Varshney et al. directly compare five widely-used annotations of active regulatory elements: stretch, super, and typical enhancers; highoccupancy target (HOT) regions; and broad domains in four human cell types. Overall, their results suggest that current...
Keywords: chromatin, gene regulation, expression QTL, chromatin QTL, GWAS
Abstract
Epigenomic signatures from histone marks and transcription factor (TF)-binding sites have been used to annotate putative gene regulatory regions. However, a direct comparison of these diverse annotations is missing, and it is unclear how genetic variation within these annotations affects gene expression. Here, we compare five widely used annotations of active regulatory elements that represent high densities of one or more relevant epigenomic marks—“super” and “typical” (nonsuper) enhancers, stretch enhancers, high-occupancy target (HOT) regions, and broad domains—across the four matched human cell types for which they are available. We observe that stretch and super enhancers cover cell type-specific enhancer “chromatin states,” whereas HOT regions and broad domains comprise more ubiquitous promoter states. Expression quantitative trait loci (eQTL) in stretch enhancers have significantly smaller effect sizes compared to those in HOT regions. Strikingly, chromatin accessibility QTL in stretch enhancers have significantly larger effect sizes compared to those in HOT regions. These observations suggest that stretch enhancers could harbor genetically primed chromatin to enable changes in TF binding, possibly to drive cell type-specific responses to environmental stimuli. Our results suggest that current eQTL studies are relatively underpowered or could lack the appropriate environmental context to detect genetic effects in the most cell type-specific “regulatory annotations,” which likely contributes to infrequent colocalization of eQTL with genome-wide association study signals.
GENOME-WIDE association studies (GWAS) have shown that most of the genetic variants associated with disease-related traits lie in nonprotein-coding regions (Hindorff et al. 2009). More importantly, these loci are specifically enriched in enhancer elements of disease-relevant cell types (Maurano et al. 2012; ENCODE Project Consortium 2012; Parker et al. 2013; Trynka et al. 2013; Corradin et al. 2014; Pasquali et al. 2014; Quang et al. 2015). This suggests that the majority of disease-associated genetic variants modulate regulatory elements that can influence gene expression. Therefore, it is essential to identify and understand the genetic signatures and molecular function(s) of gene regulatory regions.
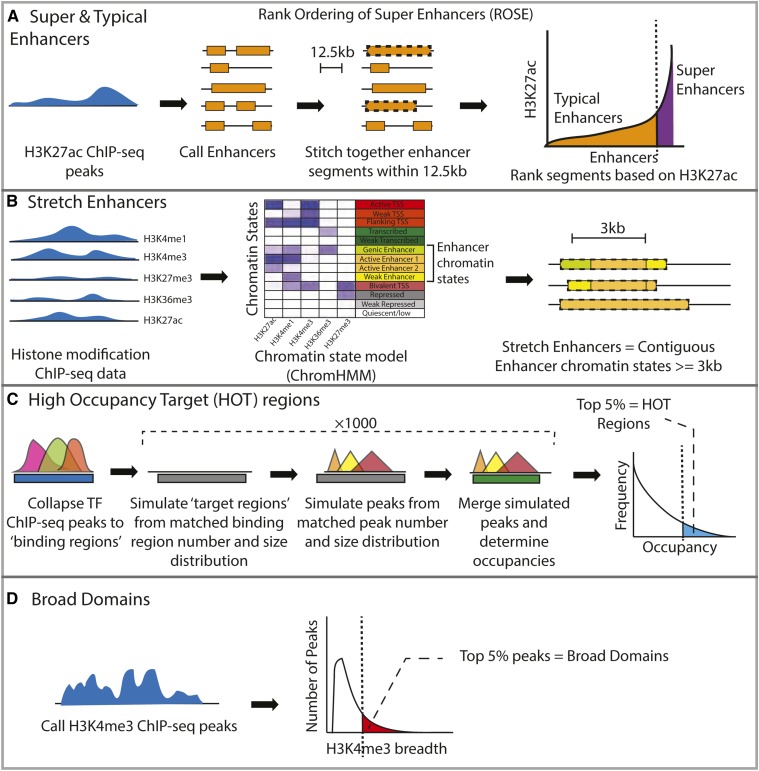
Epigenomic profiling, such as chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), of histone modifications or transcription factors (TFs) that can indicate regulatory activity in vivo has been effectively used to predict the regulatory function of genomic regions. For example, super enhancers have been defined in multiple cell types as regions with high levels of the histone H3 lysine 27 acetylation (H3K27ac) mark (Hnisz et al. 2013). Putative enhancer elements were identified from ChIP-seq peaks, and elements within 12.5 kb of each other were stitched together. After ranking these stitched regions based on the enhancer-associated ChIP-seq signal (Figure 1A), a small number (∼3%) of identified regions that contained a large fraction (>40%) of the ChIP-seq signal, observable as a steep rise in the ChIP-seq signal curve (geometrical inflection point, Figure 1A) (Whyte et al. 2013), were termed super enhancers. These elements were at least an order of magnitude larger in size than the remaining nonsuper enhancer elements (i.e., typical enhancers). This signal-based approach has been generalized as the rank ordering of super enhancers (ROSE) algorithm (Lovén et al. 2013; Whyte et al. 2013) (Figure 1A). Super enhancers are thought to encompass multiple constituent enhancer elements that collectively have high regulatory potential and drive high expression of cell identity regions (Whyte et al. 2013; Hnisz et al. 2013).
Figure 1.
Description of the regulatory annotation calling procedures. (A) Super/typical enhancers are called by using the H3K27ac mark ChIP-seq to assign enhancer elements, stitching elements within 12.5 kb and ranking the stitched segments based on H3K27ac levels. (B) Stretch enhancer calling procedure involves analyzing patterns of multiple histone marks and assigning chromatin state segmentations using ChromHMM, followed by identifying contiguous enhancer chromatin state segments longer than 3 kb. (C) HOT regions are defined as regions with higher TF-binding occupancies than expected. (D) Broad domains are defined as the top 5% of the H3K4me3 ChIP-seq peaks by length. ChIP-seq, chromatin immunoprecipitation-sequencing; HMM, hidden Markov model; HOT, high-occupancy target; ROSE, rank ordering of super enhancers; TF, transcription factor.
In another approach, ChIP-seq data for multiple histone modifications were used to annotate the genome. A hidden Markov model (HMM)-based approach identified distinct and recurrent patterns in the ChIP-seq data, and segmented the genome into chromatin states (Ernst et al. 2011; Ernst and Kellis 2012). Analyzing chromatin states across diverse cell types and tissues, the authors identified that the longest 10% of contiguous enhancer chromatin states (enhancers ≥ 3 kb) were highly cell type-specific, occurred near to genes with highly cell type-specific gene ontology terms, and were enriched for cell type-relevant disease and trait-associated variants (Parker et al. 2013). These regions were referred to as stretch enhancers (Parker et al. 2013) (Figure 1B) and represent substantially large regions of enhancer-associated chromatin.
Regulatory annotations have also been defined from TF ChIP-seq profiling. Analysis of such data sets across cell types revealed that >50% of TF-bound sites occurred in highly occupied clusters that were not randomly distributed across the genome (Moorman et al. 2006; modENCODE Consortium et al. 2010; ENCODE Project Consortium 2012; Boyle et al. 2014). To identify regions where TF occupancies were higher than expected by chance, one study first collapsed ChIP-seq peaks for multiple TFs as observed binding regions (Figure 1C, blue bar). The expected regions of TF-binding or “target regions” (Figure 1C, gray bars), and individual TF-binding sites within these regions (Figure 1C, colored triangles), were then randomly sampled 1000 times, while keeping the number and size distributions equivalent to those observed. Occupancies were scored based on observed and expected collapsed binding sites (Figure 1C, blue and green blocks, respectively); regions with the top 5% occupancies were classified as high-occupancy target (HOT) regions (Figure 1C).
The histone H3 lysine 4 trimethyl (H3K4me3) mark is associated with active and poised promoters (Bernstein et al. 2006; Mikkelsen et al. 2007; Adli et al. 2010). Unusually large regions of the H3K4me3 mark have been observed in multiple cell types across humans, mice, and other species, often spanning up to ∼60 kb (Adli et al. 2010; Benayoun et al. 2014; Chen et al. 2015). Importantly, the broadest 5% of H3K4me3 domains were found to mark genes with cell type-specific functions (Benayoun et al. 2014; Thibodeau et al. 2017). These regions have been termed broad domains (Figure 1D).
These diverse methodologies identify genomic regions with substantially high densities of epigenomic marks known to be associated with gene regulation. These regions denote important classes of regulatory elements, which show cell type specificity, transcriptional activity in reporter assays, and disease relevance based on GWAS SNP enrichments (Kvon et al. 2012; Hnisz et al. 2013, 2015; Parker et al. 2013; Benayoun et al. 2014; Boyle et al. 2014; Blinka et al. 2016; Lin et al. 2016; Dave et al. 2017). Few studies have compared the characteristics for subsets of these annotations, showing some degree of overlap between HOT regions and super enhancers (Li et al. 2016), and chromatin interactions between broad domains and super enhancers (Thibodeau et al. 2017). However, the functional differences among these annotations, especially how genetic variation in these elements affects target gene expression, are unclear. To fill this gap, we compared diverse characteristics of super, typical, and stretch enhancers, HOT regions, and broad domains (hereafter collectively referred to as regulatory annotations) in the only four matched human cell types for which they are available: the lymphoblastoid cell line (LCL) GM12878, human embryonic stem cell (hESC) line H1, leukemia cell line K562, and hepatic carcinoma cell line HepG2. We used previously published annotations as these were rigorously generated by the respective authors and are widely used. Collectively, these regulatory annotations represent the computational and statistical integration of 245 ChIP-seq data sets (an average of 61 ChIP-seq data sets per cell type). We report annotation summary statistics and the proportion of overlap with diverse chromatin states in these regions. We measure enrichment for proximity to genes that are expressed in a cell type-specific manner, and integrate genetic regulatory data to measure enrichment for expression quantitative trait loci (eQTL). Finally, as measures of strength of gene and chromatin accessibility regulation, we compare the effect sizes of loci associated with gene expression (eQTL), DNase I hypersensitivity site (dsQTL), and allelic bias in ATAC-sequencing (ATAC-seq) data. Comparisons using these metrics allow us to quantify the biological properties of these regulatory annotations.
Materials and Methods
Regulatory annotation sources
Regulatory annotations for the GM12878, H1 hESC, and HepG2 cell types were downloaded from previously published studies for HOT regions (Boyle et al. 2014), broad domains (Benayoun et al. 2014), stretch enhancers (Varshney et al. 2017), and super and typical enhancers (Hnisz et al. 2013).
Summary statistics and overlaps between annotations, chromatin states, and ATAC-seq peaks
Summary statistics, such as the number of features in each annotation, segment size distribution, and percent genome coverage (Figure 2, A–C), were calculated using custom scripts (see GitHub). To compute overlap fractions between all pairs of annotations shown in Figure 2, D and E, we calculated the base pair-level overlap between each pair using BEDtools intersect (Quinlan and Hall 2010). For each pair of annotation sets, we then calculated the Jaccard statistic by dividing the total length of the intersection region with the total length of the union region. To calculate the fraction of regulatory annotation overlap with chromatin states in Supplemental Material, Figure S2, we used chromatin states previously defined in the four cell types considered (Varshney et al. 2017) and used BEDtools intersect. Stretch enhancer annotations were also obtained from this previous study (Varshney et al. 2017).
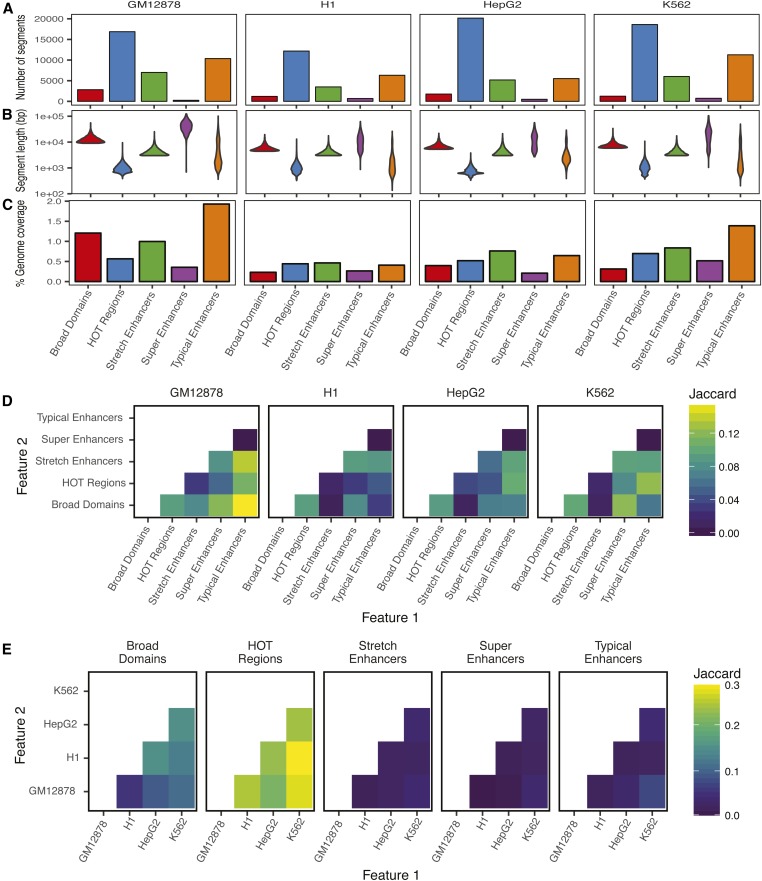
Figure 2.
Summary statistics and overlaps demonstrate differences in characteristics of regulatory annotations. For each annotation in each cell type considered, shown are number of annotation segments (A), length distribution of segment annotations (B), and percent genomic coverage (C). Jaccard statistic (base pair-level intersection/union) between each pair of annotations is shown within a cell type (D) and across cell types (E). HOT, high-occupancy target.
Enrichment for overlap between each pair of regulatory annotations in Figure S1 was calculated using the Genomic Association Tester (GAT) tool (Heger et al. 2013). To ask if two sets of regulatory annotations overlap more than that expected by chance, GAT randomly samples segments of one regulatory annotation set from the genomic workspace (hg19 chromosomes) and computes the expected overlaps with the second regulatory annotation set. We used 10,000 GAT samplings for each regulatory annotation. The observed overlap between segments and annotation is divided by the expected overlap, and an empirical P-value is obtained.
Chromatin state information content analysis
We first compiled the average posterior probabilities of a regulatory annotation segment to be called an enhancer or promoter chromatin state. We utilized the previously published 13-chromatin state ChromHMM model (also used to define stretch enhancers) (Varshney et al. 2017), which also outputs posterior probabilities for each 200-bp genomic segment to be called each of the 13 states in each of the four cell types. We considered the sum of active enhancer 1 and 2, weak enhancer, and genic enhancer posterior probabilities to represent enhancer states, and averaged these values over all the 200-bp tiles overlapping each annotation segment. We considered active, weak, and flanking transcription start site (TSS) states to denote promoter chromatin states. For example, for a segment in GM12878 broad domains, we obtained the average posterior probabilities for the region being an enhancer or promoter state in a cell for To calculate the information content, we first calculated the relative average posterior probabilities,
Next, we calculated entropy of the segment as:
We know that entropy is maximized with all segments have equal relative probabilities, or: for
We then compared with . While high posterior probabilities for enhancer or promoter states indicate preference for that state, high information content indicates cell type specificity of that chromatin state preference. When plotting Figure 3, to have the same x-axis ranges for all facets for easier comparison (stretch enhancers only show high mean posterior probabilities for enhance r states and low posterior probabilities for promoter states due to their definition), we added one pseudocount in each corner for all facets.
Figure 3.
Enhancer and promoter chromatin state information content shows cell type specificity of regulatory annotations. Mean posterior probability for an annotation segment to be called an enhancer (A) or promoter (B) chromatin state vs. the information content of that feature in GM12878 cell type. The information content is calculated by comparing the mean posterior probabilities across the four cell types. HOT, high-occupancy target.
Distance to nearest gene
We downloaded the Gencode V19 gene annotations from ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_human/release_19/gencode.v19.annotation.gtf.gz and obtained the TSS coordinates for protein-coding genes. For each segment in each annotation, we computed the distance to nearest protein-coding gene TSS using BEDtools closest (Quinlan and Hall 2010).
Enrichment of genetic variants in genomic features
Enrichment for GWAS variants for different traits and eQTL identified in the LCL in regulatory annotations was calculated using the genomic regulatory elements and GWAS overlap algorithm (GREGOR) GREGOR (version 1.2.1) (Schmidt et al. 2015). Since the causal SNP(s) for the traits are not known, GREGOR allows consideration of the input lead SNP along with SNPs in high linkage disequilibrium (LD) (based on the provided R2THRESHOLD parameter) while computing overlaps with genomic features (regulatory annotations). Therefore, as input to GREGOR, we supplied SNPs that were not in high LD with each other. We pruned the list of SNPs using the PLINK (v1.9) tool (Purcell et al. 2007; Chang et al. 2015) –clump option and 1000 genomes phase 3 vcf files (downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502) as reference. For each input SNP, GREGOR selects ∼500 control SNPs that match the input SNP for minor allele frequency (MAF), distance to the nearest gene, and the number of SNPs in LD. Fold enrichment is calculated as the number of loci at which an input SNP (either lead SNP or SNP in high LD) overlaps the feature over the mean number of loci at which the matched control SNPs (or SNPs in high LD) overlap the same features. This process accounts for the length of the features, as longer features will have more overlap by chance with control SNP sets.
Specific parameters for the GWAS enrichment were: GWAS variants for various traits were obtained from the National Human Genome Research Institute-European Bioinformatics Institute catalog (https://www.ebi.ac.uk/gwas/). Table S2 lists the individual GWAS studies for each disease/trait. We used the following parameters: pruning to remove SNPs with r2 > 0.2 for European population; GREGOR: r2 threshold = 0.8; LD window size = 1 Mb; minimum neighbor number = 500; and population = European.
Specific parameters for the LCL eQTL enrichment were: LCL eQTL data from the genotype tissue expression (GTEx V7) study was downloaded from the GTEx website (https://www.gtexportal.org/home/datasets, filename GTEx_Analysis_v7_eQTL.tar.gz). We used the following parameters: pruning to remove SNPs with r2 > 0.8 for European population; GREGOR: r2 threshold = 0.99; LD window size = 1 Mb; minimum neighbor number = 500; and population = European.
We used different r2 thresholds for GWAS (r2 = 0.8) vs. eQTL (r2 = 0.99) enrichment analyses because eQTL analyses measure a molecular feature instead of a complex phenotype, and therefore have higher resolution to identify the more likely causal variants.
Analysis of LCL-specific expression
We used an information theory approach (Schug et al. 2005; He et al. 2014) to score genes based on LCL expression level and specificity relative to the panel of 50 diverse GTEx tissues, each of which had RNA-seq data for >25 samples. We downloaded RNA-seq data from the GTEx V7 study from the website https://www.gtexportal.org/home/datasetsfilenameGTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_median_tpm.gct.gz. These data were in the form of median transcripts per million (TPM) for each gene in each tissue. We considered protein-coding genes and removed those that were lowly expressed in LCL (median TPM > 0.15) to avoid potential artifacts. We calculated the relative expression of each gene (g) in LCL compared to all 50 tissues (t) as p:
We next calculated the entropy for expression of each gene across all 50 tissues as H:
Following previous studies (Schug et al. 2005; He et al. 2014), we defined LCL tissue expression specificity (Q) for each gene as:
To aid in interpretability, we divided Q for each gene by the maximum observed Q and subtracted this value from 1, and refer to this new score as the LCL expression specificity index (LCL-ESI):
LCL-ESI scores near zero represent lowly and/or ubiquitously expressed genes, and scores near 1 represent genes that are highly and specifically expressed in LCL.
Enrichment for distance to genes based on gene expression specificity in LCL
We binned the protein-coding genes into quintiles based on LCL-ESI, such that bin 5 included the most LCL-specific genes. Each quintile bin contained N = 2753 protein-coding genes. We then used BEDtools closest to calculate the distance to the nearest protein-coding gene TSS for each bin, obtaining empirical cumulative distribution functions (ECDFs) for each regulatory annotation in each cell type. Since the regulatory annotations vary in the number of segments, and will therefore have different probabilities of occurring near a TSS, the distance to the nearest protein-coding gene TSS ECDFs cannot be directly compared. Therefore, we obtained the expected distance to the nearest protein-coding gene TSS ECDF for each annotation by randomly sampling N = 2753 genes from across the five bins 10,000 times and calculating the distance to nearest gene. We then calculated the TSS proximity enrichment for each annotation by dividing the observed with the mean expected ECDF. Enrichment therefore denotes the fold change in the observed fraction of annotation segments within a certain distance of protein-coding gene TSSs in a specific LCL-ESI bin over the mean fraction of segments at the same distance from the randomly sampled genes. The 95% C.I.s for the enrichment values were calculated as observed / (mean ± 1.96 * SE), where SE = SEM expected fraction.
Enrichment to overlap eQTL based on expression specificities of genes
We sorted the eQTL SNPs into quintiles based on the LCL-ESI of the associated genes (eGene) and grouped them into five equally sized bins, resulting in 585 eQTL in each bin. Bin numbers represent eQTL that correspond to increasingly LCL-specific genes, where bin 1 represents the least LCL-specific and bin 5 represents the most LCL-specific genes. We calculated the enrichment for each eQTL set to overlap regulatory annotations using GREGOR with the same parameters as described above for the bulk set of LCL eQTL. To quantify the trend of LCL eQTL enrichment with LCL eGene expression specificity, we calculated the Spearman correlation of the enrichment effect size expressed as log2(fold enrichment) with the eQTL bin number using the cor() function from the stats package (v3.5.1) in R (R Core Team 2015).
Gene expression and chromatin accessibility QTL effect sizes in regulatory annotations
We used the β values or the slope of the linear regression as the effect size of LCL and blood eQTL (GTEx V7), and dsQTL (Degner et al. 2012). All of these QTL studies used inverse rank-based normalization steps on the molecular features, which enables direct comparison of the effect sizes across the genome. Because low-MAF SNPs have low statistical power to be detected as significant QTL at low effect sizes, these SNPs are biased to have large QTL effect sizes. Therefore, we removed QTL SNPs with MAF < 0.2. We pruned the QTL SNPs to retain SNPs with r2 < 0.8 after sorting by P-value of association as described above using PLINK (Purcell et al. 2007; Chang et al. 2015). Since the causal SNP for the QTL signal is unknown, we also considered SNPs in high LD at r2 > 0.99 with the lead QTL SNPs, which were obtained using vcftools (Danecek et al. 2011) and the 1000 genomes phase 3 reference vcf specified above. We observed higher eQTL enrichment in annotations with increasing the r2 thresholds, which is indicative of a higher signal-to-noise ratio. A previous study analyzing LCL eQTL also showed that functional enrichment decreased rapidly from the best eQTL toward lower ranked eQTL (Lappalainen et al. 2013). We compared the absolute QTL effect sizes of loci (QTL index SNP or SNP with r2 > 0.99 with the index SNP) that overlapped each GM12878 annotation. We used the Wilcoxon rank sum test to identify significant differences between effect sizes of eQTL overlapping each annotation.
To test if there may be confounding from other genomic properties, such as the distance between the eQTL eSNP to eGene, and whether the number of SNPs in high LD with the lead SNP could also influence the eQTL effect size, we calculated the contribution of the underlying regulatory annotation on the effect size while accounting for these factors. We modeled the eQTL effect size in a linear regression using the Python statsmodels library, where we included a regulatory annotation indicator variable-encoding eQTL overlap by a stretch enhancer or HOT region annotation, and the following two covariates: (1) absolute distance of the eQTL lead SNP to its corresponding eGene TSS and (2) total number of SNPs in high LD (r2 > 0.99 with the lead SNP) that overlapped the annotation. eQTL that overlapped both annotations were not considered. Summary statistics of this regression model are presented in Table S1.
To calculate the statistical power for eQTL analysis after Bonferroni correction based on a linear regression, we used the powerEQTL.SLR function from the “powerEQTL” R package (Dong et al. 2017) (v0.1.3; https://rdrr.io/cran/powerEQTL/). For eQTL overlapping each annotation, we used the eQTL effect sizes representing the 10th to 90th percentile values and calculated power by using the following parameters: MAF = 0.2, type I error rate = 0.0005, total number of tests = 1,000,000, SD of the error term = 0.4, and sample size N = 250.
Comparison of allelic bias effect sizes in annotations
To determine SNP allelic bias in GM12878 ATAC-seq data, we used the publicly available data (Buenrostro et al. 2013) listed in Table S3. Adapters were trimmed using cta (v. 0.1.2; https://github.com/ParkerLab/cta) and reads mapped to hg19 using bwa mem (Li 2013) (default options except for the -M flag; v. 0.7.15-r1140). Bam files were filtered for high-quality autosomal read pairs using samtools (Li et al. 2009) view (-f 3 -F 4 -F 8 -F 256 -F 2048 -q 30; v. 1.3.1). WASP (van de Geijn et al. 2015) (version 0.2.1, commit 5a52185; using python version 2.7.13) was used to adjust for reference mapping bias; for remapping the reads as part of the WASP pipeline, we used the same mapping and filtering parameters described above for the initial mapping and filtering. Duplicates were removed using WASP’s rmdup_pe.py script. We used the phased GM12878 VCF file downloaded from ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/NA12878_HG001/NISTv3.3.1/GRCh37/HG001_GRCh37_GIAB_highconf_CG-IllFB-IllGATKHC-Ion-10X-SOLID_CHROM1-X_v.3.3.1_highconf_phased.vcf.gz. To avoid potential artifacts associated with double-counting alleles, overlapping read pairs were clipped using bamUtil clipOverlap (v. 1.0.14; http://genome.sph.umich.edu/wiki/BamUtil: clipOverlap). The bam files from the samples in Table S3 were then merged to create a single GM12878 bam file using samtools merge. We filtered for heterozygous autosomal SNPs with minimum coverage of 30. Since the power to detect allelic bias depends upon the read coverage at the SNP, SNPs with lower coverage are biased toward having higher effect sizes at any given level of statistical significance. To prevent this type of bias, we randomly downsampled reads at each heterozygous SNP to a total of 30 reads with base quality of at least 20. We then counted the number of reads containing each allele. We used a two-tailed binomial test that accounted for reference allele bias to evaluate the significance of the allelic bias at each SNP [as described previously (Varshney et al. 2017); implemented in a custom perl script]. We did not test SNPs in regions blacklisted by the ENCODE Consortium because of poor mappability (wgEncodeDacMapabilityConsensusExcludable.bed and wgEncodeDukeMapabilityRegionsExcludable.bed). We then selected SNPs that show significant allelic bias at a nominal threshold of binomial test P-value < 0.05 and used BEDtools intersect to identify the set of nominally significant SNPs overlapping each annotation. We defined the effect size of allelic bias as the absolute deviation from expectation, given by the absolute difference between the observed and expected fraction of reads mapping to the reference allele. We also compared the allelic bias effect sizes while only considering SNPs with MAF > 0.2.
Data availability
Workflows for analyses as described below were run using Snakemake (Köster and Rahmann 2012). All analysis steps and code to facilitate reproducibility of this work are openly shared at the GitHub repository: https://github.com/ParkerLab/regulatoryAnnotations_comparisons. Static version of scripts and all processed data are deposited at Zenodo: https://zenodo.org/record/1413623#.W8f2x1JRfpB. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7527773.
Results
Genomic distribution, coverage, and overlap of diverse regulatory annotations
To catalog super, typical, stretch enhancers, HOT regions, and broad domain regulatory annotations, we computed the number of distinct segments marked by each annotation, the length distribution of these segments, and the percentage of the genome that is covered by each annotation across the four cell types (Figure 2, A–C). Across all cell types, HOT regions comprised the greatest number of segments (Figure 2A). However, they were smaller in size (Figure 2B). Super enhancers comprised the longest segments among all annotations across the studied cell types (Figure 2B), likely due to stitching together H3K27ac peaks that are separated by ≤12.5 kb. All pairwise comparisons between segment lengths for annotations were significant (adjusted P < 2.2 × 10−06) in each of the cell types according to the Wilcoxon rank sum test followed by Bonferroni correction, highlighting the differences across annotations. While the percent genome covered by each annotation varied across cell types, these regions consistently covered <2% of the genome (Figure 2C).
Next, we calculated the fraction of overlap between all pairs of regulatory annotations. We report the Jaccard statistic (base pair-level intersection/union) for overlap between two annotations (Figure 2, D and E). We compare overlaps between different annotations within a cell type (Figure 2D) and between a single annotation (e.g., broad domains) across cell types (Figure 2E). Despite their relatively low genomic coverage (0.5% of the genome), super enhancer segments show considerable overlaps with stretch enhancer segments in the same cell type (Figure 2D), which are significantly enriched (P = 0.0001, Figure S1). This is in agreement with both of these annotations representing large domains of active enhancers marked with H3K27ac. HOT regions show extensive overlaps across cell types (Figure 2E), indicating that these regions are less cell type-specific. Broad domains display a similar pattern, though to a less pronounced degree (Figure 2E). Conversely, stretch, super, and typical enhancers show low overlaps across cell types, which indicates a higher degree of cell type specificity (Figure 2E).
Regulatory annotations comprise distinct chromatin states
Most regulatory annotations are defined using histone modification ChIP-seq profiles. However, the differences in their underlying chromatin landscape are unclear. We compared each regulatory annotation with previously reported chromatin state segmentations across all four cell types (Varshney et al. 2017) (Figure S2). Such comparisons are informative because the chromatin states (ChromHMM states) have been generated from an integrative analysis of ChIP-seq data for five diverse histone marks (H3K4me1, H3K4me3, H3K27ac, H3K36me3, and H3K27me3) resulting in 13 chromatin states encompassing active promoter (regions enriched for H3K4me3 and H3K27ac marks), enhancer (regions enriched for H3K4me1 and H3K27ac marks), transcribed (regions enriched for H3K36me3), repressed (regions enriched for H3K27me3 marks), and quiescent states (regions lacking marks) (Varshney et al. 2017). Different enhancer states, such as active enhancer 1 and 2, represent states with different levels of H3K4me1 and H3K27ac mark enrichment, and have different genomic coverage (Varshney et al. 2017). For each regulatory annotation in a particular cell type, we computed the fraction of overlap with chromatin states in the corresponding cell type and across the other three cell types (Figure S2). Generally, HOT regions and broad domains overlap with promoter-related chromatin states consistently across all four cell types, irrespective of which cell type they were called in (Figure S2, facets a1-4, b1-4). In contrast, stretch, super, and typical enhancers show a higher fraction of overlap with enhancer-related chromatin states in the corresponding cell type. Notably, stretch/super/typical enhancer regions defined in one cell type constitute mostly nonenhancer chromatin states in other cell types (Figure S2, facets c1-4, d1-4, e1-4), which further reinforces the cell type-specific nature of these annotations.
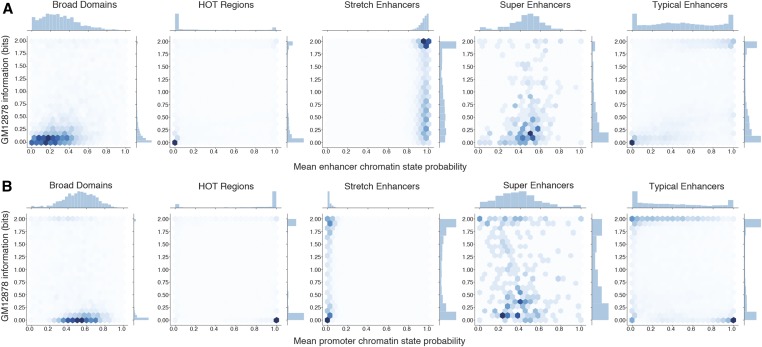
We then sought to quantify the cell type specificity of enhancer and promoter chromatin states in each regulatory annotation. For each segment of a regulatory annotation, we computed the ChromHMM posterior probabilities of being called an enhancer or active promoter state averaged over 200-bp intervals, denoting the chromatin state preference of that segment in each cell of the four cell types. We then computed the information content encoded by these probabilities across cell types (see Materials and Methods). High information content indicates high specificities of chromatin states. We observe that stretch enhancers constitute high information and a high-probability enhancer chromatin state (Figure 3A showing GM12878 annotations and Figure S3 showing annotations in all cell types), whereas HOT regions constitute low information and a high-probability promoter state (Figure 3B showing GM12878 annotations and Figure S4 showing annotations in all cell types). These analyses highlight the differences in the underlying chromatin context and cell type specificities for these annotations.
Regulatory annotations exhibit distinct cell type specificity of gene regulatory function
Regulatory annotations have been linked to common diseases based on their enrichment to overlap GWAS variants. We directly compared GWAS SNP enrichments for diseases that are relevant to the cell types represented here—such as Crohn’s disease, rheumatoid arthritis, and other autoimmune traits (relevant for LCL GM12878), and metabolic traits such as body mass index (BMI) and T2D (relevant for liver hepatocyte cell line HepG2)—in each regulatory annotation. Super and stretch enhancers in GM12878 (LCL) were generally the most enriched for autoimmune-related trait GWAS SNPs (Figure S5), whereas stretch enhancers and broad domains in HepG2 were enriched for BMI and T2D GWAS SNPs (Figure S5).
We next assessed the gene regulatory potential for these annotations using several diverse comparisons. We first measured the distance to the nearest protein-coding gene from the ends of each annotation segment, and found that broad domain and super enhancer segments tend to occur in closer proximity to gene TSSs relative to other annotations (Figure S6). Because a regulatory element does not always target the nearest gene, we next utilized cis-eQTL, which unambiguously identifies target genes by associating genetic variation (SNPs) with gene expression. We asked if regulatory annotations overlapped cis-eQTL, which were previously identified in LCLs in the genotype tissue expression (GTEx) project (GTEx Consortium 2017). HOT regions in the LCL GM12878 showed the highest enrichment to overlap LCL eQTL (Figure S7), likely because these represent active promoter regions with high TF-binding activity and lie close to protein-coding genes (Figure S6). However, HOT regions in control cell types (i.e., non-LCL) were similarly enriched to overlap LCL eQTL, which highlights the similarity of HOT regions across cell types.
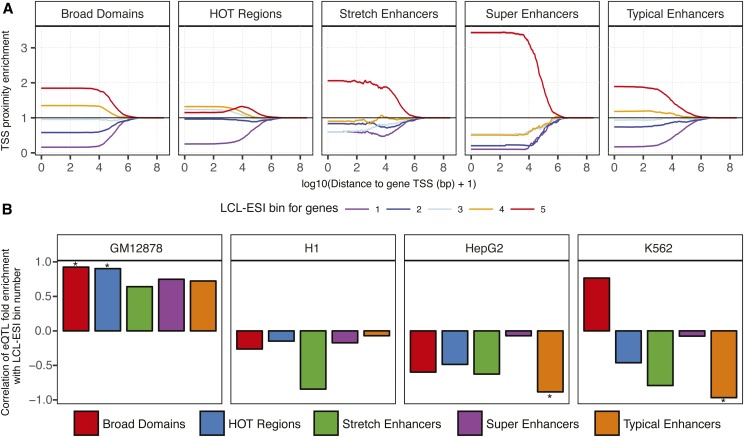
We hypothesized that significant enrichment of LCL eQTL in regulatory annotations of unrelated cell types is largely driven by eQTL for more ubiquitously expressed genes. To test this hypothesis, we classified protein-coding genes by their specificity of expression in LCLs using RNA-seq data for 50 diverse tissues from the GTEx project (GTEx Consortium 2017) and an information theory approach (Schug et al. 2005; He et al. 2014; Scott et al. 2016; Varshney et al. 2017). We calculated the expression specificities of genes by comparing the relative expression of each gene in LCLs with the entropy of the gene across all 50 tissues in the panel. We defined the LCL expression specificity index (LCL-ESI), which ranges from 0 (i.e., low or ubiquitously expressed genes) to 1 (i.e., highly and specifically expressed genes in LCL). We binned the genes into quintiles based on this LCL-ESI measure, such that bin 5 represents genes with the highest LCL-ESI scores (Figure S8). We then asked which regulatory annotations occurred closer to cell type-specific genes. We calculated the distance to the nearest TSS for genes in each LCL-ESI bin, which revealed that annotation segments occur closer to genes with higher LCL-ESI (Figure S9, colored lines). To control for the different number of segments in each annotation, we constructed a null expectation by randomly sampling genes from across the five LCL-ESI bins and calculating the distribution of distances to the nearest gene TSS (Figure S9, black). We then normalized the observed distance distribution for each LCL-ESI bin gene set with that from the null set and used this as a controlled measure of TSS proximity enrichment (Figure 4A). We observed that all regulatory annotations are depleted from occurring close to nonspecific genes (LCL-ESI bin 1) and enriched to occur closer to highly specific genes (LCL-ESI bin 5). Notably, super, stretch, and typical enhancers and broad domains were more enriched to occur near the most cell type-specific genes than HOT regions (Figure 4A). As expected, enrichments for all annotations to occur within larger distances to TSSs (order of mega bases) converge to 1 (Figure 4A), indicating a properly controlled proximity enrichment test.
Figure 4.
Proximity to protein-coding genes and enrichment for eQTL highlight functions of regulatory annotations. (A) Enrichment for regulatory annotation elements in GM12878 to lie within distances (x-axis) of TSSs of protein-coding genes binned by gene expression specificity in LCLs (LCL-ESI). Enrichment calculated in comparison to 10,000 random samplings, 95% C.I.s shown. (B) Pearson correlation of LCL-ESI gene quintile bin numbers (increasing LCL specificity) with the fold enrichment of eQTL of these genes in regulatory annotations. Positive correlation shows that the eQTL for more LCL-specific genes are more enriched in annotations. Significant (P < 0.05) correlations are marked with “*.” eQTL, expression QTL; ESI, expression specificity index; HOT, high-occupancy target; LCL, lymphoblastoid cell line; TSS, transcription start site.
We next asked which regulatory annotations were more enriched to overlap eQTL of more cell type-specific genes. We obtained sets of LCL eQTL (GTEx Consortium 2017) for genes in each LCL-ESI bin and calculated the enrichment of each eQTL set in the regulatory annotations. Indeed, we observed that GM12878 regulatory annotations were increasingly enriched to overlap eQTL for highly LCL-specific genes (Figure S10) and that the fold enrichment for eQTL in a bin is positively correlated with the LCL-ESI bin number (Figure 4B, GM12878 facet). Notably, stretch enhancers and, in some instances, typical enhancers in non-LCL cell types showed strong negative correlations of LCL eQTL fold enrichment with LCL-ESI bin number (Figure 4B), indicating higher cell type specificity for stretch enhancers. This is consistent with the previous histone modification-based chromatin state analyses (Figure 3 and Figures S2–S5), which also highlight the cell type specificity of stretch enhancers. HOT regions in non-LCL cell types show high enrichments for eQTL in less cell type-specific LCL-ESI bins 1–3 (Figure S10). This analysis shows that high enrichments of LCL eQTL in non-LCL annotations (Figure S7) were driven by eQTL for more ubiquitously expressed genes. These analyses further emphasize the differences in the cell type specificities of these regulatory annotations.
Patterns of expression and chromatin QTL effect sizes in annotations suggest regulatory buffering
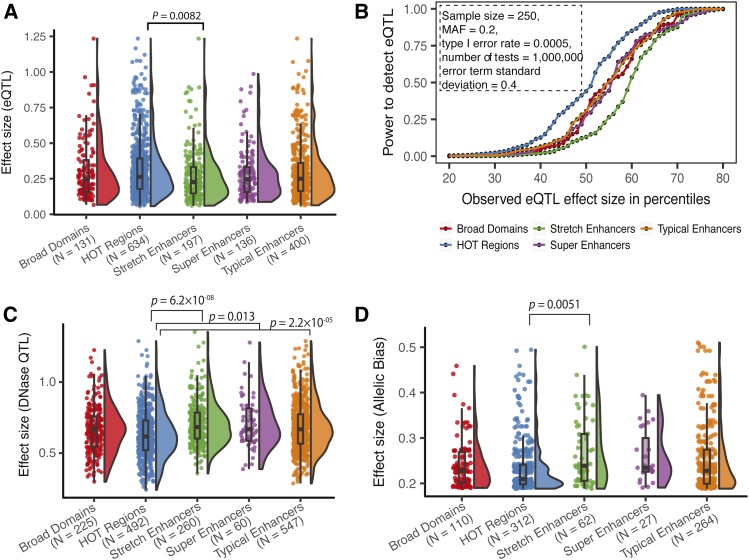
While enriched overlap with eQTL demonstrates genetic regulatory potential for each annotation (Figure 4B and Figures S7 and S10), this analysis does not distinguish the strength of these genetic effects on gene expression. To understand this, we compared the absolute effect sizes (β values from the linear regression models) of LCL eQTL overlapping different GM12878 regulatory annotations. We excluded SNPs with MAF < 0.2, since these SNPs have substantially reduced statistical power and are therefore biased to be detected as eQTL only with higher effect sizes (Figure S11). We observed that LCL eQTL in GM12878 stretch enhancers have nominally significantly lower (P = 0.032) effect sizes than GM12878 HOT regions; however, this comparison does not survive a Bonferroni correction accounting for 10 pairwise tests (Figure S12A). To achieve higher power for such an analysis, we utilized the larger GTEx blood eQTL data set and compared effect sizes in annotations of the blood relevant leukemia cell line K562. Consistent with the LCL analysis, we observed that effect sizes of blood eQTL in K562 stretch enhancers were significantly lower than those of HOT regions (Bonferroni corrected P = 0.0082, Figure 5A). We note that the differences in effect sizes for LCL and blood eQTL are largely due to different sample sizes, and therefore power to detect eQTL. To further control for potential sources of bias in this analysis, we next asked if this effect size difference was driven by distance to the eQTL target gene’s TSS or the number of SNPs in high LD with the index eQTL SNP. We modeled the eQTL absolute effect size using linear regression, including these additional two covariates along with an indicator variable encoding stretch enhancer or HOT region annotation (eQTL overlapping both annotations were not considered). We observed a significant effect on the indicator variable (P = 0.005, regression coefficient = −0.0521, Table S1), which confirms the smaller effect size of eQTL in stretch enhancers, independent of TSS distance and LD structure.
Figure 5.
Gene expression and chromatin QTL effect size differences in regulatory annotations suggest regulatory buffering. (A) Distribution of eQTL effect sizes for blood eQTL (GTEx v7, 10% FDR) in K562 regulatory annotations. (B) Power to detect eQTL after Bonferroni correction at effect sizes corresponding with the 10th through the 90th percentile observed for each annotation [shown in (A)]. Other constant parameters for the power calculation are shown in box. (C) Distribution of effect sizes for LCL DNase QTL in GM12878 regulatory annotations. (D) Distribution of effect sizes (deviation from expectation) for SNPs with significant allelic bias in GM12878 ATAC-seq (P < 0.05, minimum coverage at SNP = 30, reads downsampled to 30, see Materials and Methods) in GM12878 regulatory annotations. P-values from Wilcoxon rank sum tests, after a Bonferroni correction accounting for 10 pairwise tests. Number of QTL/allelic biased SNPs overlapping each regulatory annotation is shown in parentheses in (A, C, and D). ATAC-seq, Assay for transposase accessible chromatin-sequencing; eQTL, expression quantitative trait loci; FDR, false discovery rate; HOT, high-occupancy target; LCL, lymphoblastoid cell line; MAF, minor allele frequency.
Differences in effect sizes of eQTL in stretch enhancers compared to HOT regions directly translates to differences in the statistical power to detect eQTL residing in these regulatory annotations, which have remarkably distinct cell type specificities. To quantify this, we performed a power calculation for the 10th through the 90th percentiles of the eQTL effect size distribution observed in each annotation, keeping other parameters such as sample size, MAF, type 1 error rate, number of tests, and the SD of the error term constant. We show that variants in stretch enhancers have nearly uniform lower power to be detected as eQTL across the effect size distribution (Figure 5B and Figure S12B). Indeed, stretch enhancers showed lower enrichment to overlap eQTL than HOT regions (Figure S7). Therefore, identifying eQTL in cell type-specific stretch enhancers will require larger sample sizes.
Among other mechanisms, eQTL SNPs can influence gene expression in vivo by modulating TF binding. TFs can either bind in nucleosome-depleted regions or bind and displace nucleosomes (pioneer factors) (Gross and Garrard 1988; Wang et al. 2012; Buenrostro et al. 2013). Therefore, QTL analysis of chromatin accessibility using dsQTL can assess variant effects on regulatory element activity. Interestingly, we found that LCL dsQTL (Degner et al. 2012) in stretch enhancers have significantly higher effect sizes than those in HOT regions (Bonferroni corrected P = 6.2 × 10−08, Figure 5C), which is the opposite of what we observed for eQTL effects (Figure 5A). dsQTL in super enhancers and typical enhancers also have higher effect sizes than those in HOT regions (Bonferroni adjusted P = 0.013, 2.2 × 10−05, respectively). To examine the effect of genetic variation on open chromatin at the resolution of an individual sample, we quantified allelic bias in the assay for transposase-accessible chromatin followed by sequencing (ATAC-seq) data available in GM12878 (Buenrostro et al. 2013). Allelic bias measured by quantifying the ATAC-seq signal over each of the two alleles at a heterozygous site is an indicator of allelic differences in chromatin accessibility at a specific locus. To control for different power to detect allelic bias, we uniformly downsampled all SNPs to 30× coverage. We included all SNPs from the full range of MAFs with nominally significant allelic bias (P < 0.05) since the SNP MAF does not affect the power to detect allelic bias in an individual sample. Consistent with the dsQTL results, we observed that SNPs in stretch enhancers show a significantly larger allelic bias effect size (see Materials and Methods) compared to HOT regions (Bonferroni corrected P = 0.0051, Figure 5D). This trend remains after removing SNPs with MAF < 0.2, similar to the dsQTL analyses above (Figure S13), indicating that SNP MAF does not confound this analysis. No other pairwise tests were significant. Collectively, these observations show that stretch enhancers harbor variants that have strong genetic effects on chromatin changes, but these are buffered at the level of transcription.
Discussion
We performed a comparative analysis of five regulatory annotations, all based on diverse epigenomic signatures, to better understand their regulatory capacity and downstream transcriptional effects. We observed that stretch, super, and typical enhancers overlap enhancer chromatin states in the corresponding cell type, but overlap nonenhancer chromatin states in unrelated cell types, supporting the cell type specificity of these regulatory elements. These observations highlight H3K27ac as a good proxy for cell type-specific regulatory function. Annotations based on the H3K4me3 mark (broad domains) and TF binding (HOT regions) show a large fraction (>40%) of overlaps with promoter chromatin states across different cell types. Consistent with our observations, a recent study in the fly reported that regions bound by large numbers of TFs (such as HOT regions) are less cell type-specific (Kudron et al. 2017). While the diverse ChIP-seq data used to define regulatory annotations comes from different individuals, we note that future studies using ChIP-seq data from the same individual might have even higher power to detect cell type-specific differences.
Analysis of genetic effects on the gene regulatory function of annotations revealed that blood eQTL in K562 stretch enhancers have significantly lower effect sizes compared to HOT regions. Stretch/super enhancers are known to regulate more cell type-specific genes for which the expression levels may be tightly controlled under basal conditions. Multiple studies have observed redundancy in gene regulation by individual components of super enhancers (Hay et al. 2016; Shin et al. 2016; Moorthy et al. 2017; Xie et al. 2017). Such studies then contested the notion of super/stretch enhancers as a distinct entity, arguing that these annotations are no different from other enhancers. However, here we offer an alternative explanation: that enhancer buffering, which results from functional redundancy, could be a mechanism for tighter control of gene expression under basal conditions and would explain the low observed eQTL effect sizes. These regions could encode regulatory plasticity, allowing critical genes to respond to multiple (patho)physiologic stimuli. This would lead to smaller effects in the steady state, whereas each component could contribute to tight but pliable regulation by different signaling pathways. Therefore, the outcome of perturbing enhancer components might be different in response to different environmental stimuli, and existing studies that probe basal conditions would not detect such effects.
In contrast, genetic variants associated with open chromatin in stretch enhancers show significantly higher effects than those in HOT regions, both within a single sample (allelic bias in ATAC-seq) and across multiple samples (dsQTL). Our results present an apparent discrepancy in that genetic variants in stretch enhancers display higher chromatin QTL effect sizes and slightly but significantly lower basal expression QTL effect sizes when compared to HOT regions. It is possible that the large constellation of TFs bound in HOT regions (ENCODE Project Consortium 2012; Kudron et al. 2017) maintain more constitutively open chromatin, which would be less susceptible to effects of individual genetic variants. This concept of buffering has been demonstrated previously, where a smaller fraction of SNPs in strong DNase peaks showed significant allelic bias compared to those in weak DNase peaks (Maurano et al. 2015). We reason that chromatin accessibility, which influences TF binding, could be a molecular feature of the initial response cascade to propagate gene expression changes under stimulatory conditions. We hypothesize that the larger genetic effects on stretch enhancer chromatin accessibility will propagate to gene expression effects under specific environmental conditions. Under this hypothesis, we expect that many dsQTL will be associated with gene expression under specific stimuli (or response-specific eQTL) rather than steady state (basal eQTL). In support of this, a recent study in the macrophage model system (Alasoo et al. 2018) showed that ∼60% of eQTL that manifest upon stimulation are chromatin QTL in the basal state. Unfortunately, currently available response expression or chromatin QTL data sets are underpowered for a comparison of effect sizes in the regulatory annotations analyzed here, owing to low sample sizes.
Our observations could help reconcile why many cis-eQTL are shared across cell types and infrequently colocalize with GWAS signals (GTEx Consortium 2017; Huang et al. 2017; Liu et al. 2017). We have shown that while stretch enhancers are enriched to overlap GWAS loci for cell type-relevant traits, variants in these regions are underpowered to be identified as eQTL. Current eQTL studies are biased to identify eQTL for more broadly expressed genes. Our results suggest that larger sample sizes will be needed to identify cell type-specific eQTL. Additionally, our results suggest the need to perform response eQTL studies under carefully selected environmental conditions.
Acknowledgments
We thank the Parker laboratory members for their feedback. We acknowledge funding from an American Association for University Women International Doctoral Fellowship, a Barbour Doctoral Scholarship, and the University of Michigan Rackham Predoctoral Fellowship (awarded to A.V.), as well as National Institute of Diabetes and Digestive and Kidney Diseases grant R00 DK-099240, American Diabetes Association Pathway to Stop Diabetes grant 1–14-INI-07 and grant R01 DK-117960 (awarded to S.C.J.P.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7527773.
Communicating editor: E. Hauser
Literature Cited
- Adli M., Zhu J., Bernstein B. E., 2010. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat. Methods 7: 615–618. 10.1038/nmeth.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A. J., Mann A. L., et al. , 2018. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50: 424–431. 10.1038/s41588-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun B. A., Pollina E. A., Ucar D., Mahmoudi S., Karra K., et al. , 2014. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688 [corrigenda: Cell 163: 1281–1286 (2015)] 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., et al. , 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Blinka S., Reimer M. H., Pulakanti K., Rao S., 2016. Super-enhancers at the nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep. 17: 19–28. 10.1016/j.celrep.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P., Araya C. L., Brdlik C., Cayting P., Cheng C., et al. , 2014. Comparative analysis of regulatory information and circuits across distant species. Nature 512: 453–456. 10.1038/nature13668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Chen Z., Wu D., Zhang L., Lin X., et al. , 2015. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 47: 1149–1157. 10.1038/ng.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., et al. , 2014. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24: 1–13. 10.1101/gr.164079.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave K., Sur I., Yan J., Zhang J., Kaasinen E., et al. , 2017. Mice deficient of Myc super-enhancer region reveal differential control mechanism between normal and pathological growth. eLife 6: e23382 10.7554/eLife.23382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Pai A. A., Pique-Regi R., Veyrieras J.-B., Gaffney D. J., et al. , 2012. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482: 390–394. 10.1038/nature10808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X., T.-W. Chang, S. T. Weiss, and W. Qiu, 2017 powerEQTL: power and sample size calculation for eQTL analysis. https://CRAN.R-project.org/package=powerEQTL [DOI] [PMC free article] [PubMed]
- ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M., 2012. ChromHMM: automating chromatin state discovery and characterization. Nat. Methods 9: 215–216. 10.1038/nmeth.1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., et al. , 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T., 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159–197. 10.1146/annurev.bi.57.070188.001111 [DOI] [PubMed] [Google Scholar]
- GTEx Consortium , 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D., Hughes J. R., Babbs C., Davies J. O. J., Graham B. J., et al. , 2016. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 48: 895–903. 10.1038/ng.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Chen C., Teng L., Tan K., 2014. Global view of enhancer–promoter interactome in human cells. Proc. Natl. Acad. Sci. USA 111: E2191–E2199. 10.1073/pnas.1320308111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A., Webber C., Goodson M., Ponting C. P., Lunter G., 2013. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29: 2046–2048. 10.1093/bioinformatics/btt343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., et al. , 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106: 9362–9367. 10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., et al. , 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Schuijers J., Lin C. Y., Weintraub A. S., Abraham B. J., et al. , 2015. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58: 362–370. 10.1016/j.molcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Fang M., Jostins L., Mirkov M. U., Boucher G., et al. , 2017. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547: 173–178. 10.1038/nature22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S., 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Kudron M. M., Victorsen A., Gevirtzman L., Hillier L. W., Fisher W. W., et al. , 2017. The modERN resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics 208: 937–949. 10.1534/genetics.117.300657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E. Z., Stampfel G., Yáñez-Cuna J. O., Dickson B. J., Stark A., 2012. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 26: 908–913. 10.1101/gad.188052.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T., Sammeth M., Friedländer M. R., ’t Hoen P. A., Monlong J., et al. , 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv: 13033997 [q-bio.GN].
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu F., Ren C., Bo X., Shu W., 2016. Genome-wide identification and characterisation of HOT regions in the human genome. BMC Genomics 17: 733 10.1186/s12864-016-3077-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Y., Erkek S., Tong Y., Yin L., Federation A. J., et al. , 2016. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530: 57–62. 10.1038/nature16546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Finucane H. K., Gusev A., Bhatia G., Gazal S., et al. , 2017. Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am. J. Hum. Genet. 100: 605–616. 10.1016/j.ajhg.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Hoke H. A., Lin C. Y., Lau A., Orlando D. A., et al. , 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Haugen E., Sandstrom R., Vierstra J., Shafer A., et al. , 2015. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 47: 1393–1401 [corrigenda: Nat. Genet. 48: 101 (2016)] 10.1038/ng.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., et al. , 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Roy S., Ernst J., Kharchenko P. V., Kheradpour P., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C., Sun L. V., Wang J., de Wit E., Talhout W., et al. , 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 12027–12032. 10.1073/pnas.0605003103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy S. D., Davidson S., Shchuka V. M., Singh G., Malek-Gilani N., et al. , 2017. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27: 246–258. 10.1101/gr.210930.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S. C. J., Stitzel M. L., Taylor D. L., Orozco J. M., Erdos M. R., et al. , 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA 110: 17921–17926. 10.1073/pnas.1317023110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L., Gaulton K. J., Rodríguez-Seguí S. A., Mularoni L., Miguel-Escalada I., et al. , 2014. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46: 136–143. 10.1038/ng.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D. X., Erdos M. R., Parker S. C. J., Collins F. S., 2015. Motif signatures in stretch enhancers are enriched for disease-associated genetic variants. Epigenetics Chromatin 8: 23 10.1186/s13072-015-0015-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Schmidt E. M., Zhang J., Zhou W., Chen J., Mohlke K. L., et al. , 2015. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 31: 2601–2606. 10.1093/bioinformatics/btv201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J., Schuller W.-P., Kappen C., Salbaum J. M., Bucan M., et al. , 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6: R33 10.1186/gb-2005-6-4-r33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J., Erdos M. R., Huyghe J. R., Welch R. P., Beck A. T., et al. , 2016. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat. Commun. 7: 11764 10.1038/ncomms11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. Y., Willi M., Yoo K. H., Zeng X., Wang C., et al. , 2016. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 48: 904–911. 10.1038/ng.3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau A., Márquez E. J., Shin D.-G., Vera-Licona P., Ucar D., 2017. Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin. Sci. Rep. 7: 14466 10.1038/s41598-017-14389-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Sandor C., Han B., Xu H., Stranger B. E., et al. , 2013. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45: 124–130. 10.1038/ng.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Geijn B., McVicker G., Gilad Y., Pritchard J. K., 2015. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 12: 1061–1063. 10.1038/nmeth.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney A., Scott L. J., Welch R. P., Erdos M. R., Chines P. S., et al. , 2017. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl. Acad. Sci. USA 114: 2301–2306. 10.1073/pnas.1621192114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhuang J., Iyer S., Lin X., Whitfield T. W., et al. , 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22: 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., et al. , 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Duan J., Li B., Zhou P., Hon G. C., 2017. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66: 285–299.e5. 10.1016/j.molcel.2017.03.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Workflows for analyses as described below were run using Snakemake (Köster and Rahmann 2012). All analysis steps and code to facilitate reproducibility of this work are openly shared at the GitHub repository: https://github.com/ParkerLab/regulatoryAnnotations_comparisons. Static version of scripts and all processed data are deposited at Zenodo: https://zenodo.org/record/1413623#.W8f2x1JRfpB. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7527773.