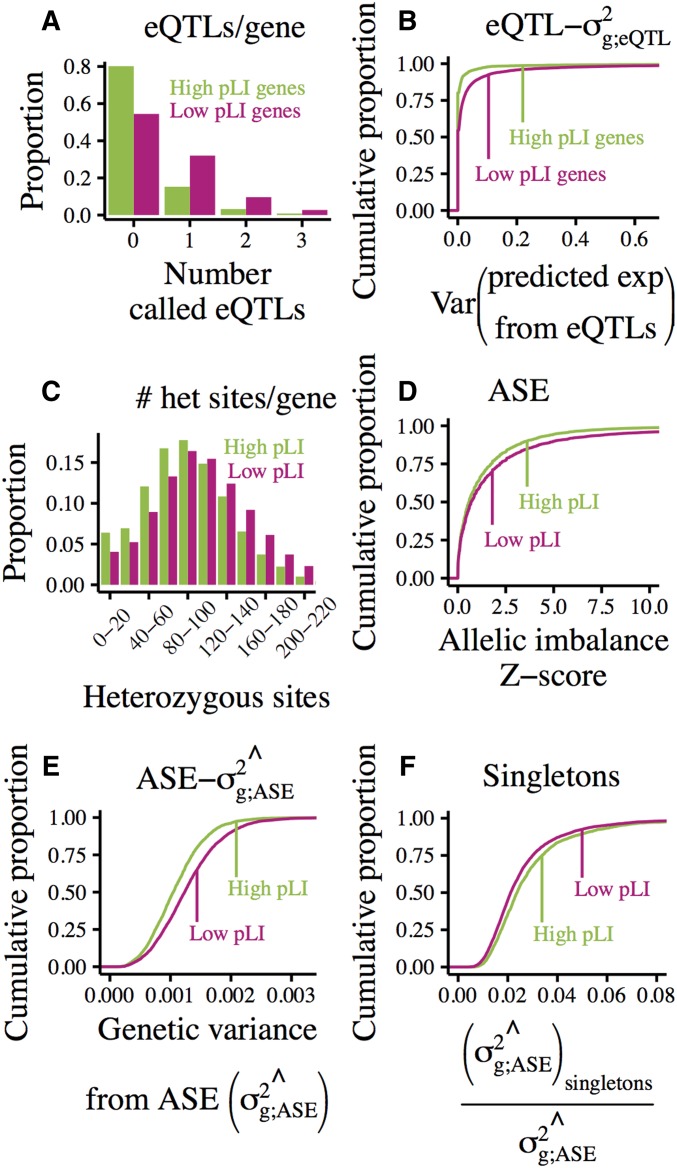
In total, 4401 eQTLs were mapped around 3103 unique low-pLI genes (of 6813 genes tested), and 688 eQTLs were mapped around 511 high-pLI genes (of 2576 genes tested). We detected fewer eQTLs per gene for dosage sensitive, high-pLI genes than for less constrained, low-pLI genes (Figure 9A). This is consistent with prior analysis of eQTLs called in GTEx (Lek et al. 2016).
Figure 9.
Regulatory genetic variation across gene sets with varying levels of constraint on gene expression as predicted by pLI class. (A) Histogram of the number of cis-eQTLs called per gene by pLI class. (B) Cumulative distributions of eQTL-estimated genetic variance by pLI class. For each gene in each individual, expression levels were predicted using genotypes and effect size estimates of called eQTLs. Genetic variance was then estimated as the variance of eQTL-predicted expression levels across individuals. (C) Histogram of the number of cis-heterozygous sites (per individual-gene pair) within 50 kb of the TSS by pLI class. (D) Cumulative distributions of allelic imbalance, measured in whole blood, by pLI class. (E) Cumulative distributions of the genetic variance of allelic imbalance captured by our model, estimated using combined-tissue ASE and allele frequencies binned according to GTEx, by pLI class. (F) Cumulative distributions of the proportion of the genetic variance of ASE attributable to singletons by pLI class.
These differences are reflected in the amount of genetic variance captured by eQTLs for low- and high-pLI genes (Figure 9B; by two-sided Kolmogorov-Smirnov test). Based on genotypes at called eQTLs, we predicted the genetically regulated component of expression for each individual in the DGN dataset. We then estimated genetic variance as the variance of predicted expression across individuals.
We also used these eQTLs to predict expression and estimate in 122 European individuals from GTEx. The differences in between gene sets are consistent across the two datasets (Figure S10).
To quantify these differences in genetic variance, we compared the median eQTL-estimated genetic variance for genes in each pLI class. We assumed log-normally distributed expression values to estimate ranges of gene expression in the population.
Across genes with at least one called eQTL, we estimate that 95% of individuals possess genetic variation resulting in expression levels between the population mean. However, for the median low-pLI gene, these ranges are and, for the median high-pLI gene, they are This may suggest that constraint on expression removes more regulatory variation around dosage-sensitive high-pLI genes than less sensitive low-pLI genes.
This observation could also be explained by differences in background selection. High-pLI genes experience greater coding sequence constraint than low-pLI genes. We might therefore expect background selection to decrease the amount of genetic variation (regulatory or not) around high-pLI genes.
As shown in Figure S2A, there are fewer common SNPs in cis to low-pLI genes (MAF>0.05, within 50 kb of the TSS) than to high-pLI genes. SNPs around low-pLI genes also tend to be more common than those around high-pLI genes (Figure S2C). Finally, low-pLI eQTLs tend to be closer to the TSS (Figure S2, B and D). These trends suggest that background selection shapes patterns of genetic variation; however, they are unlikely to explain the striking differences in eQTL-based genetic variance across gene sets.
Variation in genetic variance across genes can also be seen in allele-specific expression. As high-pLI genes tend to be more highly expressed and have more sampled reads than low-pLI genes (read depth information in Figure S8), we might expect to more confidently call ASE (i.e., higher Z-score) in high-pLI genes. However, we observed the opposite pattern; ASE Z-scores for high-pLI genes tended to be lower than those for low-pLI genes (Figure 9D).
In blood, 18.6% of 73,246 individual-gene pairs involving low-pLI genes have an ASE Z-score >3. This level of allelic imbalance is unlikely to arise from read sampling. We infer that, in these individual-gene pairs, the two haplotypes are differently cis-regulated. By contrast, only 13.3% of 36,457 individual-gene pairs involving high-pLI genes have Z-scores >3 (significantly different by binomial test, ). Given the difference in average read depth between high- and low-pLI genes, this likely under-represents the difference in expression variance between gene classes.
These differences in expression variance are reflected in genetic variation; high-pLI genes tend to have fewer cis-heterozygous sites than low-pLI genes (Figure 9C). This suggests a link between expression variance and genetic variation.
We also detected differences across gene sets in the genetic variance captured by our ASE-model (calculated according to Calculating genetic variance of ASE in Materials and Methods). The median estimated genetic variance for high-pLI genes is for low-pLI genes it is ( by Wilcoxon rank sum test). However, a larger proportion of the genetic variance of high-pLI genes is explained by singletons (median 0.053 for high-pLI genes, 0.045 for low-pLI genes; by Wilcoxon rank sum test). These observations suggest that selection reduces variance in the expression of tightly constrained genes both by reducing the number of segregating cis-regulatory sites and by keeping larger-effect regulatory variants more rare.
However, it remains unclear how much of the genome-wide signal of constraint on regulatory variants can be attributed to strong constraint on a subset of genes, like high-pLI genes, and how much is due to weak constraint subtly shifting the joint distribution of allele frequency and regulatory effect genome-wide. The extremely small number of genes with large contributions from rare variants (88% of genes have <10% genetic variance explained by singletons, 98% have <20%) may suggest that the bulk of the relationship between allele frequency and effect size is driven by weak selection on many genes, but further work is required to corroborate this interpretation. In particular, larger datasets of paired whole genome and RNA sequencing will increase the sampled genetic diversity between individuals at a locus and provide additional insight into the landscape of selection on gene expression.
Discussion
Our analyses of eQTLs and ASE suggest that selection on gene expression has detectable, but weak, effects on regulatory variants. However, much work remains to understand the connection between expression, complex traits, and fitness.
A central limitation of our approach is that we assume all heterozygous sites in an allele frequency bin have the same effect on allelic imbalance, regardless of the gene they regulate or their position relative to the TSS. In other words, we estimate the genome-wide average effect size of variants at a given allele frequency.
As a result, our model does not capture heterogeneity across regulatory variants at similar frequencies, nor does it reflect heterogeneity in regulation across genes. For example, the expression of a specific gene may be affected by a rare variant of very large effect. However, if most rare variants have negligible effects on the expression of their target genes, the average effect estimated for rare variants will remain moderate.
Our approach cannot, therefore, accurately estimate the effect of a particular variant on gene expression. Nor can we accurately estimate the proportion of genetic variance explained by singletons for a particular gene. What we can conclude is that, in general, much of the variation in gene expression is explained by common variants.
Obtaining per-variant, or average per-gene effect size estimates (rather than genome-wide averages per allele frequency bin) would provide additional insight into the gene-regulatory landscape and the effects of selection on gene expression. However, we show that statistical limitations decrease the applicability of per-variant effect size estimates from eQTL mapping to questions of constraint. New approaches and larger whole-genome and transcriptome datasets will be required to achieve this additional resolution.
Further, in seeming contrast to our findings regarding eQTLs and ASE, analyses of loss-of-function and CNVs suggest that selection on expression changes of 1.5 × is, at least in some cases, sufficient to alter patterns of expression and regulatory variation. These results indicate that gene expression has notable fitness consequences.
These observations can be reconciled with the weak selection observed here by arguing that the 1.2 × expression changes caused by the average eQTL and the 1.5 × changes in expression caused by a gene duplication have very different effects on fitness. One might also argue that the effects seen in loss-of-function and copy number variant analyses are driven by a small set of tightly constrained genes, or that loss-of-function and CNVs have effects on fitness that are unrelated to their effects on gene expression.
These arguments might lead us to conclude that gene expression has limited downstream effects on human traits and on fitness. However, if gene regulation is highly polygenic and/or pleiotropic, selection may be unable to cause dramatic changes in the frequencies of regulatory variants (regardless of the fitness consequences of expression changes).
Rampant polygenicity is likely to decrease the effectiveness of selection on individual regulatory variants. If many variants affect the expression of a gene, a new mutation with a large effect may arise on a background with existing, compensatory genetic variation. Such background-effects may explain how the expression of common CNVs is similar between individuals with two and three gene copies.
Additionally, if gene regulation is highly polygenic, an eQTL may not reflect a single causal variant with the effect estimated during mapping. If multiple causal variants with concordant effects are in linkage disequilibrium in a sample, their effects may be aggregated into a single “synthetic” eQTL with a large effect [for more on synthetic associations, see Dickson et al. (2010)]. If the variants that comprise such a synthetic eQTL are not in linkage disequilibrium in the population, their effect sizes may individually be sufficiently small as to be nearly neutral. Future work is required to determine to what degree eQTLs represent groups of concordant, small-effect variants.
This challenge of polygenicity is not limited to the regulation of a single gene. If a fitness-relevant trait is affected by the expression of many genes, the fitness consequence of a variant that regulates a single gene is likely to be small. On the other hand, if a variant regulates multiple genes, and the resulting expression changes each affect fitness, the selection experienced by the variant will be a combination of the selection on each expression trait.
Such regulatory pleiotropy may also arise if the expression of a single gene affects multiple fitness-relevant traits. For example, altered gene expression may have different effects in different tissues. Therefore, a variant that affects the expression of a single gene may have multiple (and possibly opposing) effects on fitness. Interestingly, as the polygenicity of fitness-relevant traits (e.g., the number of genes whose expression modulates the trait) increases, the likelihood that a single regulatory variant causes pleiotropic effects on multiple traits also increases.
In summary, a complex, pleiotropic gene expression network may make it difficult for selection to precisely alter the allele frequency of a single regulatory variant, even when its effects might be large and deleterious from the perspective of one gene expression trait in one tissue context. While there is much work to be done to connect gene expression to the one, or many, phenotypic traits seen by selection, we conclude that constraint has subtle, but detectable, effects on the genetic architecture of gene regulation.
Acknowledgments
We thank the members of the Pritchard Laboratory for spirited conversations regarding this work. A.H. was funded in part by a fellowship from the Stanford Center for Computational, Evolutionary and Human Genomics (CEHG). This work was supported by National Institutes of Health (NIH) grants HG008140 and HG009431.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7396673.
Communicating editor: N. Wray
Literature Cited
- Battle A., Mostafavi S., Zhu X., Potash J. B., Weissman M. M., et al. , 2014. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 24: 14–24. 10.1101/gr.155192.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. A., Li Y. I., Pritchard J. K., Gordon S., Henders A., et al. , 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D., Soumillon M., Necsulea A., Julien P., Csárdi G., et al. , 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. 10.1038/nature10532 [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B. K., Loh P.-R., Finucane H. K., Ripke S., Yang J., et al. , 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47: 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busing F. M. T. A., Meijer E., Leeden R. V. D., 1999. Delete-m jackknife for unequal m. Stat. Comput. 9: 3–8. 10.1023/A:1008800423698 [DOI] [Google Scholar]
- Chan E. T., Quon G. T., Chua G., Babak T., Trochesset M., et al. , 2009. Conservation of core gene expression in vertebrate tissues. J. Biol. 8: 33 10.1186/jbiol130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Swofford R., Johnson J., Cummings B. B., Rogel N., et al. , 2018. A quantitative model for characterizing the evolutionary history of mammalian gene expression. Genome Res. 10.1101/gr.237636.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson S. P., Wang K., Krantz I., Hakonarson H., Goldstein D. B., 2010. Rare variants create synthetic genome-wide associations. PLOS Biol. 8: e1000294 10.1371/journal.pbio.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. M., Tahmasbi R., Vrieze S. I., Abecasis G. R., Das S., et al. , 2018. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 50: 737–745. 10.1038/s41588-018-0108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T., Wen X., Pritchard J., Stephens M., 2013. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9: e1003486 10.1371/journal.pgen.1003486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D. J., Veyrieras J.-B., Degner J. F., Pique-Regi R., Pai A. A., et al. , 2012. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13: R7 10.1186/gb-2012-13-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E. R., Wheeler H. E., Shah K. P., Mozaffari S. V., Aquino-Michaels K., et al. , 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47: 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C., Vukcevic D., Schadt E. E., Franke L., Hingorani A. D., et al. , 2014. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10: e1004383 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring H. H., Terwilliger J. D., Blangero J., 2001. Large upward bias in estimation of locus-specific effects from genomewide scans. Am. J. Hum. Genet. 69: 1357–1369. 10.1086/324471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium , 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Lee S. H., Trynka G., Finucane H., Vilhjálmsson B. J., et al. , 2014. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95: 535–552. 10.1016/j.ajhg.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Ko A., Shi H., Bhatia G., Chung W., et al. , 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48: 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker R. E., Korn J. M., Nemesh J., McCarroll S. A., 2011. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet. 43: 269–276. 10.1038/ng.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker R. E., Van Doren V., Berman J. R., Genovese G., Kashin S., et al. , 2015. Large multiallelic copy number variations in humans. Nat. Genet. 47: 296–303. 10.1038/ng.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Uricchio L. H., Hartman K., Ye J., Dahl A., et al. , 2017. Singleton variants dominate the genetic architecture of human gene expression. bioRxiv. DOI: 10.1101/219238. [Google Scholar]
- Hormozdiari F., van de Bunt M., Segrè A. V., Li X., Joo J. W. J., et al. , 2016. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99: 1245–1260. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z., Ford M. J., Cusanovich D. A., Mitrano A., Pritchard J. K., et al. , 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342: 1100–1104. 10.1126/science.1242379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella R. J., Kahari A., Haider S., Zamora J., Proctor G., et al. , 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011: bar030 10.1093/database/bar030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukurba K. R., Parsana P., Balliu B., Smith K. S., Zappala Z., et al. , 2016. Impact of the X Chromosome and sex on regulatory variation. Genome Res. 26: 768–777. 10.1101/gr.197897.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X., Pritchard J. K., 2016. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science 352: 1009–1013. 10.1126/science.aad8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R. M., Chiang C., Quinlan A. R., Hall I. M., 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15: R84 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., et al. , 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau A., Santoni F. A., Bonilla X., Sailani M. R., Gonzalez D., et al. , 2014. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508: 345–350. 10.1038/nature13200 [DOI] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Battle A., Karczewski K. J., Zappala Z., Knowles D. A., et al. , 2014. Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants. Am. J. Hum. Genet. 95: 245–256. 10.1016/j.ajhg.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Kim Y., Tsang E. K., Davis J. R., Damani F. N., et al. , 2017. The impact of rare variation on gene expression across tissues. Nature 550: 239–243. 10.1038/nature24267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Pearce C. L., Pike M., Lander E. S., Hirschhorn J. N., 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33: 177–182. 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Battle A., Zhu X., Potash J. B., Weissman M. M., et al. , 2014. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol. Psychiatry 19: 1267–1274. 10.1038/mp.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae D. L., Gamazon E., Zhang W., Duan S., Eileen Dolan M., et al. , 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6: e1000888 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B., Herrero J., Fitzgerald S., Beal K., Flicek P., et al. , 2008. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 18: 1829–1843. 10.1101/gr.076521.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Marioni J. C., Pai A. A., Degner J. F., Engelhardt B. E., et al. , 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772. 10.1038/nature08872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer D. M., Hamamsy T., Lek M., Karczewski K. J., Kavanagh D., et al. , 2016. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 48: 1107–1111. 10.1038/ng.3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y. B., Bullaughey K., Hudson R. R., Sella G., 2018. A population genetic interpretation of GWAS findings for human quantitative traits. PLoS Biol. 16: e2002985 10.1371/journal.pbio.2002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D., Hemani G., Johnson M., Balding D., 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91: 1011–1021. 10.1016/j.ajhg.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E., Nica A. C., Forrest M. S., Dimas A., Bird C. P., et al. , 2007. Population genomics of human gene expression. Nat. Genet. 39: 1217–1224. 10.1038/ng2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J., Zhou X., Alberts S. C., Stephens M., Gilad Y., 2015. The genetic architecture of gene expression levels in wild baboons. eLife 4: e04729 10.7554/eLife.04729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras J.-B., Kudaravalli S., Kim S. Y., Dermitzakis E. T., Gilad Y., et al. , 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4: e1000214 10.1371/journal.pgen.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler H. E., Shah K. P., Brenner J., Garcia T., Aquino-Michaels K., et al. , 2016. Survey of the heritability and sparse architecture of gene expression traits across human tissues. PLoS Genet. 12: e1006423 10.1371/journal.pgen.1006423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Lloyd-Jones L. R., Holloway A., Marigorta U. M., Metspalu A., et al. , 2017. Constraints on eQTL fine mapping in the presence of multisite local regulation of gene expression. G3 (Bethesda) 7: 2533–2544. 10.1534/g3.117.043752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., de Vlaming R., Wu Y., Robinson M. R., Lloyd-Jones L. R., et al. , 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 50: 746–753. 10.1038/s41588-018-0101-4 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Wang G., Yang E., Ji G., Brinkmeyer-Langford C. L., et al. , 2015. Aberrant gene expression in humans. PLoS Genet. 11: e1004942 10.1371/journal.pgen.1004942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Akinsanmi I., Arafat D., Cradick T. J., Lee C. M., et al. , 2016. A burden of rare variants associated with extremes of gene expression in human peripheral blood. Am. J. Hum. Genet. 98: 299–309. 10.1016/j.ajhg.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing and genotype data used in eQTL calling were accessed by application through the NIMH Center for Collaborative Genomic Studies on Mental Disorders. Instructions for requesting access to data can be found at https://www.nimhgenetics.org/access_data_biomaterial.php. Inquiries should reference the “Depression Genes and Networks study (D. Levinson, PI).”
Gene expression measurements (RPKM) across 12 tissues for healthy individuals with CNVs were obtained from version 4 of the GTEx Project (GTEx Consortium 2017, dbGaP accession phs000424.v4.p1). RNA sequencing data for twins discordant for trisomy 21 were accessed from the Gene Expression Omnibus (GEO) data repository (accession GSE55426).
The 1000 Genomes phase 3 data, used in polarizing genotypes to the human ancestral allele, were obtained from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ALL.wgs.phase3_shapeit2_mvncall_integrated_v5b.20130502.sites.vcf.gz. Details regarding the determination of human-chimp ancestral alleles are available from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/README_vcf_info_annotation.20141104.
Allele specific expression tables as well as genotype calls from whole genome sequencing, used in estimating the effects of rare regulatory variants on gene expression, were obtained from version 6 of the GTEx Project (dbGAP accession phs000424.v6.p1).
Allele frequencies from the gnomAD dataset (Lek et al. 2016) were obtained from VCFs available through the gnomAD browser http://gnomad.broadinstitute.org/downloads. Supplemental material is available at Figshare: https://doi.org/10.25386/genetics.7396673.