Abstract
Developmental transitions of germ cells are often regulated at the level of post-transcriptional control of gene expression. In the Caenorhabditis elegans germline, stem and progenitor cells exit the proliferative phase and enter meiotic differentiation to form gametes essential for fertility. The RNA binding protein GLD-1 is a cell fate regulator that promotes meiosis and germ cell differentiation during development by binding to and repressing translation of target messenger RNAs. Here, we discovered that some GLD-1 functions are promoted by binding to DLC-1, a small protein that functions as an allosteric regulator of multisubunit protein complexes. We found that DLC-1 is required to regulate a subset of GLD-1 target messenger RNAs and that DLC-1 binding GLD-1 prevents ectopic germ cell proliferation and facilitates gametogenesis in vivo. Additionally, our results reveal a new requirement for GLD-1 in the events of oogenesis leading to ovulation. DLC-1 contributes to GLD-1 function independent of its role as a light chain component of the dynein motor. Instead, we propose that DLC-1 promotes assembly of GLD-1 with other binding partners, which facilitates formation of regulatory ribonucleoprotein complexes and may direct GLD-1 target messenger RNA selectivity.
Keywords: germline, post-transcriptional regulation, RNA binding protein, tumor
THE germ cells of Caenorhabditis elegans proceed through a precisely orchestrated developmental program to generate sperm and oocytes (Pazdernik and Schedl 2013; Voronina and Greenstein 2016). The C. elegans gonad is structured as an assembly line, where the proliferating cells including stem cells are found in the most distal region and the differentiated gametes are at the proximal end of the germline (Figure 1A). Developmental transitions in C. elegans germline are largely controlled by the post-transcriptional regulation achieved by RNA binding proteins specifically recognizing their target messenger RNAs (mRNAs) and affecting their fate (Nousch and Eckmann 2013). The importance of post-transcriptional control is reflected in the large number of RNA binding proteins required for the normal development and function of the germline.
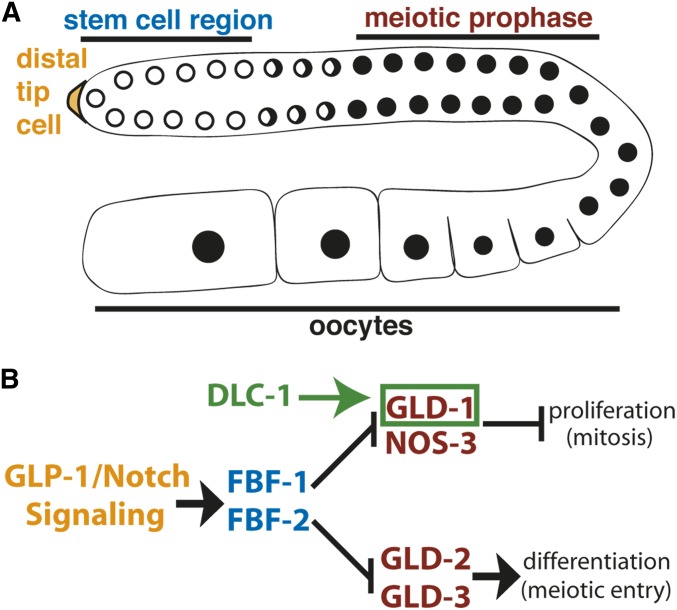
Figure 1.
(A) Schematic of C. elegans adult germline. The stem and progenitor cell region resides at the distal end followed by the transition zone where cells switch from mitosis to meiosis. Next is the pachytene region where germ cells undergo meiosis and the oocytes are located at the proximal end. (B) The regulatory network that controls the decision between stem cell proliferation and differentiation (mitotic and meiotic cell cycles). GLP-1/Notch signaling activated by a somatic distal tip cell (yellow) promotes mitotic divisions in the stem cell region of the worm germline (blue). FBF-1 and FBF-2 proteins bind and repress mRNAs that would initiate meiosis or differentiation. The GLD-1/NOS-3 and GLD-2/3 pathways promote entry into meiosis (maroon). We hypothesize that DLC-1 (green) promotes GLD-1 function.
The developmental regulatory network controlling the switch between germ cell proliferation and differentiation is well established (Kimble and Seidel 2008). GLP-1/Notch signaling from the somatic niche promotes self-renewal through the activation of Pumilio-family RNA binding proteins FBF-1 and FBF-2 (Zhang et al. 1997; Crittenden et al. 2002). Constitutive Notch signaling as observed in the strong gain-of-function allele glp-1(oz112gf) results in failure of meiotic entry and formation of a tumor (Berry et al. 1997; Hansen et al. 2004a). Weak glp-1 gain-of-function mutations display two types of ectopic proliferation: distal and proximal. The “late-onset tumor” is defined as a distal proliferative zone extending beyond the wild-type range of ∼20 cell diameters and the proximal proliferation (Pro phenotype) occurs when the most proximal germ cells fail to enter meiosis and continue proliferation, while the more distal cells are still able to enter meiosis (Berry et al. 1997; Pepper et al. 2003).
The transition to the meiotic cell cycle is promoted by three GLD (GermLine development Defective) proteins and NOS-3 (Kadyk and Kimble 1998; Eckmann et al. 2004; Hansen et al. 2004a). GLD-1 is a translational repressor (Lee and Schedl 2001; Biedermann et al. 2009), NOS-3 promotes GLD-1 accumulation (Brenner and Schedl 2016), and the complex of GLD-2 and GLD-3 forms a cytoplasmic poly(A) polymerase that promotes translation (Wang et al. 2002; Suh et al. 2006). These proteins form two main pathways (GLD-1/NOS-3 and GLD-2/GLD-3) that function redundantly to promote entry into meiosis and differentiation; however, if the activity of one gene from each pathway is simultaneously removed, a germline tumor forms. For example, in the single mutants of both gld-1(-) and gld-2(-), germ cells enter meiosis normally; however, the gld-2(-) gld-1(-) double mutant shows overproliferation due to a meiotic entry defect (Kadyk and Kimble 1998; Hansen et al. 2004a). This type of tumor is referred to as a “synthetic tumor.” The additional regulators gld-3 and nos-3 (homolog of Drosophila Nanos) (Kraemer et al. 1999; Subramaniam and Seydoux 1999) function in the GLD-2 and GLD-1 pathways, respectively, and disruption of gld-3 and nos-3 leads to synthetic tumors with null mutants in the parallel pathway (Eckmann et al. 2004; Hansen et al. 2004b).
GLD-1, a STAR domain RNA binding protein, is expressed during meiotic prophase (Figure 1A) where it promotes meiosis, gametogenesis, and germ cell identity maintenance by repressing translation of diverse mRNAs (Francis et al. 1995a; Jones et al. 1996; Marin and Evans 2003; Mootz et al. 2004; Wright et al. 2011). In addition to synthetic phenotypes with gld-2 uncovering the role of gld-1 in meiotic entry, diverse functions of GLD-1 in regulating the progression of meiotic prophase are revealed by gld-1 mutations (Francis et al. 1995a). In gld-1 null hermaphrodites, germ cells are able to enter meiosis, but exit meiotic prophase prematurely, fail to undergo oogenesis, and proliferate leading to formation of a proximal tumor (Francis et al. 1995a,b; Jones et al. 1996) (Supplemental Material, Figure S1B). Other, partial loss-of-function alleles produce phenotypes such as pachytene arrest, apoptosis in the female germline, formation of abnormal oocytes, and failure of spermatogenesis (Francis et al. 1995a,b; Schumacher et al. 2005). These multiple functions of GLD-1 reflect the broad range of its mRNA targets, although the molecular details of how the partial loss-of-function mutations affect GLD-1–dependent RNA regulation are still unknown.
The function of post-transcriptional regulators is often influenced by association with coregulators or cofactors. We recently identified a small protein DLC-1 as a cofactor of a C. elegans RNA binding protein FBF-2 (Wang et al. 2016). Direct binding between DLC-1 and FBF-2 is important for FBF-2 localization and function. DLC-1, an LC8-family protein, was originally identified as a component of the dynein motor complex (King and Patel-King 1995; Wilson et al. 2001). More recently, LC8 proteins have emerged as general cofactors promoting protein complex assembly through interactions with short linear peptides of their binding partners (Rapali et al. 2011b). Interestingly, FBF-2-DLC-1 cooperation does not require dynein motor function. We hypothesize that the dynein-independent role of DLC-1 in post-transcriptional regulation of gene expression is relevant to a broad array of RNA regulators.
In this study, we report that GLD-1 regulatory function in the germline requires GLD-1 interaction with DLC-1. Similar to the FBF-2-DLC-1 interaction, the cooperation between GLD-1 and DLC-1 is separate from the dynein motor function. Interestingly, DLC-1 is required for regulation of only a subset of GLD-1 targets in the germline. These results suggest a specific role for DLC-1 in facilitating select aspects of GLD-1–mediated regulation including prevention of ectopic germ cell proliferation.
Materials and Methods
Nematode strains and culture
Standard procedures for culture and genetic manipulation of C. elegans strains were followed (Brenner 1974). Nematodes were cultured at 20°, except as noted below for temperature-sensitive and GFP-expressing strains. A list of all strains used in this study is provided in Table S1.
Immunofluorescence
Gonads were dissected on slides treated with poly-L-lysine, frozen on dry ice, and then fixed in ice-cold 100% methanol for 1 min. Next, slides were fixed in 2% paraformaldehyde/100 mM K2HPO4 (pH 7.2) for 5 min and blocked in PBS/0.1% BSA/0.1% Tween-20 (PBS-T/BSA) for 30 min at room temperature. Samples were incubated with primary antibody diluted in PBS-T/BSA overnight at 4°. After washes, samples were incubated with secondary antibody diluted in PBS-T/BSA for 2 hr at room temperature and then 10 μl Vectashield with DAPI (Vector Laboratories, Burlingame, CA) was added to each sample before cover-slipping. Primary antibodies were mouse anti-PGL-1 (5.2 μg/ml, K76; Developmental Studies Hybridoma Bank), phospho-Histone H3 pSer10 6G3 (1:400; Cell Signaling Technology), rabbit anti-REC- 8 (catalog no. 29470002, 1:500; Novus Biologicals), and affinity-purified rabbit anti-GLD-1 antibody (gift from T. Schedl; Jones et al. 1996; 1:200). Secondary antibodies were Alexa Fluor 488-conjugated goat anti-rabbit (1:200; Jackson ImmunoResearch), Alexa Fluor 594-conjugated goat anti-mouse IgG (H+L) (1:500; Jackson ImmunoResearch), and Alexa Fluor 594-conjugated goat anti-mouse IgM (1:200; Jackson ImmunoResearch). Images were acquired with a Leica DFC300G camera attached to a Leica DM5500B microscope and stitched together using Adobe Photoshop CS3. To quantitate GLD-1 levels in either N2 vs. dlc-1(tm3153) mutant or gld-1wt::ollas vs. gld-1ndb::ollas, images were taken using identical exposure settings for each comparison. GLD-1 signal in meiotic pachytene was quantified using LAS-X software (Leica) and background signal was subtracted. Pixel intensity values obtained for experimental [dlc-1(tm3153) or gld-1ndb::ollas] samples were normalized to control values (N2 or gld-1wt::ollas, respectively). Images of perinuclear P granules were acquired using a Zeiss 880 confocal microscope and GLD intensity in cytoplasm vs. P granule was quantitated using ImageJ32 software after subtracting the background signal.
Ectopic proliferation assay
Single, double, and triple mutant worm strains (Table 1) were synchronized by bleaching and then grown at 20° for 4 days (1 day post L4 stage) except for strains encoding dynein heavy chain mutant alleles. Synchronous cultures of dhc-1(js121) worms fed either empty vector control or gld-2(RNAi) starting at L1 stage were cultured for 3 days at 24° and the hypomorphic temperature-sensitive dhc-1(or195) and dhc-1(or195); gld-3(q730) mutants were cultured at 26° for 3 days after synchronization at L1 stage. Synchronous L1 cultures of gld-1wt::ollas and gld-1ndb::ollas transgenic animals (Table 3) were fed either empty vector control, gld-2 or gld-3(RNAi) for 3 days at 24°. To assess proliferation, young adult hermaphrodites were fixed and immunostained as described above. Mitotically dividing cells were detected with mouse monoclonal antibody to phospho-Histone H3 and rabbit anti-REC-8 that preferentially detects stem and progenitor cells (Hansen et al. 2004a). Proximal germlines containing cells that stained positive for both REC-8 and phospho-histone H3 were scored as positive for proximal tumor formation. To define late-onset tumor formation, the size of single mutant or control RNA interference (RNAi) proliferative zones was established by counting the number of stem and progenitor cell rows stained positive for REC-8 antibody. Next, the size of double mutant, or gld-2 or gld-3(RNAi) proliferative zones was recorded in the same way. Germlines with proliferative zones that extended beyond the relevant controls were scored as positive for late onset tumor formation.
Table 1. Genetic interactions between dlc-1, dhc-1, and genes that function in the regulation of meiotic entry.
| Genotype | % Pro | n |
|---|---|---|
| nos-3(q650)/mT1; dlc-1(tm3153)/mT1 | 0 | 33 |
| gld-3(q730)/mT1; dlc-1(tm3153)/mT1 | 0 | 36 |
| gld-2(q497)/hT2; dlc-1(tm3153)/hT2 | 0 | 43 |
| dlc-1(tm3153) | 0 | 27 |
| nos-3(q650) | 0 | 19 |
| gld-3(q730) | 0 | 28 |
| gld-2(q497) | 0 | 44 |
| dhc-1(or195) | 0 | 48 |
| dhc-1(js121); control RNAi | 0 | 55 |
| nos-3(q650); dlc-1(tm3153) | 0 | 24 |
| gld-3(q730); dlc-1(tm3153) | 37 | 75 |
| gld-2(q497); dlc-1(tm3153) | 0 | 58 |
| dhc-1(or195); gld-3(q730) | 0 | 38 |
| dhc-1(js121); gld-2(RNAi) | 0 | 80 |
n indicates number of germlines scored for proximal tumor. Scoring was 1 day after L4 stage.
Table 3. Ectopic proliferation in gld-1ndb::ollas transgenic animals.
| Genotype and RNAi treatment | % Pro | n |
|---|---|---|
| gld-1(q485); gld-1::ollas(wt); control RNAi | 0 | 39 |
| gld-1(q485); gld-1::ollas(wt); gld-3(RNAi) | 0 | 19 |
| gld-1(q485); gld-1::ollas(wt); gld-2(RNAi) | 0 | 68 |
| gld-1(q485); gld-1::ollas(ndb); control RNAi | 0 | 39 |
| gld-1(q485); gld-1::ollas(ndb); gld-3(RNAi) | 0 | 75 |
| gld-1(q485); gld-1::ollas(ndb); gld-2(RNAi) | 7 | 132 |
n indicates number of germlines scored. Scoring was 1 day after L4.
3′ Untranslated region–based reporter analysis
Reporter transgene constructs used in this assay contained the pie-1 promoter, green fluorescent protein (GFP) fused to Histone H2B and the 3′ untranslated region (UTR) of the following GLD-1-associated mRNAs: mex-3, mes-3, cye-1, puf-5, and spn-4 (Merritt et al. 2008). These transgene constructs were chosen for analysis because their expression is repressed in meiotic pachytene. RNAi treatment was conducted as previously published (Wang et al. 2016). Briefly, synchronous cultures of worms were fed HT115 Escherichia coli expressing double-stranded RNA targeting dlc-1, gld-1, dhc-1, or empty vector control starting at L1 larval stage for 3 days at 24°. The identity of RNAi constructs was confirmed by sequencing and the efficiency of the RNAi treatments was established by visually confirming 100% sterility. Adult hermaphrodites were washed in M9, gonads dissected and fluorescent images were acquired with a Leica DFC300G camera attached to a Leica DM5500B microscope. Images were taken with identical exposure settings for each transgenic strain and composite images of full germlines were assembled using Adobe Photoshop CS3. Nuclear GFP signal was quantified using LAS-X software (Leica) and background signal was subtracted. Pixel intensity values obtained for experimental (dlc-1, gld-1 or dhc-1) RNAi-treated samples were normalized to control RNAi pixel intensity values.
GST pulldown assay
His6-GLD-1 expression vector was generated by recombination of the pDONR201 entry clone that encodes full-length GLD-1 (amino acids 1–463) with the pDEST17 destination vector using the Gateway system (Thermo Fisher Scientific). Mutagenesis of the DLC-1 binding site on GLD-1 was performed using a Q5 Site-Directed Mutagenesis Kit (New England BioLabs, Beverly, MA). All wild-type and mutant GLD-1 constructs were verified by sequencing and transformed into BL21 (DE3) cells for recombinant protein expression. Expression of His6-GLD-1 constructs was induced with 0.2 mM IPTG at 15° for 18 hr. Expression construct containing GST-DLC-1 was already present in the laboratory and GST pulldown assays were conducted as previously described (Wang et al. 2016). Briefly, GST alone or GST-tagged DLC-1 was bound to glutathione beads and then incubated with His6-GLD-1. Unbound lysate was removed, the beads were washed and protein eluted. To test if the DLC-1/GLD-1 binding interaction is RNA-dependent, 50 μg/ml RNase was added to His6-GLD-1 lysate before incubation with GST alone or GST-DLC-1. Protein samples from pulldown assays were separated using Mini-PROTEAN TGX 4–20% precast gels (BioRad, Hercules, CA) and visualized using either Coomassie (GST and GST-DLC-1) or by Western blotting (His6-GLD-1). Monoclonal anti-polyHistidine (mouse IgG2a isotype) primary antibody (1:2000; Sigma-Aldrich, St. Louis, MO) and peroxidase-conjugated goat anti-mouse IgG2a specific secondary antibody (1:1000; SouthernBiotech) was used to visualize His6-GLD-1 protein.
Western blot analysis
To determine the effect of DLC-1 on GLD-1 levels and to quantitate GLD-1wt::OLLAS or GLD-1ndb::OLLAS levels in cells, 50 worms were individually picked from synchronous cultures [N2, dlc-1(tm3153), gld-1(q485); gld-1wt::ollas or gld-1(q485); gld-1ndb::ollas] deposited in SDS-PAGE sample buffer and boiled for 30 min prior to SDS-PAGE gel electrophoresis. Proteins from worm lysate (50 worms per lane) were separated on 7.5% gel and transferred to a 0.2 μm PVDF membrane (EMD Millipore). After transfer membranes were blocked in TBS/0.1% Tween 20/5% milk powder and the blots probed with affinity purified rabbit anti-GLD-1 (T. Schedl, 1:250), monoclonal rat anti-OLLAS (1:500; Novus), and monoclonal mouse anti-α-tubulin (1:300; Sigma) primary antibodies diluted in blocking solution. Mouse anti-MYO-3 (5 μg/ml; Developmental Studies Hybridoma Bank) primary antibody was diluted in blocking solution plus 10% normal goat serum. Peroxidase-conjugated goat anti-rabbit (1:5000; Jackson ImmunoResearch), peroxidase-conjugated goat anti-rat (1:10,000; ThermoFisher Scientific), and peroxidase-conjugated goat anti-mouse IgG (1:5000; Jackson ImmunoResearch) secondary antibodies were used. Membranes were developed using Luminata Crescendo Western HRP substrate (EMD Millipore) and visualized using BioRad ChemiDoc MP Imaging System. Band intensities were quantitated using Image Lab software version 5.1.
Construction of transgenes and generation of transgenic animals
The gld-1wt::ollas and gld-1ndb::ollas transgene constructs include 1.1 kb upstream and 900 base pairs downstream of the GLD-1 coding region. gld-1 promoter, 3′UTR, and GLD-1 coding sequences were amplified from N2 Bristol DNA and recombined with the Gateway donor vectors to make entry clones. Entry clones were then recombined into pCFJ150 destination vector (Frøkjaer-Jensen et al. 2008) using Multisite Gateway technology (Thermo Fisher Scientific). Transgene constructs were injected into unc-119(ed3) worms and single-copy insertions of GLD-1wt::OLLAS and GLD-1ndb::OLLAS were generated by homologous recombination into a universal Mos1 insertion site on chromosome II and chromosome V, respectively, after Cas9-induced double-stranded break (Wang et al. 2016). Transgene insertion was confirmed by PCR spanning homology region.
DAPI-staining
Gonads were dissected on slides treated with poly-L-lysine, frozen on dry ice, and then fixed in ice cold 80% methanol/3% formaldehyde/6 mM KH2PO4. Next, slides were washed in PBS/0.1% BSA/0.1% Tween-20 for 10 min at room temperature and chromatin was stained using Vectashield with DAPI (Vector Laboratories).
Data availability
Strains (see Table S1) and reagents are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361234.
Results
dlc-1/gld-1 cooperation facilitates meiotic entry and prevents ectopic germline proliferation
Our previous research suggested that DLC-1/LC8 facilitates the function of an RNA binding protein FBF-2 in a dynein motor–independent manner. To uncover additional RNA binding proteins that might rely on DLC-1 for their function, we performed a genetic interaction assay to test if DLC-1 contributes to the regulatory network that controls the proliferation vs. differentiation decision in the C. elegans germline (Figure 1B). If activities of both GLD-1/NOS-3– and GLD-2/3–dependent pathways are disrupted, then meiotic entry is affected and the result is a germline tumor (Hansen and Schedl 2013). Thus, if DLC-1 promotes either GLD-1 or NOS-3 function, then in dlc-1(tm3153) null mutant worms [abbreviated dlc-1(-)] the GLD-1/NOS-3 pathway would be compromised. Additionally, if either gld-2 or gld-3 activity is removed in dlc-1(-) worms, then both pathways would be disrupted and a germline tumor should form.
In wild-type animals, proliferative cells are restricted to the distal end of the gonad and cells undergoing mitosis can be identified by immunostaining using an anti-phospho-histone H3 antibody. Additionally, proliferative cells in the C. elegans germline stain positively for the REC-8 antigen (Hansen et al. 2004a) (Figure 2A and Table 1). Proliferative cells were also restricted to the distal tip in dlc-1(-), gld-3(-), and nos-3(-) null single-mutant worms (Figure 2, B–E and Table 1). Although phospho-histone H3–positive cells were observed at the proximal end of gld-2(-) single mutant germline, these cells appeared to be sperm by DAPI staining and were not REC-8 positive (Figure 2D).
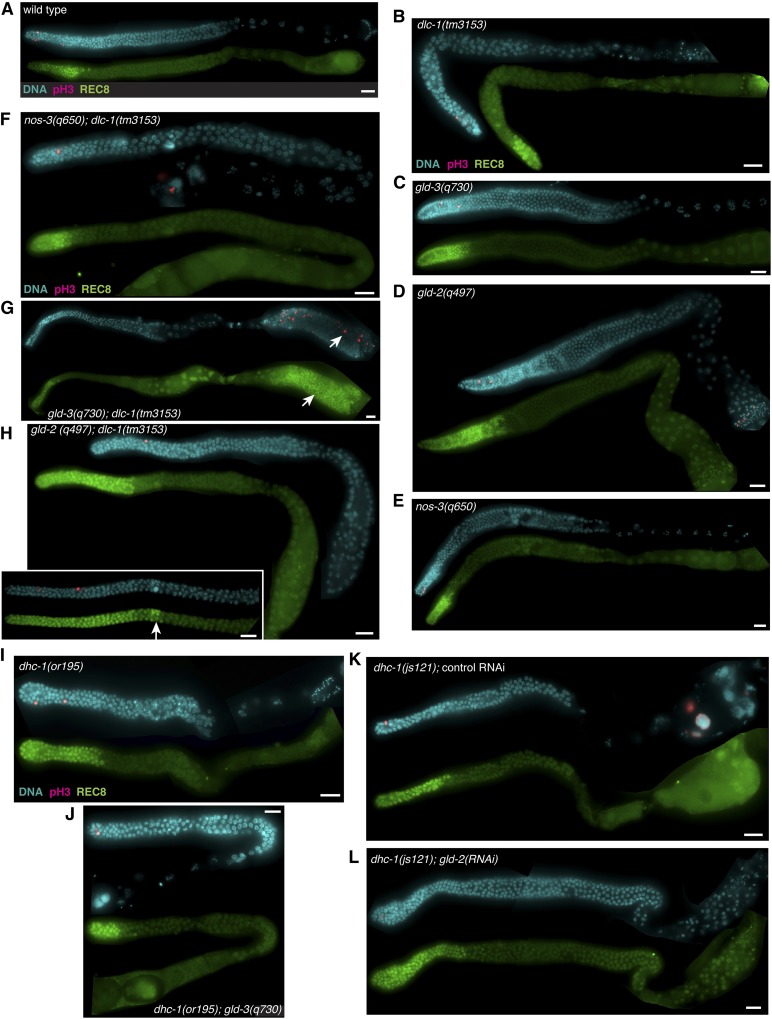
Figure 2.
Genetic evidence that dlc-1 functions with gld-1. Dissected gonads of the indicated genotypes were stained for the M-phase marker phosphohistone H3 (pink) and DNA (blue). Stem and progenitor cells are detected by anti-REC-8 staining (green). (A) Wild-type control cultured at 20°. Single mutant animals including (B) dlc-1(tm3153), (C) gld-3(q730), (D) gld-2(q497), and (E) nos-3(q650) show no ectopic germ cell proliferation. Aberrant proliferation (white arrows) is observed in (H) gld-2(q497); dlc-1(tm3153) and (G) gld-3(q730); dlc-1(tm3153) double mutant animals (inset depicts a gld-2(q497); dlc-1(tm3153) mutant germline with >35 stem and progenitor cell rows), but not in (F) nos-3(q650); dlc-1(tm3153). No ectopic proliferation is detected in (I) dhc-1(or195) single mutant animal, (J) dhc-1(or195); gld-3(q730) double mutant animal, (K) dhc-1(js121) mutant treated with control RNAi, or (L) dhc-1(js121); gld-2(RNAi). Efficacy of gld-2(RNAi) was determined by scoring sterility and embryonic lethality of dhc-1(js121)/hT2; gld-2(RNAi) treated worms. dhc-1(js121)/hT2; gld-2(RNAi) treated worms exhibited 20 ± 6% sterility and for worms with progeny the embryonic lethality was 98 ± 0.9%. Fluorescence micrographs are representative images of data collected from at least two independent experiments and 19–80 worms were scored for each genotype (see Table 1). Bar, 10 μm.
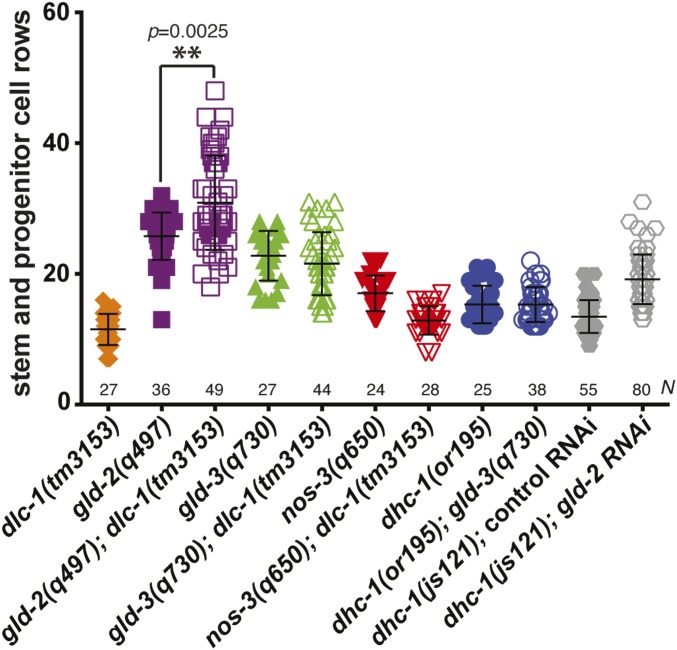
To test for synthetic phenotypes, we created double mutant strains of dlc-1(-) and either nos-3(-), gld-2(-), or gld-3(-). When nos-3(-); dlc-1(-) worms were immunostained for mitotically dividing cells using anti-phospho-histone H3 and anti-REC-8 antibodies, no aberrant cell proliferation was observed (Figure 2F and Table 1). By contrast, 37% of gld-3(-); dlc-1(-) worms exhibited Pro tumor formation (Figure 2G and Table 1). No mitotically dividing cells were observed at the proximal end of gld-2(-); dlc-1(-) double mutant worms; however, 36% of gld-2(-); dlc-1(-) worms exhibited excess proliferation at the distal end of the gonad compared to gld-2(-) control (P = 0.0025; Figure 3). The distal proliferative zone of gld-2(-); dlc-1(-) double mutant worms was up to 16 cell rows longer than gld-2(-) control and this phenotype is characteristic of late-onset tumor formation (Figure 2H and Figure 3). Since the ectopic proliferation observed in gld-2(-); dlc-1(-) and gld-3(-); dlc-1(-) double mutants is less severe than previously reported for gld-2(-) gld-1(-) and gld-1(-); gld-3(RNAi) worms (Kadyk and Kimble 1998; Eckmann et al. 2004), we conclude that absence of DLC-1 likely results in partial loss of GLD-1 function.
Figure 3.
Length of mitotic region in dlc-1, dhc-1 and gld mutant germlines. To identify defects in the transition from mitosis to meiosis, the number of REC-8–positive stem and progenitor cell rows from the distal end of the germline to the transition zone boundary were counted. A statistically significant increase in the lengths of the distal proliferative zones was observed in gld-2(q497); dlc-1(tm3153) germlines compared to gld-2(q497) control (indicated by **; P = 0.0025 by Kolmogorov–Smirnov test). No significant changes in mitotic zone length were detected in gld-3(q730); dlc-1(tm3153) vs. gld-3(q730) germlines (P > 0.25 by Kolmogorov–Smirnov test). Data were collected from at least two independent experiments and 24–80 germlines were scored for each genotype.
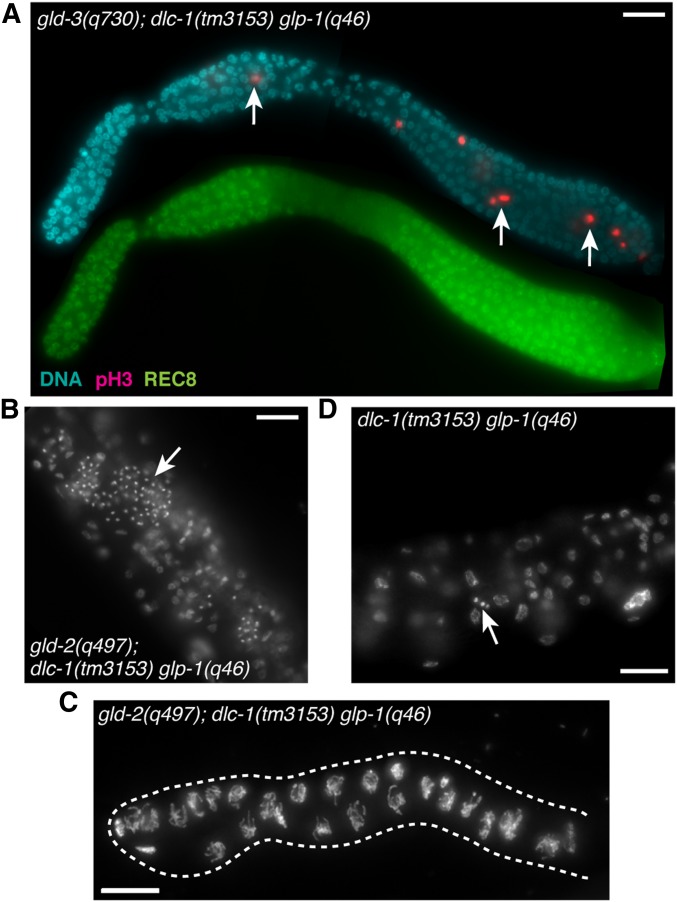
Previous research suggested that dlc-1 counteracts the activity of GLP-1/Notch signaling that promotes germline proliferation (Dorsett and Schedl 2009). To determine whether the dlc-1(-) overproliferation phenotype with gld-3(-) and gld-2(-) was dependent on glp-1 function, we examined both double mutants in the absence of glp-1 activity. We found that gld-3(-); dlc-1(-) glp-1(-) triple mutant animals were tumorous (Figure 4A). Thus, dlc-1 cannot solely function as a negative regulator of Notch signaling, and must in addition function downstream of GLP-1 to promote meiotic entry or inhibit proliferation. However, gld-2(-); dlc-1(-) glp-1(-) triple mutant germlines only supported limited proliferation before either differentiating to form sperm in early generations postcross or remaining as undifferentiated germ cells in later generations (Figure 4, B and C). The extent of germ cell proliferation in gld-2(-); dlc-1(-) glp-1(-) triple mutant was substantially greater than that of the dlc-1(-) glp-1(-) double mutant that only produces a few spermatocytes (Figure 4D; Austin and Kimble 1987). However, germ cell proliferation was only twofold (or ∼1 cell division) greater than gld-2(-); glp-1(-) double mutant previously reported to form 10–16 undifferentiated germ cells (Kadyk and Kimble 1998). We conclude that the increased capacity for mitosis found in gld-2(-); dlc-1(-) double mutant is not observed in the triple mutant lacking glp-1 activity. The reason for the observed difference between gld-2(-) and gld-3(-) synthetic mutant phenotypes and the nature of generation-dependent disruption of spermatogenesis in gld-2(-); dlc-1(-) glp-1(-) triple mutant is unknown.
Figure 4.
dlc-1(-) synthetic overproliferation phenotype is not dependent on glp-1 activity. (A) Fluorescence micrograph of dissected gonad from triple mutant animal gld-3(q730); dlc-1(tm3153) unc-32(e189) glp-1(q46) immunostained for mitotically dividing cells (pink), stem and progenitor cells (green), and DNA (blue). White arrows indicate aberrant cell proliferation. (B) Fluorescence micrograph of gld-2(q497); dlc-1(tm3153) unc-32(e189) glp-1(q46) whole worm stained with DAPI show the presence of many sperm (white arrows). (C) The ability of gld-2(q497); dlc-1(tm3153) unc-32(e189) glp-1(q46) mutant germ cells to differentiate into sperm is gradually lost after several generations, as seen in a dissected gonad stained with DAPI. (D) dlc-1(tm3153) unc-32(e189) glp-1(q46) whole worm stained with DAPI. White arrow indicates sperm. Bar, 10 μm.
To further test if dlc-1 facilitates gld-1 function we assessed whether removing dlc-1 activity would enhance a weak partial loss of function gld-1(op236) allele that appears superficially wild type at 20° (Figure S2A). At the restrictive temperature (25°) gld-1(op236) germline exhibits enhanced cell death phenotype and only a small number of cells are able to exit pachytene and produce oocytes (Schumacher et al. 2005). At 20°, dlc-1(-) mutant worms are sterile and the predominant phenotypes in the germline are enlarged germ cell nuclei and rapid deterioration of forming oocytes at the proximal end of the gonad resulting in only a few diplotene nuclei observed (Figure S2B; Dorsett and Schedl 2009; Day et al. 2018). A subset of both dlc-1(-) and gld-1(op236); dlc-1(-) double mutant worms exhibit a disorganized germline phenotype where some cells that fail to exit pachytene are interspersed with cells forming oocytes at 20° (Figure S2, B–D). Since the frequency of the disorganized germline phenotype is similar in dlc-1(-) vs. gld-1(op236); dlc-1(-) double mutant worms (38% vs. 40%, respectively), it indicates that gld-1(op236) mutation might act in the same pathway as that affected by the dlc-1(-) mutation. We have not evaluated whether failure of pachytene exit in dlc-1(-) mutant was dependent on the dynein motor function.
Together, these data suggest that dlc-1 facilitates the GLD-1/NOS-3 arm of the regulatory network that controls meiotic entry and differentiation. We hypothesized that the tumor formation observed in gld-2(-); dlc-1(-) and gld-3(-); dlc-1(-) germlines might be due to GLD-1 target mRNA misregulation and next asked if DLC-1 is involved in repressing translation of GLD-1 target mRNAs.
DLC-1 is required for regulation of a subset of GLD-1 target mRNAs
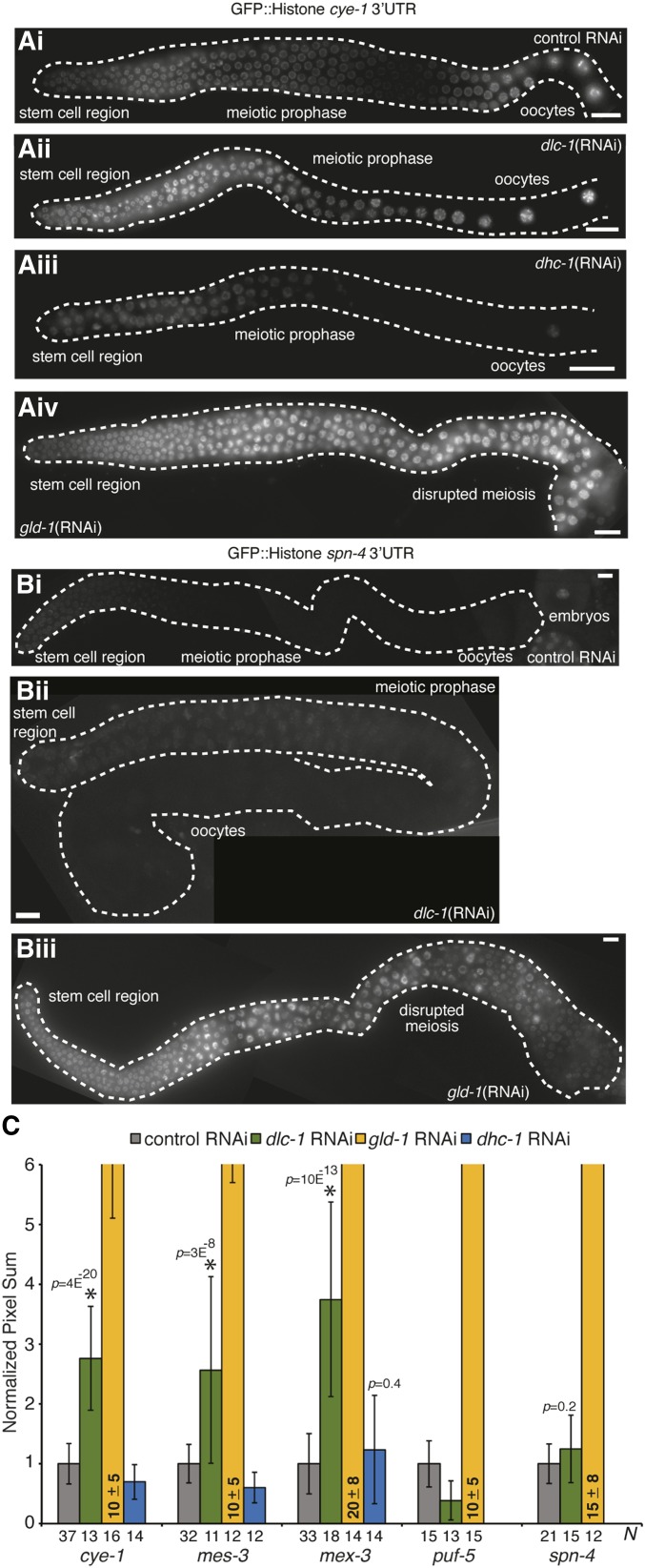
Many genes expressed in the C. elegans germline are regulated at a post-transcriptional level through the 3′UTR of the message (Merritt et al. 2008). RNA binding proteins (such as GLD-1) control the pattern of gene expression by recognizing and binding to cis-regulatory sequences present in the 3′UTR of mRNA. Therefore, we used a 3′UTR-based reporter assay to determine whether DLC-1 is involved in regulating the translation of the GLD-1 target mRNAs. The reporter transgene constructs used in this assay contain the pie-1 promoter to direct expression of the transgene to germ cells. GFP fused to histone H2B localizes to the nucleus and the GLD-1 target mRNA 3′UTR specifies the pattern of transgene expression in the germline. To examine the contribution of dlc-1 to translational regulation of the reporters, we analyzed transgenic protein expression following dlc-1 knockdown by RNAi. GLD-1 target mRNAs selected for this analysis were cye-1, mes-3, mex-3, puf-5, and spn-4 (Merritt et al. 2008; Biedermann et al. 2009). These mRNAs encode proteins that are cell cycle regulators and genes involved in chromatin modification, RNA regulation, and cell division. The results showed that DLC-1 is required for translational control of cye-1, mes-3, and mex-3 mRNA in the worm germline, as dlc-1(RNAi) leads to activation of the reporters in the meiotic prophase region of the germline (Figure 5, Ai and Aii, Figure 5C, and Figure S3, A and B). Derepression of cye-1 3′UTR reporter upon dlc-1(RNAi) was consistently observed in two independently generated transgenic strains (Figure S3, C and D). However, only minor changes in GFP intensity were observed in control vs. dlc-1(RNAi) treated puf-5 and spn-4 transgenic reporter worms (Figure 5, Bi and Bii and Figure 5C). Although all strains of transgenic reporter worms were sterile after treatment with dlc-1(RNAi) it is possible that small residual amounts of DLC-1 protein might be sufficient to maintain repression of puf-5 and spn-4 transgenic reporters. To test this possibility, we crossed endogenously tagged GFP::3xFLAG::PUF-5 and SPN-4::GFP::3xFLAG worms into the dlc-1(-) null mutant background and observed the pattern of tagged protein expression. Consistent with the data obtained using dlc-1(RNAi), tagged PUF-5 and SPN-4 protein expression was repressed in meiotic pachytene in the absence of DLC-1 (Figure S4, A–D and Table S2). Finally, to ensure that expression of the cye-1, mes-3, mex-3, puf-5, and spn-4 transgenic reporters is responsive to changes in GLD-1 levels, the worms were treated with gld-1(RNAi) and GFP signal in the germline was assessed. When transgenic reporter worms were treated with gld-1(RNAi), GFP signal was strongly derepressed (Figure 5, Aiv and Biii and Figure 5C). Since DLC-1 is required for appropriate expression of cye-1, mes-3, and mex-3 but not puf-5 or spn-4 reporters, we conclude that DLC-1 likely only contributes to some of GLD-1 functions as it is required for regulating a subset of GLD-1 mRNA targets.
Figure 5.
DLC-1 helps regulate a subset of GLD-1 target mRNAs in a dynein motor independent manner. Fluorescence micrographs of dissected gonads expressing GFP-tagged Histone H2B under the control of either (A) cye-1 3′UTR or (B) spn-4 3′UTR after the indicated RNAi treatments. Bar, 10 μm. (C) Quantitation of reporter expression in the meiotic pachytene region of C. elegans germline after the following RNAi treatments: control (gray), dlc-1 (green), gld-1 (yellow), and dhc-1 (blue). Transgenes are identified along the x-axis, and the number of germlines scored (N) is indicated for each treatment. Efficiencies of dlc-1 and dhc-1(RNAi) treatments were established by confirming 100% sterility. Nuclear GFP signal in meiotic pachytene germ cells was quantified using LAS-X software (Leica). Signal intensities following experimental RNAi treatments (dlc-1, gld-1, or dhc-1) were normalized to respective control values. Results are representative of at least three independent experiments and error bars represent SD from the mean. Student’s unpaired t-test was used to calculate P-values for dlc-1 and dhc-1(RNAi) treatments compared to control. * indicates statistically significant differences.
DLC-1 mRNA regulatory function likely does not require dynein motor activity
Since DLC-1 is a multifunctional protein and interacts with a wide variety of cellular proteins, DLC-1 may promote the function of GLD-1 either by facilitating formation of GLD-1 RNPs or through a dynein motor–related function. To determine if the dynein motor–related function of DLC-1 plays a role in mRNA regulation, we used RNAi to deplete the dynein motor subunit (DHC-1) in cye-1, mes-3, and mex-3 transgenic reporter strains. GFP intensity in worms treated with dhc-1(RNAi) vs. empty vector control was assessed and only small differences in transgenic protein expression were observed (Figure 5Aiii and Figure 5C). To ensure that the negative RNAi result was not due to the presence of residual amounts of DHC-1 protein, we crossed endogenously tagged GFP::3xFLAG::MEX-3 and transgenic cye-1 3′UTR reporter into dhc-1(js121) mutant background. GFP::3xFLAG::MEX-3 protein expression was restricted to the distal tip and oocytes in both wild-type and dhc-1(js121) mutant background (Figure S4, E and F and Table S2). Similarly, cye-1 reporter expression was repressed in the meiotic prophase region in dhc-1(js121) mutant background, which is consistent with the data obtained using dhc-1(RNAi) (Figure S4, G and H and Table S2). Additionally, we tested if the GLD-1 function of preventing ectopic germ cell proliferation is compromised by the absence of dhc-1 by combining a null gld-3 mutant with the hypomorphic temperature-sensitive mutant allele dhc-1(or195ts). Synchronous cultures of dhc-1(or195ts) and dhc-1(or195ts); gld-3(-) embryos were grown at 26° until worms reached adulthood and then stained for mitotically dividing cells using anti-phospho-histone H3 and anti-REC-8 antibodies. Although dissected germlines from dhc-1(or195ts) mutant worms appear relatively normal (Figure 2I; compare to a wild-type germline at 26°, Figure S1E), this mutant exhibited an embryonic lethal phenotype when cultured at 26° (no live progeny observed for 100% adults; n = 172). No ectopic or excessive proliferation was observed in either dhc-1(or195ts) control or dhc-1(or195ts); gld-3(-) worms (Figure 2, I and J, Figure 3, and Table 1). We also treated dhc-1(js121) mutant worms with either control or gld-2(RNAi), immunostained dissected germlines with anti-phospho-histone H3 and anti-REC-8 antibodies, and observed no aberrant cell proliferation (Figure 2, K and L, Figure 3, and Table 1). Since disruption of the dynein motor function does not affect either GLD-1–dependent translational control or cell proliferation, we conclude that DLC-1 contribution to GLD-1 activity is likely independent of its role as a light chain component of the dynein motor.
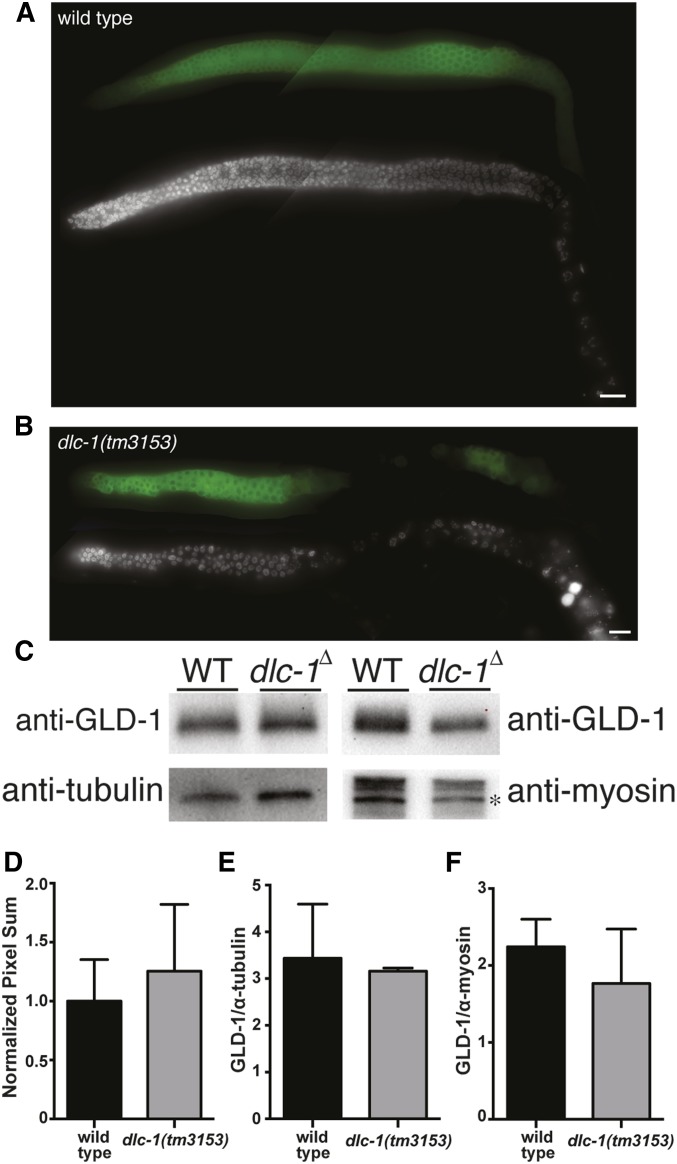
DLC-1 does not regulate GLD-1 protein levels
To test whether DLC-1 promotes the functions of GLD-1 by regulating GLD-1 levels in germ cells, wild-type (N2) and dlc-1(-) worm germlines were dissected and immunostained using anti-GLD-1 antibody (Jones et al. 1996). In wild-type worms, GLD-1 protein expression is low in the distal region of the germline where cells undergo mitosis, high in meiotic cells, and then levels decrease as cells exit pachytene (Figure 6A). GLD-1 levels are also highest in the meiotic prophase region of dlc-1(-) worm germline and low in the distal and proximal ends (Figure 6B). In the dlc-1(-) germlines that show proximal germline disorganization, GLD-1 expression pattern is altered, and aberrant patches of cells unable to exit pachytene are still expressing GLD-1 (Figure 6B). To determine if the level of GLD-1 expression is different in dlc-1(-) germline compared to the wild-type control, the staining intensity in the meiotic prophase region of the germline was measured and no significant difference was observed (P > 0.17 by the Student’s t-test; Figure 6D). Anti-GLD-1 antibody was also used to quantitate the total level of GLD-1 in wild-type and dlc-1(-) worms by Western blot analysis. The amount of GLD-1 present in the lysate of 50 worms of each genotype was normalized to either tubulin or myosin and no significant differences in GLD-1 protein levels were observed in dlc-1(-) vs. wild-type control (GLD-1/anti-tubulin P > 0.7; GLD-1/anti-myosin P > 0.4; Figure 6, C, E, and F). Thus, DLC-1 does not promote GLD-1 function by regulating GLD-1 protein levels. This result suggests that DLC-1 facilitates GLD-1 (as opposed to NOS-3) activity because previous research established that NOS-3 promotes GLD-1 accumulation (Hansen et al. 2004b; Brenner and Schedl 2016).
Figure 6.
GLD-1 protein expression pattern and abundance is similar in wild-type vs. dlc-1(-) worms. Fluorescence micrographs of (A) wild-type and (B) dlc-1(tm3153) dissected gonads immunostained for GLD-1 protein using anti-GLD-1 antibody (Jones et al. 1996). Bar, 10 μm. (C) Western blot analysis of GLD-1 protein levels in the lysate of 50 wild-type or dlc-1(tm3153) whole worms probed with anti-GLD-1, anti-tubulin, and anti-MYO-3 antibodies. * indicates 210 kDa myosin heavy chain band. (D) Quantitation of GLD-1 protein expression in meiotic pachytene germ cells using LAS-X software (Leica). Wild type, N = 14; dlc-1(tm3153), N = 13; P > 0.17. Quantitation of GLD-1 protein level normalized to (E) tubulin and (F) myosin. Error bars represent SD from the mean. No significant differences in GLD-1 protein levels were observed in dlc-1(-) vs. wild-type control (GLD-1/anti-tubulin: P > 0.7, N = 3 of each genotype; GLD-1/anti-myosin: P > 0.4, N = 3 of each genotype). Student’s unpaired t-test was used to calculate P values.
DLC-1 binds the N-terminal domain of GLD-1
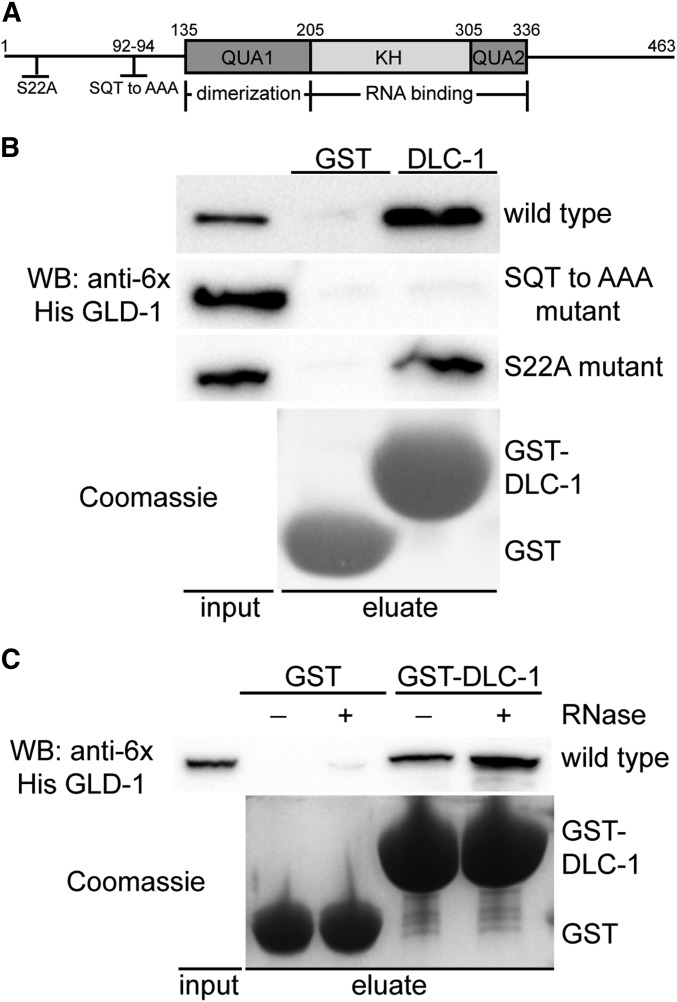
Since genetic interactions may be direct or indirect we asked whether DLC-1 and GLD-1 proteins interact. To determine if DLC-1 binds directly to GLD-1 in vitro we used a GST pulldown assay with recombinant GST-tagged DLC-1 and His-tagged GLD-1. We found that wild-type GLD-1 binds specifically to DLC-1 and not the GST control (Figure 7B). DLC-1/LC8 family proteins interact with linear peptides that conform to a weak (D/S)KX(T/I/V)Q(T/V)(D/E) consensus sequence (Rapali et al. 2011b). We identified a putative DLC-1 binding site (YSQT) in the N-terminal domain of GLD-1 (Figure 7A) that appears conserved among the nematode orthologs of GLD-1 (ME, unpublished data). Mutating three residues of the putative DLC-1 binding site (SQT to AAA) abolished DLC-1-GLD-1 binding in vitro (Figure 7B). By contrast, mutating a conserved serine residue present on the N-terminal domain of GLD-1 to alanine did not disrupt the DLC-1-GLD-1 interaction (Figure 7, A and B). These results show that DLC-1 recognizes and binds to a specific amino acid sequence present on the N-terminal domain of GLD-1. Since GLD-1 regulates mRNA expression by binding to the 3′UTR of its mRNA targets and forming ribonucleoprotein complexes (RNPs), we next asked if the DLC-1-GLD-1 binding is RNA dependent. We found that the DLC-1-GLD-1 association is not RNA-dependent, as the presence of RNase did not disrupt the interaction between DLC-1 and GLD-1 (Figure 7C).
Figure 7.
DLC-1 binds the N-terminal domain of GLD-1 in vitro. (A) Schematic diagram showing the domain structure of GLD-1. GLD-1 forms homodimers through interactions in the QUA1 domain and the KH and QUA2 domains bind RNA. Mutations to the N-terminal domain are indicated. (B) GST pulldown assay to test if GST alone or GST-DLC-1 (detected by Coomassie) binds either wild-type His6 GLD-1, His6 GLD-1 that has a mutated DLC-1 binding site (SQT to AAA mutant), or His6 GLD-1 that has a conserved serine mutated to alanine (S22A mutant). His6 GLD-1 constructs were detected by Western blot. (C) GST and GST-DLC-1 were assayed for the ability to bind wild-type His6 GLD-1 in the presence or absence of RNase.
Interaction with DLC-1 facilitates GLD-1 function in vivo
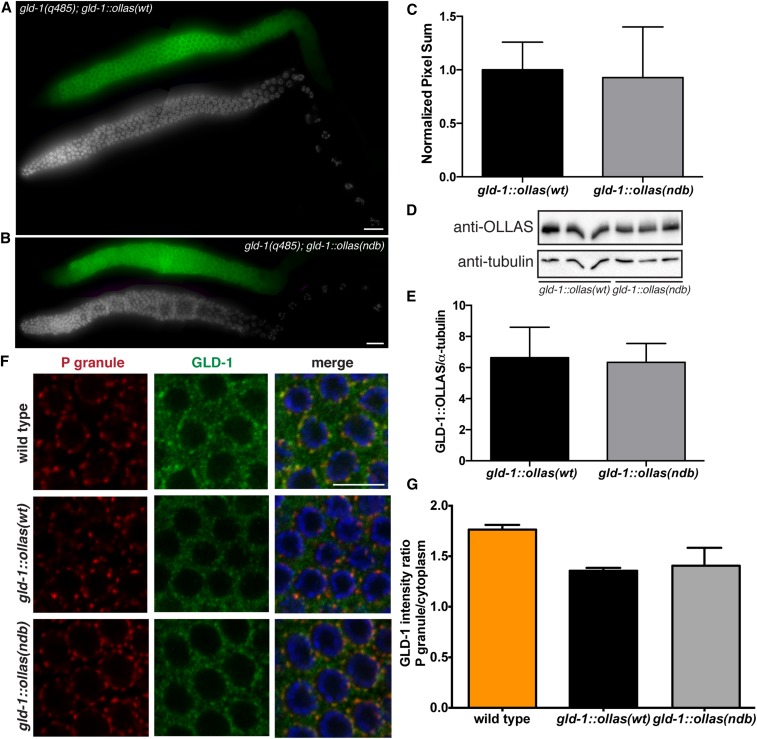
To test whether the DLC-1-GLD-1 binding identified in vitro is important for GLD-1 function in vivo, we generated mutated OLLAS-tagged GLD-1 transgene unable to bind DLC-1 (SQT to AAA mutant, abbreviated further as GLD-1ndb::OLLAS) and crossed this transgene into gld-1(q485) null mutant background. Additionally, we generated a wild-type OLLAS-tagged transgene of GLD-1 (GLD-1wt::OLLAS), which rescues gld-1(-). We will refer to the wild-type and mutant transgenes that have been crossed into a gld-1 null mutant background as gld-1wt::ollas and gld-1ndb::ollas throughout the manuscript. Tables and figures contain the full genotype of the transgenic worms (gld-1(q485); gld-1wt::ollas and gld-1(q485); gld-1ndb::ollas). To document GLD-1::OLLAS protein expression, gld-1wt::ollas and gld-1ndb::ollas worm gonads were immunostained with anti-GLD-1 antibody. As previously reported, GLD-1 expression is highest in the central meiotic pachytene region of the germline and low in the distal and proximal ends (Jones et al. 1996). Similar patterns and levels of transgenic protein expression were observed in both gld-1wt::ollas and gld-1ndb::ollas worms (P > 0.6 by Student’s t-test; Figure 8, A–C). Western blot analysis with anti-OLLAS antibody was used to test whether similar levels of GLD-1wt::OLLAS and GLD-1ndb::OLLAS protein are present in whole-worm lysate. GLD-1::OLLAS protein levels were quantitated, normalized to tubulin, and the results showed that GLD-1wt::OLLAS and GLD-1ndb::OLLAS proteins are expressed at similar levels (P > 0.7; Figure 8, D and E).
Figure 8.
GLD-1ndb mutation does not affect protein expression and localization in vivo. GLD-1wt::OLLAS and GLD-1ndb::OLLAS protein expression (green) in (A) gld-1(q485); gld-1wt::ollas and (B) gld-1(q485); gld-1ndb::ollas transgenic worm gonads. GLD-1::OLLAS is highly expressed in the meiotic pachytene region of the germline. DNA was stained using DAPI (gray) for reference. Bar, 10 μm. (C) Quantitation of GLD-1::OLLAS immunostaining intensity levels in meiotic pachytene cells using LAS-X software (Leica). No significant difference in expression was observed using Student’s unpaired t-test (P > 0.6; gld-1wt::ollas N = 12, gld-1ndb::ollas N = 13). (D) Western blot analysis of GLD-1wt::OLLAS and GLD-1ndb::OLLAS protein levels in whole lysates of transgenic worms. Tubulin is used as a loading control. (E) Quantitation of GLD-1wt::OLLAS and GLD-1ndb::OLLAS protein level in 50 transgenic whole worm lysate normalized to tubulin. No significant difference in expression was detected by Student’s unpaired t-test (P > 0.7; N = 5 for each genotype). (F) Confocal images of meiotic pachytene region of wild-type, gld-1(q485); gld-1wt::ollas, and gld-1(q485); gld-1ndb::ollas germlines co-immunostained for P granule component PGL-1 (red) and GLD-1::OLLAS (green). Bar, 10 μm. (G) Quantitation of GLD-1, GLD-1wt::OLLAS and GLD-1ndb::OLLAS protein enrichment in P granules from confocal images. No significant difference in enrichment of GLD-1wt::OLLAS and GLD-1ndb::OLLAS was detected by Student’s unpaired t-test (P > 0.6; N = 3 for each genotype).
Our previous research established that DLC-1 is a specific cofactor of the RNA binding protein FBF-2 (Wang et al. 2016). The DLC-1-FBF-2 interaction is required for FBF-2 to localize to perinuclear P granules and function as a translational repressor. P granules are RNPs found exclusively in germ cells and required for fertility (Voronina 2013). GLD-1 was reported to partially localize to P granules in the embryo (Jones et al. 1996), but its localization to germline P granules was not assessed. Since our current data suggest DLC-1 is required for GLD-1 function, we next asked if GLD-1 is enriched in germline P granules and whether DLC-1 binding to GLD-1 is important for GLD-1 subcellular localization. Immunostaining showed that endogenous GLD-1 protein as well as GLD-1::OLLAS protein from transgenic lines is expressed in the cytoplasm of the meiotic pachytene cells and revealed the presence of some granular structures (Figure 8F). To compare the distribution of GLD-1 protein to P granules, we calculated the ratio of GLD-1 intensity in P granules to cytoplasm. The ratio in wild-type, gld-1wt::ollas, and gld-1ndb::ollas worms was 1.7, 1.3, and 1.4, respectively. This result suggests that GLD-1 is slightly enriched in P granules and that the OLLAS tag does not cause the observed enrichment of GLD-1::OLLAS protein in transgenic lines. Since the P granule enrichment of GLD-1 was not significantly different between gld-1wt::ollas and gld-1ndb::ollas when evaluated using Student’s unpaired t-test (P > 0.6), we conclude that DLC-1 binding does not affect subcellular localization of GLD-1 (Figure 8, F and G).
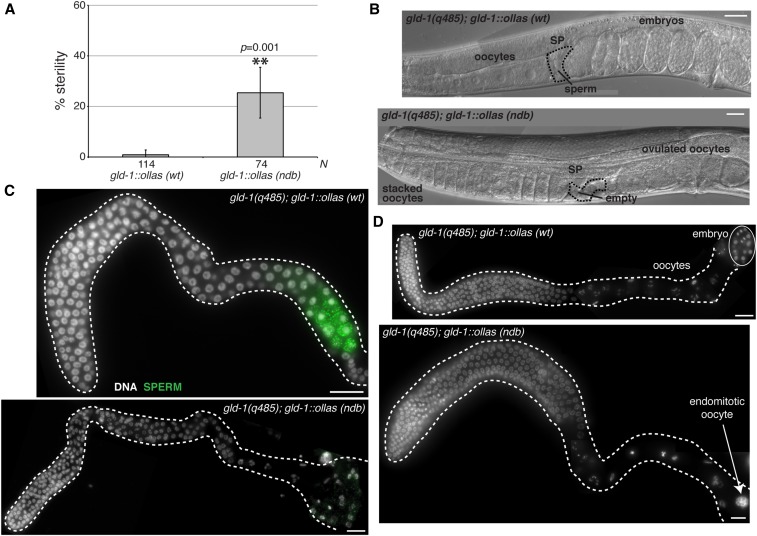
Finally, we compared the fertility of gld-1wt::ollas to gld-1ndb::ollas transgenic worms in the gld-1(-) background by isolating worms at the fourth larval stage of development (L4) and quantitating the percentage of worms unable to produce offspring. We found a statistically significant increase in sterility in gld-1ndb::ollas compared to gld-1wt::ollas transgenic worms (25% vs. <1%) when cultured at 24° (Figure 9A). Sterile hermaphrodites appeared to accumulate excessive oocytes and occasionally ovulated unfertilized oocytes (Figure 9B). Accumulation of arrested oocytes is often linked to depletion of available sperm (McCarter et al. 1999). By immunostaining for a sperm antigen, we found that 8% of gld-1ndb::ollas fail to produce sperm during larval development and exhibit feminized germline phenotype (Figure 9C and Table 2). Germline feminization is consistent with disruption of GLD-1 protein function (Francis et al. 1995b) and we conclude that interaction with DLC-1 facilitates GLD-1 function in vivo. Staining of gld-1ndb::ollas sterile germlines with DAPI further showed that in a subset (27%) of gld-1ndb::ollas germlines oocytes undergo meiotic maturation in absence of ovulation, as evidenced by the presence of endomitotic oocyte DNA at the proximal end of the gonad (Figure 9D and Table 2). Interestingly, endomitotic oocyte phenotype has not been associated with previously described gld-1 mutants. These results suggest a new requirement for GLD-1 in regulation of oocyte maturation or ovulation.
Figure 9.
DLC-1/GLD-1 interaction promotes GLD-1 function in vivo. (A) Analysis of sterility phenotype of gld-1(q485); gld-1wt::ollas and gld-1(q485); gld-1ndb::ollas worms at 24°. Sterile worms were identified by the inability to produce viable offspring. L4 larvae from synchronous cultures of either gld-1(q485); gld-1wt::ollas or gld-1(q485); gld-1ndb::ollas worms were isolated and their ability to produce viable offspring assessed. Percent sterility from four independent experiments was calculated and error bars represent SD from the mean. Number of animals scored for each genotype (N) is indicated below the chart. ** indicates statistically significant difference in percent sterility determined by Student’s paired t-test (P = 0.001). (B) Images of gld-1(q485); gld-1wt::ollas and gld-1(q485); gld-1ndb::ollas whole worms acquired using Nomarski DIC microscopy. Morphological landmarks include oocytes, spermatheca (SP; black dashed outline), sperm and embryos. gld-1(q485); gld-1ndb::ollas worms accumulate oocytes, ovulate unfertilized oocytes and spermatheca appears empty. (C) Fluorescence micrograph of gld-1(q485); gld-1wt::ollas or gld-1(q485); gld-1ndb::ollas gonads dissected at L4 stage and immunostained for sperm (green) using anti-MSP antibody. A subset (8%) of gonads from gld-1(q485); gld-1ndb::ollas worms lack sperm and exhibit feminized germline phenotype (Table 2). (D) Dissected gonads from either gld-1(q485); gld-1wt::ollas or sterile gld-1(q485); gld-1ndb::ollas worms stained with DAPI to reveal DNA morphology. Gonads from gld-1(q485); gld-1ndb::ollas mutant worms exhibit endomitotic oocyte phenotype (white arrow) (27%; Table 2). Bar, 10 μm.
Table 2. Phenotype of gld-1ndb::ollas transgenic animals.
| Genotype | Endomitotic oocyte (%) | n | Spermatogenesis defect (%) | n |
|---|---|---|---|---|
| gld-1(q485); gld-1::ollas (wt) | 0 | 63 | 0 | 10 |
| gld-1(q485); gld-1::ollas (ndb mutant) | 27 | 52 | 8 | 52 |
Is DLC-1 binding GLD-1 important for GLD-1 target mRNA regulation?
We next asked whether disruption of gametogenesis observed upon the loss of GLD-1 interaction with DLC-1 was associated with disruption of GLD-1 target translational repression. To test this we crossed gfp::3xflag::mex-3 into gld-1(q485) null mutant background in the presence of gld-1wt::ollas or gld-1ndb::ollas and observed the pattern of tagged GFP::FLAG::MEX-3 protein expression. MEX-3 expression was restricted to the distal mitotic region and proximal oocytes in gld-1wt::ollas genetic background and brightfield microscopy revealed normal germlines (Figure S5A and Table S2). In a subset of gld-1ndb::ollas worms that exhibited germline feminization and accumulation of arrested oocytes (Figure 9B and Table 2), we observed MEX-3 localizing to large granular structures in the oocytes (Figure S5B). This is consistent with previously described accumulation of MEX-3 in large ribonucleoprotein granules in the arrested oocytes (Schisa et al. 2001; Jud et al. 2008). However, the extent of MEX-3 repression in meiotic prophase region of the germline in gld-1wt::ollas vs. gld-1ndb::ollas worms appeared similar (Figure S5, A and B and Table S2). It is possible that activation of mex-3 transgenic reporter in pachytene observed after dlc-1(RNAi) was due to disrupted function of several RNA binding proteins including GLD-1 in the absence of DLC-1. These results also suggest that disruption of gametogenesis in the gld-1ndb::ollas background is due to misregulation of GLD-1 targets other than mex-3.
DLC-1 binding to GLD-1 facilitates meiotic entry and prevents ectopic germline proliferation
Since genetic redundancy may have influenced the ability to determine if DLC-1 binding GLD-1 is important for RNA regulation in vivo, we next asked whether DLC-1 binding GLD-1 is required to prevent ectopic germline proliferation in a sensitized genetic background. We used RNAi to knock down either gld-2 or gld-3 in gld-1(q485); gld-1wt::ollas and gld-1(q485); gld-1ndb::ollas animals and assessed germ cell proliferation by immunostaining with anti-phospho-histone H3 and anti-REC-8 antibody as previously described. As expected, no ectopic proliferation was observed in gld-1wt::ollas and gld-1ndb::ollas gonads treated with control RNAi (Figure 10, A and C and Table 3). Interestingly, 7% of gld-1ndb::ollas gonads developed tumors at the proximal end of the germline when fed gld-2(RNAi) (Figure 10D and Table 3). By contrast, no Pro tumors formed in gld-1wt::ollas; gld-2(RNAi), gld-1wt::ollas; gld-3(RNAi) or gld-1ndb::ollas; gld-3(RNAi) worms (Figure 10B and Table 3). Treatment of gld-1wt::ollas with either gld-2(RNAi) or gld-3(RNAi) increased the length of the distal proliferative zone compared to control RNAi (Figure 10E). This result is consistent with the longer mitotic zones observed in gld-2(q497) and gld-3(q730) mutant worms (Figure 3; Eckmann et al. 2004). Following gld-2(RNAi), there were no differences in the distal proliferative zones of gld-1wt::ollas and gld-1ndb::ollas worms (P > 0.4 by Kolmogorov–Smirnov test; Figure 10E). A subset of gld-1ndb::ollas; gld-3(RNAi) germlines exhibited an extended distal proliferative zone (up to 14 cell rows longer than the control) and the cumulative distribution of mitotic zone lengths was significantly different in gld-1ndb::ollas; gld-3(RNAi) germlines compared to gld-1wt::ollas; gld-3(RNAi) control (P = 0.024 by Kolmogorov–Smirnov test; Figure 10E). Together, these data show that mutation of the DLC-1 binding site on GLD-1 disrupts the function of GLD-1 in suppression of ectopic proliferation in vivo and that the phenotypes observed in gld-2(-); dlc(-) and gld-3(-); dlc-1(-) double mutants (Figure 2, G and H) are likely due to the failure of DLC-1-GLD-1 interaction.
Figure 10.
GLD-1 requires interaction with DLC-1 to prevent ectopic germline proliferation in vivo. Gonads were immunostained for mitotically dividing cells (pink), stem and progenitor cells (green), and DNA (blue) as in Figure 2. Dissected germlines from gld-1(q485); gld-1wt::ollas transgenic animals treated with either (A) empty vector control or (B) gld-2(RNAi). Dissected gonads from gld-1(q485); gld-1ndb::ollas mutant worms treated with either (C) control or (D) gld-2(RNAi). Efficacy of gld-2(RNAi) was determined by scoring sterility and embryonic lethality. gld-1(q485); gld-1wt::ollas transgenic animals fed gld-2(RNAi) exhibited 100% sterility and gld-1(q485); gld-1ndb::ollas mutant worms exhibited on average 86% sterility and the embryonic lethality of worms with eggs ranged from 43 to 100%. White arrow indicates aberrant cell proliferation. Images represent data collected from two independent experiments and 19–132 worms were scored for each genotype (see Table 3). Bar, 10 μm. (E) Mitotic region length was measured by counting REC-8 positive stem and progenitor cell row number of gld-1(q485); gld-1wt::ollas (blue) and gld-1(q485); gld-1ndb::ollas (green) transgenic animals fed either control, gld-2(RNAi), or gld-3(RNAi), respectively. A total of 22–80 germlines were scored for each genotype (N, indicated below the chart). A statistically significant difference in distribution of mitotic zone lengths was observed between gld-1(q485); gld-1wt::ollas and gld-1(q485); gld-1ndb::ollas worms treated with gld-3(RNAi) (indicated by *; P = 0.024) but not gld-1(q485); gld-1wt::ollas vs. gld-1(q485); gld-1ndb::ollas worms treated with gld-2(RNAi) (P > 0.4). Kolmogorov–Smirnov test was used to calculate P-values. Embryonic lethality of gld-1(q485); gld-1wt::ollas; gld-3(RNAi) worms was 69 ± 25% and gld-1(q485); gld-1ndb::ollas; gld-3(RNAi) was 54 ± 11%.
Discussion
In this study, we show that direct association with DLC-1 is required for the function of germline post-transcriptional regulator GLD-1. Similar to the previously characterized interaction of DLC-1 and FBF-2, the DLC-1-GLD-1 cooperation is independent of DLC-1’s function in the context of the dynein motor. These results suggest a widespread role for DLC-1 as a cofactor in post-transcriptional regulation of gene expression.
DLC-1 cooperation with GLD-1 is consistent with allosteric effect
The genetic and molecular analysis presented here suggests that DLC-1 promotes GLD-1 functions in supporting meiosis and gametogenesis in the C. elegans germline. These findings extend the previously proposed role for DLC-1 in the inhibition of proliferative cell fate (Dorsett and Schedl 2009). Previous research suggested that DLC-1 antagonized GLP-1/NOTCH activity in part through promoting accumulation and nuclear import of methyltransferase METT-10, yet genetic analysis suggested additional METT-10–independent mechanisms (Dorsett and Schedl 2009). Our data suggests that dlc-1 also functions downstream of GLP-1, since overproliferation of gld-3(-); dlc-1(-) mutant was independent of glp-1 activity (Figure 4A). Formation of synthetic tumors in gld-2(-); dlc-1(-) and gld-3(-); dlc-1(-) backgrounds suggests that DLC-1 likely acts in the GLD-1 pathway that promotes entry into meiosis. Cooperation of DLC-1 with GLD-1 may represent one of METT-10–independent mechanisms suggested by the prior studies. Although the previous genetic analysis suggested that DLC-1 was functioning in the context of the dynein motor as mutations of the motor subunit dhc-1 exhibited similar genetic interactions, our results suggest that cooperation of DLC-1 and GLD-1 does not involve dynein motor (Figure 2, I–L, Figure 3, Figure 5Aiii, Figure 5C, Figure S4, E–H, Table 1, and Table S2).
LC8-type proteins may affect accumulation or subcellular distribution of their binding partners (Moseley et al. 2007; Dorsett and Schedl 2009; Rapali et al. 2011b; Wang et al. 2016). Additionally, LC8 proteins can function as allosteric regulators of protein networks by binding to intrinsically disordered regions of proteins and facilitating structural organization and dimerization (Lightcap et al. 2009; Rapali et al. 2011a). Here, we tested the potential mechanisms that could be used by DLC-1 to promote GLD-1 function.
We find that DLC-1 does not appear to influence the stability of GLD-1 since neither dlc-1(-) mutation nor mutagenesis of DLC-1 binding site in GLD-1 affect GLD-1 protein levels (Figure 6 and Figure 8, A–E). Since our previous data suggested DLC-1 was important for recruiting the RNA binding protein FBF-2 to P granules (Wang et al. 2016), we tested whether GLD-1 was similarly enriched in P granules and if interaction with DLC-1 contributed to this localization. We find that GLD-1 is slightly enriched in P granules compared to the surrounding cytoplasm (Figure 8, F and G), although the extent of the enrichment varies between the individual germlines. This parallels enrichment of vertebrate STAR domain proteins SAM68 and KSRP in the chromatoid body of mouse early spermatids (Messina et al. 2012; Zhang et al. 2017). Mutation of DLC-1 binding site in GLD-1 causes a small increase in GLD-1 accumulation in P granules; however, this effect is not statistically significant (Figure 8, F and G). These results suggest that there must be multiple mechanisms targeting P granule–associated RNA binding proteins to these cytoplasmic organelles, and interaction with DLC-1 is only one of the potential localization determinants. Since DLC-1 is widely present in both P granules and the cytoplasm (Wang et al. 2016), it may promote GLD-1’s function in either location. If association with DLC-1 does not affect accumulation or localization of GLD-1, the remaining possibility explaining the effect on GLD-1 is that association with DLC-1 changes GLD-1 conformation. Although previous research suggests that DLC-1/LC8 dimers promote dimerization of their binding partners (Barbar and Nyarko 2015), it is unclear whether this could be relevant to GLD-1 because the RNA binding domain of GLD-1 dimerizes very effectively in vitro without requiring any cofactors (Ryder et al. 2004). More likely, the conformational change induced by DLC-1 binding would change the assembly of GLD-1 with other molecular partners.
DLC-1 binding facilitates GLD-1’s role in spermatogenesis, oocyte development leading to maturation and ovulation, and regulating germ cell proliferation
Mutation of DLC-1 binding site on GLD-1 compromises several aspects of GLD-1 function. It is possible that in addition to disrupting the interaction with DLC-1, gld-1ndb mutation affects GLD-1 interaction with other protein partners or general folding. Since gld-1ndb mutation shows similar synthetic phenotypes as dlc-1(-), we conclude that disruption of GLD-1ndb function is mainly due to a loss of interaction with DLC-1. A subset (8%; Table 2) of gld-1ndb::ollas mutant worms exhibits feminization of the germline (Fog). Feminized gonads fail to make sperm but oogenesis is not affected and our observed phenotype is similar to the previously described class D partial loss-of-function gld-1 mutant allele (Francis et al. 1995a). A larger subset (27%) of gld-1ndb::ollas mutant worms exhibit an endomitotic oocyte phenotype where oocytes fail to arrest in diakinesis, undergo maturation, and start DNA replication prior to ovulation. This phenotype has never been described for a gld-1 mutant allele and analysis of gld-1ndb::ollas mutant worms reveals a new requirement for GLD-1 in regulating oocyte development, maturation, or ovulation.
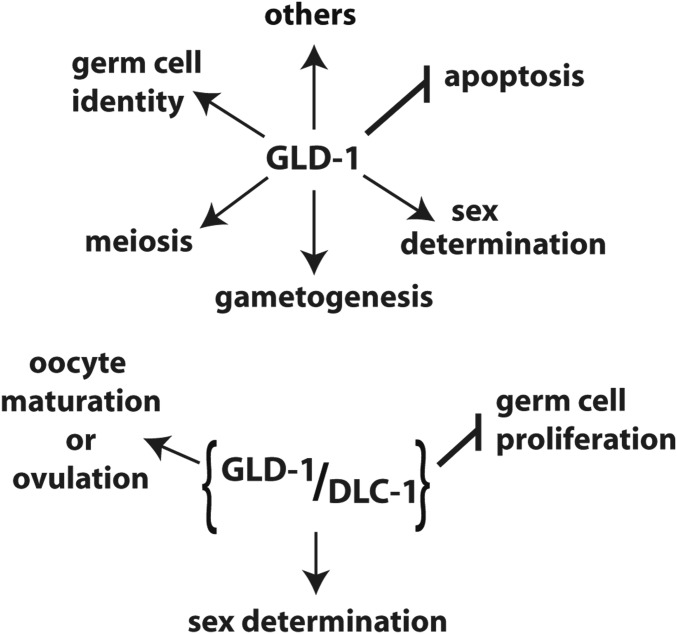
Disruption of GLD-1 activity is also associated with overproliferation in the C. elegans germline. Both gld-2(-) gld-1(-) and gld-1(-); gld-3(RNAi) worms fail to enter meiosis and germlines contain only mitotically dividing cells (Kadyk and Kimble 1998; Eckmann et al. 2004). Our results show that 37% of gld-3(-); dlc-1(-) double mutant worms exhibit proximal (Pro) tumor formation where the germ cells fail to produce oocytes and return to the mitotic cell cycle (Figure 2G and Table 1). A subset of gld-2(-); dlc-1(-) worms exhibited late onset tumor formation, which suggests a defect in meiotic entry (Figure 2H and Figure 3). Additionally, 7% of gld-1ndb::ollas; gld-2(RNAi) worms exhibit Pro tumor formation (Figure 10D and Table 3). The fact that the ectopic proliferation observed in gld-2(-); dlc-1(-) and gld-3(-); dlc-1(-) double mutants and gld-1ndb::ollas; gld-2(RNAi) worms is less severe than previously reported for gld-2(-) gld-1(-) and gld-1(-); gld-3(RNAi) worms (Kadyk and Kimble 1998; Eckmann et al. 2004) suggests that abolishing GLD-1-DLC-1 binding results in partial loss of GLD-1 function and that GLD-1-DLC-1 binding may play a secondary role in promoting entry into meiosis. Collectively, these data suggest that DLC-1 facilitates a subset of GLD-1 functions (Figure 11).
Figure 11.
GLD-1 is a key regulator of germline development that affects multiple aspects of germ cells function. DLC-1 facilitates a subset of GLD-1 functions.
DLC-1 is needed for regulation of a subset of GLD-1 targets
One mechanistic explanation for DLC-1 facilitating select GLD-1 functions is that only some GLD-1 target mRNAs require DLC-1 for efficient translational control. Consistent with this possibility, analysis of the in vivo reporters representing mRNA targets of GLD-1 regulation suggests that DLC-1 is required for repression of some, but not all reporters in the meiotic pachytene cells (Figure 5). A likely reason for this selectivity is that GLD-1 forms several regulatory RNPs depending on the specific target mRNA, only some of which contain DLC-1 and rely on DLC-1 for their function. The other GLD-1 RNPs are capable of regulating their specific targets independent of DLC-1. In support of this model, a subset of GLD-1 target mRNAs recovered through DLC-1 immunoprecipitation includes cye-1 and mes-3 mRNAs, but excludes puf-5 and spn-4 mRNAs (Day et al. 2018). In the future, it would be of interest to identify GLD-1 target mRNAs that are misregulated in the gld-1ndb::ollas mutant worms leading to disruption of gametogenesis and oocyte cell cycle control or ovulation.
A role for DLC-1 in post-transcriptional regulation of gene expression
RNA binding proteins control many post-transcriptional steps of gene expression for large and diverse sets of mRNA targets. Although binding specificity in terms of the nucleotide sequence recognized by a particular RNA binding protein plays a large role in target specificity and regulatory output, other factors such as structural accessibility of the nucleic acid molecule or allosteric modulation of molecules to enable formation of higher order RNPs are also important. Our previous research showed that a small protein DLC-1 helped regulate gene expression in the distal region of the worm gonad by functioning as a cofactor to the RNA binding protein FBF-2. Since DLC-1 was required for FBF-2 localization to perinuclear P granules, we concluded that DLC-1 could facilitate gene regulation by helping its binding partner sort to the appropriate RNP regulatory complex. Here, we expand upon our previous studies by characterizing DLC-1 interaction with another RNA regulator, the STAR domain RNA binding protein GLD-1. Our results show that DLC-1 is required for GLD-1 regulation of a subset (but not all) GLD-1 target mRNAs and this suggests that DLC-1 might regulate post transcriptional gene expression by directing GLD-1 target mRNA selectivity. Interestingly, disruption of DLC-1 binding motif in gld-1ndb mutant produced a weaker effect on one of GLD-1 targets (mex-3) than dlc-1(RNAi) (Figure 5C and Figures S3A and S5). This suggests that DLC-1 affects germline gene regulation by facilitating the function of other RNA binding proteins in addition to GLD-1. Our results also support the idea that DLC-1 uses different mechanisms to affect the regulatory activity of its binding partners. Elucidating the molecular details of how DLC-1 promotes GLD-1 functions in translational control and preventing ectopic proliferation is an exciting new direction for future research.
Acknowledgments
We thank the members of Voronina laboratory for helpful discussions. Several nematode strains were provided by Caenorhabditis Genetics Center, funded by the National Institutes of Health (NIH) (grant P40OD010440). The K76, anti-MSP, and anti-MY0-3 antibodies were obtained from the Developmental Studies Hybridoma Bank (National Institute of Child Health and Human Development, The University of Iowa). The anti-GLD-1 antibody was a gift from Tim Schedl. Confocal microscopy was performed in the University of Montana BioSpectroscopy Core Research Laboratory, which is operated with support from NIH Center of Biomedical Research Excellence Award P20GM103546 to the Center for Biomolecular Structure and Dynamics and from the Vice President of Research and Creative Scholarship at the University of Montana, and the S10OD021806 NIH award for acquisition of confocal microscope. This work was supported by the NIH grant GM109053 to E.V., a University of Montana Research Award to M.E., and a University of Montana Undergraduate Research Award to E.O.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361234.
Communicating editor: D. Greenstein
Literature Cited
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. 10.1016/0092-8674(87)90128-0 [DOI] [PubMed] [Google Scholar]
- Barbar E., Nyarko A., 2015. Polybivalency and disordered proteins in ordering macromolecular assemblies. Semin. Cell Dev. Biol. 37: 20–25. 10.1016/j.semcdb.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry L. W., Westlund B., Schedl T., 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124: 925–936. [DOI] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. 10.1016/j.devcel.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Brenner J. L., Schedl T., 2016. Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics 202: 1085–1103. 10.1534/genetics.115.185678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. 10.1038/nature754 [DOI] [PubMed] [Google Scholar]
- Day N. J., Ellenbecker M., Voronina E., 2018. Caenorhabditis elegans DLC-1 associates with ribonucleoprotein complexes to promote mRNA regulation. FEBS Lett. 592: 3683–3695. 10.1002/1873-3468.13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett M., Schedl T., 2009. A role for dynein in the inhibition of germ cell proliferative fate. Mol. Cell. Biol. 29: 6128–6139. 10.1128/MCB.00815-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann C. R., Crittenden S. L., Suh N., Kimble J., 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160. 10.1534/genetics.104.029264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T., 1995a. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T., 1995b. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. 10.1007/978-1-4614-4015-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Hubbard E. J., Schedl T., 2004a. Multi-pathway control of the proliferation vs. meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268: 342–357. 10.1016/j.ydbio.2003.12.023 [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004b. Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. 10.1242/dev.00916 [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Jud M. C., Czerwinski M. J., Wood M. P., Young R. A., Gallo C. M., et al. , 2008. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 318: 38–51. 10.1016/j.ydbio.2008.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kimble J., Seidel H., 2008. C. elegans germline stem cells and their niche in StemBook. Harvard Stem Cell Institute, Cambridge, MA. [PubMed] [Google Scholar]
- King S. M., Patel-King R. S., 1995. The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270: 11445–11452. 10.1074/jbc.270.19.11445 [DOI] [PubMed] [Google Scholar]
- Kraemer B., Crittenden S., Gallegos M., Moulder G., Barstead R., et al. , 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. 10.1016/S0960-9822(99)80449-7 [DOI] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420. 10.1101/gad.915901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightcap C. M., Kari G., Arias-Romero L. E., Chernoff J., Rodeck U., et al. , 2009. Interaction with LC8 is required for Pak1 nuclear import and is indispensable for zebrafish development. PLoS One 4: e6025 10.1371/journal.pone.0006025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin V. A., Evans T. C., 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130: 2623–2632. 10.1242/dev.00486 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina V., Meikar O., Paronetto M. P., Calabretta S., Geremia R., et al. , 2012. The RNA binding protein SAM68 transiently localizes in the chromatoid body of male germ cells and influences expression of select microRNAs. PLoS One 7: e39729 10.1371/journal.pone.0039729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D., Ho D. M., Hunter C. P., 2004. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131: 3263–3272. 10.1242/dev.01196 [DOI] [PubMed] [Google Scholar]
- Moseley G. W., Roth D. M., Dejesus M. A., Leyton D. L., Filmer R. P., et al. , 2007. Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 18: 3204–3213. 10.1091/mbc.e07-01-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Eckmann C. R., 2013. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757: 205–247. 10.1007/978-1-4614-4015-4_8 [DOI] [PubMed] [Google Scholar]
- Pazdernik N., Schedl T., 2013. Introduction to germ cell development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 1–16. 10.1007/978-1-4614-4015-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S., Killian D. J., Hubbard E. J., 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P., García-Mayoral M. F., Martínez-Moreno M., Tárnok K., Schlett K., et al. , 2011a. LC8 dynein light chain (DYNLL1) binds to the C-terminal domain of ATM-interacting protein (ATMIN/ASCIZ) and regulates its subcellular localization. Biochem. Biophys. Res. Commun. 414: 493–498. 10.1016/j.bbrc.2011.09.093 [DOI] [PubMed] [Google Scholar]
- Rapali P., Szenes Á., Radnai L., Bakos A., Pál G., et al. , 2011b. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278: 2980–2996. 10.1111/j.1742-4658.2011.08254.x [DOI] [PubMed] [Google Scholar]
- Ryder S. P., Frater L. A., Abramovitz D. L., Goodwin E. B., Williamson J. R., 2004. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11: 20–28. 10.1038/nsmb706 [DOI] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. 10.1016/j.cell.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G., 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871. [DOI] [PubMed] [Google Scholar]
- Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J., 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103: 15108–15112. 10.1073/pnas.0607050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E., 2013. The diverse functions of germline P-granules in Caenorhabditis elegans. Mol. Reprod. Dev. 80: 624–631. 10.1002/mrd.22136 [DOI] [PubMed] [Google Scholar]
- Voronina E., Greenstein D., 2016. Germ cell fate determination in C. elegans. in eLS John Wiley & Sons, Ldt: Chichester: 1–8. [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. 10.1038/nature01039 [DOI] [PubMed] [Google Scholar]
- Wang X., Olson J. R., Rasoloson D., Ellenbecker M., Bailey J., et al. , 2016. Dynein light chain DLC-1 promotes localization and function of the PUF protein FBF-2 in germline progenitor cells. Development 143: 4643–4653. 10.1242/dev.140921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. J., Salata M. W., Susalka S. J., Pfister K. K., 2001. Light chains of mammalian cytoplasmic dynein: identification and characterization of a family of LC8 light chains. Cell Motil. Cytoskeleton 49: 229–240. 10.1002/cm.1036 [DOI] [PubMed] [Google Scholar]
- Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E., et al. , 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 30: 533–545. 10.1038/emboj.2010.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. 10.1038/37297 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang G., Liu L., Liang X., Lin Y., et al. , 2017. KH-type splicing regulatory protein is a new component of chromatoid body. Reproduction 154: 723–733. 10.1530/REP-17-0169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains (see Table S1) and reagents are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361234.