Abstract
Background:
For over a decade, Sca-1+ cells within the mouse heart have been widely recognized as a stem cell population with multipotency that can give rise to cardiomyocytes, endothelial cells and smooth muscle cells in vitro and after cardiac grafting. However, the developmental origin and authentic nature of these cells remain elusive.
Methods:
Here, we used a series of high-fidelity genetic mouse models to characterize the identity and regenerative potential of cardiac resident Sca-1+ cells.
Results:
With these novel genetic mouse models, we found that Sca-1 does not label cardiac precursor cells during early embryonic heart formation. Postnatal cardiac resident Sca-1+ cells are in fact a pure endothelial cell population. They retain endothelial properties and exhibit minimal cardiomyogenic potential during development, normal aging and upon ischemic injury.
Conclusions:
Our study provides definitive insights into the nature of cardiac resident Sca-1+ cells. The observations challenge the current dogma that cardiac resident Sca-1+ cells are intrinsic stem cells for myocardial development, renewal and repair and suggest that the mechanisms of transplanted Sca-1+ cells in heart repair need to be reassessed.
Keywords: Sca-1, cardiac stem cells, heart failure, heart regeneration, heart repair
Introduction
Whether mammalian hearts harbor a population of intrinsic stem cells for myocardial renewal and repair and, if they do, how to identify these cells for use in cell-based therapy for heart failure are central questions in cardiac regenerative medicine1–5. In the past two decades, tremendous efforts have been made to search for such cells, and many forms of cardiac stem cells (CSCs) have been identified1, 2. Recently, the nature of c-Kit+ CSCs and their function in heart repair were questioned6–10. Three independent groups coincidentally indicated that cardiac c-Kit+ cells lack myogenic potential during heart development and repair11–13.
Murine stem cell antigen-1 (Sca-1) is a member of the Ly-6 gene family (gene name Ly6a)14, 15. Sca-1 encodes a cell surface protein widely used to enrich hematopoietic stem cells (HSCs) from the bone marrow (BM)14, 16. With this perception, Sca-1 has been persistently thought to be a marker to identify adult stem cells in multiple organs17–21. Cardiac Sca-1+ cells were one of the first putative CSCs identified in the adult mouse heart22–26, and are found distributed in diverse CSC subtypes in mice (e.g., cardiospheres, side populations, and cardiac colony-forming unit fibroblasts)27–32. The human equivalent of the murine Sca-1 ortholog has not been identified. However, Sca-1+-like cells were isolated from the adult human heart using an anti-mouse Sca-1 antibody and showed cardiomyogenic potential when cultured in vitro33. Importantly, a phase I clinical trial (CADUCEUS) was performed in which autologous Sca-1-related cardiosphere-derived cells were administered to patients with myocardial infarction (MI). Reduced scar size with improved cardiac function was observed in the patients34.
Despite these findings, questions have been raised regarding the mechanisms of Sca-1+ cells in heart repair. In transplantation of exogenously expanded Sca-1+ cells, the number of identifiable engrafted cells has been found to be extremely low (<0.5%), and thus, they are unlikely to contribute to functional heart repair through myocardial differentiation35. The myogenic potential of engrafted Sca-1+ cells may also require further investigation because the conclusions are mainly based on immunostaining with potential microscopic artifacts36, 37. In addition, previous reports determining the myogenic potential of cardiac Scal-1+ cells have largely relied on in vitro cardiomyogenic differentiation culture procedures and that may not represent the nature of endogenous Scal-1+ cells23, 30. Furthermore, the developmental origin of cardiac Sca-1+ cells remains largely unknown. These questions raise doubts about whether Sca-1 expression marks bona fide embryonic and/or adult CSCs38. In summary, there is an urgent need to define the authentic identity of Sca-1+ cells in the developing and adult hearts, to provide definitive answers as to whether Sca-1 expression represents a true and applicable CSC population for heart repair.
Methods
The data, analytical methods, and study materials will be made available to other researchers for the purposes of reproducing the results or replicating the procedure upon reasonable request. Inquiries can be directed to the corresponding author.
Mouse models
All mouse experiments were conducted in accordance with an approved IACUC protocol at the Icahn School of Medicine at Mount Sinai and were in compliance with institutional and governmental regulations (PHS Animal Welfare Assurance A3111–01). Nkx2.5H2B-GFP/+, cTNTH2B-GFP/+, c-KitH2B-GFP/+, and ROSA26RtdTomato/+ mouse lines were described previously12, 39–41. PDGFRαH2B-GFP/+ mice were obtained from Dr. Philippe Soriano42.
Three cassettes (LoxP-4XployA-LoxP-H2B-tdTomato-FRT-Neo-FRT, LoxP-nLacZ-4XPloyA-LoxP-H2B-GFP-FRT-Neo-FRT and MerCreMer-FRT-Neo-FRT) were inserted into the start codon of the Sca-1 (Ly6a) locus to generate Sca-1H2B-tdTomato/+, Sca-1nLacZ-H2B-GFP/+ and Sca-1MerCreMer/+ knock-in mouse models, respectively. The cassettes were flanked by a 5.0 kb 5ʹ homologous arm and a 4.0 kb 3ʹ homologous arm in the targeting constructs. The constructs were linearized and electroporated into mouse embryonic stem (ES) cells. Positive ES cells were identified by long-range PCR (Roche) with two pairs of primers (P1+P2 and P3+P4). The primer sequences are as follows: P1, 5-ATGAATAGTTGACCCCCACATGCT-3; P2, 5-CAGGGTGGACCTGCTTCAGAACCT-3 (Sca-1STOP-H2B-tdTomato/+); P2, 5-GGATGTGCTGCAAGGCGATTAAGT-3 (Sca-1nLacZ-H2B-GFP/+); P2, 5-GTTCAGCATCCAACAAGGCACTGA-3 (Sca-1MerCreMer/+); P3, 5-AGAGCTTGGCGGCGAATGGGCTGACCG-3; and P4, 5-TGACAACCATCAAGGTTATGATCT-3. The PCR fragments were further subcloned and verified by DNA sequencing. Targeted ES cells were microinjected into blastocysts to generate chimeric mice. The chimeric mice were crossed with C57BL/6 and Black Swiss mice to obtain germline transmission mice. The Neo cassette was removed by crossing with Flippase deleter mice. Sca-1H2B-tdTomato/+ mice were obtained by crossing Sca-1LoxP−4XPloyA-LoxP-H2B-tdTomato/+ with Protamine-Cre mice.
Tamoxifen (Sigma, cat. T5648) was injected intraperitoneally into the mice at a dose of 0.12 mg/g body weight.
X-gal staining
Mouse tissues were isolated in ice-cold PBS and fixed in 4% paraformaldehyde for 30 min at 4°C. After fixation, the tissues were washed three times with PBS and incubated in X-gal solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, and 1 mg/ml X-gal) overnight at room temperature (RT). For section staining, after fixation, the tissues were treated with 30% sucrose overnight at 4°C and embedded in optimal cutting temperature (OCT) compound (Tissue-Tek, 4583) on dry ice. Then, 10-μm sections were cut and post-fixed in 4% paraformaldehyde for 5 min. The sections were stained with X-gal solution overnight at 37°C.
Immunofluorescence
Mouse tissues were perfused with 4% paraformaldehyde, dehydrated with sucrose and embedded in OCT. Embedded tissues were cut into 10-μm sections. Sections were blocked with 10% donkey serum (Sigma, D9663) in PBS for 1 h at RT and incubated with primary antibodies overnight at 4°C. The primary antibodies were goat anti-PECAM (CD31) (1:50, R&D Systems, AF3628) and rat anti-Sca-1 (1:200, BD Biosciences, 553333). Sections were then incubated with secondary antibodies for 1 h at RT. The secondary antibodies used were donkey anti-goat Alexa Fluor 488 (1:500; Invitrogen) and donkey anti-rat Alexa Fluor 594 (1:500; Invitrogen). Stained sections were mounted with Vectashield mounting medium with DAPI (Vector Laboratories). Immunofluorescence images were obtained using a Leica fluorescence microscope.
A TSA plus Fluorescein System (Perkin Elmer, NEL741001KT) was used to amplify Sca-1 antibody fluorescent signals when necessary. After primary antibody incubation, an HRP-conjugated secondary antibody was applied for 1 h at RT. HRP-conjugated donkey anti-rat IgG (1:1000; Invitrogen, A18745) was used as the secondary antibody. Sections were washed three times in TNT buffer and amplified with TSA Plus Working Solution for 10 min at RT.
Myocardial infarction
MI was induced by ligation of the LAD coronary artery as previously described43. Briefly, 8- to 16-week-old mice were anesthetized with 4% isoflurane before intubation. The left thoracic region was trimmed with an electric razor and sterilized with 70% isopropanol. After thoracotomy, an 8–0 nylon suture was placed to ligate the LAD. The ribcage and muscle layers were closed with 5–0 polypropylene sutures. Excess air and blood was removed from the chest cavity, and the skin was closed with 4–0 polypropylene sutures. The intubation tube was removed, and mice were housed with moist food and a water bottle.
Heart perfusion and flow cytometry
The procedure for preparing single non-myocardial cells from murine hearts was described previously12. Before surgery, mice were injected with heparin. Animals were anesthetized by isoflurane inhalation. Hearts were perfused with Ca2+-free collagenase type II solution. Atria and connective tissues were removed. Ventricles were cut into small pieces and gently dissociated into single cells with a Pasteur pipette. Dissociated single cells were transferred into a 50 ml Falcon tube and centrifuged at 10×g for 5 min. Cardiomyocytes formed a cell pellet on the bottom. Non-cardiomyocytes were harvested and transferred into a new tube without disturbing the cardiomyocyte pellets. The cells were centrifuged at 300×g for 5 min and resuspended and incubated in 5–10 ml of 1X RBC (Red Blood Cell) lysis buffer at RT for 10 min to remove red blood cells. The cells were collected, washed twice, and resuspended in PBS with 0.5% BSA for flow cytometry analysis.
Bone marrow flow cytometry
Flow cytometric analyses were performed as we previously described44. Mice (8 weeks old) were sacrificed for BM cell collection. Single-cell suspensions derived from the BM were stained with panels of fluorochrome-conjugated antibodies (Sca-1, BD Biosciences 553108; c-Kit, BD Biosciences 560557). Dead cells were excluded by DAPI staining. Analyses were performed using an LSRII flow cytometer. All data were analyzed using FlowJo7.6 software.
Statistical Analysis
Results are shown as mean±SEM. Statistical analysis was performed in Student t test to compare data from individual experimental groups. For each group, at least 3–5 animals or tissue samples were collected for experimentation.
Results
The new Sca-1H2B-tdTomato reporter mouse recapitulates endogenous Sca-1 expression
To characterize the nature of cardiac Sca-1+ cells, we first generated a Sca-1H2B-tdTomato/+ knock-in mouse model by inserting an H2B-tdTomato cassette into the start codon of Sca-1 (Ly6a) through homologous recombination (Figure S1 A). In this model, Sca-1 genomic sequences are preserved. Nuclear tdTomato (H2B-tdTomato) expression is under the control of complete Sca-1 regulatory elements, thereby providing a sensitive, robust genetic tool to identify endogenous Sca-1+ cells in developing and adult mouse hearts. All Sca-1H2B-tdTomato/+ mice were viable and exhibited completely normal development into adulthood (>12 months).
To confirm the fidelity of the knock-in allele, we compared tdTomato with endogenous Sca-1 expression in various organs of Sca-1H2B-tdTomato/+ mice at postnatal day (P) 60 to 90 (P60–90). Sca-1H2B-tdTomato expression co-localized with a Sca-1 antibody in multiple cell types within the kidney, intestine and lung (Figure S1 C-F). With this reporter line, we also detected Sca-1+ cells in the spleen and thymus (data not shown), consistent with previous findings that Sca-1 is expressed in these organs45–49. Sca-1 is a cell surface marker that is widely used along with c-Kit in the identification of HSCs16, and thus, Sca-1 and tdTomato expression should overlap in the c-Kit+ BM cell population from Sca-1H2B-tdTomato/+ mice. We tested tdTomato expression on c-Kit+ HSCs of Sca-1H2B-tdTomato/+ mice via flow cytometry (Figure S1 G1). Indeed, Sca-1H2B-tdTomato and Sca-1 expression largely overlapped (Figure S1 G4). These results further demonstrated that H2B-tdTomato signals in Sca-1H2B-tdTomato/+ mice recapitulate endogenous Sca-1 expression.
Heterogeneous Sca-1+ cell populations in postnatal mouse hearts
Next, we used Sca-1H2B-tdTomato/+ mice to identify Sca-1+ cells in the heart. Cardiac Sca-1+ cells were found at all postnatal stages inspected (P0, P7, P14, and P30–360), with a low number and expression level (based on the brightness of H2B-tdTomato) at birth. The number and expression level progressively increased as the heart grew. By P30, Sca-1H2B-tdTomato-positive cells were dispersed in all cardiac chambers at high density (Figure 1).
Figure 1. Heterogeneity of cardiac resident Sca-1+ cells.
(A) Diagram of the Sca-1H2B-tdTomato/+;c-KitH2B-GFP/+ double heterozygous alleles. (B-C) Longitudinal sections of Sca-1H2B-tdTomato/+;c-KitH2B-GFP/+ mouse hearts at P30 (B) and P120 (C). Partial Sca-1H2B-tdTomato cells and c-KitH2B-GFP cells showed co-localization (yellow, arrows in B2 and C2). (D) Flow cytometry analysis of 4-month-old Sca-1H2B-tdTomato/+;c-KitH2B-GFP/+ mouse heart cells. (E) Diagram of the Sca-1H2B-tdTomato/+;PDGFRaH2B-GFP/+ double heterozygous alleles. (F-G) Longitudinal sections of Sca-1H2B-tdTomato/+;PDGFRaH2B-GFP/+ mouse hearts at P60 (F) and P240 (G). Some Sca-1H2B-tdTomato cells and PDGFRaH2B-GFP cells showed co-localization (yellow, arrows in F2 and G2). (H) Flow cytometry analysis of 4-month-old Sca-1H2B-tdTomato/+;PDGFRaH2B-GFP/+ mouse heart cells. (I) Diagram of the Sca-1H2B-tdTomato/+;Nkx2.5H2B-GFP/+ double heterozygous alleles. (J, K) Longitudinal sections of Sca-1H2B-tdTomato/+;Nkx2.5H2B-GFP/+ mouse hearts at P30 (M) and P120 (N). Sca-1H2B-tdTomato cells (red, arrows in J2 and K2) and NKx2.5H2B-GFP-positive cells (green, arrowheads in M2 and N2) were not co-localized. (L) Diagram of the Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+ double heterozygous alleles. (M, N) Longitudinal sections of Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+ mouse hearts at P30 (J) and P120 (K). No Sca-1H2B-tdTomato cells (red, arrows in M2 and N2) were cTnTH2B-GFP-positive (green, arrowheads in J2 and K2). LV, left ventricle; RV, right ventricle. n=3 for each stage. Scale bar, 100 μm.
Sca-1+ CSCs were initially described as a population of cells lacking the HSC marker c-Kit22. However, later studies showed that Sca-1 is widely co-expressed with c-Kit in many CSC subtypes (e.g., cardiospheres, side populations, and cardiac colony-forming unit fibroblasts)27–32. To obtain a definitive answer regarding cardiac Sca-1 cell identity related to c-Kit, we crossed Sca-1H2B-tdTomato/+ mice with c-KitH2B-GFP/+ knock-in mice (Figure 1 A). Bright, nuclear-localized tdTomato and GFP signals facilitated identification of co-localization. Hearts from the compound heterozygotes (Sca-1H2B-tdTomato/+;c-KitH2B-GFP/+) were collected at P30–120. By directly inspecting sections under a microscope, we found that a substantial number of Sca-1H2B-tdTomato-positive cells were also c-KitH2B-GFP-positive (Figure 1 B, C). By flow cytometry, we estimated that ~51.8% of Sca-1 H2B-tdTomato-positive cells express c-Kit in a 4-month-old heart (Figure 1 D).
Recently, pro-epicardial origin cardiac resident CFU-Fs (colony-forming units–fibroblasts) were identified in the adult mouse heart. These cardiac CFU-Fs express platelet-derived growth factor receptor α (PDGFRα) and Sca-1 and exhibit mesenchymal stem cell properties with multipotency (including cardiomyogenic potential) when cultured in vitro32. A new study also showed that PDGFRα+/Sca-1+ side population cells from the adult mouse heart are clonogenic and have the capacity to produce cardiomyocytes, endothelial cells, and smooth muscle cells after cardiac grafting50. To further characterize cardiac Sca-1+ cells and their relationship with PDGFRα, we crossed PDGFRα H2B-GFP/+ knock-in mice with Sca-1H2B-tdTomato/+ mice (Figure 1 E). Cryosections of Sca-1H2B-tdTomato/+;PDGFRαH2B-GFP/+ mouse hearts were examined at P60–240. Sca-1H2B-tdTomato/PDGFRaH2B-GFP double-positive cells were widely observed in all cardiac chambers (Figure 1 F, G). Flow cytometry revealed that ~49.3% of Sca-1 H2B-tdTomato cells express PDGFRα in a 3-month-old heart (Figure 1 H).
Exogenously expanded cardiac Sca-1+ cells were shown to express early cardiomyogenic markers, including Nkx2.5, Gata4 and Mef2c, upon treatment with oxytocin23. Lineage tracing with a Sca-1 transgenic mouse model also showed that Sca-1+ cells continuously contribute to myocardial turnover during physiological aging at adulthood51. If any subset of cardiac resident Sca-1+ cells acts as intrinsic stem cells that provide a progenitor pool for myocardial growth during heart maturation after birth or myocardial turnover during aging at adulthood, we speculate that these Sca-1+ cells may transiently express the cardiomyogenic marker Nkx2.5 during progenitor to cardiomyocyte conversion. Therefore, we attempted to determine whether any cardiac Sca-1+ cells simultaneously express Nkx2.5 in postnatal hearts. Nkx2.5H2B-GFP/+ knock-in mice were crossed with Sca-1H2B-tdTomato/+ mice (Figure 1 I), and cardiac tissues of the compound heterozygous animals (Nkx2.5H2B-GFP/+;Sca-1H2B-tdTomato/+) were rigorously examined at P30–180 (15–30 day intervals between stages). Surprisingly, we did not find any Sca-1H2B-tdTomato and Nkx2.5H2B-GFP double-positive cells at any of the stages examined (Figure 1 J, K).
Cardiac troponin T (cTnT) is an indicator of differentiated cardiomyocytes. Stable fluorescence of the H2B-tdTomato fusion protein from Sca-1H2B-tdTomato/+ mice may allow short-term cell lineage tracing52, 53 and detection of Sca-1 and cTnT double-positive cells when Sca-1+ progenitor cells differentiate into cardiomyocytes. To determine whether Sca-1 is expressed in cardiomyocytes, cTnTH2B-GFP/+ mice were crossed with Sca-1H2B-tdTomato/+ mice (Figure 1 L). Cardiac sections of the compound heterozygous mice (Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+) at P30–180 (15–30-day interval between stages) were thoroughly examined. However, no Sca-1H2B-tdTomato and cTnTH2B-GFP double-positive cells were found at any of the stages examined (Figure 1 M, N).
Cardiac resident Sca-1+ cells are of the Tie2 endothelial lineage
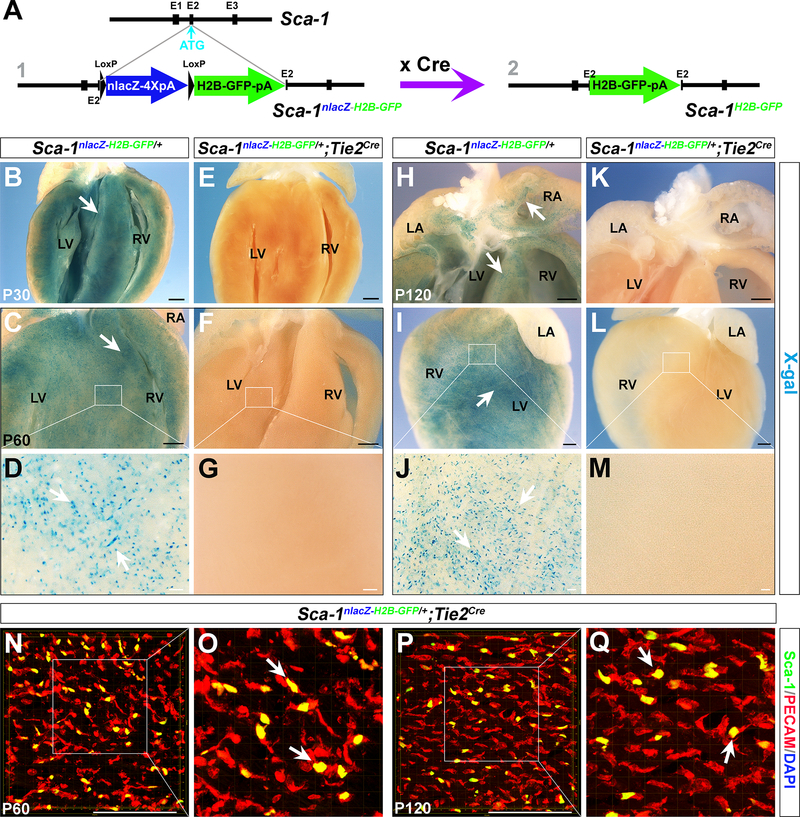
To further determine the identity of Sca-1+ cells throughout heart formation, we generated a dual-reporter mouse line Sca-1nLacZ-H2B-GFP/+ in which a LoxP-nLacZ-4XPolyA-LoxP-H2B-GFP cassette was inserted into the start codon of Sca-1 (Ly6a) through homologous recombination (Figure S2 A, B). The nLacZ cassette was flanked by two LoxP sites, and thus, Sca-1H2B-GFP expression is initiated when the nlacZ cassette is removed by Cre excision. We performed whole-mount X-gal staining on various tissues/organs from Sca-1nLacZ-H2B-GFP/+ mice. nLacZ signals were detected in the kidney, lung, spleen, thymus, intestine, and stomach, consistent with previous reports45–49 and observations of Sca-1H2B-tdTomato/+ mice (Figure S2 C-J).
We crossed Sca-1nLacZ-H2B-GFP/+ mice with endothelial-specific Tie2Cre mice54 to determine endothelial identity and to ascertain how many Sca-1+ cells are of the cardiac endothelium30, 35, 37, 55. X-gal staining was performed on the hearts of Sca-1nLacZ-H2B-GFP/+;Tie2Cre mice as well as Sca-1nLacZ-H2B-GFP/+ control littermates at P30–120 (15–30-day interval between stages) (Figure 2 B-M). In Sca-1nLacZ-H2B-GFP/+ hearts, we detected a vast number of Sca-1nLacZ-positive cells. However, hardly any (nearly zero) X-gal+ cells were found in Sca-1nLacZ-H2B-GFP/+;Tie2Cre hearts at any of the stages detected. We performed immunostaining on Sca-1nLacZ-H2B-GFP/+;Tie2Cre cardiac tissues with an anti-PECAM (CD31) antibody. Sca-1H2B-GFP-positive cells generated by Tie2Cre excision were co-localized with PECAM (Figure 2 N-Q). These results conclusively suggest that cardiac Sca-1+ cells are purely of the Tie2 endothelial lineage.
Figure 2. Cardiac resident Sca-1+ cells are of the Tie2 lineage.
(A) Diagram of the Sca-1nLacZ-H2B-GFP/+ reporter allele. Sca-1H2B-GFP is expressed when the nlacZ cassette is removed by Cre excision. (B-M) X-gal staining of hearts from Sca-1nLacZ-H2B-GFP/+ and Sca-1nLacZ-H2B-GFP/+;Tie2Cre/+ littermate mice at P30 (B, E), P60 (C, F) and P120 (H-L). D, G, J and M are high-magnification photomicrographs corresponding with the areas outlined in C, F, I and L (white rectangle). Numerous Sca-1nLacZ-positive cells were observed in Sca-1nLacZ-H2B-GFP/+ hearts (arrows in B, C D, H, I and J), but no X-gal-positive cells were seen in Sca-1nLacZ-H2B-GFP/+;Tie2Cre/+ hearts. (N-Q) Z-stack images of immunostaining with an anti-PECAM antibody (red) of Sca-1nLacZ-H2B-GFP/+;Tie2Cre/+ hearts at P60 (N, O) and P120 (P, Q). Sca-1H2B-GFP-positive cells co-localized with PECAM (yellow, arrows in O and Q). LA, left atria; LV, left ventricle; RA, right atria; RV, right ventricle. n=3 for each stage. Black scale bar, 1 mm. White scale bar, 100 μm.
Sca-1 does not label any cardiac precursor cells during early embryonic heart formation
Currently, the developmental origin of cardiac Sca-1+ cells remains unknown. If resident Sca-1+ cells represent a population of CSCs for myocardial renewal and repair in adulthood51, we speculate that Sca-1 could possibly be expressed in early cardiac precursors at mid-late gestation, during which cardiac progenitors from the first and second heart field, pro-epicardium/epicardium, and cardiac neural crest progressively migrate and differentiate to form a four-chambered heart with great arteries56–58. Therefore, we performed X-gal staining to search for Sca-1+ cells in E7.0-P0 hearts of Sca-1nLacZ-H2B-GFP/+ mice. In fact, Sca-1 expression was not detected in any of the early cardiogenic regions (including cardiac crescent at E7.0–7.5, first and second heart field at E8.0–9.5, pro-epicardium/epicardium at E8.0–16.5, and cardiac neural crest cells at E8.5–11.5) (Figure S3 A-H). The earliest stage with Sca-1nLacZ-positive cells was E17.5 (Figure S3 I-J), consistent with observations of Sca-1H2B-tdTomato/+ mice, in which we detected a few Sca-1H2B-tdTomato-positive cells at E17.5 but no Sca-1-expressing cells at or before E16.5 (Figure S3 M-P).
Sca-1+ cells minimally differentiate into cardiomyocytes during homeostasis and aging
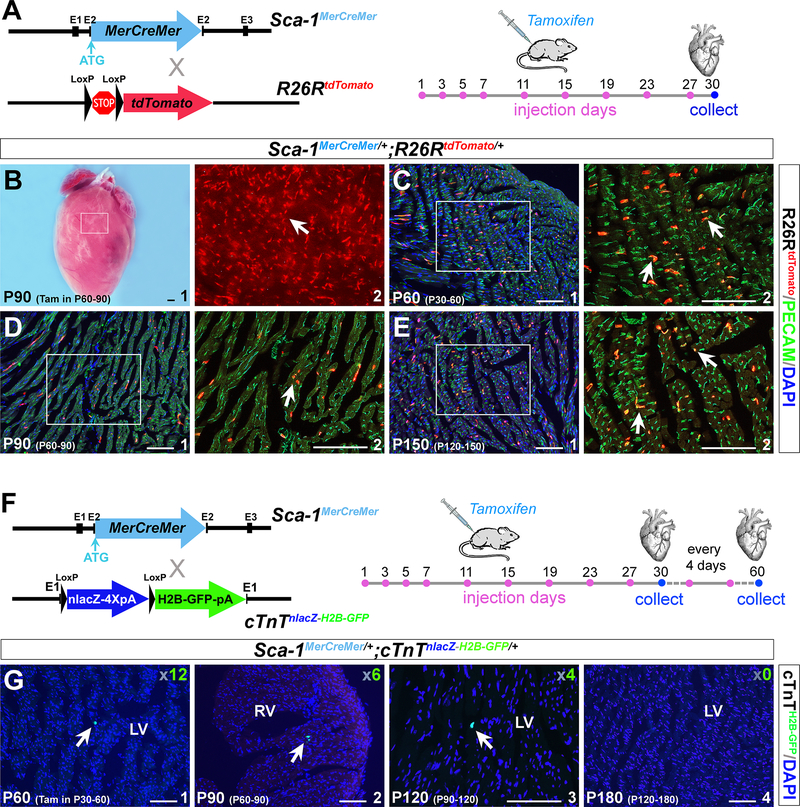
To further investigate the differentiation potential of cardiac resident Sca-1+ cells, we generated a third knock-in mouse model, Sca-1MerCreMer/+, by inserting an inducible MerCreMer cassette into the Ly6a start codon (Figure S4). Sca-1MerCreMer/+ mice were crossed with the ROSA26RtdTomato reporter41 to obtain Sca-1MerCreMer/+;ROSA26RtdTomato/+ double heterozygous mice (Figure 3 A). To examine the potential occurrence of Sca-1MerCreMer leakiness, we examined 10-month-old Sca-1MerCreMer/+;ROSA26RtdTomato/+ mouse hearts without tamoxifen induction, and tdTomato signals were not detected. Next, we treated Sca-1MerCreMer/+;ROSA26RtdTomato/+ mice with tamoxifen at P30, P60 and P120 for 1 month (on days 1, 3, 5, 7, 11, 15, 19, 23 and 27) (Figure 3 A), and ROSA26RtdTomato cells were detected throughout the hearts (Figure 3 B). Immunostaining with an anti-PECAM antibody showed that tdTomato cells are PECAM+ (Figure 3 C-E), further confirming the endothelial identity of cardiac Sca-1+ cells.
Figure 3. Sca-1+ cells in the adult heart have minimal myogenic potential.
(A) Diagram of the Sca-1MerCreMer/+ allele. Sca-1MerCreMer/+ mice were crossed with the ROSA26RtdTomato reporter to obtain Sca-1MerCreMer/+;ROSA26RtdTomato/+ double heterozygous mice. Tamoxifen was administered 9 times in one month (days 1, 3, 5, 7, 11, 15, 19, 23 and 27) to induce Sca-1MerCreMer expression. (B) A representative Sca-1MerCreMer/+;ROSA26RtdTomato/+ heart showed substantial ROSA26RtdTomato-positive cells present after tamoxifen treatment (red, arrows in B2). (C-E) Immunostaining with an anti-PECAM antibody (green) of Sca-1MerCreMer/+;ROSA26RtdTomato/+ hearts at P30, P60 and P90 after 1 month of tamoxifen treatment. ROSA26RtdTomato cells co-localized with PECAM staining in these hearts (yellow, arrows in C2, D2 and E2). (F) Diagram of the Sca-1MerCreMer/+;cTnTnLacZ-H2B-GFP/+ double heterozygous alleles. cTnT2H2B-GFP was expressed when Cre activity was specifically induced in cardiomyocytes. Hearts were collected after 1 month of tamoxifen treatment. (G) All cryosections (10 μm) of Sca-1MerCreMer/+;cTnTnLacZ-H2B-GFP/+ mouse hearts at P60, P90, P120 and P180 were examined under a microscope. Very few GFP cells were detected (arrows in G), and the total number was determined and is shown in the corner. LV, left ventricle; RV, right ventricle. Black scale bar, 1 mm. White scale bar, 100 μm.
Recent lineage tracing of cardiac Sca-1 cells in a transgenic mouse model carrying a 14-kb Sca-1 regulatory element showed that cardiac Sca-1-derived cardiomyocytes continuously contribute to myocardial replacement during aging, although the frequency is relatively low (an average of 2–5% of total cardiomyocytes at 2–18 months)51. Whether the transgenic mouse model utilized in the study represents the endogenous activity of Sca-1+ cells is unknown. Therefore, we introduced a super-sensitive cardiomyocyte-specific reporter mouse model, cTnTnlacZ-H2B-GFP/+ and crossed it with Sca-1MerCreMer/+ mice. The LoxP-nLacZ-4XPloyA-LoxP-H2B-GFP cassette was inserted into the cTnT start codon in cTnTnlacZ-H2B-GFP/+ mice40. Bright nuclear GFP (cTnTH2B-GFP) was expressed when Cre activity was present in the myocardium or myocardial precursor cells (Figure 3 F). In the absence of tamoxifen, no GFP+ cells were observed in Sca-1MerCreMer/+;cTnTnlacZ-H2B-GFP/+ hearts (data not shown). We injected tamoxifen into Sca-1MerCreMer/+;cTnTnlacZ-H2B-GFP/+ mice at P30, P60, P90 and P120. After 1–2 months of induction, cardiac tissues were collected at P60, P90, P120 and P180 (Figure 3 F). Very few cTnTH2B-GFP-positive cells were found in the cardiac sections examined: only 12, 6, 4 and 0 cells were detected in whole hearts at P60, P90, P120 and P180, respectively (Figure 3 G). To increase Sca-1MerCreMer effectiveness, we administered tamoxifen for 3 months. However, the number of cTnTH2B-GFP-positive cells did not significantly change (altogether <15 cells in the whole heart, ~0.0012% of total cardiomyocytes). These results suggest that the myogenic potential of cardiac resident Sca-1+ cells is exceedingly low or non-existent.
Sca-1+ cells retain their endothelial identity and do not differentiate into cardiomyocytes after injury
A previous report showed that transplanted cardiac Sca-1+ cells with telomerase activity have the ability to migrate to injured areas of the myocardium22. Lineage tracing in the Sca-1 transgenic mouse model also revealed that cardiac Sca-1+ cells exhibited increased cardiomyocyte differentiation potential under pathological conditions51. These observations may need further evaluation because the transplanted Sca-1+ cells and the transgenic allele-labeled Sca-1+ cells may not mimic the characteristics of endogenous cardiac Sca-1+ cells.
To investigate the differentiation potential of resident Sca-1+ cells upon injury, we ligated the left anterior descending (LAD) coronary artery of Sca-1H2B-tdTomato/+;Nkx2.5H2B-GFP/+ mice and Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+ mice at P60–150 (Figure 4 A-G). These sensitive genetic tools allowed us to precisely locate Sca-1-derived cardiomyogenic progenitors (Sca-1+/Nkx2.5+) and cardiomyocytes (Sca-1+/cTnT+) when they were present in the injured zone. Longitudinal sections of the injured heart showed decreased ventricular wall and septum thickness, suggesting acute MI (Figure 4 A). All cardiac sections of Sca-1H2B-tdTomato/+;Nkx2.5H2B-GFP/+ mice at 2, 5, 8 and 15 days post-surgery (dps) were examined, and Sca-1H2B-tdTomato-positive cells were found in the infarcted zone. However, no Sca-1H2B-tdTomato/Nkx2.5H2B-GFP double-positive cells were found (Figure 4 B-D). Moreover, we did not find any Sca-1H2B-tdTomato/cTnTH2B-GFP double-positive cells at 5, 8 and 15 dps in the injured area of Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+ hearts (Figure 4 E-G).
Figure 4. Resident Sca-1+ cells do not convert into cardiomyocytes upon injury.
(A) Longitudinal sections of Sca-1H2B-tdTomato/+ mouse hearts showing that Sca-1+ cells (arrow) are present in the injured area at 5 days post-surgery (dps). Sections were counterstained with DAPI. The infarcted region is indicated by asterisks. (B-D) Sca-1H2B-tdTomato/+;Nkx2.5H2B-GFP/+ mouse hearts were collected at 2 dps (B), 5 dps (C) and 8 dps (D). No Sca-1H2B-tdTomato (arrows in B, C and D) and Nkx2.5H2B-GFP (arrowheads in B, C and D) double-positive cells were found in the infarcted area. (E-G) Sca-1H2B-tdTomato/+;cTnTH2B-GFP/+ mouse hearts were collected at 5 dps (E), 8 dps (F) and 15 dps (G). Sca-1H2B-tdTomato cells in the injured area (arrows in E, F and G) were not co-localized with cTnTH2B-GFP cells (arrowheads in E, F and G). (H) Diagram of time points for LAD surgery and tamoxifen administration for Sca-1MereCreMer/+;cTnTnlacZ-H2B-GFP/+;ROSA26RtdTomato/+ triple heterozygous mice. (I-J) Representative whole-mount view of Sca-1MereCreMer/+;cTnTnlacZ-H2B-GFP/+;ROSA26RtdTomato/+ mouse heart at 60 dps (I). ROSA26RtdTomato-positive cells were detected throughout the heart, including the infarcted region (arrows in J3). J2 and J3 are high-magnification images of the border zone (J2) and infarcted area (J3) in J1. Asterisks in J1–3 indicate the infarcted region. (K-M) Longitudinal sections of Sca-1MereCreMer/+;cTnTnlacZ-H2B-GFP/+;ROSA26RtdTomato/+ hearts at 30 dps (K), 60 dps (L) and 120 dps (M). ROSA26RtdTomato cells were detected in the infarcted region (arrows in K-M). No cTnTH2B-GFP-positive cells were detected in the infarcted region at 30, 60 and 120 dps. M2 is the green fluorescence filter image of M1 and shows the absence of cTnTH2B-GFP-positive cells. (N-O) Sca-1H2B-GFP-positive endothelial cells in the infarcted region of Sca-1nLacZ-H2B-GFP/+;Tie2Cre hearts at 30 dps. N2 and O2 are high-magnification photomicrographs corresponding to the areas outlined in N1 and O1. Black scale bar, 1 mm. White scale bar, 100 μm.
Furthermore, we performed LAD ligation in Sca-1MerCreMer/+;cTnTnlacZ-H2B-GFP/+;ROSA26RtdTomato/+ triple heterozygous mice (3–5 months old). Tamoxifen was administered from 1 dps to 30–120 dps for continuous labeling of the Sca-1+ cells (Figure 4 H), and ROSA26RtdTomato signals were detected throughout the injured heart, including the infarcted regions, in these animals (indicating efficient induction), and exhibited enhanced density in the border zone (Figure 4 J1–2). Examination of Sca-1MerCreMer/+;cTnTnlacZ-H2B-GFP/+;ROSA26RtdTomato/+ hearts revealed a very limited number of cTnTH2B-GFP-positive cells (Figure 4 K-M). Only ~10 cells (~0.001% of total cardiomyocytes) were found in the entire heart at 30, 60, and 120 dps, and none were located in the injured area (Figure 4 K-M). These observations confirm the extremely low myogenic potential of cardiac resident Sca-1+ cells and suggest that the myogenic potential of these cells (if any) is not spontaneously stimulated under pathological conditions. In addition, we also performed LAD ligation in Sca-1nLacZ-H2B-GFP/+;Tie2Cre mice (3–6 months old) and found that Sca-1H2B-GFP-positive cells were distributed in the border zone and infarcted area at 3–30 dps, indicating the Sca-1+ cells maintain their endothelial identity upon injury (Figure 4 N-Q).
Discussion
In this study, we employed a series of new mouse models to define the nature of cardiac resident Sca-1+ cells. These high-fidelity genetic tools avoid potential artifacts from immunostaining, and provide definitive conclusions regarding the identity and potency of cardiac resident Sca-1+ cells. With these models, we determined that Sca-1 is not expressed in early cardiac precursors during embryonic heart formation. Although cardiac Sca-1+ cells are heterogeneous, they belong to the Tie2 endothelial lineage exclusively, with minimal cardiomyogenic potential under both physiological and pathological conditions. These observations challenge the long-standing dogma that resident Sca-1+ cells are intrinsic CSCs for myocardial development, renewal and repair.
Previous lineage tracing with a transgenic mouse model carrying a 14-kb Sca-1 genomic sequence detected many more Sca-1-derived myocardial cells in the adult mouse heart (~2–5% of total cardiomyocytes) than we observed in this study using a Sca-1MereCreMer mouse model carrying a complete Sca-1 genomic sequence (~0.001% of total cardiomyocytes). We presume that a partial Sca-1 genomic fragment does not recapitulate endogenous Sca-1 expression. Use of partial promoter fragments has also confounded previous attempts to lineage label c-Kit+ cells59. The extremely low number of Sca-1-derived cardiomyocytes may not even be functionally essential to the heart. These cells may arise from rare, sporadic, and transient Sca-1 expression barely detected in the Sca-1 reporter mice (Sca-1H2B-tdTomato).
Previous studies have also suggested that two types of Sca-1+ cells exist in the mouse heart: Sca-1+/CD31− and Sca-1+/CD31+30, 35, 37, 55. With Sca-1nLacZ-H2B-GFP/+;Tie2Cre mice, we found that all the cardiac resident Sca-1+ cells are of the Tie2 lineage. Endothelial cells are known to be heterogeneous with mixed expression of distinct endothelial markers60–62. Whether Tie2 labels a slightly larger cardiac endothelial population than CD31 in the adult mouse heart remains unknown. This requires further investigation in the future.
Over the past 15 years, reports from various groups have repeatedly demonstrated the myogenic potency of cardiac Sca-1+ cells in vitro. Although our study conclusively suggests that endogenous cardiac Sca-1+ cells do not primarily convert to cardiomyocytes, our results are not in opposition to the multipotency of these cells when they are cultured in vitro. We speculate that exogenous expansion of cardiac Sca-1+ cells with specific media and factors may “reprogram” them and significantly change their nature. The induced multipotency of exogenously expanded Sca-1+ cells does not confirm their cardiomyogenic potential in vivo. Indeed, a study that used transplantation of Sca-1+ cells to the injured heart showed very low efficiency of myocardial conversion35. Recent study by Ye et al. in which various types of cells (including BM cells, cardiospheres, cardiosphere-derived Sca-1+/CD45− cells, and human embryonic stem cell-derived cardiomyocytes) were delivered to ischemic hearts revealed that all these cells, regardless of their type or origin, almost equivalently reduced infarct size and improved cardiac function63. Based on these observations and the endothelial nature of cardiac Sca-1+ cells, we believe the major beneficial effects of transplanted Sca-1+ cells to injured hearts are not due to cardiomyogenic potential. In the future, it will be interesting to investigate whether and how the paracrine effects and/or neovascularization of Sca-1+ cells contribute to heart repair given their endothelial identity.
Supplementary Material
Clinical Perspective:
What is new?
We show that Sca-1 does not label cardiac stem cells in the embryonic or adult mouse hearts.
Cardiac Sca-1+ cells are purely of the Tie2 endothelial lineage.
Resident Sca-1+ cells rarely contribute to cardiomyocytes during normal aging and after injury.
What are the clinical implications?
The identity of cardiac Sca-1+ cells is endothelium.
Mechanisms of transplanted Sca-1+ cells in heart repair need to be reevaluated
Acknowledgments:
The authors thank Dr. Kevin Kelly in the Transgenic Core at Mount Sinai for generating mouse models, Sakib Ahmed and Nadia Hossain for their help in histology experiments. C.L.C. and L.Z. designed the study and wrote the paper; L.Z., N.S., J.Y., F.Y., F.C., E.C., Q.D., M.X, and L.B. performed the primary experiments and analyzed the data; L.Z. (Zangi) helped to perform the experiments and provided technical assistance.
Sources of Funding: C.L.C. is supported by the National Institutes of Health (1R01HL131735, 1R56HL129807, 1R01HL095810 and 1R01HL137036) and American Heart Association (15GRNT25710153 and 0855808D). J.Y. is supported by the National Natural Science Foundation of China (81470488, 81770280 and 81728004).
Footnotes
Conflicts of Interest Disclosures: None.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Passier R, van Laake LW and Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. [DOI] [PubMed] [Google Scholar]
- 2.Segers VF and Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. [DOI] [PubMed] [Google Scholar]
- 3.Laflamme MA and Murry CE. Heart regeneration. Nature. 2011;473:326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ptaszek LM, Mansour M, Ruskin JN and Chien KR. Towards regenerative therapy for cardiac disease. Lancet. 2012;379:933–942. [DOI] [PubMed] [Google Scholar]
- 5.van Berlo JH and Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med. 2014;20:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B and Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. [DOI] [PubMed] [Google Scholar]
- 7.Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD, Henning BJ, Stirparo GG, Papait R, Scarfo M, Agosti V, Viglietto G, Condorelli G, Indolfi C, Ottolenghi S, Torella D and Nadal-Ginard B. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. [DOI] [PubMed] [Google Scholar]
- 8.Jesty SA, Steffey MA, Lee FK, Breitbach M, Hesse M, Reining S, Lee JC, Doran RM, Nikitin AY, Fleischmann BK and Kotlikoff MI. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci U S A. 2012;109:13380–13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaruba MM, Soonpaa M, Reuter S and Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molkentin JD and Houser SR. Are resident c-Kit+ cardiac stem cells really all that are needed to mend a broken heart? Circ Res. 2013;113:1037–1039. [DOI] [PubMed] [Google Scholar]
- 11.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E and Molkentin JD. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultana N, Zhang L, Yan J, Chen J, Cai W, Razzaque S, Jeong D, Sheng W, Bu L, Xu M, Huang GY, Hajjar RJ, Zhou B, Moon A and Cai CL. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun. 2015;6:8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Yang R, Huang X, Zhang H, He L, Zhang L, Tian X, Nie Y, Hu S, Yan Y, Zhang L, Qiao Z, Wang QD, Lui KO and Zhou B. Genetic lineage tracing identifies in situ Kit-expressing cardiomyocytes. Cell Res. 2016;26:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Rijn M, Heimfeld S, Spangrude GJ and Weissman IL. Mouse hematopoietic stem-cell antigen Sca-1 is a member of the Ly-6 antigen family. Proc Natl Acad Sci U S A. 1989;86:4634–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palfree RG and Hammerling U. Biochemical characterization of the murine activated lymphocyte alloantigen Ly-6E.1 controlled by the Ly-6 locus. J Immunol. 1986;136:594–600. [PubMed] [Google Scholar]
- 16.Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y and Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- 17.Xin L, Lawson DA and Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:6942–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asakura A, Seale P, Girgis-Gabardo A and Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clayton E and Forbes SJ. The isolation and in vitro expansion of hepatic Sca-1 progenitor cells. Biochem Biophys Res Commun. 2009;381:549–553. [DOI] [PubMed] [Google Scholar]
- 20.McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ and Bertoncello I. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells. 2009;27:623–633. [DOI] [PubMed] [Google Scholar]
- 21.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM and Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. [DOI] [PubMed] [Google Scholar]
- 22.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML and Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100:12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H and Komuro I. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. [DOI] [PubMed] [Google Scholar]
- 24.Matsuura K, Honda A, Nagai T, Fukushima N, Iwanaga K, Tokunaga M, Shimizu T, Okano T, Kasanuki H, Hagiwara N and Komuro I. Transplantation of cardiac progenitor cells ameliorates cardiac dysfunction after myocardial infarction in mice. J Clin Invest. 2009;119:2204–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tateishi K, Ashihara E, Takehara N, Nomura T, Honsho S, Nakagami T, Morikawa S, Takahashi T, Ueyama T, Matsubara H and Oh H. Clonally amplified cardiac stem cells are regulated by Sca-1 signaling for efficient cardiovascular regeneration. J Cell Sci. 2007;120:1791–1800. [DOI] [PubMed] [Google Scholar]
- 26.Takamiya M, Haider KH and Ashraf M. Identification and characterization of a novel multipotent sub-population of Sca-1(+) cardiac progenitor cells for myocardial regeneration. PLoS One. 2011;6:e25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J, Boyle A, Shih H, Sievers RE, Zhang Y, Prasad M, Su H, Zhou Y, Grossman W, Bernstein HS and Yeghiazarians Y. Sca-1+ cardiosphere-derived cells are enriched for Isl1-expressing cardiac precursors and improve cardiac function after myocardial injury. PLoS One. 2012;7:e30329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, Goetsch SC, Gallardo TD and Garry DJ. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. [DOI] [PubMed] [Google Scholar]
- 29.Nagai T, Matsuura K and Komuro I. Cardiac side population cells and Sca-1-positive cells. Methods Mol Biol. 2013;1036:63–74. [DOI] [PubMed] [Google Scholar]
- 30.Pfister O, Mouquet F, Jain M, Summer R, Helmes M, Fine A, Colucci WS and Liao R. CD31- but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. [DOI] [PubMed] [Google Scholar]
- 31.Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, Takahashi T, Goto M, Mikami Y, Yasuda N, Akazawa H, Uezumi A, Takeda S and Komuro I. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, Rashidianfar A, Biben C, Zoellner H, Colvin EK, Pimanda JE, Biankin AV, Zhou B, Pu WT, Prall OW and Harvey RP. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 2011;9:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smits AM, van Vliet P, Metz CH, Korfage T, Sluijter JP, Doevendans PA and Goumans MJ. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4:232–243. [DOI] [PubMed] [Google Scholar]
- 34.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G and Marban E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Hu Q, Nakamura Y, Lee J, Zhang G, From AH and Zhang J. The role of the sca-1+/CD31- cardiac progenitor cell population in postinfarction left ventricular remodeling. Stem Cells. 2006;24:1779–1788. [DOI] [PubMed] [Google Scholar]
- 36.Laflamme MA and Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. [DOI] [PubMed] [Google Scholar]
- 37.Liang SX, Tan TY, Gaudry L and Chong B. Differentiation and migration of Sca1+/CD31- cardiac side population cells in a murine myocardial ischemic model. Int J Cardiol. 2010;138:40–49. [DOI] [PubMed] [Google Scholar]
- 38.Valente M, Nascimento DS, Cumano A and Pinto-do OP. Sca-1+ cardiac progenitor cells and heart-making: a critical synopsis. Stem Cells Dev. 2014;23:2263–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Nomura-Kitabayashi A, Sultana N, Cai W, Cai X, Moon AM and Cai CL. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev Biol. 2014;390:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J, Zhang L, Sultana N, Oh JG, Wu B, Hajjar RJ, Zhou B and Cai CL. A series of robust genetic indicators for definitive identification of cardiomyocytes. J Mol Cell Cardiol. 2016;97:278–285. [DOI] [PubMed] [Google Scholar]
- 41.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES and Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton TG, Klinghoffer RA, Corrin PD and Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT and Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC and Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blake PG, Madrenas J and Halloran PF. Ly-6 in kidney is widely expressed on tubular epithelium and vascular endothelium and is up-regulated by interferon gamma. J Am Soc Nephrol. 1993;4:1140–1150. [DOI] [PubMed] [Google Scholar]
- 46.Flanagan K, Modrusan Z, Cornelius J, Chavali A, Kasman I, Komuves L, Mo L and Diehl L. Intestinal epithelial cell up-regulation of LY6 molecules during colitis results in enhanced chemokine secretion. J Immunol. 2008;180:3874–3881. [DOI] [PubMed] [Google Scholar]
- 47.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT and Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. [DOI] [PubMed] [Google Scholar]
- 48.Hanson P, Mathews V, Marrus SH and Graubert TA. Enhanced green fluorescent protein targeted to the Sca-1 (Ly-6A) locus in transgenic mice results in efficient marking of hematopoietic stem cells in vivo. Exp Hematol. 2003;31:159–167. [DOI] [PubMed] [Google Scholar]
- 49.Holmes C and Stanford WL. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. [DOI] [PubMed] [Google Scholar]
- 50.Noseda M, Harada M, McSweeney S, Leja T, Belian E, Stuckey DJ, Abreu Paiva MS, Habib J, Macaulay I, de Smith AJ, al-Beidh F, Sampson R, Lumbers RT, Rao P, Harding SE, Blakemore AI, Jacobsen SE, Barahona M and Schneider MD. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat Commun. 2015;6:6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida S, De Gaspari P, Kostin S, Jenniches K, Kilic A, Izumiya Y, Shiojima I, Grosse Kreymborg K, Renz H, Walsh K and Braun T. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Reports. 2013;1:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaner NC, Steinbach PA and Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. [DOI] [PubMed] [Google Scholar]
- 53.Challen GA and Goodell MA. Promiscuous expression of H2B-GFP transgene in hematopoietic stem cells. PLoS One. 2008;3:e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA and Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. [DOI] [PubMed] [Google Scholar]
- 55.Liang SX, Khachigian LM, Ahmadi Z, Yang M, Liu S and Chong BH. In vitro and in vivo proliferation, differentiation and migration of cardiac endothelial progenitor cells (SCA1+/CD31+ side-population cells). J Thromb Haemost. 2011;9:1628–1637. [DOI] [PubMed] [Google Scholar]
- 56.Vincent SD and Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. [DOI] [PubMed] [Google Scholar]
- 57.Epstein JA. Franklin H Lecture Epstein. Cardiac development and implications for heart disease. N Engl J Med. 2010;363:1638–1647. [DOI] [PubMed] [Google Scholar]
- 58.Xin M, Olson EN and Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molkentin JD and Houser SR. Response to Torella et al. Circ Res. 2014;114:e27. [DOI] [PubMed] [Google Scholar]
- 60.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorin E and Shreeve SM. Heterogeneity of vascular endothelial cells in normal and disease states. Pharmacol Ther. 1998;78:155–166. [DOI] [PubMed] [Google Scholar]
- 62.Garlanda C and Dejana E. Heterogeneity of endothelial cells. Specific markers. Arterioscler Thromb Vasc Biol. 1997;17:1193–202. [DOI] [PubMed] [Google Scholar]
- 63.Ye J and Yeghiazarians Y. Cardiac stem cell therapy: Have we put too much hype in which cell type to use? Heart Fail Rev. 2015;20:613–619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.