Abstract
Mutations are the ultimate source of all genetic variation. However, few direct estimates of the contribution of mutation to molecular genetic variation are available. To address this issue, we first analyzed the rate and spectrum of mutations in the Arabidopsis thaliana reference accession after 25 generations of single-seed descent. We then compared the mutation profile in these mutation accumulation (MA) lines against genetic variation observed in the 1001 Genomes Project. The estimated haploid single nucleotide mutation (SNM) rate for A. thaliana is 6.95 × 10−9 (SE ± 2.68 × 10−10) per site per generation, with SNMs having higher frequency in transposable elements (TEs) and centromeric regions. The estimated indel mutation rate is 1.30 × 10−9 (±1.07 × 10−10) per site per generation, with deletions being more frequent and larger than insertions. Among the 1694 unique SNMs identified in the MA lines, the positions of 389 SNMs (23%) coincide with biallelic SNPs from the 1001 Genomes population, and in 289 (17%) cases the changes are identical. Of the 329 unique indels identified in the MA lines, 96 (29%) overlap with indels from the 1001 Genomes dataset, and 16 indels (5% of the total) are identical. These overlap frequencies are significantly higher than expected, suggesting that de novo mutations are not uniformly distributed and arise at polymorphic sites more frequently than assumed. These results suggest that high mutation rate potentially contributes to high polymorphism and low mutation rate to reduced polymorphism in natural populations providing insights of mutational inputs in generating natural genetic diversity.
Keywords: mutation rate, mutation accumulation line, Arabidopsis thaliana, transposable element, natural polymorphism, indel
MUTATIONS contribute to genetic variation—the substrate of evolution. Thus, a full understanding of evolution requires knowledge of the rate of spontaneous mutation and the fitness consequences of new mutational inputs. Observable genetic variation in the real world is generated and maintained through multiple processes; for instance, genetic variation could be a balance between mutational input, natural selection, drift, and gene flow (Wright 1988; Hartl and Clark 2006). However, what exactly determines the extent of genetic diversity in different species is still unclear (Leffler et al. 2012). To understand the contribution of mutation to patterns of genetic variation, we need to know the degree to which intragenomic variation of mutational inputs contributes to the reported differences in genetic diversity along the genome. Variation in the activity and effectiveness of the molecular machinery that detects and repairs such errors directly influences genome-wide mutation rates (Hoffman et al. 2004; Turrientes et al. 2013). These, in turn, are affected by localized factors, such as heterozygous status (Yang et al. 2015), transcription (Park et al. 2012; Chen et al. 2016), and recombination (Lercher and Hurst 2002; Zhang and Gaut 2003; Yang and Gaut 2011). Ultimately, a better knowledge of mutation rates and sources of their variation will help us to understand the relative roles of mutation and selection in excesses or deficits of substitutions in the genome.
Because de novo mutations in each generation are rare, mutation rate estimates are commonly derived from comparisons over long phylogenetic distances, or by making inferences from observed genetic diversity in extant populations. However, such estimates are not only affected by selection on mutations, but they are also greatly confounded by genetic processes, including variation in recombination and mutation rates as well as gene conversion, and demographic factors such as the population size and its associated expansions and contractions. An alternative is the analysis of pedigrees over very short evolutionary time frames, by comparing parents and their direct offspring, or by experimentally studying the accumulation of mutations in inbred lineages over a limited number of generations, i.e., mutation accumulation (MA) lines. During the propagation of MA lines kept at small population size under benign conditions, the drift-dominant condition allows mutations with all but the most extreme negative effects on fitness to accumulate in the genome (Halligan and Keightley 2009).
Although precise estimates of whole genome mutation rates in multicellular organisms have been made, we are still far from understanding fine-scale intragenomic variation of mutation rates, primarily because the number of mutations observed has been small. A further limitation of several previous studies has been that they were based on early versions of Illumina short-read sequencing technology, and therefore were limited in detection of insertions and deletions (indels). Both problems apply to mutational estimates for land plants, where MA line sequences have only been analyzed for A. thaliana (Ossowski et al. 2010; Jiang et al. 2014; Exposito-Alonso et al. 2018).
Here, we use whole-genome sequencing to characterize the mutation profile from 107 Arabidopsis thaliana MA lines of the reference accession Col-0 at the 25th generation. With the analysis of this population, we increase by more than an order of magnitude the number of spontaneous mutations documented in this species. We use this information to address the following questions: what are the spectrum, frequency, and pattern of spontaneous mutations in A. thaliana? How does the distribution of spontaneous mutation in the genome compare with the pattern of genetic variation observed in the wild?
Materials and Methods
MA line sequencing
Seeds of the 24th generation A. thaliana MA lines are from an experiment described previously (Shaw et al. 2000, 2002). Briefly, the 107 MA lines were derived from a single founder of the Col-0 accession. The MA lines were maintained in the greenhouse by single-seed descent to reduce selection and maximize drift. An additional 25th generation was propagated in the University of Maryland greenhouse (Rutter et al. 2010). Fitness of these MA lines has been assessed extensively in different environments (Shaw et al. 2000, 2002; Rutter et al. 2010, 2012b; Roles et al. 2016). DNA was extracted from the leaf tissue germinated from the seeds of the 25th generation MA lines, and sequenced on the Illumina HiSeq3000 platform (2 × 150 bp) at the Max Planck Institute for Developmental Biology. The average read depth per MA line was 36×.
Identification of mutations
Sequencing reads from each MA line were mapped to the A. thaliana TAIR10 reference genome (The Arabidopsis Information Resource at www.arabidopsis.org) using NextGenMap (Sedlazeck et al. 2013), then sorted, indexed, and assigned line identification numbers with Picard Tools v1.136 (https://broadinstitute.github.io/picard/). The sequence reads were then locally realigned using GATK v3.6 (McKenna et al. 2010). Duplicate reads were marked and removed with Picard Tools. SNM and indel variants for each line were identified using GATK’s HaplotypeCaller tool. The resulting individual gVCF files were merged using GATK’s GenotypeGVCFs tool. Plastid and mitochondrion variants were searched with parameter settings for haploid. The fixed differences between the MA line ancestor and the TAIR reference genome were identified as the shared derived homozygous variants, SNMs, and indels, across all sequenced MA lines. The final mutations were filtered for the unique homozygous variants that are present in a single or multiple MA lines.
The presence of large structural variants, including deletions, duplications, insertions, inversions, and translocations, were quantified using the Delly software package (Rausch et al. 2012). The MA line specific variants that passed Delly’s quality filtering were examined using the integrated genome viewer (IGV) v2.3.82 (Robinson et al. 2011). Detection of de novo TE insertions was attempted with the jitterbug software package (Hénaff et al. 2015). Genomic locations of the mutations were annotated using the SnpEff v4.2 (Cingolani et al. 2012) with default settings.
The haploid mutation rate was estimated with the equation µ = m/(L*n*T) (Denver et al. 2009), where µ is the mutation rate per nucleotide site per generation, m is the number of identified SNMs or indels, L is the number of MA lines, n is the number of reference nucleotide sites accessible for variant calling, and T is the number of generations. To count the number of reference nucleotide sites accessible for variant calling, we used the -allSite option in GATK’s GenotypeGVCFs tool, which reports both variant and nonvariant sites. Because GATK estimates the genotype quality for variant and nonvariant sites differently, we used read depth to filter both variant and nonvariant sites. For variants, three filtering criteria were used: variant quality (QD) >30, read depth after filtering at a specific line (DP) >3, and number of called alleles (AN) >107, equivalent to one-half of the 107 diploid MA lines having called genotypes. A mutation was called if it is a homozygous alternative allele in a single MA line. For nonvariant sites, sites with read depth <3 and phred-scaled quality score (QUAL) <30 were not considered. We report estimated mutation rates from the 48 MA lines that do not share mutations with other. All other analyses were based on all 107 MA lines. The bash scripts for the analyses are included in the Supplemental material. The distribution of the number of mutations per line was tested for overdispersion using the dispersiontest function in AER package (Kleiber and Zeileis 2008) in R v.3.3.1 (R Core Team 2017). We also adjusted the number of SNMs dividing by the proportion of accessible reference sites of each line.
To validate mutations experimentally, we randomly selected 10 MA lines for PCR amplification and Sanger sequencing. We also analyzed the targeted mutations in five subfounder lines that were three generations descended from the same founder of the MA lines, with the propagation of subfounder lines having been independent from the MA lines. We identified variants in the subfounder lines using the same pipeline as for the MA lines.
To test the accuracy of our mutation calling pipelines, we performed two types of simulations. In the first simulation, we simulated 500 random point mutations throughout the genome, and investigated whether our pipeline recovered the simulated mutations (Ness et al. 2012). We simulated independent mutations in five copies of the reference genome such that simulated mutations would be positions in one reference genome but not in the other four reference genomes. Twenty mutations per chromosome, and a total of 100 mutations for each reference, were simulated. Five MA lines (lines 24, 31, 36, 62, and 109), which have different reference genome coverage depth, were chosen to test whether our pipeline can recover the simulated mutations regardless coverage depth variation. Original reads of the MA lines were individually mapped to one of the mutated reference genomes. We then called the genotype using the GATK’s GenotypeGVCFs individually for each MA line, and report the number of recovered simulated mutations and the number of accessible nonvariant sites. In the second simulation, we introduced homozygous mutations at the sequencing reads in the bam file using Bamsurgeon tool (https://github.com/adamewing/bamsurgeon; Ewing et al. 2015). We changed the basecalls of the sequencing reads in the original bam file of the same five MA lines that we selected previously for reference mutation simulation. We then call variants (GVCF files) on these five modified bam files individually using HaplotyeCaller in GATK. By combining these five GVCF files with the original GVCF files of the rest of MA lines, we call and filter SNMs jointly across all lines using GenotypeGVCFs in GATK. However, we noted that due to the modification of the bam file, HaplotypeCaller targeted regions with simulated mutations for de novo assembly and hence altered the mapping position of the modified bases, resulting in bam files that are different from the original ones, and positions that have higher than 10× coverage are less affected by the HaplotypeCaller de novo assembly. We therefore reported the number of recovered SNMs at different read depth coverage.
DNA methylation
DNA methylation data were retrieved from publicly available A. thaliana datasets GSM980986 and GSM980987 (Stroud et al. 2013). Methylation levels for cytosines at each CG, CHG, and CHH (H refers to A, T, or G) context were reported in both data sets. Cytosines with methylation levels above 0.1 in one of the data sets were considered as methylated sites.
Interaction between methylation, TE, and chromosome regions
A logistic regression framework was used to test main and interactive effects of cytosine methylation, TE, and chromosome regions on the likelihood of a given nucleotide being mutated. The analysis conducted in R v.3.3.1 (R Core Team 2017) included each genomic position as a record and whether a position underwent mutation based on the total mutations identified among all MA lines. Three categorical predictor variables were methylation (methylated or nonmethylated cytosine), TE (TE or non-TE bases), and chromosome regions (chromosome arm or pericentromeric/centromeric sites). The response variable depended on whether a given nucleotide is mutated. Akaike information criterion (AIC) scores were used to assess the fit of the three models that (1) only includes main effects, (2) includes main effects and two-way interactions, and (3) includes main effects and two and three-way interactions.
Genetic variation in natural populations
VCF variant files of the 1001 Genomes project were downloaded from the project website (http://1001genomes.org) to assess SNP positions and allele frequency. Ancestral and derived SNPs were identified based on a three-way whole-genome alignment between A. thaliana (TAIR10), A. lyrata, and Capsella rubella using progressive Cactus (Paten et al. 2011a,b). Subalignments with single sequence from each of the three species were extracted as homologous regions using the HAL tools (Hickey et al. 2013). Of the alignable regions, sites identical in the three species are considered as conserved sites in A. thaliana, and SNPs found in conserved sites are defined as derived variants. Sites that are identical between A. lyrata and C. rubella but different in A. thaliana were also extracted. SNPs found among at sites where the alternative allele is identical to C. rubella and A. lyrata were defined as ancestral variants.
Data availability
The FASTQ and BAM files of each MA line have been uploaded to NCBI Short Read Archive (SRA) with the accession number of SRP133100. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6456065.
Results
Mutation spectrum and rate
We utilized a population of MA lines that were derived from a single founder in the Col-0 reference background (Shaw et al. 2000). Because A. thaliana is self-fertilizing, and because the founder line was advanced through three generations of selfing in the laboratory before establishment of the MA lines, we expect the founder to have been highly homozygous; the genetic differences among MA lines should be due overwhelmingly to new mutations that have arisen since divergence from the founder. Lines had been advanced by single seed descent, minimizing selection on new mutations to the extent possible. We sequenced 107 of these MA lines from generation 25 of the experiment at an average read depth coverage of 36× per line, making on average 111 Mb (93%) of the reference genome accessible for variant calling (Supplemental Material, Table S1). When compared to the Col-0 reference genome, a total of 292 single nucleotide mutations (SNMs) and 792 indels (332 deletions and 460 insertions) are found in all 107 MA lines, i.e., must have already been fixed in the founder of the MA lines (File S1). These SNMs and indels are also found in the sequences of third generation subfounder lines that are descendants of the same founder, confirming the presence of these fixed differences between the MA line founder and the Col-0 reference genome.
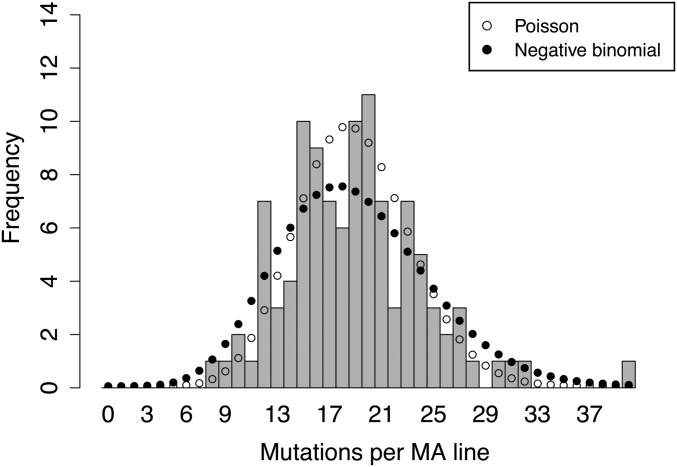
There are 1694 unique SNMs; in addition, 159 SNMs are shared by multiple MA lines, for an average of 18.9 SNMs per line (Files S1–S3). Among 107 MA lines, 59 MA lines have at least one shared mutation. Based on the 48 MA lines that do not share mutations with other lines, we estimate a haploid (or gamete) mutation rate of 6.95 × 10−9 (SE ± 2.68 × 10−10) per site per generation (Table S1). The total number of SNMs varies considerably among MA lines, ranging from 8 to 40 SNMs in a single line (Table S1). These differences do not correlate with sequence coverage (P = 0.35) but correlate with the number of accessible reference sites (P = 0.007). In addition the distribution of total SNMs per MA line fits both Poisson (χ2 = 14.0, d.f. = 13, P = 0.38) and negative binomial distributions (χ2 = 10.4, d.f. = 13, P = 0.66) where the overdispersion is not significant (dispersion test, P = 0.09) (Figure 1). The significant correlation with accessible reference sites (P = 0.02) and nonsignificant overdispersion (dispersion test, P = 0.24) are also found for the 48 MA lines that do not have shared mutations. When adjusted by the number of accessible reference sites, the distribution of unique SNMs per MA line still fits both Poisson (χ2 = 6.0, d.f. = 6, P = 0.42) and negative binomial distributions (χ2 = 2.9, d.f. = 6, P = 0.82) with nonsignificant overdispersion (dispersion test, P = 0.23).
Figure 1.
Distribution of SNM numbers in each of the 107 A. thaliana MA lines. The expected numbers for best-fit Poisson (lambda = 18.9) and negative binomial (mean = 18.9, overdispersion = 1.74) distributions are shown as open and closed circles, respectively. The distribution of SNMs per MA line fits both negative binomial (χ2 = 10.4, d.f. = 13, P = 0.66) and Poisson distributions (χ2 = 14.0, d.f. = 13, P = 0.38). For chi-square analysis, bins with few counts were combined so that every bin has at least five counts. The distribution includes 1694 unique and 159 shared SNMs.
The 356 indels (329 unique and 27 shared between lines), including 212 deletions and 144 insertions, provide an estimated indel mutation rate of 1.30 × 10−9 (±1.07 × 10−10) per site per generation (Files S1–S3). There are significantly more deletions than insertions (Fisher’s Exact test, P = 0.02). Seven deletions are larger than 100 bp, and four of these large deletions overlap open reading frames. When excluding these seven large deletions, the remaining deletions are still significantly larger (mean = 6.5 bp) than insertions (mean = 3.9 bp) (two sample t-test, t = −2.3, d.f. = 319.9, P = 0.02). None of the insertions are larger than 100 bp. We did not detect any novel transposable element (TE) insertions in the MA lines using the jitterbug software (Hénaff et al. 2015). The distributions of indels and SNMs among genomic regions are similar to each other (Figure S1). Assuming that primarily nonsynonymous SNMs, including gains and losses of stop codons, and indels in coding regions affect fitness, we estimate a diploid genomic mutation rate affecting fitness of 0.16 (±0.01) per generation.
Mutation validation
To experimentally validate mutations, we randomly selected 10 MA lines for PCR and Sanger sequencing of 96 regions with mutations. Of these 96, 92 were amplified successfully by PCR and sequenced. Sanger sequencing confirmed 78 of 79 SNMs and 15 of 17 indels, including five sets of complex/double mutations and three SNMs that are shared by two MA lines (File S1). Our confirmation rates are comparable to a recent MA line study in Chlamydomonas reinhardtii (SNMs: 98.7% vs. 98.3%; indels: 88.2% vs. 90.5%) (Ness et al. 2015) and almost the same as in a previous A. thaliana MA line study (Ossowski et al. 2010). To investigate whether the mutations we identified could be due to ancestral heterozygosity in the founder, we sequenced five third-generation subfounder lines that are descendants of the same founder of MA lines. None of the unique SNMs and indels are variants in any of the five subfounder lines.
To test the accuracy of our mutation calling pipelines, we performed two types of simulation and investigated whether our pipeline recovered the simulated mutations (Ness et al. 2012). In the first simulation, we simulated random point mutations throughout the genome. Original reads of five MA lines with different genome read depth coverage were individually mapped to one of the mutated reference genomes. For each mutated reference genome, 100 random point mutations were simulated (File S5). Based on the filtering threshold, we determined that on average 107 of the 119 Mb reference genome can be confidently called. From the 500 simulated mutations, we therefore expected to identify 450 point mutations. Our pipeline recovered 445 (98.9%) simulated point mutations (Table S2). In the second simulation, we introduced homozygous mutations at the sequencing reads. After filtering the modified positions that have higher than 10× read depth coverage, we recovered 72–90% of the homozygous point mutations (Table S3).
Mutations shared between MA lines
Our observation of mutations shared between MA lines could have multiple explanations. (1) Shared mutations are at sites that were heterozygous at the founder genotype. (2) Mapping errors cause incorrect mutation calling. (3) Shared mutations occurred independently at mutation hotspots in multiple lines. (4) Some MA lines were split into multiple lines during the MA line propagation. The first three potential explanations do not appear to be consistent with the validation results. By sequencing the subfounder lines, we show that of 186 shared mutations (159 SNMs and 27 indels), 183 cases are nonvariant in all five subfounder lines, two are heterozygous in one of the subfounder lines, and one varies among subfounder lines (File S4). We therefore conclude that, except for the last three cases, 183 out of the 186 shared mutations are not likely due to ancestral heterozygosity. The PCR and Sanger sequencing confirmed three SNMs that were shared by two MA lines (File S1), excluding the possibility of incorrect callings for those shared mutations. Mutation hotspot is less likely because the majority of the shared mutations are shared by a pair of lines rather than multiple lines (File S1), which also provides more evidence for line-splitting or outcrossing. In addition, if it were a hotspot effect, we would expect to see different mutations at the same position. Instead, we identify the same mutation at the same genomic location in different lines. We conclude it is most probable that the majority of shared SNMs are due to accidental splitting of some MA lines (mutations shared by pairs of lines) or outcrossing among a subset of MA lines (mutations shared by three or four lines) at some point during propagation, although without resequencing new plant material, we cannot rule out the possibility of contamination during sequencing library preparation. We also found that the distribution of the presence of mutation in TEs (χ2 = 0.94, d.f. = 1, P = 0.33) and the transition-to-transversion ratio (χ2 = 1.4, d.f. = 1, P = 0.24) are not significantly different between shared and unique SNMs. The shared SNMs also did not vary significantly from the unique SNMs in their distributions between chromosomes (χ2 = 7.04, d.f. = 4, P = 0.13). Thus, regardless of the reason for some mutations being shared across lines, the shared mutations likely originate from the same processes during mutation accumulation that generated the unique mutations.
Chromosome-level variation in mutation rates
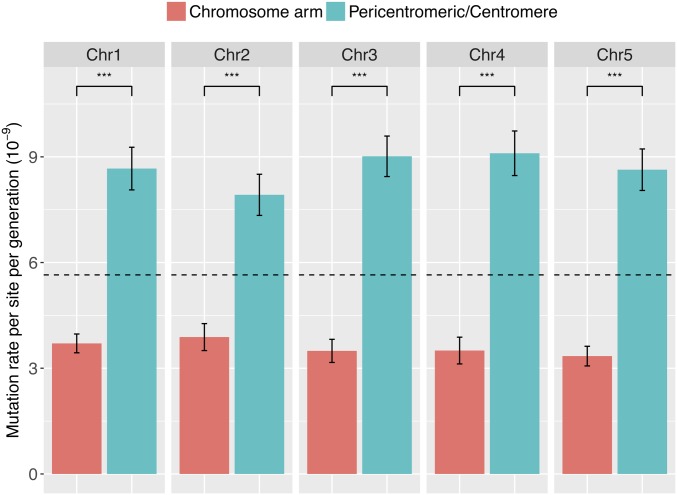
While the SNM rate does not vary significantly among the five chromosomes (χ2 = 8.14, d.f. = 4, P = 0.09), it is substantially lower on the arms of all five chromosomes than in pericentromeric and centromeric regions (χ2 = 567.4, d.f. = 2, P < 2e−16) (Figure 2).
Figure 2.
Comparison of mutation rates between chromosome arms and pericentromeric/centromere regions in 107 MA lines. The base mutation rate for chromosome arms is significantly lower than pericentromeric/centromere regions across five chromosomes (χ2 = 567.4, d.f. = 2, P < 2e−16). Dashed line indicates genome-wide average. The pericentromeric region is defined as being within 4 Mb of the centromere. Error bar is equal to one SEM. Asterisks indicate P < 0.001 (***). Mutation rates are adjusted for accessible reference genome of respective regions that pass the filtering threshold.
SNM rates observed in TEs, intergenic, and genic regions are significantly different from each other (χ2 = 837.3, d.f. = 2, P < 2e−16). The haploid mutation rate estimate at TEs is 1.36 × 10−8 per site per generation, about twofold higher than the genome-wide average estimate. The mutation rate in intergenic regions is close to the genome-wide mean, 5.75 × 10−9, while the rate in genic regions is about half that, 3.35 × 10−9 per site per generation.
Interaction between methylation, TE annotation, and chromosome region
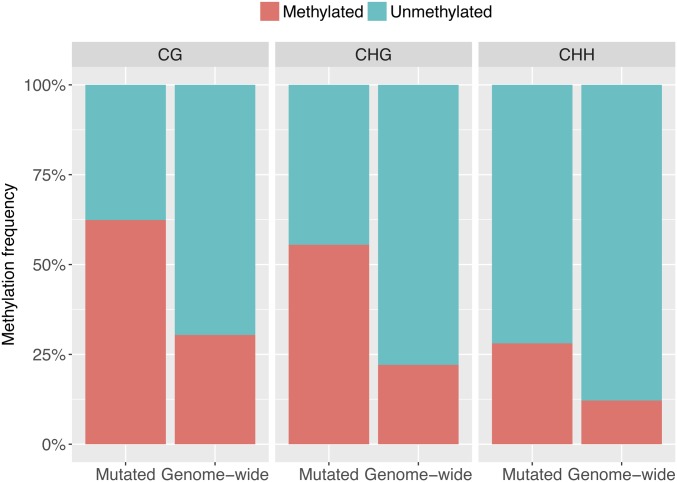
Because methylated cytosines can undergo spontaneous deamination to thymidine, mutation rates at these sites have long been known to be elevated (Bird 1980; Xia et al. 2012). We therefore investigated mutations at CG, CHG, and CHH sites (H refers to A, T, or G) that are either methylated or nonmethylated (Stroud et al. 2013). The likelihood of a cytosine occurring in any of the three methylated contexts is much higher for mutated than for nonmutated sites: CG (χ2 = 97.6, d.f. = 1, P < 2e−16), CHG (χ2 = 141.6, d.f. = 1, P < 2e−16) and CHH (χ2 = 181.7, d.f. = 1, P < 2e−16) (Figure 3). The ratio of transitions to transversions at methylated and nonmethylated cytosine in CG, CHG, and CHH contexts (Table S4), however, is not different (Fisher’s Exact test, P = 0.252).
Figure 3.
Comparison of cytosine methylation frequencies at all bases in the genome and mutated bases in 107 MA lines. H refers to A, T, or G. The methylation frequency is significantly higher at mutated bases than the genome-wide occurrence of all three contexts, CG (χ2 = 97.6, d.f. = 1, P < 2e−16), CHG (χ2 = 141.6, d.f. = 1, P < 2e−16), and CHH (χ2 = 181.7, d.f. = 1, P < 2e−16).
We used a logistic regression model to disentangle the effects of methylation, TE position and chromosome region on mutation rate, since methylation occurs mainly at TEs, and TEs are concentrated in centromeres (Kawakatsu et al. 2016; Sigman and Slotkin 2016; Underwood et al. 2017; Zhang et al. 2018). The logistic regression model with main effects and two- and three-way interactions has the lowest AIC score, 94.3 (Table S5). Alternative models including only main effects or main effects plus two-way interactions have AIC scores of 147.7 and 101.9, respectively. TEs, methylated sequences, and pericentromeric/centromeric regions all show significantly positive associations with mutations (Table S5). Methylated TE positions on chromosome arms have the highest mutation rate, whereas AT sites outside TEs on chromosome arms have the lowest mutation rates, with a 30-fold difference between the highest and lowest mutation rate (Figure 4).
Figure 4.
Effects of annotation as TE, cytosine methylation, and chromosome region on mutation rates in 107 MA lines. Log-transformed mutation rates per site per generation are shown. Mutation rates for TE and non-TE positions are highlighted in blue and red, respectively. Mutation rates for AT positions as well as methylated and nonmethylated CG positions are indicated in square, triangle, and circle, respectively. Error bars indicate one SEM. Difference in mutation rates between chromosome arms and pericentromeric/centromeric regions among six classes was assessed by Student’s t-test and significance levels of these multiple tests were adjusted by sequential Bonferroni correction. NS indicates not significant and asterisks indicate P < 0.001 (***).
Although sites in pericentromeric and centromeric regions are more likely to be mutated than those on chromosome arms, this effect is restricted to AT sites and non-TE regions (Figure 4), as indicated by a significant negative interaction between TE and distance from centromere (Table S5). Furthermore, TE, methylation status, and chromosomal position do not have additive effects on mutation rates (Figure 4 and Table S5).
Context effects and distribution of mutations along the genome
We calculated the spacing of all unique 1694 SNMs, finding that the inter-SNM distance is consistent with an exponential distribution on chromosome arms (Kolmogorov-Smirnov test, D = 0.032, P = 0.564), but not at the genome-wide scale (D = 0.055, P = 1e−4), due to deviations from the exponential distribution of spacing in pericentromeric/centromeric regions (D = 0.074, P = 0.019).
To test the effects of flanking nucleotides, we estimated mutation rates for A/T and G/C positions flanked by different nucleotides on either side, regardless of DNA strand orientation. G/C bases have a higher mutation rate than A/T bases (two sample t-test, t = 14.4, d.f. = 18.9, P = 1e−11). The nucleotide one position upstream (P = 0.96) or downstream (P = 0.97) does not have effects on mutation rate variation and of all 16 possible combinations of flanking nucleotides, after sequential Bonferroni correction, none have significantly higher or lower mutation rates within the G/C or A/T groups (Figure S2).
To examine potential nonindependence of the distribution of mutations, we examined double mutations, defined as two mutations in the same MA line within 10 bp of each other, and at complex events, defined as multiple mutations within a 50 bp region (Zhu et al. 2014; Behringer and Hall 2016). Twenty-three pairs of double mutations are found in 20 different MA lines (Table S6), representing 2.7% of all SNMs. In contrast to other SNMs, the ratio of transitions (37) to transversions (34) at double mutations is nearly one. Forty-eight complex mutations, including 17 deletions, 6 insertions, and 25 SNMs, are found in 16 different MA lines (Table S6). Complex and double mutations occur significantly more often than expected from a random distribution of mutations across the genome, as tested by 1000 random draws assuming each mutation as independent (Z-test, P = 2e−16). This observation suggests that most of the complex and double mutations are the results of single mutational events, consistent with the finding that multi-nucleotide mutations are common in eukaryotes (Schrider et al. 2011).
Comparison of de novo mutations and natural genetic variation
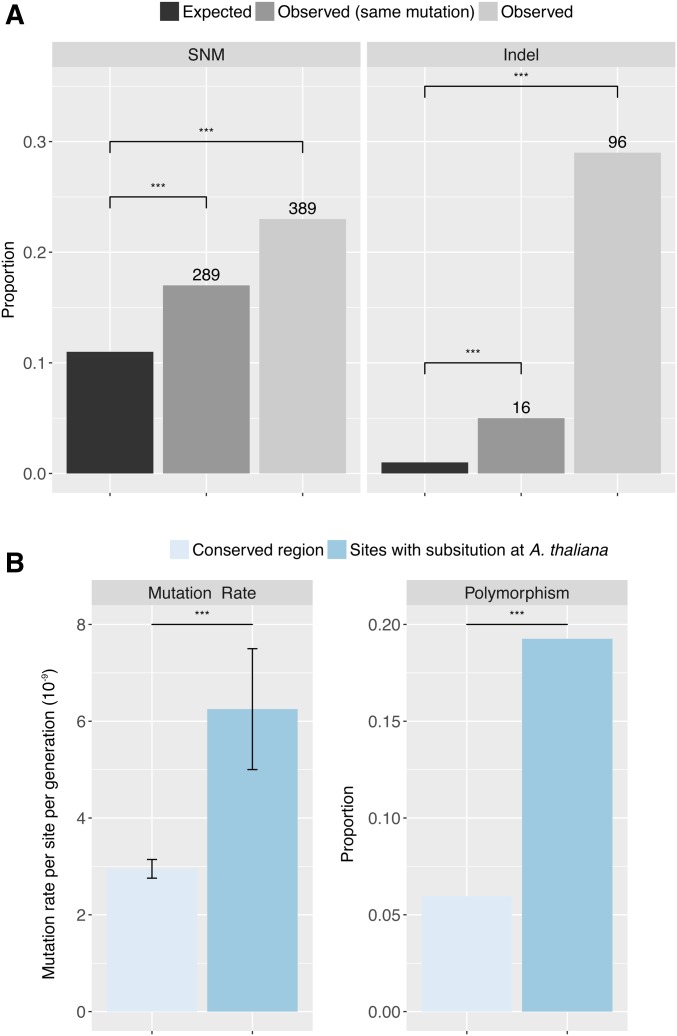
In a survey of 1135 natural accessions of A. thaliana, 10,707,430 biallelic SNPs and 1,424,879 small indels of up to 40 bp have been found, using similar approaches as used in this work (1001 Genomes Consortium 2016). These polymorphisms affect 11% and 1.5% of the reference genome, respectively. Among the 1694 unique SNMs identified in the MA lines, 389 SNMs (23%) coincide with biallelic SNPs from the 1001 Genomes population, and in 289 (17%) cases the variants are identical (Figure 5A). Of the 329 unique indels identified in the MA lines, 95 (29%) overlap with indels from the 1001 Genomes dataset, and 16 indels (5% of the total) are identical (Figure 5A). The overlap between MA-line polymorphisms and naturally occurring variants is thus highly significant compared to expectations of overlap based on a random distribution of mutations and polymorphisms (Figure 5A) (SNM: Fisher’s Exact test, P = 1e−10; indel: Fisher’s Exact test, P = 6e−5). These results are also found for the shared mutations, in which 44 of the 159 shared SNMs overlap with 1001 Genome SNPs. The proportion of the overlap with 1001 Genome SNP is not significantly different between shared or unique SNMs (χ2 = 0.88, d.f. = 1, P = 0.35). We asked whether this overlap is biased toward SNPs with low allele frequency in the 1001 Genomes population. It is not. The proportion of SNPs with allele frequency <0.01 is not significantly different between 1001 Genomes SNPs overlapping or not overlapping with MA line SNMs (χ2 = 0.02, d.f. = 1, P = 0.90).
Figure 5.
Overlap between mutations identified in 107 MA lines (SNMs) and polymorphism detected in the 1001 Genomes population (SNPs). (A) Comparison of expected and observed proportions of SNMs and indels that overlap with biallelic SNPs and indels in the 1001 Genomes population. Numbers on top of each bar indicate absolute overlap. (B) Comparison of mutation rates and proportions of polymorphism at conserved sites (identical in C. rubella, A. lyrata, and the A. thaliana reference genome) and at sites with substitutions in the A. thaliana Col-0 reference genome. The proportion of polymorphism is calculated as the number of biallelic SNPs divided by the number of reference sites. Asterisks indicate P < 0.001 (***) of Fisher’s Exact test following sequential Bonferroni correction.
To further evaluate how much mutational processes might influence patterns of intraspecific polymorphism and interspecific substitutions, we used a whole-genome alignment between the A. thaliana Col-0 reference accession and reference genomes of A. lyrata (Hu et al. 2011) and Capsella rubella (Slotte et al. 2013) to partition the A. thaliana genome into conserved sites, where the three reference genomes are identical, and lineage-specific substitutions in A. thaliana, where a site is identical between A. lyrata and C. rubella, but differs in the A. thaliana reference. We then separated the 1001 Genomes biallelic SNPs (which were initially called against the Col-0 reference) into 1,799,125 derived (SNPs occurring at conserved sites) and 219,909 ancestral variants (SNPs occurring at substitution sites of A. thaliana and having the same sequence in A. lyrata and C. rubella). The overlap between MA line SNMs and derived 1001 Genomes variants (11%) is about half the overlap between SNMs and ancestral variants (21%). We further investigated whether the lower overlap between SNMs and derived SNPs is due to low mutation rate or low polymorphism at the conserved sites. The base mutation rate (2.95 × 10−9) estimated from MA lines at these conserved sites is indeed significantly lower than the mutation rate at substitution sites (6.25 × 10−9) (χ2 = 194.3, d.f. = 1, P < 2e−16) (Figure 5B). The proportion of polymorphism at the conserved sites (6%) is also significantly lower than the polymorphism at substitution sites (20%) (Figure 5B) (χ2 = 855,260, d.f. = 1, P < 2e−16).
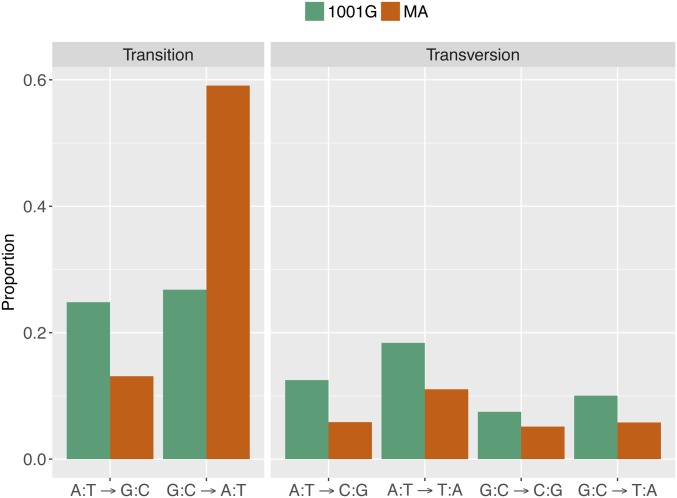
To compare the ratios of transitions to transversions, we considered only the derived variants of the 1001 Genomes population, to ensure that nucleotide changes could be polarized. The ratio of transitions to transversions is significantly higher in MA lines (2.60) than in the derived variants of the 1001 Genomes population (1.05) (Fisher’s Exact test, P < 2e−16). Among the six possible nucleotide substitutions, mutations are strongly biased toward G:C to A:T transitions in the MA lines, but such bias is not evident in the 1001 Genomes population (Figure 6) (χ2 = 431.4, d.f. = 5, P < 2e−16). The G:C to A:T transition class accounts for 59% of changes in the MA lines, but only 26% in the 1001 Genomes population. Excluding changes from G:C to A:T from the analysis greatly reduces the difference in proportion of specific nucleotide changes between the MA lines and the 1001 Genomes set (χ2 = 8.9, d.f. = 4, P = 0.06).
Figure 6.
Comparison of nucleotide changes in 107 MA lines (brown bars) and the 1001 Genomes population (green bars). Complementary mutations, such as A:T→G:C and T:A→C:G, are collapsed. The distribution of nucleotide changes in MA lines is significantly different from the polymorphisms discovered in the 1001 Genomes Project (χ2 = 431.4, d.f. = 5, P < 2e−16).
A final result from the comparison of SNMs in our MA lines and the 1001 Genomes polymorphisms reveals the distribution of SNMs and SNPs, with respect to different sequence changes and genomic regions, to differ significantly between the two sets (χ2 = 41.49, d.f. = 5, P = 7e−8) (Figure 7), with the ratio of nonsynonymous to synonymous mutations being significantly higher in MA lines (2.19 vs. 1.46) (Fisher’s Exact test, P = 2e−3) and intron SNMs being significantly less frequent than intron SNPs (Fisher’s Exact test, P = 3e−5).
Figure 7.
Comparison of the genomic locations of SNMs in 107 MA lines (brown bars) and SNPs discovered in the 1001 Genomes Project (green bars). The distributions are significantly different from each other (χ2 = 37.8, d.f. = 6, P = 1e−6). NS indicates not significant and asterisks indicate P < 0.001 (***) following sequential Bonferroni correction.
Discussion
Our whole-genome sequencing effort of 107 MA lines provides a comprehensive estimate of the spontaneous mutation profile in A. thaliana. In agreement with earlier and more limited analyses (Ossowski et al. 2010; Jiang et al. 2014; Exposito-Alonso et al. 2018), we find that mutations in A. thaliana do not occur evenly across the genome: they are biased toward G:C to A:T transitions, localize predominantly in TEs and pericentrometic/centromeric regions, and are more frequent at methylated cytosines. As far as we are aware, no other studies have documented accelerated mutation rate in TEs. By increasing the number of documented spontaneous mutations more than an order of magnitude, we show in addition that mutation rates vary as much as 30-fold across the A. thaliana genome: they are highest at methylated cytosines in TEs on chromosome arms and lowest at AT sites outside TEs on chromosome arms. Furthermore, deletions are more frequent and larger than insertions, in agreement with evidence that A. thaliana genomes are still shrinking (Hu et al. 2011). Our data demonstrate that the frequency of SNMs is independent of nearby trinucleotide contexts. Either the number of mutations does not yet provide sufficient power to detect more subtle effects on mutation rates, or mutations in Arabidopsis are indeed less affected by immediately surrounding sequence contexts than mutations in Chlamydomonas genomes (Ness et al. 2015). That the overlap between mutations and natural polymorphisms, which historically must have experienced a wide range of environments, is significantly higher than expected by chance, points to the stability of mutation rate variation within a species.
Although the total number of SNMs varies considerably among MA lines, ranging from 8 to 40 SNMs among lines, the occurrence of SNMs per MA line is consistent with a Poisson distribution, suggesting that SNMs are independently distributed among the MA lines, and that the number of SNMs across lines reflected different outcomes of a probabilistic process rather than variation in the inherent mutation rate across the lines. Indeed, the line with the highest number of mutations (MA line 32 contains 40 SNMs) did not contain any mutations in known DNA repair genes. Early mutations at DNA repair genes could cause significant variation of mutation rates among MA lines (Hoffman et al. 2004; Turrientes et al. 2013; Belfield et al. 2018). Since the overdispersion is insignificant, as expected, the distribution of SNMs per MA line also fit a negative binomial distribution with the best fit overdispersion parameter. A Poisson or negative binomial distribution for the number of SNM per MA line has also been reported in budding yeast (Saccharomyces cerevisiae) (Zhu et al. 2014) and fission yeast, Schizosaccharomyces pombe (Behringer and Hall 2016).
We have confirmed that mutation is biased toward centromeres in A. thaliana, as previously observed (Ossowski et al. 2010). The same pattern has also been reported for budding yeast, S. cerevisiae (Bensasson 2011). Centromeric chromatin contains a specific histone variant, CENH3 (Ravi et al. 2011), and Bensasson (2011) postulated that DNA might be difficult to unwind from CENH3, resulting in DNA breaks not easily accessible to the repair machinery, accounting in turn for the higher centromere mutation rates.
A high mutation rate in TEs could lead to their genetic inactivation through sequence degradation (Le Rouzic et al. 2007). Our estimated base mutation rate at TEs is 1.36 × 10−8, about twice the genome-wide average. This elevated rate, along with absence of novel TE insertion in the MA lines, could account for the observation that TE copy number is generally low in A. thaliana, even though all known plant TE families are present in the A. thaliana genome (Arabidopsis Genome Initiative 2000). TE content is strongly correlated with plant genome size (Tenaillon et al. 2010). Given that TE mutation rate strongly affects TE content, as our results imply, we would predict a negative correlation between TE mutation rate and plant genome size. Alternatively, more efficient selection against TE insertion in selfers like A. thaliana might lead to the same result (Lockton and Gaut 2010). Nevertheless, it is important to account for the elevated mutation rate of TE sequences in estimating TE age and diversification in plants.
Methylated cytosines have an increased mutation rate (Ossowski et al. 2010; Xia et al. 2012), consistent with spontaneous deamination of methylated cytosines leading to substitution with thymine (Duncan and Miller 1980). If the transition-biased mutation rate in A. thaliana MA lines was driven mainly by methylation at cytosines, we should observe an increased transition rate at methylated sites compared to nonmethylated ones. However, this increase is not significant compared to nonmethylated sites. These results suggest that in addition to methylation, other factors contribute to the high transition rate found in MA lines (see also Ossowski et al. 2010).
Genome size is directly affected by deletions and insertions. We observed a deletion bias, with deletions being not only more frequent but also larger than insertions, providing a plausible explanation for genome size shrinkage in A. thaliana relative to related species (Hu et al. 2011), with the proviso that the analysis tools we used probably are more powerful for detection of deletions compared to insertions. Natural variants also suggest a bias toward deletions (Hu et al. 2011), as do comparisons with related species (Rutter et al. 2012a). This deletion-biased mutation is consistent with the finding that the repair mechanism for DNA double-strand breaks in A. thaliana preferentially causes larger deletions (Vu et al. 2017). While deletion-biased mutation has also been observed in Drosophila (Keightley et al. 2009; Leushkin et al. 2013) and budding yeast S. cerevisiae (Zhu et al. 2014), an opposite trend of insertion-biased mutation was found in fission yeast S. pombe (Farlow et al. 2015; Behringer and Hall 2016) and nematodes (Denver et al. 2004), suggesting that DNA repair mechanisms might function differently among taxa.
The ratio of nonsynonymous to synonymous substitution is significantly lower in the 1001 Genomes data set (1.46) (1001 Genomes Consortium 2016) than in our MA lines (2.19). This suggests that the strength of selection is reduced in our MA experiments, although we cannot rule out the possibility that some mutations that affect germination are selected against during the propagation of MA lines, and that lethal or highly deleterious mutations are excluded. Our finding that intron SNPs are more frequent than intron SNMs is inconsistent with the inference in C. grandiflora that introns evolve neutrally (Williamson et al. 2014). Transitions are 2.6 times more frequent than transversions in the MA lines, with a strong bias toward G:C to A:T transitions. The base composition equilibrium due to mutations alone would be 85% AT, which is far higher than the observed 74% AT in the A. thaliana reference genome. These results imply that the base composition does not solely reflect mutation outcome.
The 1001 Genomes polymorphism set is much less skewed toward G:C to A:T substitutions than our MA lines (Figure 5), suggesting selection favoring G:C over A:T or a mechanism that compensates for AT-biased mutations in natural populations. High thermal stability may be the cause of selection for increased GC content in animal genomes (Bernardi 2000), whereas GC-biased conversion and selection for codon usage efficiency were shown to drive GC content increase in the rice genome (Muyle et al. 2011).
Assuming that only indels in coding regions and nonsynonymous SNMs affect fitness, we estimate that the diploid genomic rate of mutation rate affecting fitness is 0.16 per generation, which is very close to conclusions made with a much smaller data set (Ossowski et al. 2010). These estimates are also similar to the estimates based on fitness measures of these lines in the field (Rutter et al. 2010) and greenhouse experiments (Shaw et al. 2002). In aggregate, our findings are entirely consistent with de novo mutations in protein coding regions contributing to population level genetic variation for fitness.
Mutational processes influence the patterns of intraspecific polymorphism and interspecific variation, and a comparison between the MA lines and the 1001 Genomes population revealed that the specific sites of mutations and natural polymorphisms significantly overlap in the two datasets. Because the strength of selection is minimized in the MA experiment, the significant overlaps between SNMs and SNPs could not be due to selection alone, but indicates that polymorphic sites tend to have a higher mutation rate than monomorphic sites and are therefore also more often mutated in the MA lines. For interspecific comparisons, we found that conserved sites (identical between the A. thaliana, A. lyrata, and C. rubella reference genomes) have reduced mutation rates (2.62 × 10−9) compared to the genome-wide mutation rate (6.95 × 10−9). Our results thus strongly suggest an important role of ongoing mutation in generating current natural genetic diversity, with high mutation rates at some sites contributing to high polymorphism and low mutation rates at other sites reducing genetic variation in natural populations. Thus, variation in mutation rates across the genome must be considered when interpreting observed patterns of genetic diversity.
Conclusions
The current study provides the most comprehensive estimate of mutation spectrum and mutational inputs for a plant. Using A. thaliana MA lines, we greatly increase the evidence for a series of conclusions: (1) mutation rates vary across the genome; (2) mutations are biased toward transitions, toward sites in TEs, toward pericentromeric/centromeric regions, and toward methylated cytosines; (3) patterns of mutational distribution along the genome play a large role in contributing to patterns of genetic variation found in natural populations. We conclude that incorporating the observed genomic patterns of mutations into analyses of natural patterns of variation enhances our understanding of how natural variants have been maintained, and how populations respond to selection (Bailey and Bataillon 2016).
Acknowledgments
The authors thank the Maryland Advanced Research Computing Center (MARCC) and the Extreme Science and Engineering Discovery Environment (XSEDE) for access on high performance computers. Support was provided by the Max Planck Society (D.W.) and the National Science Foundation (DEB 0844820 and DEB 1257902 to C.B.F., DEB 0845413 and DEB 1258053 to M.T.R., and DEB 9981891 to R.G.S.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6456065.
Communicating editor: M. Hahn
Literature Cited
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Bailey S. F., Bataillon T., 2016. Can the experimental evolution programme help us elucidate the genetic basis of adaptation in nature? Mol. Ecol. 25: 203–218. 10.1111/mec.13378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer M. G., Hall D. W., 2016. Genome-wide estimates of mutation rates and spectrum in Schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda) 6: 149–160. 10.1534/g3.115.022129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield E. J., Ding Z. J., Jamieson F. J. C., Visscher A. M., Zheng S. J., et al. , 2018. DNA mismatch repair preferentially protects genes from mutation. Genome Res. 28: 66–74. 10.1101/gr.219303.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D., 2011. Evidence for a high mutation rate at rapidly evolving yeast centromeres. BMC Evol. Biol. 11: 211 10.1186/1471-2148-11-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G., 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241: 3–17. 10.1016/S0378-1119(99)00485-0 [DOI] [PubMed] [Google Scholar]
- Bird A. P., 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8: 1499–1504. 10.1093/nar/8.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang J.-R., Zhang J., 2016. Nascent RNA folding mitigates transcription-associated mutagenesis. Genome Res. 26: 50–59. 10.1101/gr.195164.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. 10.1038/nature02697 [DOI] [PubMed] [Google Scholar]
- Denver D. R., Dolan P. C., Wilhelm L. J., Sung W., Lucas-Lledó J. I., et al. , 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. USA 106: 16310–16314. 10.1073/pnas.0904895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H., 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561. 10.1038/287560a0 [DOI] [PubMed] [Google Scholar]
- Ewing A. D., Houlahan K. E., Hu Y., Ellrott K., Caloian C., et al. , 2015. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods 12: 623–630. 10.1038/nmeth.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M., Becker C., Schuenemann V. J., Reiter E., Setzer C., et al. , 2018. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14: e1007155 10.1371/journal.pgen.1007155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow A., Long H., Arnoux S., Sung W., Doak T. G., et al. , 2015. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics 201: 737–744. 10.1534/genetics.115.177329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001 Genomes Consortium , 2016. 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 162: 481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. 10.1146/annurev.ecolsys.39.110707.173437 [DOI] [Google Scholar]
- Hartl D. L., Clark A. G., 2006. Principles of Population Genetics, Ed. 4 Sinauer Associates, Inc., Sunderland, MA. [Google Scholar]
- Hénaff E., Zapata L., Casacuberta J. M., Ossowski S., 2015. Jitterbug: somatic and germline transposon insertion detection at single-nucleotide resolution. BMC Genomics 16: 768 10.1186/s12864-015-1975-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey G., Paten B., Earl D., Zerbino D., Haussler D., 2013. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29: 1341–1342. 10.1093/bioinformatics/btt128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P. D., Leonard, J. M., Lindberg, G. E., Bollmann, S. R., and Hays, J.B. 2004. Rapid accumulation of mutations during seed-to-seed propagation of mismatch-repair-defective Arabidopsis. Genes Dev. 18: 2676–2685. 10.1101/gad.1217204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J.-F., et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481. 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Mithani A., Belfield E. J., Mott R., Hurst L. D., et al. , 2014. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 24: 1821–1829. 10.1101/gr.177659.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Huang S. C., Jupe F., Sasaki E., Schmitz R. J., et al. , 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. 10.1101/gr.091231.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber C., Zeileis A., 2008. Applied Econometrics with R. Springer-Verlag, New York: 10.1007/978-0-387-77318-6 [DOI] [Google Scholar]
- Leffler E. M., Bullaughey K., Matute D. R., Meyer W. K., Ségurel L., et al. , 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10: e1001388 10.1371/journal.pbio.1001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M. J., Hurst L. D., 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340. 10.1016/S0168-9525(02)02669-0 [DOI] [PubMed] [Google Scholar]
- Le Rouzic A., Boutin T. S., Capy P., 2007. Long-term evolution of transposable elements. Proc. Natl. Acad. Sci. USA 104: 19375–19380. 10.1073/pnas.0705238104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leushkin E. V., Bazykin G. A., Kondrashov A. S., 2013. Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection. Genome Biol. Evol. 5: 514–524. 10.1093/gbe/evt021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S., Gaut B. S., 2010. The evolution of transposable elements in natural populations of self-fertilizing Arabidopsis thaliana and its outcrossing relative Arabidopsis lyrata. BMC Evol. Biol. 10: 10 10.1186/1471-2148-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A., Serres-Giardi L., Ressayre A., Escobar J., Glémin S., 2011. GC-biased gene conversion and selection affect GC content in the Oryza Genus (rice). Mol. Biol. Evol. 28: 2695–2706. 10.1093/molbev/msr104 [DOI] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. 10.1534/genetics.112.145078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Vasanthakrishnan R. B., Colegrave N., Keightley P. D., 2015. Extensive de novo mutation rate variation between individuals and across the genome of Chlamydomonas reinhardtii. Genome Res. 25: 1739–1749. 10.1101/gr.191494.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Qian W., Zhang J., 2012. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 13: 1123–1129. 10.1038/embor.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B., Earl D., Nguyen N., Diekhans M., Zerbino D., et al. , 2011a. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 21: 1512–1528. 10.1101/gr.123356.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B., Diekhans M., Earl D., John J. S., Ma J., et al. , 2011b. Cactus graphs for genome comparisons. J. Comput. Biol. 18: 469–481. 10.1089/cmb.2010.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stütz A. M., Benes V., et al. , 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Shibata F., Ramahi J. S., Nagaki K., Chen C., et al. , 2011. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 7: e1002121 10.1371/journal.pgen.1002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: Available at: https://www.R-project.org/ [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roles A. J., Rutter M. T., Dworkin I., Fenster C. B., Conner J. K., 2016. Field measurements of genotype by environment interaction for fitness caused by spontaneous mutations in Arabidopsis thaliana. Evolution 70: 1039–1050. 10.1111/evo.12913 [DOI] [PubMed] [Google Scholar]
- Rutter M. T., Shaw F. H., Fenster C. B., 2010. Spontaneous mutation parameters for Arabidopsis thaliana measured in the wild. Evolution 64: 1825–1835. 10.1111/j.1558-5646.2009.00928.x [DOI] [PubMed] [Google Scholar]
- Rutter M. T., Cross K. V., Van Woert P. A., 2012a. Birth, death and subfunctionalization in the Arabidopsis genome. Trends Plant Sci. 17: 204–212. 10.1016/j.tplants.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Rutter M. T., Roles A., Conner J. K., Shaw R. G., Shaw F. H., et al. , 2012b. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution 66: 2335–2339. 10.1111/j.1558-5646.2012.01583.x [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Hourmozdi J. N., Hahn M. W., 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21: 1051–1054. 10.1016/j.cub.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck F. J., Rescheneder P., Von Haeseler A., 2013. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29: 2790–2791. 10.1093/bioinformatics/btt468 [DOI] [PubMed] [Google Scholar]
- Shaw F. H., Geyer C. J., Shaw R. G., 2002. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56: 453–463. 10.1111/j.0014-3820.2002.tb01358.x [DOI] [PubMed] [Google Scholar]
- Shaw R. G., Byers D. L., Darmo E., 2000. Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman M. J., Slotkin R. K., 2016. The first rule of plant transposable element silencing: location, location, location. Plant Cell 28: 304–313. 10.1105/tpc.15.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F., et al. , 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. 10.1038/ng.2669 [DOI] [PubMed] [Google Scholar]
- Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E., 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 (erratum: Cell 161: 1697–1698) 10.1016/j.cell.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M. I., Hollister J. D., Gaut B. S., 2010. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15: 471–478. 10.1016/j.tplants.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Turrientes M.-C., Baquero F., Levin B. R., Martínez J.-L., Ripoll A., et al. , 2013. Normal mutation rate variants arise in a mutator (Mut S) Escherichia coli population. PLoS One 8: e72963 (erratum: PLoS One 8) 10.1371/journal.pone.0072963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood C. J., Henderson I. R., Martienssen R. A., 2017. Genetic and epigenetic variation of transposable elements in Arabidopsis. Curr. Opin. Plant Biol. 36: 135–141. 10.1016/j.pbi.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu G. T. H., Cao H. X., Reiss B., Schubert I., 2017. Deletion-bias in DNA double-strand break repair differentially contributes to plant genome shrinkage. New Phytol. 214: 1712–1721. 10.1111/nph.14490 [DOI] [PubMed] [Google Scholar]
- Williamson R. J., Josephs E. B., Platts A. E., Hazzouri K. M., Haudry A., et al. , 2014. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 10: e1004622 10.1371/journal.pgen.1004622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1988. Surfaces of selective value revisited. Am. Nat. 131: 115–123. 10.1086/284777 [DOI] [Google Scholar]
- Xia J., Han L., Zhao Z., 2012. Investigating the relationship of DNA methylation with mutation rate and allele frequency in the human genome. BMC Genomics 13: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Gaut B. S., 2011. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol. Biol. Evol. 28: 2359–2369. 10.1093/molbev/msr058 [DOI] [PubMed] [Google Scholar]
- Yang S., Wang L., Huang J., Zhang X., Yuan Y., et al. , 2015. Parent–progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523: 463–467. 10.1038/nature14649 [DOI] [PubMed] [Google Scholar]
- Zhang L., Gaut B. S., 2003. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13: 2533–2540. 10.1101/gr.1318503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lang Z., Zhu J.-K., 2018. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19: 489–506. 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–E2318. 10.1073/pnas.1323011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The FASTQ and BAM files of each MA line have been uploaded to NCBI Short Read Archive (SRA) with the accession number of SRP133100. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6456065.