Abstract
Objectives
This study evaluated and compared a variety of active learning strategies, including a novel strategy we proposed, as applied to the task of filtering incorrect semantic predications in SemMedDB.
Materials and methods
We evaluated 8 active learning strategies covering 3 types—uncertainty, representative, and combined—on 2 datasets of 6,000 total semantic predications from SemMedDB covering the domains of substance interactions and clinical medicine, respectively. We also designed a novel combined strategy called dynamic β that does not use hand-tuned hyperparameters. Each strategy was assessed by the Area under the Learning Curve (ALC) and the number of training examples required to achieve a target Area Under the ROC curve. We also visualized and compared the query patterns of the query strategies.
Results
All types of active learning (AL) methods beat the baseline on both datasets. Combined strategies outperformed all other methods in terms of ALC, outperforming the baseline by over 0.05 ALC for both datasets and reducing 58% annotation efforts in the best case. While representative strategies performed well, their performance was matched or outperformed by the combined methods. Our proposed AL method dynamic β shows promising ability to achieve near-optimal performance across 2 datasets.
Discussion
Our visual analysis of query patterns indicates that strategies which efficiently obtain a representative subsample perform better on this task.
Conclusion
Active learning is shown to be effective at reducing annotation costs for filtering incorrect semantic predications from SemMedDB. Our proposed AL method demonstrated promising performance.
Keywords: active machine learning, supervised machine learning, natural language processing, medical informatics, drug interactions, clinical medicine
BACKGROUND AND SIGNIFICANCE
As of February 2018, PubMed contains over 28 million citations. While this comprises a vast amount of valuable information, its storage as unstructured text makes it infeasible for researchers to utilize it effectively without automated assistance. Literature-based discovery (LBD) is an automatic method to discover hypotheses based on findings in the literature, and it has led to finding new potential treatments for diseases (1–3) and previously unknown drug–drug interactions (4–6). Instead of depending on co-occurrence of words, using semantic predications has demonstrated to improve LBD (7).
SemRep (8), developed by the National Library of Medicine’s Semantic Knowledge Representation Project, is a natural language processing (NLP) tool to extract semantic predications from MEDLINE. These predications are triplets subject entity, predicate, object entity where the subject and object entities are Unified Medical Language System (UMLS) concepts and the predicate is one of the 30 relationships defined in (9). For example, SemRep extracts the predication TGF-beta (C0040690), STIMULATES, IL-1Ra (C0245109) from the sentence “TGF-beta stimulates secretion of the IL-1Ra.” The output of SemRep applied to the entirety of MEDLINE citations comprises the Semantic MEDLINE Database (SemMedDB) (10), which totals over 90 million semantic predications as of December 31, 2017.
While semantic predications have been used in a variety of research efforts (4,11–13), SemRep’s precision is relatively low, reported in the range 0.42–0.58 (4). This limits the use of semantic predications in biomedical NLP systems as they are often incorrect. A previous study (5) showed that machine learning (ML) techniques can be employed to filter incorrect semantic predications from SemRep’s output, improving precision. However, training an ML model requires an expert-annotated dataset, which is costly to develop. Reducing the annotation cost of building such a model is imperative for using the output of SemRep in biomedical NLP tasks.
Active learning (AL) is a method for reducing the annotation cost for training statistical models. In AL, the learning algorithm chooses the order in which it sees the training data using an algorithm called a query strategy. The goal of this process is to query examples in an order such that the model achieves the best possible performance given the least amount of labeled training data, thereby reducing the total annotation cost.
AL has been well described in the general ML literature (14–18) and has been applied to biomedical and clinical text (19–24). However, the effectiveness of different AL methods varies widely across datasets and tasks (25,26). Previous studies investigate this variation by analyzing how AL affects the hypothesis space (15,27) as well as discussing how the nature of clinical text data affects the performance of different AL methods (22). Still, without a formal evaluation it is impossible to determine which AL methods perform well on the task of filtering semantic predications. We, therefore, provide here the first application and comparative evaluation of AL to semantic relationships extracted from biomedical literature. Moreover, we designed a novel AL method, dynamic β, without hand-tuned hyperparameters that achieves near-optimal performance on this task.
OBJECTIVES
Our preliminary work (28) showed the potential value of AL applied to semantic predications in biomedical literature. Expanding on this, the objectives of this study are 3-fold:
To assess the effectiveness of AL for reducing annotation cost for the task of filtering incorrect semantic predications.
To evaluate and compare query strategies and design a novel AL method that does not use hand-tuned parameters
To provide a comparative analysis of AL methods through visualization to better understand how different types of methods perform on this task.
Towards these objectives, we conducted simulated AL experiments on 2 datasets of semantic predications using 8 query strategies covering 3 query strategy types: uncertainty, representative, and combined; and evaluated each strategy against a baseline, passive learning. For the combined type, we developed an innovative query strategy, dynamic β, for dynamically computing the weight hyperparameter in an effort to obtain a more generalizable AL model. We also performed an error analysis of low middle, and high performing query strategies using a novel method for visualizing their query patterns and comparing them to their learning curves.
MATERIALS AND METHODS
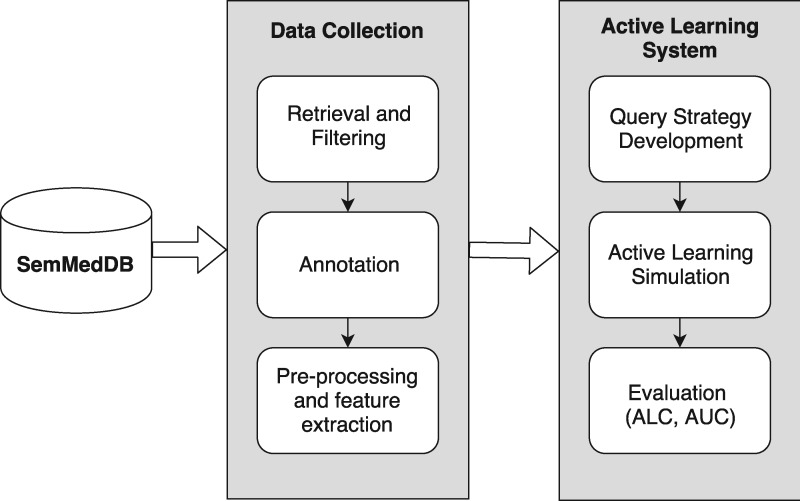
Figure 1 illustrates the development process of the AL system. We first retrieved a random subset of semantic predications from SemMedDB within the substance interactions (SI) and clinical medicine (CM) domains. These predications were annotated as either “correct” or “incorrect” by 2 health informatics experts. Features were then extracted from these examples as input to the ML algorithm. The ML task was a binary classification problem in which correct predications receive a positive label and incorrect predications receive a negative label. We used a linear support vector machine (SVM) with L2 regularization (29) as the classification algorithm, implemented using the SGDClassifier in the scikit-learn Python package (30). We then developed an AL system to simulate experiments for each query strategy. We evaluated the annotation cost of each strategy using the Area Under the Learning Curve (ALC) and the number of iterations required to reach a target Area Under the ROC Curve (AUC).
Figure 1.
An overview of the active learning system development process.
Datasets
We created 2 datasets for this study: an SI dataset and a CM dataset, each containing 3000 semantic predications. These were chosen because SI predicates describe low-level molecular phenomena, whereas CM predicates cover macro-level observable phenomena. We included the following predicates from the SemMedDB December 2016 release:
SI dataset: INTERACTS_WITH, STIMULATES, or INHIBITS. These predicates specifically describe SI according to (9). Additionally, for this group the semantic types of the subject and object entities were constrained to belong to the “Chemicals and Drugs” UMLS semantic group.
CM dataset: ADMINISTERED_TO, COEXISTS_WITH, COMPLICATES, DIAGNOSES, MANIFESTATION_OF, PRECEDES, PREVENTS, PROCESS_OF, PRODUCES, TREATS, or USES. This subset was determined to denote CM relationships by a health informatician and physician (R.R.).
For each dataset an annotation guideline was generated by the consensus of 2 annotators: a health informatician (J.V.) and a health informatician and physician (R.R.). According to this guideline, the annotators annotated a subset of 200 predications from each dataset and inter-annotator agreement was established by computing Cohen’s kappa and percentage agreement. The remaining semantic predications were then split and independently annotated to obtain the gold-standard labels for evaluation. Each semantic predication was labeled as either “correct” or “incorrect” by comparing the relation stated in the source sentence to the predication triplet and the definition of the predicate as given in the appendix of (9).
Pre-processing and feature extraction
The sentences were converted to lower case, tokenized on whitespace, and English stop words were removed. Punctuation was also removed, with the exception of hyphens in order to not split hyphenated entity names such as CCK-PZ. The features extracted consisted of tf-idf computed over the source sentences as well as the UMLS CUIs of the entities in the predication. We did not find any performance improvement using additional features such as predicate part-of-speech and argument distance. Using the ANOVA F-test, we retained the top 10% of features that explain the greatest amount of variance in the data. This resulted in 517 features for the SI dataset and 614 features for the CM dataset. This number of features was tuned to both obtain acceptable performance of the classifier and allow training and prediction to run quickly.
Active learning
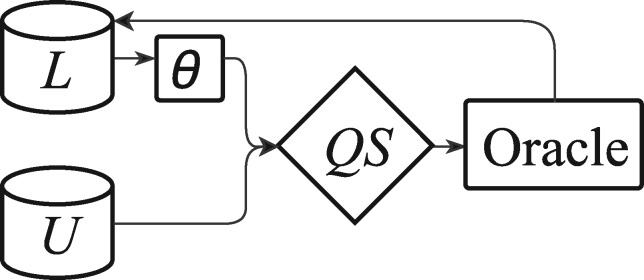
The AL system has 5 main components: a query strategy QS, an ML model θ, a pool of unlabeled data U, a pool of labeled data L, and the gold-standard or “oracle” which provides the labels for the data in U. There is also the held-out test data T upon which θ is evaluated. Training and annotation run in tandem in an iterative process in which (i) the query strategy QS chooses an example from U, (ii) the oracle is queried for the example’s label, (iii) the example is added to L, and (iv) θ is retrained on the new L. This process is illustrated in Figure 2. Additionally, θ is evaluated on T at every iteration.
Figure 2.
The active learning process. From an initial labeled set L, train the ML model θ, choose the most informative example from the unlabeled set U using the query strategy QS and the updated θ, query the oracle for its label, and update L.
We evaluated 8 query strategies covering 3 types: uncertainty, representative, and combined. These strategies are detailed below. The baseline query strategy against which each was evaluated is passive learning which, rather than making a series of informed choices as to which examples to pick from U, picks each example at random.
Uncertainty sampling
Uncertainty based query strategies operate under the assumption that the most informative examples are those closest to the decision boundary of the model θ. The uncertainty sampling methods used here are simple margin, least confidence, and least confidence with dynamic bias.
Simple margin (SM): SM sampling (15) queries the least certain example from U by measuring each example’s distance to the separating hyperplane. For this reason, simple margin is restricted to SVM models. The chosen example from U is computed by (1).
| (1) |
Where is the decision function of the SVM.
Least confidence (LC): The LC strategy (20) chooses the example from U whose posterior probability given the ML model is closest to 0.5. This is computed by (2).
| (2) |
Where is the most probable class for example under the model. As shown in (18), in the case of binary classification LC is equivalent to the other uncertainty sampling methods margin sampling and entropy sampling. For this reason, these methods are not included in this study.
Least Confidence with Dynamic Bias (LCB2): In LC the class distribution of L can become imbalanced resulting in a poor prediction model. LCB2 (19) corrects for this by introducing the term which compensates for class imbalance. Equation 2 is updated as shown in (3).
| (3) |
Where
, the linear combination of the uncertainty term and the bias term .
, the weight of uncertainty: the ratio of the size of the current labeled set L to the size of the initial unlabeled set U0.
, the weight of the bias.
is the proportion of positive to negative labels in L.
The uncertainty term represents how certain the system is that the current class distribution (represented by pp) is representative of the true class distribution. When L is large relative to U0, this certainty is high. In this case the query strategy should compensate less for any class imbalance. Thus, the influence of the bias term is inversely proportional to the progress of the AL system and diminishes as L increases.
LC and LCB2 both require posterior probabilities from the classifier. Platt scaling (31) was used to obtain posterior probabilities from the SVM for these 2 strategies.
Representative sampling
Uncertainty sampling strategies may result in a labeled set distribution that is very different from the true distribution. In other words, the system may get “stuck” modeling one area of the data. Representative strategies, on the other hand, aim to keep the distributions of the labeled and unlabeled sets similar to ensure the ML model generalizes well to the test data. They do this by using distance and similarity metrics to choose examples that are spread across the data distribution. We used Euclidean distance in all of our representative sampling experiments.
Distance to Center (D2C): The distance to center strategy (19) aims to choose from U the examples most dissimilar from those in L. It is given by (4).
| (4) |
Where ⋅) is a vector distance measure and is the mean vector across samples in L.
Density: Rather than choosing the example with the greatest distance from the average in L, as D2C does, density sampling, adapted from (17), chooses the example with the greatest average distance from every other in U. It is given by (5).
| (5) |
Density sampling thus focuses on querying examples that are representative of U, rather than examples that are not representative of L.
Min-Max: Min-Max sampling was originally developed for AL applied to semi-supervised clustering tasks (16,32). Like D2C, this method obtains a representative sample from U by choosing points that are dissimilar from those in L. The difference lies in how the dissimilarity is computed. Whereas D2C measures the distance of an example to the mean L, Min-Max sampling computes the distance between each pair of points and chooses the example from U that has the greatest minimum distance to any other point in L. This approach is given by (6).
| (6) |
Min-Max aims to obtain a representative sample quickly by ensuring that very similar points are not queried in succession. At the time of writing, this study is the first to use Min-Max sampling for fully supervised classification.
Combined sampling
Combined strategies leverage the benefits of uncertainty and representative query strategies to outweigh the pitfalls of both. A combined strategy thus aims to choose the example that is relatively uncertain while still being representative of the unlabeled set.
Information density (ID): ID sampling (17) balances informativeness and representativeness by combining the scores output by query strategies of both types into a single score. This is shown in (7).
| (7) |
Where is the uncertainty sampling score for and is the representative sampling score for . β is a hyperparameter that weights the representative sampling score. In our implementation, and are scaled to the interval [0, 1] to ensure consistent behavior of β. In this study, we used LCB2 as the uncertainty sampling strategy and Min-Max as the representative sampling strategy for the ID sampling experiments, these being the best performing strategies from each type.
Dynamic β: There are two things to note about the early stages of the AL process, when L is small and U is large: (i) it is unlikely that L is representative of U; (ii) given that L is small and unrepresentative, the prediction model trained on L is likely to be poor. Therefore, it is crucial to make L more representative early in the AL process, while later it is more important to fine-tune the decision boundary. These points motivated the development of dynamic β, which adjusts the weight of the representative sampling score in (7) according to the progress of the AL system. The definition of β in equation 7 is updated to (8).
| (8) |
Where |U| is the size of the current unlabeled set and |L| is the size of the current labeled set.
Experiments and evaluation
We used 10-fold cross validation to evaluate each query strategy. Ten percent (300) of the examples comprised the test fold and the remaining 2700 examples comprised the training fold. Ten percent (270) of the training examples were randomly selected for the initial labeled set L0 while the remaining 2430 comprised the initial unlabeled set U0. Each time L was updated and the ML model was retrained, the model was evaluated on the test data T using AUC as the performance metric. As the performance of the classifier is dependent upon how the data is initially split into L0and U0, we ran each experiment ten times with different initializations of L0and U0and averaged the AUC scores at each update of L.
The AL system was evaluated using 2 metrics: the normalized ALC as used in the active learning challenge (14) and the number of training examples required to achieve 0.80 AUC. This AUC threshold was chosen as the target because preliminary experiments found that the best performing ML classifier achieved an AUC in the 0.80–0.84 range on both datasets. Plotting the AUC as a function of the size of the labeled set produces a learning curve. The ALC is the area under this curve. The ALC is normalized using equation (9).
| (9) |
Where is the area under the learning curve given random predictions (0.5 AUC at every point on the learning curve) and is the area under the best possible learning curve (1.0 AUC at every point on the learning curve). In our experiments, ALC is computed using the full set of 2700 examples. Hereafter, ALC is taken to mean the normalized ALC in (9).
RESULTS
Inter-annotator agreement computed over 200 semantic predications for both datasets was in the “substantial agreement” range (33). Cohen’s kappa and percentage agreement on the SI and CM datasets were 0.74 and 87%, 0.72 and 91%, respectively.
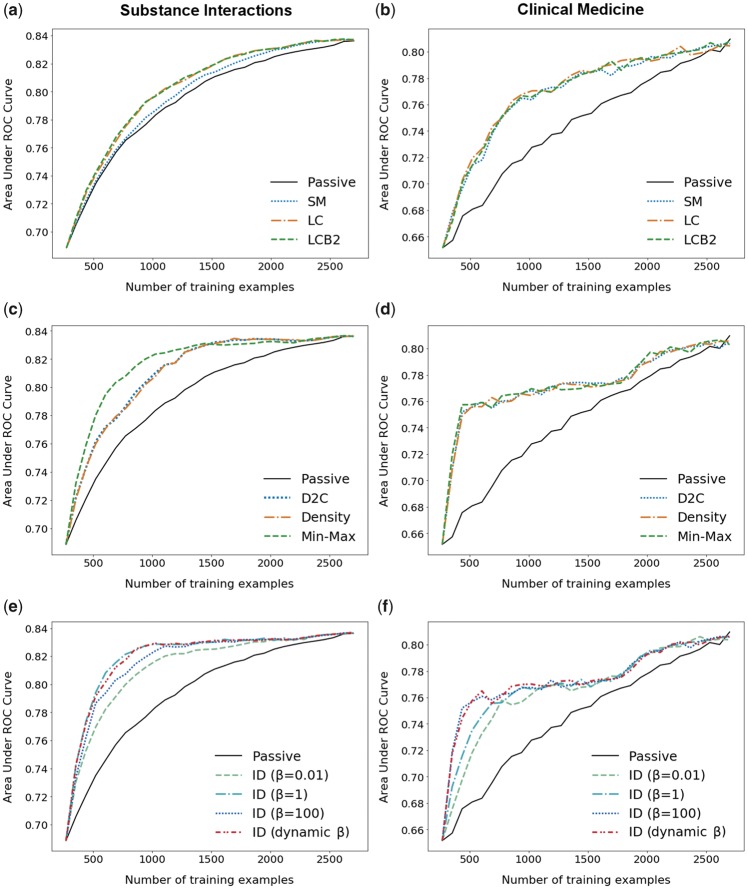
Table 1 shows the results of the simulated AL experiments on the SI and CM datasets. The learning curves for each query strategy on each dataset are given in Figure 3. All query strategies outperformed the passive learning baseline on both datasets. The representative sampling methods generally outperformed the uncertainty sampling methods in terms of ALC. However, on the CM dataset the representative-based methods required more training examples to reach 0.80 AUC than the uncertainty-based methods, largely due to a relative plateau in AUC from 500 to around 1700 training examples. The best performing query strategy on the SI dataset was ID sampling with β = 1, which outperformed the baseline by 0.052 ALC (ALC = 0.642). Additionally, the number of annotations required to reach 0.80 AUC on the SI dataset was reduced by 58% compared with the baseline. The best performing strategy on the CM datasets was tied in terms of ALC between ID sampling with β =100 and Min-Max, both of which achieved an ALC of 0.550, 0.059 greater than the baseline. Min-Max did, however, require 19 fewer annotations to reach 0.80 AUC, a reduction of 13%. Our proposed dynamic β method closely approximated (by 0.001 ALC) the learning curve of the best performing β value for both datasets, achieving comparable ALCs of 0.641 and 0.549, respectively.
Table 1.
Area under the learning curve (ALC) and number of training examples required to reach target area under the ROC curve (AUC) of the uncertainty, representative, and combined query strategies evaluated on the substance interactions and clinical medicine datasets
| Type | Query strategy | Substance interactions |
Clinical medicine |
||
|---|---|---|---|---|---|
| ALC | |L| @ 0.80 AUC | ALC | |L| @ 0.80 AUC | ||
| Baseline | Passive | 0.590 | 1295 | 0.491 | 2473 |
| SM | 0.597 | 1218 | 0.541 | 2093 | |
| Uncertainty | LC | 0.606 | 1051 | 0.543 | 2043 |
| LCB2 | 0.607 | 1060 | 0.542 | 2089 | |
| D2C | 0.623 | 891 | 0.548 | 2166 | |
| Representative | Density | 0.622 | 905 | 0.547 | 2136 |
| Min-Max | 0.634 | 657 | 0.550 | 2127 | |
| Combined | ID (β = 0.01) | 0.626 | 771 | 0.534 | 2157 |
| ID (β =1) | 0.642 | 546 | 0.542 | 2146 | |
| ID (β = 100) | 0.635 | 653 | 0.550 | 2174 | |
| ID (dynamic β)a | 0.641 | 587 | 0.549 | 2180 | |
Bold values indicate the best performing method for that metric.
Novel algorithm.
Figure 3.
Average area under the ROC curve (AUC) learning curves for the uncertainty-based, representative-based, and combined query strategy types for the substance interactions and clinical medicine datasets. Rows correspond to query strategy types. Columns correspond to the datasets.
DISCUSSION
We have shown that AL is able to reduce the number of annotations required for this task by 749 (58%) in the best case. As the annotators for this task averaged around 100 annotations per hour, this amounts to a full work-day of annotation time. Additionally, the ID strategy achieves the best ALC on both datasets and our proposed method, dynamic β, shows promising ability to approximate the learning curves of the best performing query strategy. These strategies could thus reduce annotation cost when used in other AL tasks by removing the need to manually choose the query strategy type or the β value, which the results show can dramatically influence performance. Nevertheless, it is necessary to understand how to best apply AL in order to reap its benefits. To contribute to this understanding, we present comparative visualization of the AL strategies used in this study.
Comparative analysis of query patterns
Overall, we observed that the representative and combined sampling methods outperformed the uncertainty sampling methods on both datasets, largely due to a difference in slope of the learning curves in the early stages of AL. We hypothesized that this difference is due to the data distribution and how the query strategies pick the next example from U. Uncertainty sampling methods rely entirely on the current model trained on L to compute the informativeness of the examples in U. When L is small, the prediction model is likely to be poor, yet uncertainty sampling will choose examples close to the decision boundary, reinforcing it. The result is a model that converges slowly to the optimal decision boundary for the dataset. Representative sampling, on the other hand, finds a good decision boundary quickly by ensuring that L (and the model trained on it) generalizes to U.
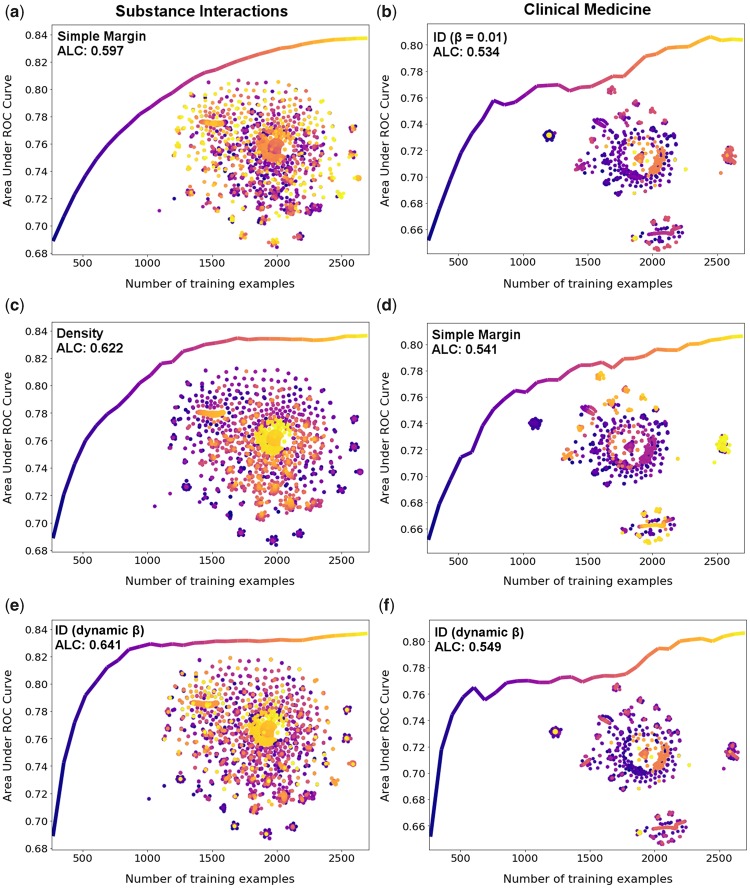
To investigate this performance discrepancy, we compared the learning curves and query patterns of three query strategies for each dataset, including dynamic β, stratified by their type and performance. We logged the orders in which examples were chosen from U by each query strategy. U was then transformed using t-Distributed Stochastic Neighbor Embedding (t-SNE) (34) to 2 dimensions for visualization. Overlaying this visualization with a heat map corresponding to the order in which examples were chosen shows how trends differ by query strategy (Figure 4).
Figure 4.
Query patterns of the low, middle, and high performing query strategies for the substance interactions and clinical medicine datasets overlaid on a visualization of U generated using t-SNE along with their corresponding learning curves. Dark blue corresponds to the first examples queried. Yellow corresponds to the last examples queried. Columns correspond to the substance interactions and clinical medicine datasets, respectively. Rows from the top correspond to the low, middle, and high performing query strategies, respectively.
The low performing strategies, SM (Figure 4a) and ID β =0.01 (Figure 4b), exhibit different trends across the datasets. Both strategies first sample data around the middle of the distribution. However, there is little improvement over the baseline on the SI dataset in the early stages. This indicates that the first points queried by SM from the SI dataset are not informative for the model, reinforcing the aforementioned point of how uncertainty sampling strategies can become “stuck” reinforcing a sub-optimal decision boundary. On the other hand, ID β =0.01 on the CM dataset achieves a large deviation from the baseline in the early stages, indicating that these points are informative for the model.
These indications are supported by the query patterns of the middle (Figure 4c and 4d) and high (Figure 4e and 4f) performing strategies. On the SI dataset, these strategies choose examples on the outer edge of the data first (density sampling, Figure 4c) or sample a relatively even spread (ID dynamic βFigure 4e) in the early stages. As these strategies obtain a large deviation from the baseline learning curve early on, we infer that the most informative examples in the SI dataset are spread around the outer portions of the distribution, rather than around the center where SM focuses. On the CM dataset, the query patterns of the middle (SM, Figure 4d) and high (ID dynamic β, Figure 4f) performing strategies are similar to the low performing strategy in the early stages, i.e. they choose examples around the center and in the far-left cluster first. As all three strategies achieve improvements over the baseline in the early stages of AL, we infer that these examples are informative for the model.
Overall, the query patterns indicate that strategies which quickly obtain an L that is representative of U perform best on this task. The middle and high performing strategies on the SI dataset (Figure 4c and 4e) obtain a representative subset by 1000 examples, whereas the low performing strategy (Figure 4a) samples the outer portions of the distribution only in the later stages of AL. Also, on the CM dataset the strategies with the steepest initial learning curves (ID β = 1 and ID β = dynamic, Figure 4b and 4f) sample from the outer portions of the distribution earlier than SM (Figure 4d).
The improvement of the ID dynamic β = dynamic strategy over the density strategy on the SI dataset is due to the efficiency in which the ID strategy obtains a representative subsample of the data. Unlike density sampling, most of the points that ID dynamic β queries last (the yellow points) lie at the center of small clusters. Given that the ID learning curve increases faster than the density curve, we conclude that these points do not greatly influence the ML model’s ability to generalize to the test data. ID dynamic β thus achieves a generalizable model by querying a representative subsample of the data more efficiently than density sampling.
Limitations and future work
Although the inter-annotator agreement was “substantial” for both datasets according to (33), we encountered issues of ambiguity during annotation. For example, take the sentence and predication “The influence of caffeine on the mitomycin C-induced chromosome aberration frequency in normal human and xeroderma pigmentosum cells” xeroderma pigmentosum, PROCESS_OF, human. Here, it is unclear whether the sentence is contrasting human cells and xeroderma pigmentosum cells or normal human cells and xeroderma pigmentosum human cells. Additionally, we noticed different levels of annotator disagreement across predicates. For example, there were 3 times more disagreements regarding the MANIFESTATION_OF and TREATS predicates than the PRODUCES and ADMINISTERED_TO predicates.
This study covered the major uncertainty and representative sampling query strategies. Still, there are numerous strategies in addition to ID sampling that aim to combine informativeness and representativeness that were not explored (25–27). Future work is to perform a more in-depth analysis of how these strategies compare and how informativeness and representativeness measures combine.
CONCLUSION
This study evaluated 8 different AL query strategies belonging to 3 different types on the task of filtering incorrect semantic predications from SemMedDB. Combined sampling methods were the most effective on both datasets, in the best case reducing the annotation cost by 58%. For the ID sampling strategy, we designed dynamic β, a method for dynamically weighting the representative sampling score, which demonstrated promising performance. We also performed a comparative analysis of the query strategies, visualizing their query patterns with respect to their learning curves and performance on this dataset.
Author’s contributions
J.V. and R.Z. conceived the study idea and design. J.V. retrieved the data and did the programming. J.V. and R.R. annotated the corpus. All authors participated in writing and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by National Center for Complementary & Integrative Health Award (#R01AT009457) (Zhang), the Agency for Healthcare Research & Quality grant (#1R01HS022085) (Melton), and the National Center for Advancing Translational Science (#U01TR002062) (Liu/Pakhomov/Jiang).
Conflict of interest statement. none declared.
ACKNOWLEDGEMENTS
The authors thank Dr. Halil Kilicoglu and Dr. Mike Cairelli for their guidance in building and annotating the datasets used in this study.
REFERENCES
- 1. Hristovski D, Kastrin A, Peterlin B, Rindflesch TC.. Combining Semantic Relations and DNA Microarray Data for Novel Hypotheses Generation. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010: 53–61. [Google Scholar]
- 2.Rastegar-Mojarad M, Elayavilli RK, Wang L, Prasad R, Liu H. Prioritizing Adverse Drug Reaction and Drug Repositioning Candidates Generated by Literature-Based Discovery. Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics (BCB ’16). New York, NY, USA: ACM; 2016: 289–296.
- 3. Kostoff RN. Literature-related discovery (LRD): introduction and background. Technol Forecast Soc Change 2008; 752: 165–185. [Google Scholar]
- 4. Zhang R, Cairelli MJ, Fiszman M, et al. Using semantic predications to uncover drug-drug interactions in clinical data. J Biomed Inform 2014; 49: 134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang R, Adam TJ, Simon G, et al. Mining biomedical literature to explore interactions between cancer drugs and dietary supplements. AMIA Jt Summits Transl Sci Proc 2015; 2015: 69–73. [PMC free article] [PubMed] [Google Scholar]
- 6. Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC.. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA Annu Symp Proc 2007; 2007: 6–10. [PMC free article] [PubMed] [Google Scholar]
- 7. Hristovski D, Friedman C, Rindflesch TC, Peterlin B.. Exploiting semantic relations for literature-based discovery. AMIA Annu Symp Proc 2006; 2006: 349–53. [PMC free article] [PubMed] [Google Scholar]
- 8. Rindflesch TC, Fiszman M.. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform 2003; 366: 462–77. [DOI] [PubMed] [Google Scholar]
- 9. Kilicoglu H, Rosemblat G, Fiszman M, Rindflesch TC.. Constructing a semantic predication gold standard from the biomedical literature. BMC Bioinformatics 2011; 12: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. SemMedDB Database Details https://skr3.nlm.nih.gov/SemMedDB/dbinfo.html Accessed January 10 2017.
- 11. Liu Y, Bill R, Fiszman M, et al. Using SemRep to label semantic relations extracted from clinical text. AMIA Annu Symp Proc 2012; 2012: 587–95. [PMC free article] [PubMed] [Google Scholar]
- 12. Rosemblat G, Resnick MP, Auston I, et al. Extending SemRep to the Public Health Domain. J Am Soc Inf Sci Technol 2013; 6410: 1963–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fathiamini S, Johnson AM, Zeng J, et al. Automated identification of molecular effects of drugs (AIMED). J Am Med Inform Assoc 2016; 234: 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guyon I, Cawley G, Dror G, Lemaire V. Results of the active learning challenge. Proceedings of Machine Learning Research - Proceedings Track, 2011; 16: 19–45. http://proceedings.mlr.press.
- 15. Kremer J, Pedersen KS, Igel C.. Active learning with support vector machines. Wires Data Mining Knowl Discov 2014; 44: 313–26. [Google Scholar]
- 16. Mallapragada PK, Jin R, Jain AK. Active query selection for semi-supervised clustering. 2008 19th International Conference on Pattern Recognition IEEE, Tampa, FL, USA; 2008: 1–4.
- 17. Settles B, Craven M. An analysis of active learning strategies for sequence labeling tasks. Presented at the Proceedings of the Conference on Empirical Methods in Natural Language Processing, Honolulu, Hawaii, 2008.
- 18. Settles B. Active learning, San Rafael, Calif.: Morgan & Claypool, 2012, pp. 1 online resource (xiii, 100 pages). http://www.morganclaypool.com/doi/abs/10.2200/S00429ED1V01Y201207AIM018. Available through Synthesis Digital Library of Engineering and Computer Science.
- 19. Chen Y, Mani S, Xu H.. Applying active learning to assertion classification of concepts in clinical text,” (in eng). J Biomed Inform 2012; 452: 265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Lasko TA, Mei Q, Denny JC, Xu H.. A study of active learning methods for named entity recognition in clinical text. J Biomed Inform 2015; 58: 11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y, Lask TA, Mei Q, et al. An active learning-enabled annotation system for clinical named entity recognition. BMC Med Inform Decis Mak 2017; 17 (S2): 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Figueroa RL, Zeng-Treitler Q, Ngo LH, Goryachev S, Wiechmann EP.. Active learning for clinical text classification: is it better than random sampling?. J Am Med Inform Assoc 2012; 195: 809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kholghi M, Sitbon L, Zuccon G, Nguyen A.. Active learning: a step towards automating medical concept extraction. J Am Med Inform Assoc 2016; 232: 289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Y, Cao H, Mei Q, Zheng K, Xu H.. Applying active learning to supervised word sense disambiguation in MEDLINE. J Am Med Inform Assoc 2013; 205: 1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du B, Wang Z, Zhang L, et al. Exploring representativeness and informativeness for active learning. IEEE Trans Cybern 2017; 471: 14–26. [DOI] [PubMed] [Google Scholar]
- 26. Huang SJ, Jin R, Zhou ZH.. Active learning by querying informative and representative examples. IEEE Trans Pattern Anal Mach Intell 2014; 3610: 1936–49. [DOI] [PubMed] [Google Scholar]
- 27. Xu Z, Yu K, Tresp V, Xu X, Wang J.. Representative Sampling for Text Classification Using Support Vector Machines. Berlin, Heidelberg: Springer Berlin Heidelberg; 2003: 393–407. [Google Scholar]
- 28. Chen X, Cairelli MJ, Sneiderman C, Rindflesch R, Pakhomov S, Melton G, Zhang R Applying active learning to semantic predications in SemMedDB. Poster presented at IEEE International Conference on Biomedical and Health Informatics, Feb 24, 2016; Las Vegas, NV, USA.
- 29. Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Presented at the Proceedings of the fifth annual workshop on Computational learning theory, Pittsburgh, Pennsylvania, USA, 1992.
- 30. Pedregosa F, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 31. Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in Large Margin Classifiers. Cambridge, MA, USA: MIT Press; 1999; 10: 61–74. [Google Scholar]
- 32. Vu V-V, Labroche N, Bouchon-Meunier B. Active learning for semi-supervised K-means clustering. Presented at the 2010 22nd IEEE International Conference on Tools with Artificial Intelligence, 2010.
- 33. Viera AJ, Garrett JM.. Understanding interobserver agreement: the kappa statistic. Fam Med 2005; 375: 360–3. [PubMed] [Google Scholar]
- 34. van der Maaten LJP, Hinton GE.. Visualizing high-dimensional data using t-SNE. J Mach Learn Res 2008; 9: 2579–605. [Google Scholar]