Abstract
The Mre11-Rad50-Xrs2 (MRX) complex acts together with the Sae2 protein to initiate resection of DNA double-strand breaks (DSBs) and to regulate a checkpoint response that couples cell cycle progression with DSB repair. Sae2 supports resistance to DNA damage and downregulates the signaling activities of MRX, Tel1, and Rad53 checkpoint proteins at the sites of damage. How these functions are connected to each other is not known. Here, we describe the separation-of-function sae2-ms mutant that, similar to SAE2 deletion, upregulates MRX and Tel1 signaling activities at DSBs by reducing Mre11 endonuclease activity. However, unlike SAE2 deletion, Sae2-ms causes neither DNA damage sensitivity nor enhanced Rad53 activation, indicating that DNA damage resistance depends mainly on Sae2-mediated Rad53 inhibition. The lack of Sae2, but not the presence of Sae2-ms, impairs long-range resection and increases both Rad9 accumulation at DSBs and Rad53–Rad9 interaction independently of Mre11 nuclease activity. Altogether, these data lead to a model whereby Sae2 plays distinct functions in limiting MRX-Tel1 and Rad9 abundance at DSBs, with the control on Rad9 association playing the major role in supporting DNA damage resistance and in regulating long-range resection and checkpoint activation.
Keywords: MRX, Rad9, Rad53, Sae2, Tel1
MECHANISMS devoted to repair DNA lesions are essential for maintaining genome integrity. Among DNA lesions, DNA double-strand breaks (DSBs) are the most severe because they have the potential to cause loss of genetic information and chromosomal rearrangements (Mehta and Haber 2014). DSBs can be repaired by homologous recombination (HR), which requires that the 5′ strands of both DSB DNA ends are nucleolytically degraded (resected) (Bonetti et al. 2018). Then, the resulting 3′-ended single-stranded DNA (ssDNA) tails can invade an undamaged homologous DNA template, like the sister chromatid or the homologous chromosome (Mehta and Haber 2014).
In both yeast and mammals, DNA end resection is a two-step process. First, the Sae2 protein (CtIP in mammals) activates a latent endonuclease activity of Mre11 within the context of the Mre11-Rad50-Xrs2 (MRX) complex to incise the 5′-terminated strands at both DNA ends (Cannavo and Cejka 2014). The resulting nick generates an entry site for the Mre11 exonuclease, which degrades back toward the DSB end in the 3′–5′ direction, and for the long-range resection nucleases Exo1 and Dna2 that degrade away from the DSB in the 5′–3′ direction (Mimitou and Symington 2008; Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010; Garcia et al. 2011; Nimonkar et al. 2011; Shibata et al. 2014; Reginato et al. 2017; Wang et al. 2017). In addition to a nucleolytic activity in the vicinity of the broken DNA ends, MRX has a structural role in recruiting and promoting the activity of Exo1 and Dna2 at DNA DSBs (Cejka et al. 2010; Nicolette et al. 2010; Niu et al. 2010; Shim et al. 2010; Cannavo et al. 2013; Gobbini et al. 2018).
The short-range resection catalyzed by MRX-Sae2 is particularly important for the processing of DNA ends that possess protein blocks at their 5′-terminated DNA strands, such as stalled topoisomerases or Spo11 in meiosis (Trujillo and Sung 2001; Neale et al. 2005; Cannavo and Cejka 2014). By contrast, it can be dispensable during the processing of endonuclease-induced DNA breaks, whose DNA ends are readily accessible to Exo1 and Dna2 nucleases (Llorente and Symington 2004).
DSB generation triggers activation of the checkpoint protein kinases Mec1 and Tel1 (ATR and ATM in mammals, respectively), which sense and signal the presence of DNA DSBs, leading to arrest of cell cycle progression (Gobbini et al. 2013; Villa et al. 2016). The lack of any MRX/MRN subunit abolishes Tel1/ATM activation by preventing its association to DSBs (Nakada et al. 2003; Falck et al. 2005; Lee and Paull 2005; You et al. 2005; Berkovich et al. 2007), indicating that MRX/MRN is required for Tel1/ATM recruitment to DSBs. By contrast, Mec1/ATR (in association with Ddc2/ATRIP) recognizes the RPA-coated ssDNA that results from resection of the DSB DNA ends (Zou and Elledge 2003; Jazayeri et al. 2006; Myers and Cortez 2006). Once activated by damaged DNA, Tel1 and Mec1 propagate the checkpoint signals through the Rad53 and Chk1 effector kinases (Chk2 and Chk1 in mammals, respectively) (Ciccia and Elledge 2010). Rad53 activation requires the BRCT-domain-containing protein Rad9 (53BP1 in mammals). Rad9 undergoes Mec1- and/or Tel1-dependent phosphorylation upon DNA damage (Emili 1998; Vialard et al. 1998), and these phosphorylation events create a binding site for Rad53, thus allowing Rad53 in-trans autophosphorylation that leads to Rad53 activation as a kinase (Sun et al. 1998; Durocher et al. 1999; Pellicioli et al. 1999; Gilbert et al. 2001; Schwartz et al. 2002; Sweeney et al. 2005; Smolka et al. 2007).
Cells lacking SAE2 are more sensitive to DNA damaging agents than mre11 nuclease-dead mutants (Bonetti et al. 2015), indicating that Sae2 has Mre11-nuclease independent roles in the DNA damage response. Consistent with this hypothesis, the lack of Sae2 enhances Tel1 signaling activity by increasing MRX and Tel1 persistence at the DSB ends (Usui et al. 2001; Lisby et al. 2004; Clerici et al. 2006, 2014). This persistent MRX-Tel1 activation in sae2∆ cells is associated with enhanced activity of the downstream checkpoint kinase Rad53, which causes a permanent cell cycle arrest (Usui et al. 2001; Clerici et al. 2006). The increased MRX-Tel1-Rad53-mediated checkpoint activation has been proposed to account for the DNA damage hypersensitivity and the resection defect of sae2∆ cells. In fact, mre11 mutant alleles that reduce MRX binding to DSBs restore DNA damage resistance and resection in sae2∆ cells (Chen et al. 2015; Puddu et al. 2015; Cassani et al. 2018). A similar effect also occurs when Tel1 function is affected by reducing either its association to DSBs or its kinase activity (Gobbini et al. 2015). Moreover, impairment of Rad53 activity by affecting either its interaction with Rad9 or its kinase activity suppresses both the hypersensitivity to DNA damage and the resection defect of sae2∆ cells (Gobbini et al. 2015).
Tel1 and Rad53 hyperactivation in sae2∆ cells leads to an increased accumulation of Rad9 at DSBs, which acts as a barrier to Sgs1-Dna2-mediated DSB resection (Bonetti et al. 2015; Ferrari et al. 2015; Gobbini et al. 2015). Furthermore, as Rad53 is known to phosphorylate and inhibit Exo1 (Morin et al. 2008), Rad9-mediated Rad53 hyperactivation in sae2∆ cells also leads to Exo1 inhibition. These findings lead to a model whereby the DNA damage hypersensitivity and the resection defect of sae2∆ cells are due to an increased MRX-Tel1 activation, which, in turn, leads to an increased Rad9 association at DSBs and Rad53 hyperactivation, thereby inhibiting Exo1 and Dna2-Sgs1 resection activity (Bonetti et al. 2018).
To better understand the contribution of MRX, Tel1, and Rad53 to the DNA damage hypersensitivity of Sae2-lacking cells, and how Sae2 modulates the signaling activities of the above factors, we searched for sae2 alleles that failed to inhibit Tel1 but retained Sae2 function in supporting DNA damage resistance. Here, we describe the hypomorphic sae2-ms allele that, similar to sae2∆ and mre11 nuclease defective alleles, increases MRX and Tel1 persistence at DSBs by affecting MRX cleavage activity. However, unlike SAE2 deletion, the Sae2-ms mutant variant is capable of supporting DNA damage resistance and long-range resection, indicating that the enhanced MRX persistence at DSBs is not responsible for the increased DNA damage sensitivity and the resection defect of sae2∆ cells. Furthermore, unlike SAE2 deletion, Sae2-ms does not enhance Rad53 activation, indicating separable functions of Sae2 in the downregulation of MRX-Tel1 and Rad53 signaling activities. Accordingly, the lack of Sae2, but not the presence of Sae2-ms, increases Rad9 association at DSBs and Rad53-Rad9 interaction independently of MRX nuclease activity. Altogether, these data indicate that Sae2 limits MRX association at DSBs and therefore Tel1 activation in a nuclease-dependent manner. Furthermore, it limits Rad9 persistence at DSBs and this inhibition plays the major role in supporting DNA damage resistance and in regulating both long-range resection and checkpoint activation.
Materials and Methods
Yeast strains
Strain genotypes are listed in Supplemental Material, Table S1. Strain YJK40.6, used to detect end-tethering, was kindly provided by D. P. Toczyski (University of California, San Francisco). Strain HS21, used to detect hairpin opening, was kindly provided by M. A. Resnick (National Institutes of Health, NC). Strains JKM139 and YMV45, used to detect DSB resection and single-strand annealing (SSA), respectively, were kindly provided by J. Haber (Brandeis University, Waltham). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone) supplemented with 2% glucose (YEPD), 2% raffinose (YEPR), or 2% raffinose and 3% galactose (YEPRG). Gene disruptions were generated by one-step PCR disruption method. All experiments were performed at 26°.
Search for sae2 mutations that suppress mec1∆ sensitivity to HU and MMS
To search for sae2 alleles that suppress mec1∆ sensitivity to HU and MMS, but that do not impair Sae2 function in DSB repair, we used low-fidelity PCR to random mutagenize the SAE2 gene. Genomic DNA from a strain carrying the LEU2 gene located 122 bp downstream of the SAE2 stop codon was used as template to amplify by low-fidelity PCR a SAE2 region spanning from position −54 to +212 bp from the SAE2 coding sequence. Thirty independent PCR reaction mixtures were prepared, each containing 5 U GoTaq G2 Flexi DNA polymerase (Promega), 10 ng genomic DNA, 500 ng each primer, 0.5 mM each dNTP (dATP, dTTP, dCTP), 0.1 mM dGTP, 0.5 mM MnCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl and 3 mM MgCl2. The resulting PCR amplification products, containing the SAE2 coding sequence and the LEU2 marker gene, were used to transform a mec1∆ sml1∆ mutant strain in order to replace the SAE2 wild type sequence with the mutagenized DNA fragments. Transformant clones were selected on synthetic medium without leucine and then assayed by drop test for increased viability in the presence of HU and MMS compared to mec1∆. Among them, the clones that, after transformation with a plasmid carrying the wild-type MEC1 gene, showed camptothecin (CPT) and phleomycin (phleo) resistance compared to sae2∆ cells were chosen for further characterization.
DSB resection and repair by SSA
DSB end resection at the MAT locus in JKM139 derivative strains was analyzed on alkaline agarose gels by using a single-stranded probe that anneals to the unresected DSB strand on one side of the break (Colombo et al. 2018). Quantitative analysis of DSB resection was performed by calculating the ratio of band intensities for ssDNA to total amount of DSB products. DSB repair by SSA was detected by Southern blot analysis using an Asp718–Sal1 fragment containing part of the LEU2 gene as a probe (Trovesi et al. 2011). Quantitative analysis of the repair product was performed by calculating the ratio of band intensities for SSA product with respect to a loading control.
Plasmid religation assay
The centromeric pRS316 plasmid digested with the BamHI restriction enzyme was transformed into the cells. Efficiency of religation was calculated by determining the number of colonies that were able to grow on medium selective for the plasmid marker and was normalized respect to the transformation efficiency for each strain. Transformation efficiency was determined after transformation with undigested pRS316 DNA.
ChIP and qPCR
Chromatin immunoprecipitation (ChIP) analysis was performed with anti-HA (12CA5) and anti-Myc antibodies (Ab32 from Abcam) as previously described (Cassani et al. 2016). Quantification of immunoprecipitated DNA was achieved by quantitative real-time PCR (qPCR) on a Bio-Rad MiniOpticon apparatus. Triplicate samples in 20 μl reaction mixture containing 10 ng of template DNA, 300 nM of each primer, 2× SsoFast EvaGreen supermix (#1725201; Bio-Rad) (2× reaction buffer with dNTPs, Sso7d-fusion polymerase, MgCl2, EvaGreen dye, and stabilizers) were run in white 48-well PCR plates Multiplate (#MLL4851; Bio-Rad). The qPCR program was as follows: step 1, 98° for 2 min; step 2, 98° for 5 sec; step 3, 60° for 10 sec; step 4, return to step 2 and repeat 30 times. At the end of the cycling program, a melting program (from 65 to 95° with a 0.5° increment every 5 sec) was run to test the specificity of each qPCR. Data are expressed as fold enrichment at the HO-induced DSB over that at the noncleaved ARO1 locus, after normalization of each ChIP signal to the corresponding input for each time point. Fold enrichment was then normalized to the efficiency of DSB induction.
Western blotting and immunoprecipitation
Protein extracts for western blot analysis were prepared by trichloroacetic acid (TCA) precipitation and separated on 10% polyacrylamide gels. Rad53 was detected by using anti-Rad53 polyclonal antibodies kindly provided by J. Diffley (The Francis Crick Institute, London, UK), while Rad9 was detected by using anti-Rad9 polyclonal antibodies kindly provided by N. Lowndes (University of Ireland, Galway, Ireland). Epitope-tagged Mre11, Tel1, Rad9, and Sae2 were detected by using anti-HA (12CA5) or anti-Myc antibodies (Ab32 from Abcam). Rad53–Rad9 coimmunoprecipitations were performed as previously described (Schwartz et al. 2002). Sae2–Sae2 coimmunoprecipitations were performed as previously described (Kim et al. 2008).
Hairpin opening assay
The rate of Lys+ recombinants was derived from the median recombination frequency determined from 10 different isolates of each strain as previously described (Lobachev et al. 2002). Three trials were performed and the mean recombination rate was calculated.
Data availability
Table S1 includes names and genotypes of each strain used in this work. Figure S1 illustrates the DNA damage sensitivity of sae2-ms and sae2-S134L cells. All the strains are available upon request. All data necessary for confirming the conclusions of the article are present within the article and the associated supplemental files. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7392671.
Results
Search for sae2 alleles that hyperactivate Tel1 but do not cause DNA damage hypersensitivity
Cells lacking Sae2 are hypersensitive to DNA damaging agents and increase MRX occupancy at DSBs, which activates a Tel1-dependent checkpoint that is accompanied by persistent Rad53 phosphorylation and prolonged cell cycle arrest (Usui et al. 2001; Lisby et al. 2004; Clerici et al. 2006).
To gain insights into the role of Sae2 in DNA damage resistance and downregulation of the checkpoint response, we searched for separation-of-function sae2 mutants that hyperactivated Tel1, similar to sae2∆ cells, but conserved Sae2 function in DNA damage resistance. We took advantage of the finding that Tel1 hyperactivation allows SAE2 deletion to suppress the hypersensitivity to hydroxyurea (HU) and methyl methanesulfonate (MMS) of cells lacking Mec1 and kept viable by SML1 deletion (Usui et al. 2001). We randomly mutagenized the SAE2 gene by low-fidelity PCR, followed by transformation of mec1∆ sml1∆ cells with the linear SAE2 PCR products obtained, in order to replace the corresponding SAE2 wild-type sequence with the mutagenized DNA fragments. Transformant clones were first chosen based on their increased viability in the presence of HU and MMS compared to mec1∆ cells. Among them, we selected for further characterization clones that were more resistant to camptothecin (CPT) and phleomycin (phleo) compared to sae2∆ cells after transformation with a plasmid carrying the wild-type MEC1 gene.
By the above analysis we identified the sae2-ms allele, DNA sequencing of which revealed three missense mutations leading to replacement of Ser134 with Leu, Pro217 with Thr, and Ala230 with Val, respectively. Similar to sae2∆ mec1∆ cells, sae2-ms mec1∆ cells showed increased viability in the presence of HU or MMS compared to mec1∆ cells (Figure 1A), indicating that the sae2-ms allele compensates for Mec1 deficiency under genotoxic treatments. Unlike SAE2 deletion, which, by itself, causes hypersensitivity to HU, MMS, CPT, and phleo, sae2-ms cells did not lose viability in the presence of any of the above tested drugs (Figure 1B), indicating that Sae2-ms maintains Sae2 function in DNA damage resistance.
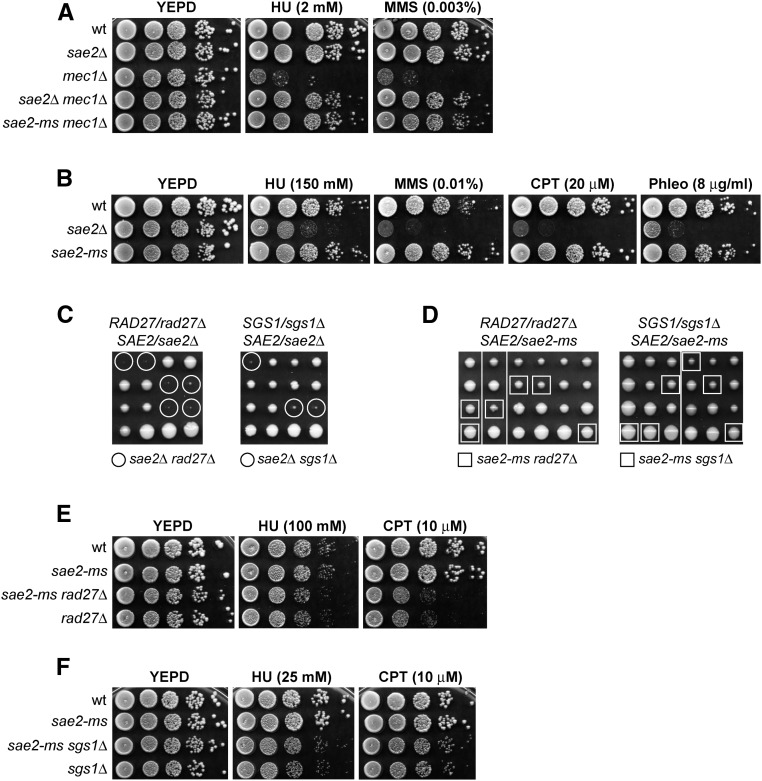
Figure 1.
Sae2-ms suppresses the hypersensitivity to HU and MMS of mec1∆ cells. (A, B, E, and F) Exponentially growing cells were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without HU, MMS, CPT, or phleo at the indicated concentrations. All strains in (A) carried SML1 deletion that kept mec1∆ cells viable. (C and D) Meiotic tetrads were dissected on YEPD plates that were incubated at 25°, followed by spore genotyping.
Sae2-ms supports viability of rad27∆ and sgs1∆ cells
Synthetic lethality/sickness is observed when SAE2 deletion is combined with deletion of the RAD27 gene, which encodes a nuclease involved in Okazaki fragment processing during lagging strand DNA synthesis (Tishkoff et al. 1997), suggesting that Sae2 is required for the processing of DNA lesions generated in a rad27Δ background (Moreau et al. 1999; Debrauwère et al. 2001). A similar synthetic effect is also seen when SAE2 is deleted in cells lacking the helicase Sgs1, possibly due to defective DSB resection and excessive telomere shortening (Mimitou and Symington 2008; Hardy et al. 2014).
To determine whether Sae2-ms maintains the Sae2 functions mentioned above, diploid cells heterozygous for both rad27∆ and sae2-ms or sgs1∆ and sae2-ms were generated, and, after sporulation, tetrads were dissected to determine whether viable rad27Δ sae2-ms or sgs1Δ sae2-ms spores could be obtained. As expected, rad27∆ sae2∆ and sgs1∆ sae2∆ spores were unviable or grew so slowly that they could not be further propagated (Figure 1C). By contrast, the rad27∆ sae2-ms and sgs1∆ sae2-ms spores grew remarkably well (Figure 1D). Furthermore, sae2-ms did not exacerbate the hypersensitivity to HU and CPT of rad27∆ (Figure 1E) and sgs1∆ cells (Figure 1F). These findings indicate that Sae2-ms maintains Sae2 function in supporting cell viability in the absence of Rad27 or Sgs1 both in the presence and in the absence of DNA damage.
Sae2-ms maintains Sae2 functions in end-tethering and long-range resection
Sae2 promotes DSB repair by supporting DNA-end resection and by maintaining the DSB ends adjacent to each other (Clerici et al. 2005). The lack of Sae2 affects not only short-range resection by abrogating Mre11 nuclease activity, but also reduces the efficiency of long-range resection by increasing the association of Rad9 at DSBs, which directly and indirectly inhibits the resection activity of Dna2-Sgs1 and Exo1, respectively (Morin et al. 2008; Bonetti et al. 2015; Ferrari et al. 2015; Gobbini et al. 2015).
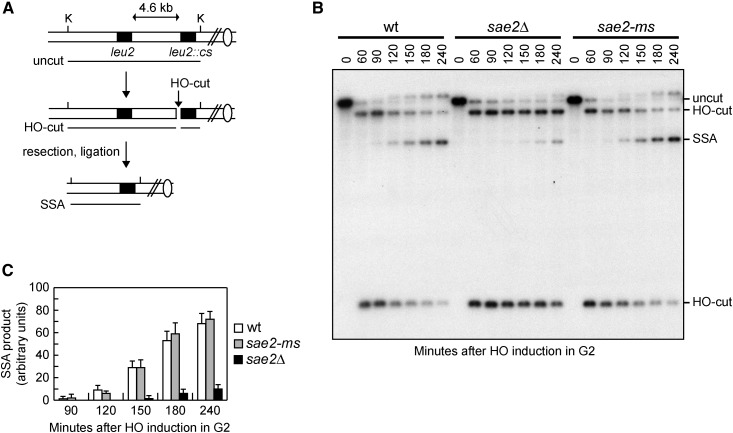
The lack of Sae2 leads to severe defects in repairing a DSB by SSA (Clerici et al. 2005). This mechanism repairs a DSB flanked by direct DNA repeats when sufficient resection exposes the complementary DNA sequences, which can then anneal to each other, resulting in deletion of the DNA region between the repeats (Fishman-Lobell et al. 1992; Ivanov et al. 1996). To assess whether Sae2-ms affects DSB repair by SSA, we introduced the sae2-ms allele in the YMV45 strain, which carries two direct sequence repeats of the LEU2 gene on chromosome III separated by 4.6 kb. An HO endonuclease cleavage site was inserted at the junction between one of the leu2 repeats and the intervening sequence (Vaze et al. 2002) (Figure 2A). This strain also carries a GAL-HO construct that provides galactose-inducible HO expression. Accumulation of the SSA repair products after HO induction was reduced in sae2∆ cells compared to wild type, whereas it occurred with almost wild-type kinetics in sae2-ms cells (Figure 2, B and C), indicating that Sae2-ms does not affect DSB repair by SSA.
Figure 2.
Sae2-ms is proficient in SSA-mediated DSB repair. (A) Map of the YMV45 chromosome III region where the HO-cut site is flanked by homologous leu2 sequences that are 4.6 kb apart. HO-induced DSB formation results in generation of 12 and 2.5 kb DNA fragments (HO-cut) that can be detected by Southern blot analysis of KpnI-digested genomic DNA with a LEU2 probe. DSB repair by SSA generates a product of 8 kb (SSA). K, KpnI. (B) Exponentially growing YEPR cell cultures were arrested in G2 with nocodazole and transferred to YEPRG in the presence of nocodazole at time zero to induce HO expression. Southern blot analysis of KpnI-digested genomic DNA with a LEU2 probe. (C) Densitometric analysis. The experiment as in (B) was repeated independently and the mean values are represented with error bars denoting SD (n = 3).
The SSA-mediated DSB repair defect in sae2∆ cells has been attributed to the lack of Sae2 function in both DNA-end tethering and long-range resection (Clerici et al. 2005). To assess more directly the ability of Sae2-ms to support end tethering, we used a strain where Lac repressor binding site (LacO) arrays were inserted at a distance of 50 kb on opposite sides of an irreparable HO-inducible cut site, and can be visualized by the binding of a constitutively expressed LacI-GFP fusion protein (Lobachev et al. 2002). HO expression was induced by galactose addition to cell cultures that were arrested and kept blocked in G2 by nocodazole treatment to ensure that all cells would remain arrested in metaphase. Most wild-type and sae2-ms cells showed a single LacI-GFP focus after HO induction, indicating their ability to maintain the broken DNA ends together, whereas sae2∆ cells showed an increase of cells with two LacI-GFP spots after HO induction (Figure 3A) (Clerici et al. 2005). Altogether, these findings indicate that Sae2-ms does not impair DNA end-tethering.
Figure 3.
Sae2-ms is proficient in end-tethering and long-range resection. (A) DSB end-tethering. Exponentially growing YEPR cell cultures were arrested in G2 with nocodazole and transferred to YEPRG in the presence of nocodazole at time zero; 200 cells for each strain were analyzed to determine the percentage of cells showing two LacI-GFP foci. Plotted values are the mean values with error bars denoting SD (n = 3). * P < 0.05 (Student’s t-test). (B) Map of the JKM139 chromosome III region. 5′–3′ resection progressively eliminates SspI sites, producing larger SspI fragments (r1 through r6). S, SspI. (C) DSB resection. YEPR exponentially growing cultures of JKM139 derivative strains were arrested in G2 with nocodazole and transferred to YEPRG in the presence of nocodazole at time zero to induce HO expression. SspI-digested genomic DNA was separated on alkaline agarose gel and hybridized with a single-stranded MAT probe that anneals to the unresected 3′ end at one side of the break. (D) Densitometric analysis. The experiment as in (C) was repeated independently and the mean values are represented with error bars denoting SD (n = 3). (E) ChIP analysis. HO was induced by galactose addition at time zero in exponentially growing JKM139 derivative cells. Relative fold enrichment of Rad9-HA protein at the indicated distance from the HO cleavage site was determined after ChIP with anti-HA antibodies and qPCR analysis. Plotted values are the mean values with error bars denoting SD (n = 3). * P < 0.05 (Student’s t-test). (F) Exponentially growing cells were spotted out onto YEPD plates with or without CPT. (G) Plasmid religation assay. Data are expressed as percentage of religation relative to wild type that was set up at 100% after normalization to the corresponding transformation efficiency. Plotted values are the mean values with error bars denoting SD (n = 3). * P < 0.05 (Student’s t-test).
The lack of Sae2 was shown to reduce long-range resection of an endonuclease-induced DSB by increasing the amount of Rad9 bound DSBs, which, in turn, acts as a barrier to Sgs1-Dna2-mediated resection and inhibits Exo1 activity by activating Rad53 (Morin et al. 2008; Bonetti et al. 2015; Ferrari et al. 2015). To test more directly the ability of sae2-ms cells to support resection by Sgs1-Dna2 and Exo1, we monitored ssDNA formation and Rad9 persistence at the HO-induced DSB generated at the MAT locus in JKM139 derivative strains expressing the HO gene from the galactose-inducible GAL1 promoter (Lee et al. 1998). The HML and HMR loci were deleted in these strains to prevent DSB repair by gene conversion. Resection of the HO-induced DSB renders the DNA sequence flanking the HO break resistant to cleavage by restriction enzymes, resulting in the appearance of resection intermediates that can be detected by Southern blot with a probe that anneals to the 3′ end at one side of the break (Figure 3B). As expected, sae2∆ cells showed a slight defect in resection of the HO-induced DSB compared to wild type, whereas the resection products accumulated with wild-type kinetics in sae2-ms cells (Figure 3, C and D). Furthermore, the amount of Rad9 bound at the HO-induced DSB, which was increased in sae2∆ cells compared to wild-type cells, was similar in both wild type and sae2-ms cells (Figure 3E). The increased Rad9 association at DSBs in sae2∆ cells is not due to altered Mre11 nuclease activity, as cells carrying the nuclease-dead mre11-H125N allele did not increase Rad9 persistence at the HO-induced DSB (Figure 3E).
Consistent with the ability of Sae2-ms to support long-range resection by Dna2 and Exo1, the sae2-ms mutation did not exacerbate the DNA damage hypersensitivity of exo1∆ cells, as it did SAE2 deletion possibly because of a more severe resection defect of sae2∆ exo1∆ double mutant compared to each single mutant (Figure 3F) (Zhu et al. 2008). Furthermore, sae2-ms cells did not increase the efficiency of ligation by NHEJ of a self-replicating plasmid, which was instead increased in sae2∆ cells likely because the reduced ssDNA generation increases the ability of NHEJ repair events to occur (Figure 3G). Altogether, these findings indicate that Sae2-ms supports both DNA end-tethering and the activity of the long-range resection nucleases.
Suppression of Mec1 deficiency by Sae2-ms requires Tel1, Rad9, and Rad53
Tel1 promotes activation of the downstream effector kinase Rad53 in response to DNA damage, and this activation requires Rad9 (Gobbini et al. 2013). To assess whether suppression of the DNA damage hypersensitivity of mec1∆ cells by Sae2-ms is due to hyperactivation of a Tel1-mediated checkpoint response, we asked whether mec1∆ suppression by sae2-ms requires Tel1, Rad9, and/or Rad53. The sae2-ms allele failed to suppress the HU hypersensitivity of tel1∆ mec1∆ cells, which lose viability dramatically even in the absence of DNA damage compared to each single mutant (Figure 4A), possibly due to excessive telomere shortening and premature senescence (Ritchie et al. 1999). Similarly, sae2-ms did not restore HU resistance of mec1∆ cells carrying either RAD9 deletion (Figure 4B) or the kinase defective rad53-K227A allele (Figure 4C). These findings indicate that the bypass of Mec1 function by Sae2-ms requires Tel1, Rad9, and Rad53 checkpoint proteins. Consistent with a Tel1 involvement, suppression of the HU sensitivity of mec1∆ sae2-ms double mutant cells was unaffected by the lack of Ddc1 (Figure 4D), which interacts with Mec3 and Rad17 to form a heterotrimeric complex that stimulates Mec1 kinase activity but not Tel1 kinase activity (Gobbini et al. 2013).
Figure 4.
Sae2-ms requires Tel1, Rad53, and Rad9 for suppression of Mec1 deficiency. (A–E) Exponentially growing cells were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without HU or MMS at the indicated concentrations. All strains carried SML1 deletion that kept mec1∆ cells viable.
The sae2-S134L mutation is responsible for suppression of Mec1 deficiency
The Sae2-ms mutant variant carries the three amino acid substitutions S134L, P217T, and A230V. We asked which substitution(s) was responsible for the suppression of mec1∆ hypersensitivity to DNA damage by constructing strains expressing the sae2-S134L or the sae2-P217T, A230V allele. Comparison analysis revealed that the sae2-S134L allele restored resistance of mec1∆ cells to HU and MMS to a level similar to that observed in sae2-ms mec1∆ cells, whereas the sae2-P217T, A230V allele did not (Figure 4E). Thus, effective mec1∆ suppression appears to be due exclusively to the S134L aminoacid substitution. Similar to sae2-ms cells, sae2-S134L cells were not hypersensitive to DNA damaging agents (Figure S1).
The Sae2 S134 residue was shown to be phosphorylated by Cdk1 (Huertas et al. 2008; Fu et al. 2014), prompting us to test the effect of substituting this residue with either the nonphosphorylatable alanine residue or aspartic acid, which mimics constitutive phosphorylation. We found that the sae2-S134A allele suppressed the HU and MMS sensitivity of mec1∆ cells as efficiently as sae2-S134L (Figure 4E). However, the S134D aminoacid substitution also restored resistance of mec1∆ cells to HU and MMS (Figure 4E), suggesting that the negative charge associated with the phosphorylation event of S134 is not relevant for Sae2 function in bypassing Mec1 deficiency. Consistent with this hypothesis, substitution of the E131 residue, which is located close to S134, with valine suppressed the sensitivity to MMS of mec1∆ cells without causing DNA damage hypersensitivity by itself (Kim et al. 2008), suggesting that the region of the protein surrounding the S134 residue, rather than its phosphorylation, is important for the bypass of Mec1 function.
Sae2-S134L and Sae2-ms reduce hairpin cleavage and increase MRX and Tel1 association at DNA DSBs
Previous work has established that SAE2 deletion leads to increased MRX persistence at DSBs, which can account for enhanced Tel1 activation and bypass of Mec1 deficiency (Usui et al. 2001; Lisby et al. 2004; Clerici et al. 2006). Thus, we measured Mre11 and Tel1 association at the HO-induced DSB by ChIP and qPCR. Association to DNA DSBs of both Mre11 (Figure 5A) and Tel1 (Figure 5B) was more robust and persisted longer, not only in sae2∆ cells but also in sae2-ms and sae2-S134L cells, indicating that Sae2-ms and Sae2-S134L increase the amount of MRX and Tel1 bound at DSBs. The increased Mre11 and Tel1 association was not due to increased amounts of Mre11 or Tel1, as similar levels of Mre11 (Figure 5C) and Tel1 (Figure 5D) proteins were detected in protein extracts from wild type, sae2-ms, and sae2-S134L cells.
Figure 5.
Sae2-ms and Sae2-S134L enhance Mre11 and Tel1 association to DSBs and reduce hairpin cleavage. (A) ChIP analysis. HO was induced by galactose addition at time zero in exponentially growing JKM139 derivative cells. Relative fold enrichment of Mre11-Myc protein at the indicated distances from the HO cleavage site was determined after ChIP with anti-Myc antibodies and subsequent qPCR analysis. Plotted values are the mean values with error bars denoting SD (n = 3). * P < 0.05 (Student’s t-test). (B) As in (A), but showing relative fold enrichment of Tel1-HA after ChIP with anti-HA antibodies. (C and D) Western blot analysis with anti-Myc or anti-HA antibodies of protein extracts prepared from exponentially growing cells. The same amount of extracts was probed with anti-Pgk1 antibodies as loading control. (E) Recombination frequency of strains with the lys2-AluIR and lys2-Δ5’ ectopic recombination system. The rate of Lys+ recombinants was derived from the median recombination frequency. The reported values are the mean values with SD indicated in brackets (n = 3). (F) Exponentially growing cells were serially diluted (1:10) and spotted out onto YEPD plates with or without HU or MMS. All strains carried SML1 deletion that kept mec1∆ cells viable. (G) Protein extracts prepared from exponentially growing cells were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (Input) or after immunoprecipitation (IP) with anti-HA antibodies. *indicates a cross-hybridization signal.
The Sae2-dependent Mre11 endonucleolytic activity is essential to initiate resection at DNA ends that are not directly accessible to Exo1 and Dna2-Sgs1 because they are capped by hairpin DNA structures or bound proteins (Trujillo and Sung 2001; Lobachev et al. 2002; Cannavo and Cejka 2014). mre11-H125N allele, which specifically eliminates Mre11 nuclease activity, increases Mre11 and Tel1 persistence at DSBs (Figure 5, A and B) (Lisby et al. 2004; Clerici et al. 2006), suggesting that this activity can contribute to MRX displacement from DSBs. Thus, we investigated whether the sae2-ms and sae2-S134L mutations might specifically reduce Mre11 nuclease activity. As the Mre11 nuclease activity and Sae2 are required to open DNA hairpin structures both in vitro and in vivo (Trujillo and Sung 2001; Lobachev et al. 2002), we used a genetic assay to measure hairpin resolution in sae2-ms and sae2-S134L cells. Inverted Alu elements inserted in the LYS2 gene on chromosome III form a hairpin-capped end, whose opening by the MRX nuclease and Sae2 stimulates recombination with a truncated lys2 gene on chromosome II to generate Lys+ cells (Lobachev et al. 2002). As expected, sae2∆ and the nuclease defective mre11-H125N cells showed decreased rates of Lys+ recombinants compared to wild-type cells (Figure 5E). Interestingly, the rates of Lys+ prototrophs were reduced also in sae2-ms and sae2-S134L cells, although to lower extents than in sae2∆ and mre11-H125N cells (Figure 5E). These findings suggest that Sae2-ms and Sae2-S134L can impair MRX removal from the sites of DNA damage by altering Mre11 nuclease activity.
Although Mre11-H125N persisted longer at DNA DSBs (Figure 5A) and caused increased Tel1 association at DSBs (Figure 5B), it did not suppress the hypersensitivity to HU of mec1∆ cells and only slightly suppressed their hypersensitivity to MMS (Figure 5F). This finding suggests that upregulation of MRX and Tel1 in the presence of the Mre11-H125N mutant variant is not sufficient to bypass Mec1 deficiency.
It has been shown that Sae2 oligomerization is important for Sae2 function in the DNA damage response (Kim et al. 2008; Cannavo et al. 2018). A region of Sae2 spanning from 120 to 170 amino acids (and therefore containing the S134 residue) was shown to be important for Sae2 self-interaction (Kim et al. 2008), prompting us to test the self-association properties of Sae2-ms. Sae2 was immunoprecipitated with anti-HA antibodies from protein extracts of SAE2-HA/SAE2-MYC, sae2-ms-HA/SAE2-MYC and sae2-ms-HA/sae2-ms-MYC diploid cells. The amount of either Sae2-Myc or Sae2-ms-Myc detected by anti-Myc antibodies in immunoprecipitates of Sae2-ms-HA was similar to that of wild-type Sae2-Myc detected in immunoprecipitates of Sae2-HA (Figure 5G). Thus, the sae2-ms mutation does not affect Sae2 self-interaction.
Sae2 plays distinct functions in downregulation of MRX-Tel1 and Rad53 activities
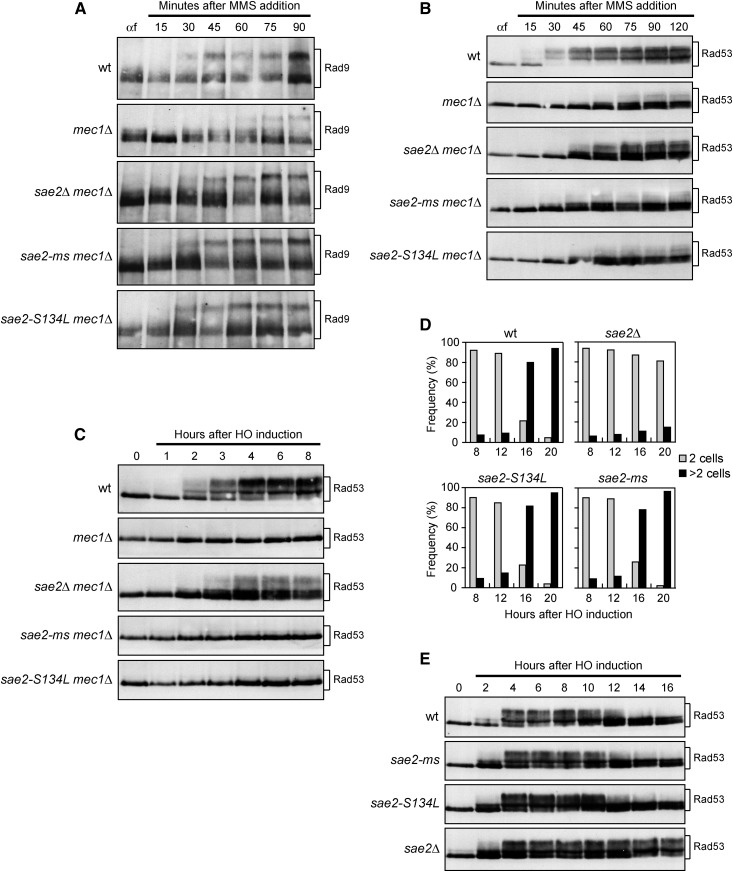
Activation of Rad53 requires its interaction with the adaptor Rad9 that is phosphorylated by Mec1/Tel1 (Emili 1998; Sun et al. 1998; Vialard et al. 1998; Durocher et al. 1999; Pellicioli et al. 1999; Gilbert et al. 2001; Schwartz et al. 2002; Sweeney et al. 2005; Smolka et al. 2007). To better understand the effects of Sae2-ms and Sae2-S134L on Tel1-mediated Rad53 activation, we analyzed Rad9 and Rad53 phosphorylation, detected as electrophoretic mobility shifts, in mec1∆, sae2∆ mec1∆, sae2-ms mec1∆, and sae2-S134L mec1∆ cells arrested in G1 and then released into the cell cycle in the presence of MMS. As expected, MMS-treated mec1∆ cells showed a decrease of both Rad9 (Figure 6A) and Rad53 phosphorylation (Figure 6B) compared to wild type cells. Consistent with the finding that the sae2∆, sae2-ms, and sae2-S134L alleles increase Tel1 signaling activity, Rad9 phosphorylation was increased in MMS-treated sae2∆ mec1∆, sae2-ms mec1∆, and sae2-S134L mec1∆ cells compared to mec1∆ cells (Figure 6A). However, while sae2∆ mec1∆ cells showed also enhanced Rad53 phosphorylation compared to mec1∆ cells, sae2-ms mec1∆, and sae2-S134L mec1∆ cells did not (Figure 6B). The inability of sae2-ms mec1∆ and sae2-S134L mec1∆ cells to hyperactivate Rad53 compared to sae2∆ mec1∆ cells is not due to a more efficient DNA repair, as sae2-ms and sae2-S134L cells did not show Rad53 hyperactivation also in response to a single irreparable DSB (Figure 6C). In fact, when cultures of JKM139 derivative strains were transferred to galactose to induce HO, sae2∆ mec1∆ cells showed an increased amount of Rad53 phosphorylation compared to mec1∆ cells, while neither sae2-ms mec1∆ nor sae2-S134L mec1∆ cells did (Figure 6C). These findings indicate that Sae2-ms and Sae2-S134L mutant variants are defective in the downregulation of MRX-Tel1 signaling, but not of Rad53 signaling.
Figure 6.
Sae2-ms and Sae2-S134L do not enhance Rad53 phosphorylation. (A and B) Exponentially growing cells were arrested in G1 with α-factor (αf) and released into the cell cycle in the presence of MMS (0.03%). Western blot analysis with anti-Rad9 (A) and anti-Rad53 antibodies (B). (C) Exponentially growing YEPR cultures of JKM139 derivative strains were transferred to YEPRG at time zero to induce HO. Western blot analysis with anti-Rad53 antibodies. (D) Adaptation assay. YEPR G1-arrested cell cultures were plated on galactose-containing plates (time zero). At the indicated time points, 200 cells for each strain were analyzed to determine the frequency of large budded cells (two cells) and of cells forming microcolonies of more than two cells. (E) Exponentially growing YEPR cell cultures were transferred to YEPRG at time zero to induce HO. Western blot analysis with anti-Rad53 antibodies.
Cells carrying a single irreparable DSB undergo checkpoint-mediated cell cycle arrest, but then they adapt to this checkpoint, decreasing Rad53 activation and re-entering the cell cycle (Toczyski et al. 1997; Pellicioli et al. 2001). The heightened Rad53 activation in sae2∆ cells prevents the turning off of the checkpoint triggered by a single irreparable DSB (Clerici et al. 2006). To assess further that Tel1/MRX upregulation by Sae2-ms and Sae2-S134L does not increase Rad53 activation, we analyzed the ability of sae2-ms and sae2-S134L cells to adapt to a single irreparable DSB. When G1-arrested cell cultures of JKM139 derivative strains were spotted on galactose-containing plates to induce HO, most sae2Δ cells were still arrested at the two-cell dumbbell stage after 20 hr, whereas wild type, sae2-ms, and sae2-S134L cells over-rode the checkpoint-mediated cell cycle arrest within 16 hr, producing microcolonies with more than two cells (Figure 6D). Moreover, when galactose was added to exponentially growing cell cultures of the same strains, Rad53 phosphorylation decreased in wild type, sae2-ms, and sae2-S134L cells 12–14 hr after galactose addition, while it persisted throughout the experiment in sae2Δ cells (Figure 6E). Altogether, these findings indicate that Sae2-ms and Sae2-S134L mutant variants are specifically defective in downregulating Tel1 activation but not Rad53 activation, indicating that Sae2 plays distinct functions in the inhibition of MRX-Tel1 and Rad53 activities.
Sae2 inhibits the interaction between Rad9 and Rad53
Activation of Rad53 in vivo requires its interaction with Rad9, which acts both as an adaptor mediating the interaction between Mec1 and Rad53, and as a scaffold facilitating the concentration of Rad53 molecules at the sites of damage. In fact, Rad9 phosphorylation by Mec1 or Tel1 creates a binding site for Rad53 interaction (Sun et al. 1998; Durocher et al. 1999; Schwartz et al. 2002). Mec1 and Tel1 subsequently phosphorylate Rad53, which is associated with Rad9 (Sweeney et al. 2005; Smolka et al. 2007), followed by Rad53 in trans autophosphorylation and full activation of the kinase (Pellicioli et al. 1999; Gilbert et al. 2001).
The finding that Sae2-ms and Sae2-S134L are capable of inducing Rad9 hyperphosphorylation but not Rad53 hyperphosphorylation suggests that Sae2 can inhibit Rad53 activation by limiting Rad53–Rad9 interaction and/or Rad53 autophosphorylation. We therefore immunoprecipitated HA epitope-tagged Rad9 from cell extracts prepared from undamaged exponentially growing cells. A basal level of Rad53 binding to Rad9 was detected in wild-type cells even in the absence of DNA damage, and this interaction increased when Rad9 was immunoprecipitated from sae2∆ cells (Figure 7A). By contrast, both sae2-ms and sae2-S134L cells showed a level of Rad53 binding to Rad9 similar to that observed in wild-type cells (Figure 7A). These findings indicate that Sae2 inhibits Rad53–Rad9 interaction and that Sae2-ms and Sae2-S134L maintain this function. This inhibition does not depend on Sae2 stimulation of MRX nuclease activity, as Rad53 binding to Rad9 in nuclease defective mre11-H125N cells was similar to that of wild-type cells or even lower (Figure 7B).
Figure 7.
Sae2 inhibits Rad9-Rad53 interaction. (A and B) Protein extracts prepared from exponentially growing cells were analyzed by western blotting with anti-HA (Rad9) and anti-Rad53 antibodies either directly (Input) or after immunoprecipitation (IP) with anti-HA antibodies. *indicates a cross-hybridization signal. (C) Western blot analysis with anti-HA antibodies of protein extracts prepared from exponentially growing cells. The same amount of extracts was stained with Coomassie Blue as loading control. (D) ChIP analysis. HO was induced by galactose addition at time zero in exponentially growing JKM139 derivative cells. Relative fold enrichment of Sae2-HA protein at the indicated distance from the HO cleavage site was determined after ChIP with anti-HA antibodies and subsequent qPCR analysis. Plotted values are the mean values with error bars denoting SD (n = 3). * P < 0.05 (Student’s t-test).
Sae2 overproduction was shown to decrease Rad53 phosphorylation and activation independently of DSB repair (Clerici et al. 2006). The ability of Sae2-ms and Sae2-S134L to downregulate Rad53 activation is not due to increased production or binding to the sites of damage of the corresponding mutant proteins. In fact, similar amounts of Sae2, Sae2-ms, and Sae2-S134L were detected in protein extracts from wild type, sae2-ms, and sae2-S134L cells (Figure 7C). Furthermore, the amount of Sae2-ms and Sae2-S134L bound at an HO-induced DSB was similar, or even lower, than that of wild-type Sae2 (Figure 7D).
Discussion
SAE2 deletion causes DNA damage hypersensitivity and enhances Tel1 and Rad53 signaling activities (Usui et al. 2001; Lisby et al. 2004; Clerici et al. 2006). The persistent Tel1- and Rad53-mediated checkpoint activation in sae2∆ cells requires the function of MRX, whose association at DSBs is increased in sae2∆ cells (Lisby et al. 2004; Clerici et al. 2006). Reducing either MRX association to DSBs or Rad53/Tel1 signaling restores DNA damage resistance in Sae2-deficient cells (Chen et al. 2015; Gobbini et al. 2015; Puddu et al. 2015; Cassani et al. 2018), suggesting that the DNA damage hypersensitivity of sae2∆ cells is due to a failure to downregulate MRX/Tel1 and/or Rad53 activities.
To better understand the function of Sae2 in DNA damage resistance and in the regulation of MRX association to DSBs and of Tel1 and Rad53 activation, we searched for sae2 alleles that hyperactivate Tel1, but that do not cause DNA damage hypersensitivity by themselves. This screen allowed us to identify the Sae2-ms mutant variant, which restores resistance of mec1∆ cells to HU and MMS in a Tel1-, Rad9- and Rad53-dependent manner. Sae2-ms carries three amino acid substitutions, with S134L being responsible for mec1∆ suppression.
Similar to SAE2 deletion, both Sae2-ms and Sae2-S134L increase Tel1 signaling activity by enhancing MRX and Tel1 association to DNA ends, and are defective in hairpin cleavage, which is known to depend on Mre11 endonucleolytic activity (Lobachev et al. 2002). This finding suggests that the MRX-Sae2-mediated cleavage activity contributes to eliminate MRX bound to DNA ends and this MRX displacement limits Tel1 signaling activity. Consistent with a role of Mre11 endonuclease in MRX removal, abolition of Mre11 nuclease activity by the H125N substitution increases the amount of MRX and Tel1 bound at DSBs to an extent similar to that caused by SAE2 deletion.
Upregulation of MRX-Tel1 in sae2∆ cells is accompanied by enhanced DSB-induced Rad53 phosphorylation and activation. Although sae2∆, sae2-ms, and sae2-S134L cells show equivalent increase of MRX and Tel1 association to DSBs, Sae2-ms and Sae2-S134L do not cause persistent Rad53 activation as the absence of Sae2. The inability of sae2-ms and sae2-S134L cells to hyperactivate Rad53 compared to sae2∆ cells does not appear to be due to different amounts of MRX-Tel1 bound to DSBs and/or residual Mre11 clipping activity. In fact, nuclease defective mre11-H125N cells, which increase MRX-Tel1 association at DSBs and reduce hairpin cleavage to an extent similar to sae2∆ cells, fail to hyperactivate Rad53. These findings suggest that Sae2 plays distinct functions in dampening Tel1 and Rad53 signaling activities.
Mec1 is known to play two distinct roles in Rad53 activation. First, Mec1 phosphorylates multiple Rad9 residues (Schwartz et al. 2002), and phosphorylated Rad9 recruits Rad53 to DNA lesions (Sun et al. 1998; Vialard et al. 1998; Durocher et al. 1999; Schwartz et al. 2002). Then, Mec1 phosphorylates Rad53 bound to Rad9 on multiple sites (Sweeney et al. 2005; Smolka et al. 2007), and this phosphorylation of Rad53 presumably contributes to the relief of catalytic autoinhibition, allowing Rad53 autophosphorylation and activation (Pellicioli et al. 1999; Gilbert et al. 2001). Consistent with an upregulation of Tel1 activity, both the lack of Sae2 and the presence of Sae2-ms or Sae2-S134L increase DSB-induced Rad9 phosphorylation in cells lacking Mec1. However, only the lack of Sae2, but neither the presence Sae2-ms nor Sae2-S134L, increases the interaction between Rad53 and Rad9 even in the absence of DNA lesions. Since Rad53 autophosphorylation and activation requires Mec1/Tel1-dependent phosphorylation of Rad53 molecules that are bound to Rad9 (Sweeney et al. 2005), we propose that Sae2 limits Rad53 activation by inhibiting Rad53–Rad9 interaction, and that Sae2-ms and Sae2-S134L maintain this function.
How does Sae2 limit Rad9-Rad53 interaction? Sae2-mediated inhibition of Rad53 activation does not require Sae2 function in promoting MRX nuclease activity, as Rad53–Rad9 interaction is not enhanced in mre11-H125N cells. We have previously shown that Rad9 persistence is the primary cause of the DNA damage hypersensitivity and the resection defect of sae2∆ cells (Gobbini et al. 2015). Interestingly, neither Sae2-ms nor Mre11-H125N increases Rad9 persistence at DSBs. This finding suggests that the Mre11 nuclease activity does not limit Rad9 accumulation at DSBs and that Sae2 by itself can directly interfere with Rad9 persistence at DNA ends. As Rad9 is required to activate Rad53, a robust Rad9 accumulation at DSBs in sae2∆ cells can account for the increased Rad9–Rad53 interaction and therefore Rad53 hyperactivation. However, since Sae2 interacts with Rad53 (Liang et al. 2015), and defective Rad53 kinase activity bypasses Sae2 function in DNA damage resistance and resection by decreasing the amount of Rad9 bound at DSBs (Gobbini et al. 2015), it is also possible that Sae2 directly inhibits Rad9–Rad53 interaction, and that the lack of this function leads to Rad53 hyperactivation, which in turn increases Rad9 association to DSBs in a positive feedback loop. In any case, the finding that sae2-ms and sae2-S134L are proficient in long-range resection, and are DNA damage resistant, indicates that the increased Rad9 accumulation at DSBs is responsible for the DNA damage hypersensitivity and the impaired long-range resection of sae2∆ cells.
Although Rad53 is not hyperactivated in both sae2-ms and sae2-S134L cells as in sae2∆ cells, both Sae2-ms and Sae2-S134L are capable of compensating for Mec1 deficiency in a Rad53-dependent manner. This finding suggests that upregulation of MRX/Tel1 signaling by these Sae2 mutant variants increases Rad53 activation to a level that is sufficient to compensate for Mec1 deficiency. However, as Rad9 persistence at DSBs is not enhanced in these cells compared to sae2∆ cells, the retained ability of Sae2-ms and Sae2-S134L to limit Rad9 accumulation at DSBs does not allow to reach the extent of Rad53 activation that is responsible for the persistent DNA damage-induced cell cycle arrest of sae2∆ cells.
Although the nuclease defective Mre11-H125N variant increases MRX and Tel1 accumulation at DSBs, unlike Sae2-ms and Sae2-S134L, it is not capable to compensate for Mec1 deficiency. Interestingly, the Mre11-H125N mutant variant was shown to increase the amount of Sae2 bound at DSBs (Lisby et al. 2004). As Sae2 overproduction decreases Rad53 phosphorylation and activation (Clerici et al. 2006), the increased Sae2 persistence at DSBs in mre11-H125N cells may limit Rad53 activation, and, therefore, the ability of Mre11-H125N to compensate for Mec1 deficiency despite an increased MRX-Tel1 signaling. As Rad9 and Rad53 limit DSB resection by inhibiting Sgs1/Dna2 and Exo1 (Morin et al. 2008; Bonetti et al. 2015; Ferrari et al. 2015; Gobbini et al. 2015), downregulation of both Rad9 persistence at DSBs and Rad53 activation can also explain why mre11-H125N cells are proficient in long-range resection and are considerably less sensitive to DNA damaging agents than sae2Δ cells.
In summary, our findings support a model whereby Sae2 has two distinct functions in checkpoint downregulation. On the one hand, it removes MRX and Tel1 from DNA ends by promoting Mre11 endonuclease activity; on the other, it limits Rad9 accumulation to DSBs independently of Mre11 nuclease activity. Both these Sae2 functions contribute to downregulate Rad53 activation, with control of Rad9 association playing the major role, providing different layers of regulation of the checkpoint response in the maintenance of genome stability.
Acknowledgments
We thank G. Lucchini for critical reading of the manuscript. We also thank J. Haber, M. A. Resnick, and D.P. Toczyski for yeast strains, and J. Diffley and N. Lowndes for antibodies. The research leading to these results has received funding from Associazione Italiana per la Ricerca sul Cancro (AIRC) under IG 2017 – ID. 19783 project – P.I. Longhese Maria Pia, and Progetti di Ricerca di Interesse Nazionale (PRIN) 2015 to M.P.L.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7392671.
Communicating editor: O. Cohen-Fix
Literature Cited
- Berkovich E., Jr. Monnat R. J., Kastan M. B., 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9: 683–690. 10.1038/ncb1599 [DOI] [PubMed] [Google Scholar]
- Bonetti D., Villa M., Gobbini E., Cassani C., Tedeschi G., et al. , 2015. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 16: 351–361. 10.15252/embr.201439764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D., Colombo C. V., Clerici M., Longhese M. P., 2018. Processing of DNA ends in the maintenance of genome stability. Front. Genet. 9: 390 10.3389/fgene.2018.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Cejka P., 2014. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514: 122–125. 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- Cannavo E., Cejka P., Kowalczykowski S. C., 2013. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. USA 110: E1661–E1668. 10.1073/pnas.1305166110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Johnson D., Andres S. N., Kissling V. M., Reinert J. K., et al. , 2018. Regulatory control of DNA end resection by Sae2 phosphorylation. Nat. Commun. 9: 4016 10.1038/s41467-018-06417-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani C., Gobbini E., Wang W., Niu H., Clerici M., et al. , 2016. Tel1 and Rif2 regulate MRX function in end-tethering and repair of DNA double-strand breaks. PLoS Biol. 14: e1002387 10.1371/journal.pbio.1002387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani C., Gobbini E., Vertemara J., Wang W., Marsella A., et al. , 2018. Structurally distinct Mre11 domains mediate MRX functions in resection, end-tethering and DNA damage resistance. Nucleic Acids Res. 46: 2990–3008. 10.1093/nar/gky086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., et al. , 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116. 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Donnianni R. A., Handa N., Deng S. K., Oh J., et al. , 2015. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc. Natl. Acad. Sci. USA 112: E1880–E1887. 10.1073/pnas.1503331112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S. J., 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40: 179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M. P., 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280: 38631–38638. 10.1074/jbc.M508339200 [DOI] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M. P., 2006. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7: 212–218. 10.1038/sj.embor.7400593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Trovesi C., Galbiati A., Lucchini G., Longhese M. P., 2014. Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 33: 198–216. 10.1002/embj.201386041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo C. V., Menin L., Clerici M., 2018. Alkaline denaturing southern blot analysis to monitor double-strand break processing. Methods Mol. Biol. 1672: 131–145. 10.1007/978-1-4939-7306-4_11 [DOI] [PubMed] [Google Scholar]
- Debrauwère H., Loeillet S., Lin W., Lopes J., Nicolas A., 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98: 8263–8269. 10.1073/pnas.121075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Henckel J., Fersht A. R., Jackson S. P., 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4: 387–394. 10.1016/S1097-2765(00)80340-8 [DOI] [PubMed] [Google Scholar]
- Emili A., 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2: 183–189. 10.1016/S1097-2765(00)80128-8 [DOI] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P., 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611. 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- Ferrari M., Dibitetto D., De Gregorio G., Eapen V. V., Rawal C. C., et al. , 2015. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 11: e1004928 10.1371/journal.pgen.1004928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J. E., 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12: 1292–1303. 10.1128/MCB.12.3.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Chow J., Bernstein K. A., Makharashvili N., Arora S., et al. , 2014. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol. Cell. Biol. 34: 778–793 (erratum: Mol. Cell Biol. 34: 4213) 10.1128/MCB.00963-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Phelps S. E., Gray S., Neale M. J., 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. 10.1038/nature10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C. S., Green C. M., Lowndes N. F., 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8: 129–136. 10.1016/S1097-2765(01)00267-2 [DOI] [PubMed] [Google Scholar]
- Gobbini E., Cesena D., Galbiati A., Lockhart A., Longhese M. P., 2013. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair (Amst.) 12: 791–799. 10.1016/j.dnarep.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Gobbini E., Villa M., Gnugnoli M., Menin L., Clerici M., et al. , 2015. Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 11: e1005685 10.1371/journal.pgen.1005685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbini E., Cassani C., Vertemara J., Wang W., Mambretti F., et al. , 2018. The MRX complex regulates Exo1 resection activity by altering DNA end structure. EMBO J. 337: e98701 10.15252/embj.201798588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J., Churikov D., Géli V., Simon M. N., 2014. Sgs1 and Sae2 promote telomere replication by limiting accumulation of ssDNA. Nat. Commun. 5: 5004 10.1038/ncomms6004 [DOI] [PubMed] [Google Scholar]
- Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692. 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Sugawara N., Fishman-Lobell J., Haber J. E., 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., et al. , 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8: 37–45. 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Vijayakumar S., Reger M., Harrison J. C., Haber J. E., et al. , 2008. Functional interactions between Sae2 and the Mre11 complex. Genetics 178: 711–723. 10.1534/genetics.107.081331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T., 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554. 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., et al. , 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. 10.1016/S0092-8674(00)81482-8 [DOI] [PubMed] [Google Scholar]
- Liang J., Suhandynata R. T., Zhou H., 2015. Phosphorylation of Sae2 mediates forkhead-associated (FHA) domain-specific interaction and regulates its DNA repair function. J. Biol. Chem. 290: 10751–10763. 10.1074/jbc.M114.625293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Llorente B., Symington L. S., 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24: 9682–9694. 10.1128/MCB.24.21.9682-9694.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Gordenin D. A., Resnick M. A., 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. 10.1016/S0092-8674(02)00614-1 [DOI] [PubMed] [Google Scholar]
- Mehta A., Haber J. E., 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6: a016428 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Ferguson J. R., Symington L. S., 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. 10.1128/MCB.19.1.556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin I., Ngo H. P., Greenall A., Zubko M. K., Morrice N., et al. , 2008. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 27: 2400–2410. 10.1038/emboj.2008.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. S., Cortez D., 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281: 9346–9350. 10.1074/jbc.M513265200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17: 1957–1962. 10.1101/gad.1099003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M. J., Pan J., Keeney S., 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057. 10.1038/nature03872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette M. L., Lee K., Guo Z., Rani M., Chow J. M., et al. , 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17: 1478–1485. 10.1038/nsmb.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., et al. , 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25: 350–362. 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., et al. , 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., et al. , 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561–6572. 10.1093/emboj/18.22.6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lee S. E., Lucca C., Foiani M., Haber J. E., 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7: 293–300. 10.1016/S1097-2765(01)00177-0 [DOI] [PubMed] [Google Scholar]
- Puddu F., Oelschlaegel T., Guerini I., Geisler N. J., Niu H., et al. , 2015. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 34: 1509–1522. 10.15252/embj.201590973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato G., Cannavo E., Cejka P., 2017. Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 31: 2325–2330. 10.1101/gad.308254.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B., Mallory J. C., Petes T. D., 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065–6075. 10.1128/MCB.19.9.6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., et al. , 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9: 1055–1065. 10.1016/S1097-2765(02)00532-4 [DOI] [PubMed] [Google Scholar]
- Shibata A., Moiani D., Arvai A. S., Perry J., Harding S. M., et al. , 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53: 7–18 (erratum: Mol. Cell 53: 361) 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., et al. , 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29: 3370–3380. 10.1038/emboj.2010.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H., 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104: 10364–10369. 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Hsiao J., Fay D. S., Stern D. F., 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274. 10.1126/science.281.5374.272 [DOI] [PubMed] [Google Scholar]
- Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., et al. , 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15: 1364–1375. 10.1016/j.cub.2005.06.063 [DOI] [PubMed] [Google Scholar]
- Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D., 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263. 10.1016/S0092-8674(00)81846-2 [DOI] [PubMed] [Google Scholar]
- Toczyski D. P., Galgoczy D. J., Hartwell L. H., 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90: 1097–1106. 10.1016/S0092-8674(00)80375-X [DOI] [PubMed] [Google Scholar]
- Trovesi C., Falcettoni M., Lucchini G., Clerici M., Longhese M. P., 2011. Distinct Cdk1 requirements during single-strand annealing, crossover, and noncrossover recombination. PLoS Genet. 7: e1002263 10.1371/journal.pgen.1002263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K. M., Sung P., 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276: 35458–35464. 10.1074/jbc.M105482200 [DOI] [PubMed] [Google Scholar]
- Usui T., Ogawa H., Petrini J. H., 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7: 1255–1266. 10.1016/S1097-2765(01)00270-2 [DOI] [PubMed] [Google Scholar]
- Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., et al. , 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10: 373–385. 10.1016/S1097-2765(02)00593-2 [DOI] [PubMed] [Google Scholar]
- Vialard J. E., Gilbert C. S., Green C. M., Lowndes N. F., 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17: 5679–5688. 10.1093/emboj/17.19.5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa M., Cassani C., Gobbini E., Bonetti D., Longhese M. P., 2016. Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell. Mol. Life Sci. 73: 3655–3663. 10.1007/s00018-016-2262-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Daley J. M., Kwon Y., Krasner D. S., Sung P., 2017. Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31: 2331–2336. 10.1101/gad.307900.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Chahwan C., Bailis J., Hunter T., Russell P., 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25: 5363–5379. 10.1128/MCB.25.13.5363-5379.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 includes names and genotypes of each strain used in this work. Figure S1 illustrates the DNA damage sensitivity of sae2-ms and sae2-S134L cells. All the strains are available upon request. All data necessary for confirming the conclusions of the article are present within the article and the associated supplemental files. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7392671.