Abstract
A gene drive biases the transmission of one of two copies of a gene such that it is inherited more frequently than by random segregation. Highly efficient gene drive systems were recently developed in insects, which leverage the sequence-targeted DNA cleavage activity of CRISPR/Cas9 and endogenous homology directed repair mechanisms to convert heterozygous genotypes to homozygosity1–4. If implemented in laboratory rodents, similar systems would enable the rapid assembly of currently impractical genotypes that involve multiple homozygous genes (e.g., to model multigenic human diseases). However, such a system has not yet been demonstrated in mammals. Here, we utilize an “active genetic” element that encodes a guide RNA embedded in the mouse Tyrosinase gene to evaluate whether targeted gene conversion can occur when CRISPR/Cas9 is active in the early embryo or in the developing germline. Although Cas9 efficiently induces double strand DNA breaks in the early embryo and male germline, these breaks are not resolved by homology directed repair. However, Cas9 expression limited to the female germline forms double strand breaks that are resolved by homology directed repair, which copies the active genetic element from the donor to the receiver chromosome and increases its rate of inheritance in the next generation. These results demonstrate feasibility of CRISPR/Cas9-mediated systems that bias inheritance in mice, which have potential to transform the use of rodent models in basic and biomedical research.
A cross of mice that are heterozygous for each of three unlinked genes must produce 146 offspring for a 90% probability that one will be triple homozygous mutant. The likelihood decreases further if any of the three mutations are genetically linked but on opposite homologous chromosomes, since recombination events that combine alleles onto the same chromosome would be very infrequent. The cost, time, and requirement for a large number of mice to obtain a few individuals of the desired genotype are therefore prohibitive for certain complex models of multigenic evolutionary traits or human diseases, like arthritis and cancer.
Recently, CRISPR/Cas9-mediated gene drive systems were developed in Drosophila and Anopheline mosquitoes that increase the frequency of inheritance of desired alleles1–4. These utilized genetic elements, which we refer to broadly as “active genetic elements”, that can carry transgenes or orthologous sequences from other species5. Critically, an active genetic element includes a guide RNA (gRNA) and is inserted into the genome at the precise location that is targeted for cleavage by the encoded gRNA. In a heterozygous animal that also expresses the Cas9 nuclease, the gRNA targets cleavage of the wild type homologous chromosome. Genomic sequences that flank the active genetic element then resolve the double strand break (DSB) by homology directed repair (HDR), which copies the active genetic element from donor to receiver chromosome and converts the heterozygous genotype to homozygosity. The frequency of transmitting the active genetic element to the next generation is therefore greater than expected by random segregation of heterozygous alleles and is referred to as “super-Mendelian”. In addition to the potential to surmount the obstacles of assembling complex genotypes in laboratory rodents, variations of a CRISPR/Cas9-mediated system have been proposed that might suppress invasive rodent populations and/or reduce the impacts of rodent-borne disease6,7.
Despite the high efficiency observed in insects, divergence over 790 million years since their last common ancestor with mammals presents two potential obstacles to implementation of active genetics in mice; the frequency of DSB formation using genetically encoded Cas9 and gRNA and/or of HDR may preclude efficient gene conversion. The alternative DSB repair pathway, non-homologous end joining (NHEJ), frequently generates small insertions or deletions, known as “indels”, that make CRISPR/Cas9 an effective means of mutating specific sites in the genome. Although HDR of CRISPR/Cas9 induced DSBs does occur in vitro and in vivo in mammalian cells and embryos, usually from a plasmid or single stranded DNA template, NHEJ is the predominant mechanism of DSB repair in somatic cells8,9.
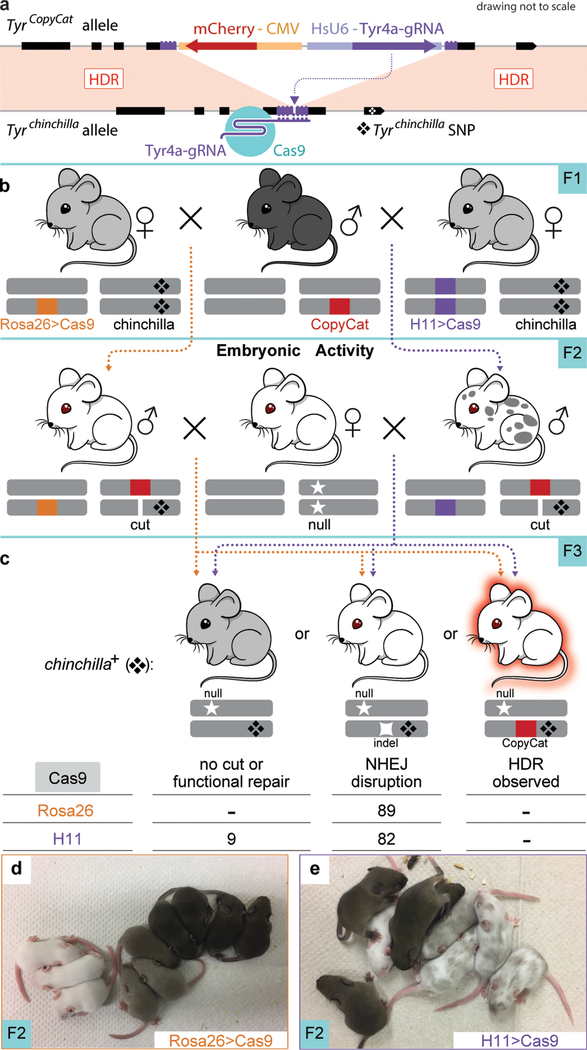
To assess the feasibility of active genetic systems in mice, we designed a “CopyCat” element10 that differs from the genetic element used initially in insects in that it cannot self-propagate because it encodes a gRNA but not the Cas9 protein (Fig. 1a). We designed our strategy to disrupt the Tyrosinase gene (Tyr), because of the obvious albino phenotype of homozygous loss-of-function animals11 and to make use of a previously characterized Tyr gRNA with high activity12. Extended Data Fig. 1 illustrates the insertion of this element precisely into the gRNA cut site in exon 4 of Tyr to obtain the TyrCopyCat knock-in allele (Supplementary Methods and Supplementary Fig. 1, 2). Briefly, the Tyr gRNA is transcribed from a constitutive human RNA polymerase III U6 promoter13. On the reverse strand of DNA, to minimize possible transcriptional conflict, mCherry is ubiquitously expressed from the human cytomegalovirus (CMV) immediate-early promoter and enhancer14. Since the 2.8 kb insert disrupts the Tyr open reading frame, TyrCopyCat is a functionally null (albino) allele that is propagated by Mendelian inheritance in absence of Cas9. Crossing mice that carry the TyrCopyCat element to transgenic mice that express Cas9 allows us to test whether it is possible to observe super-Mendelian inheritance of the TyrCopyCat allele. For this analysis, we assessed eight different genetic strategies using existing tools that provide spatial and temporal control of Cas9 expression in the early embryo and in the male and female germlines.
Figure 1 |. Embryonic Cas9 activity does not copy the TyrosinaseCopyCat allele from the donor to the receiver chromosome.
(a) The genetically encoded TyrCopyCat element, when combined with a transgenic source of Cas9, is expected to induce a DSB in the Tyrchinchilla-marked receiver chromosome, which could be repaired by inter-homologue HDR. (b) Breeding strategy to unite TyrCopyCat with a constitutive Cas9 transgene followed by test cross to Tyrnull. (c) Sum of F3 test cross offspring from five independent families for each Rosa26-Cas9 and H11-Cas9 genotype. (d) A representative of six Rosa26-Cas9 F2 litters. Black mice did not inherit TyrCopyCat. Grey mice inherited TyrCopyCat but not Cas9. White mice inherited both transgenes. (e) A representative of five F2 litters in which all offspring inherited H11-Cas9. The mosaic mice also inherited TyrCopyCat.
We employed two “constitutive” Cas9 transgenic lines, Rosa26-Cas915 and H11-Cas916, that reportedly express Cas9 in all tissues that have been assessed. Each is driven by a ubiquitous promoter and is placed in the respective Rosa26 or H11 “safe harbor” locus. In order to track inheritance of the chromosome that is targeted for gene conversion, we bred the chinchilla allele of Tyrosinase (Tyrc-ch, here simplified to Tyrch) into each Cas9 transgenic line. Tyrch encodes a hypomorphic point mutation in exon 5 that is tightly linked to the gRNA target site in exon 4. Tyrch homozygotes or heterozygotes complementing a null allele have a grey coat color, and the G to A single nucleotide polymorphism can be scored with certainty by PCR followed by DNA sequencing17 (Supplementary Fig. 4).
Female Rosa26-Cas9; Tyrch/ch and H11-Cas9; Tyrch/ch mice were each crossed to TyrCopyCat/+ males to unite the gRNA and Cas9 protein in the early embryo (Fig. 1b). In absence of a loss-of-function mutation in exon 4 of the receiver chromosome, TyrCopyCat/ch mice should appear grey (see Cas9-; TyrCopyCat/ch mice in Fig. 1d). However, we did not observe any grey Cas9+; TyrCopyCat/ch mice in the F2 offspring of either cross. Instead, all 17 of the Rosa26-Cas9; TyrCopyCat/ch mice were entirely white. Among H11-Cas9; TyrCopyCat/ch mice, 21 of the F2 progeny (87.5%) were a mosaic mixture of grey and white fur, and three mice (12.5%) were entirely white (Fig. 1d, e and Extended Data Table 1). The prevalence of mosaicism in the H11-Cas9 strategy, compared to the all-white mice produced by Rosa26-Cas9, suggests that there may be a difference in the level and/or timing of Cas9 expression driven by these two transgenes.
Our next goal was to determine what type of repair events (NHEJ mutations or gene conversions by HDR) were transmitted to the next generation. In order to assess inheritance in many offspring, we crossed each F2 male Rosa26-Cas9; TyrCopyCat/ch and H11-Cas9; TyrCopyCat/ch mouse to multiple albino CD-1 females (Fig. 1c), which carry a loss-of-function point mutation in Tyr exon 1 (Tyrc, here designated as Tyrnull)11,17. We then genotyped F3 offspring of this test cross by PCR and DNA sequencing exon 5 to identify those that inherited the Tyrch-marked receiver chromosome (Supplementary Fig. 4). In the absence of gene conversion, effectively none of these chromosomes would be predicted to also carry the TyrCopyCat allele due to separation of Tyr exons 4 and 5 by only ~9.1 kb.
Tyrch/null animals should appear grey due to partial activity of the hypomorphic Tyrch allele. However, among F3 offspring with this genotype, 100% in the Rosa26-Cas9 lineage and 90.4% in the H11-Cas9 lineage were completely white, indicating frequent transmission of a CRISPR/Cas9 induced loss-of-function mutation on the receiver chromosome and consistent with the primarily albino coat color of the F2 parents (Fig. 1c and Extended Data Table 1, 2). If the induced null alleles resulted from inter-homologue HDR to copy the TyrCopyCat allele from the donor to the Tyrch-marked receiver chromosome, these white animals should also express the fluorescent mCherry marker. In these experiments, however, none of the F3 offspring that inherited the receiver chromosome in either the Rosa26-Cas9 or H11-Cas9 lineages expressed mCherry. PCR amplification of Tyr exon 4 confirmed that the TyrCopyCat element was not present in white Tyrch/null F3 progeny (Extended Data Table 2).
The different propensities to yield full albino versus mosaic coat color patterns in the Rosa26-Cas9 and H11-Cas9 lineages were paralleled by differences in the number of unique NHEJ mutations that we identified on receiver chromosomes in individuals of each genotype. Sequenced PCR products from Rosa26-Cas9; TyrCopyCat F2 tails (somatic tissues comprised of both ectodermal and mesodermal derivatives) and from individual F3 outcross offspring (representing the germline) typically exhibited only two unique NHEJ mutations suggesting that many of these Cas9 induced mutations may have been generated in 2–4 cell stage embryos (average 2.4 alleles among offspring of five families; Extended Data Fig. 2 and Extended Data Table 3). In contrast, H11-Cas9; TyrCopyCat F2 tails and F3 offspring harbored significantly more unique NHEJ mutations (average 4.6 alleles in five families; two-tailed Student’s t-test, p=0.041), consistent with the hypothesis that Cas9 is expressed at a later embryonic stage and/or at lower levels in this lineage.
We considered two explanations for the observation that TyrCopyCat was not copied to the receiver chromosome in the early embryo. The first possibility is that homologous chromosomes are not aligned for inter-homologue HDR to repair DSBs. Second, the DNA repair machinery in somatic cells typically favors NHEJ over HDR8,9. A possible solution to overcome both potential obstacles is to restrict CRISPR/Cas9 activity to coincide with meiosis in the developing germline. During meiosis, recombination of the maternal and paternal genomes is initiated by the formation of DSBs that are repaired by exchanging regions of homologous chromosomes that are physically paired during Meiosis I18. Indeed, the molecular mechanisms of NHEJ are actively repressed to favor HDR during meiosis in many species, including mice19.
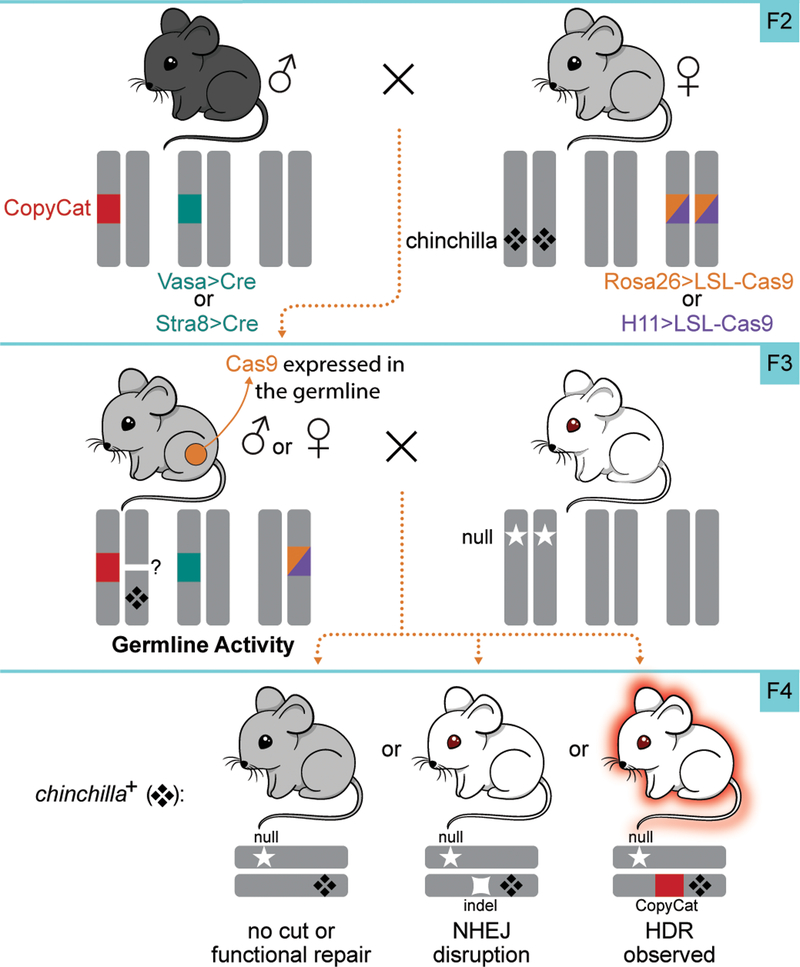
In order to test the hypothesis that CRISPR/Cas9 activity will convert a heterozygous active genetic element to homozygosity during meiosis, we designed a crossing scheme to initiate Cas9 expression during germline development in TyrCopyCat/ch mice. Since no currently available transgenic mice express Cas9 under direct control of a germline-specific promoter, we combined conditional Rosa26- or H11-LSL-Cas9 transgenes, each with a Lox-Stop-Lox preceding the Cas9 translation start site15,16, with available Vasa-Cre or Stra8-Cre germline transgenic mice. Vasa-Cre is expressed later than the endogenous Vasa transcript in both male and female germ cells20 while Stra8-Cre is limited to the male germline and is initiated in early stage spermatogonia21. Although oogonia and spermatogonia are pre-meiotic, and spermatogonia are in fact mitotic, we reasoned that Cre protein must first accumulate before Cas9 can be expressed from the LSL-Cas9 conditional allele. The possible time delay may require initiation of Cre expression before the onset of meiosis so that Cas9-induced DSBs can be resolved by inter-homologue HDR prior to segregation of homologous chromosomes at the end of Meiosis I. We generated each combination of these Cre and conditional Cas9 lines in case the timing or levels of Cas9 expression are critical variables in these crosses. We also assessed males and females of the Vasa strategies in case there are sex-dependent differences in animals that inherit the same genotype.
Males heterozygous for TyrCopyCat and the Vasa-Cre transgene were crossed to females homozygous for both the Tyrch allele and one of the two conditional Cas9 transgenes (Fig. 2). We avoided the reverse cross using female Vasa-Cre mice, since Cre protein maternally deposited in the egg20 might induce recombination of the conditional Cas9 allele and induce mutations in the early embryo similar to what we observed in the experiments using constitutive Cas9 transgenes. Introducing the Vasa-Cre transgene by inheritance from the male instead resulted in most offspring that were entirely grey, due to the TyrCopyCat/ch genotype, and a few mosaic animals (Supplementary Table 4). The presence of any mosaicism suggests that this conditional approach to restrict Cas9 expression to the germline resulted in some spurious cleavage of the Tyr locus in somatic tissues.
Figure 2 |. Breeding strategy to produce TyrCopyCat/chinchilla mice with a conditional Cas9 transgene and a germline restricted Cre transgene.
F3 offspring were test crossed to Tyrnull animals to assess F4 phenotypes and genotypes. Quantification of observed outcomes in F4 test cross offspring are presented in Table 1.
We first tested whether Cas9 in the female germline could promote copying of the TyrCopyCat element onto the receiver chromosome by crossing F3 female mice of each Vasa-Cre lineage to CD-1 (Tyrnull) males. In each cross, we identified F4 offspring that inherited the Tyrch-marked chromosome (Fig. 2). As in the crosses to assess the effects of embryonic Cas9 expression, we expected Tyrch/null mice without a loss-of-function mutation in exon 4 of the receiver chromosome would be grey. Mice with a CRISPR/Cas9 induced NHEJ mutation in exon 4 should be white. Similarly, mice carrying a CRISPR/Cas9 induced mutation that was repaired by inter-homologue HDR should also be white, but should additionally fluoresce red due to transmission of the mCherry-marked TyrCopyCat active genetic element.
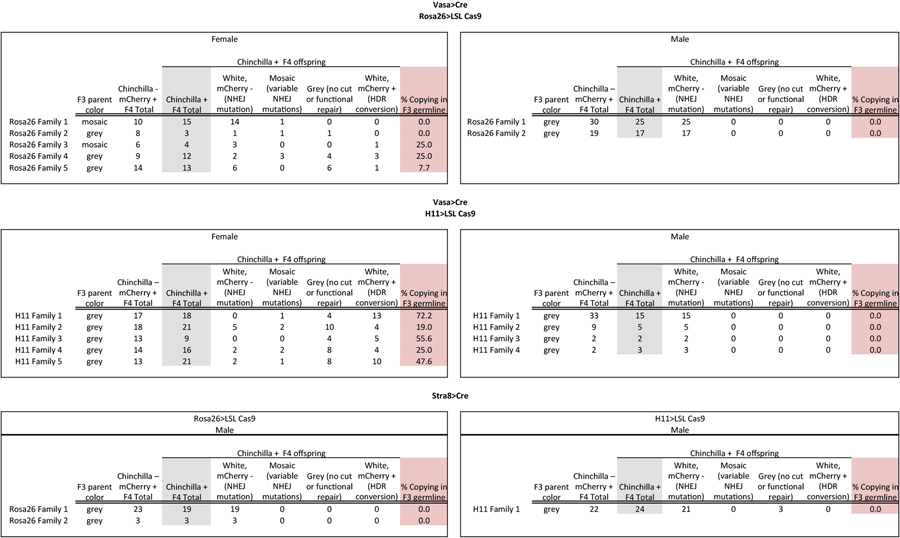
Table 1 summarizes the results of these crosses that demonstrate gene conversion upon Cas9 expression in the female germline. In contrast with early embryonic expression of Cas9, we observed that the TyrCopyCat transgene was copied to the Tyrch-marked receiver chromosome in both Vasa-Cre; Rosa26-LSL-Cas9 and Vasa-Cre; H11-LSL-Cas9 lineages. However, the observed efficiency differed between genotypes; three out of five females of the Vasa-Cre; Rosa26-LSL-Cas9 lineage and all five of five females of the Vasa-Cre; H11-LSL-Cas9 lineage transmitted a Tyrch-marked chromosome containing a TyrCopyCat insertion to at least one offspring. Although there was considerable variation among females with the same genotype, the highest observed frequency of gene conversion (72.2%) within a germline produced 13 out of 18 Tyrch offspring with a TyrCopyCat insertion in the Vasa-Cre; H11-LSL-Cas9 lineage (Table 1 and Extended Data Table 4). The probability of obtaining an animal with this genotype by natural meiotic recombination mechanisms is very low (4.7×10−5) due to ultra-tight linkage between the TyrCopyCat and Tyrch alleles. Although it seems likely that inter-homologue HDR of Cas9-induced DSBs utilizes the same DNA repair machinery that is active during meiotic recombination, these copying events cannot be explained by an increased incidence of chromosomal crossover, since all animals that inherited the donor chromosome lacking the Tyrch marker expressed mCherry (Extended Data Table 4).
Table 1:
Observed F4 outcomes of germline Cas9 strategies.
| Cre | Cas9 | F3 parent (F)emale or (M)ale |
No cut or functional repair |
NHEJ disruption |
HDR conversion |
%HDR conversion observed |
|
|---|---|---|---|---|---|---|---|
| Vasa | Rosa26 | F | 1 | - | 15 | - | - |
| 2 | 1 | 2 | - | - | |||
| 3 | - | 3 | 1 | 25% | |||
| 4 | 4 | 5 | 3 | 25% | |||
| 5 | 6 | 6 | 1 | 8% | |||
| M | 1 | - | 25 | - | - | ||
| 2 | - | 17 | - | - | |||
| H11 | F | 1 | 4 | 1 | 13 | 72% | |
| 2 | 10 | 7 | 4 | 19% | |||
| 3 | 4 | - | 5 | 56% | |||
| 4 | 8 | 4 | 4 | 25% | |||
| 5 | 8 | 3 | 10 | 48% | |||
| M | 1 | - | 15 | - | - | ||
| 2 | - | 5 | - | - | |||
| 3 | - | 2 | - | - | |||
| 4 | - | 3 | - | - | |||
| Stra8 | Rosa26 | F | 1 | - | 19 | - | - |
| 2 | - | 3 | - | - | |||
| H11 | M | 1 | 3 | 21 | - | - | |
In contrast to the 41 copying events we observed out of 132 total Tyrch/null offspring of female mice, we observed no copying in a total of 113 offspring of males where conditional Cas9 expression was induced by either Vasa-Cre or Stra8-Cre (Table 1 and Extended Data Table 4). It is possible, however, that the number of families and of offspring in each family, which was limited by unexplained low male fertility, was insufficient to detect low efficiency copying in each of four genetic strategies. If there is indeed a difference between males and females in the efficiency of TyrCopyCat copying, we can conceive of two potential explanations. First, despite equivalent genotypes in the male and female Vasa-Cre lineages, Cre, Cas9, and/or gRNA may not be well expressed in the male germline. However, the high frequency of white Tyrch/null offspring suggests that DSB formation is very efficient in males. Second, spermatogonia continually undergo mitosis and produce new primary spermatocytes throughout the life of a male in mammals22. In contrast, oogonia directly enlarge without further mitosis to form all of the primary oocytes in the embryo23. The difference in the observed efficiency of inter-homologue HDR between females and males at this locus may therefore reflect a requirement for the precise timing of CRISPR/Cas9 activity to coincide with meiosis (Fig. 3). NHEJ indels in males may result from DSB repair that occurs prior to the alignment of homologous chromosomes in Meiosis I. Similarly, the higher observed efficiency of inter-homologue HDR in females of the H11-LSL-Cas9 conditional strategy may relate to the lower or delayed Cas9 expression from the H11 locus, compared to Rosa26, that was evident in the constitutive crosses (Fig. 1d, e and Extended Data Table 1 and 3). Thus, in the Vasa-Cre; H11-LSL-Cas9 mice, DSB formation may have been fortuitously delayed to fall within a more optimal window during female meiosis.
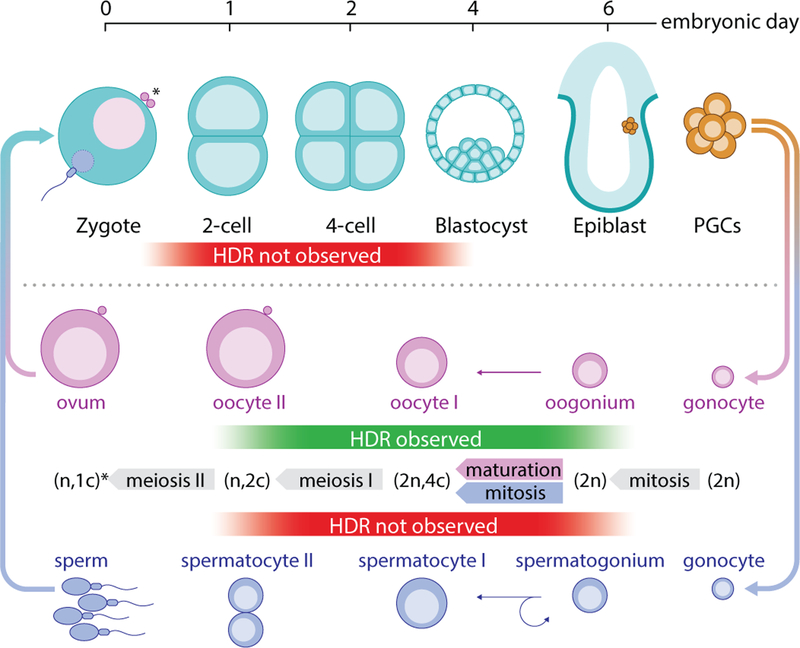
Figure 3 |. Gene conversion by an active genetic element was observed in the female germline and not in the male germline or in the early embryo.
Schematic representation of early embryonic and male and female germline development overlaid with the presence or absence of observed HDR. [PGCs: primordial germ cells, n: number of homologous chromosomes, c: chromosome copy number. Asterisk indicates the difference between male sperm (n, 1c) and female ovum, which remains (n, 2c) until second polar body extrusion after fertilization.]
In summary, we demonstrate that the fundamental mechanism of a CRISPR/Cas9-mediated gene drive is feasible in mice. However, our comparison of eight different genetic strategies indicates that precise timing of Cas9 expression may present a greater challenge in rodents than in insects in order to restrict DSB formation to a window when breaks can be efficiently repaired by the endogenous meiotic recombination machinery. These data are therefore critical to the ongoing discussion about whether CRISPR/Cas9-mediated gene drives could be used to reduce invasive rodent populations, since it appears that both the optimism and concern are likely premature. Further optimization to increase the frequency of gene conversion in both males and females and to reduce the prevalence of drive-resistant alleles (NHEJ indels that alter the gRNA target site) would be necessary to achieve rapid and sustained suppression of wild populations24–29.
Nevertheless, the copying efficiencies we observed here would be more than sufficient for a broad range of laboratory applications. For example, the average observed copying rate of 44% using the most efficient genetic strategy in females (Vasa-Cre; H11-LSL-Cas9) combined ultra-tightly linked Tyrosinase mutations such that 22.5% of all offspring inherited a chromosome with both alleles, an impossibility by Mendelian inheritance. This observed average copying rate would also be expected to increase the inheritance of a single desired allele from 50% to 72%, and the highest rate of gene conversion we observed (72.2%) would result in an 86% frequency of transmitting a desired allele. If multiple genes could be simultaneously converted to homozygosity, such high transmission frequencies that bypass the onerous constraint of genetic linkage stand to greatly accelerate the production of rodent models for a variety of complex genetic traits.
Extended Data
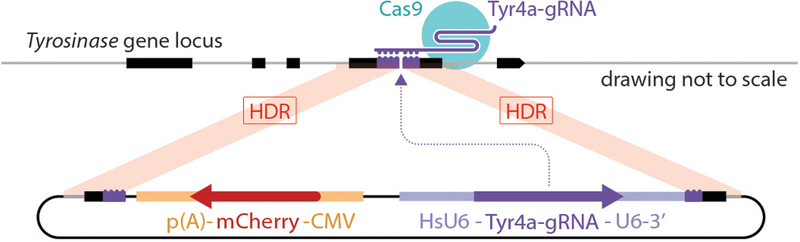
Extended data figure 1. Knock-in strategy using the TyrCopyCat targeting vector.
The U6-Tyr4a-gRNA and CMV-mCherry were inserted into the cut site of the Tyr4a-gRNA by homology directed repair after CRISPR/Cas9 DSB formation targeted by the Tyr4a-gRNA. See Supplemental Methods and Supplementary Fig. 1 and 2 for additional details.
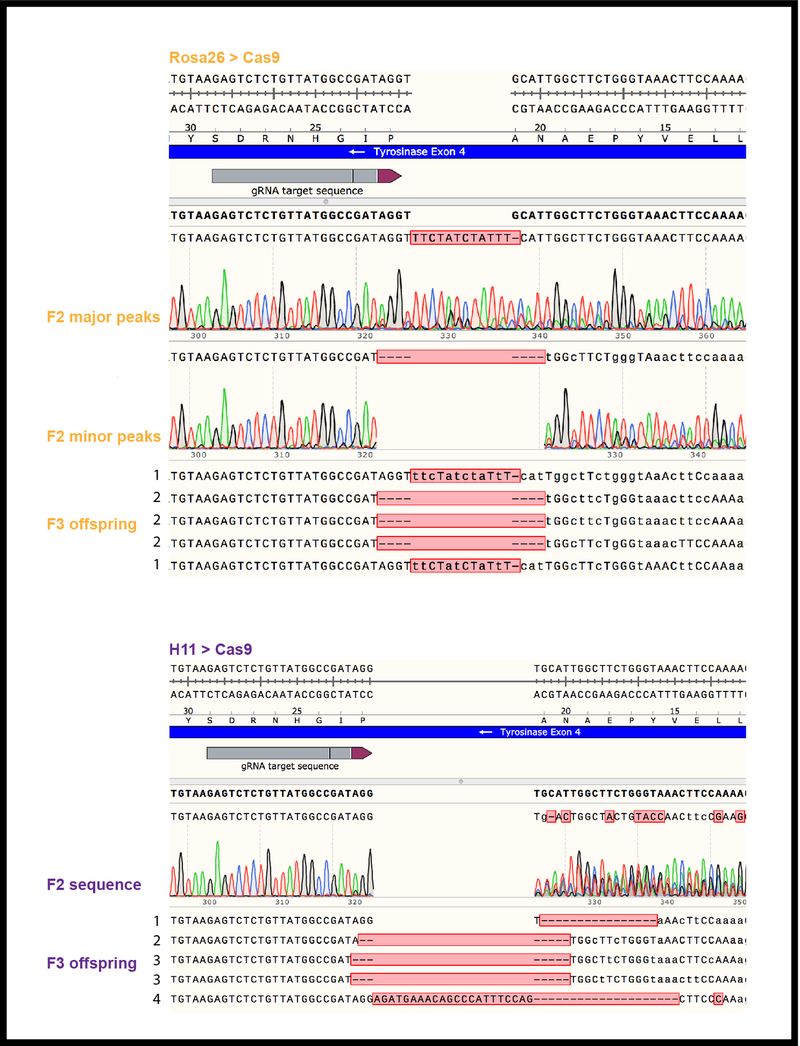
Extended data figure 2. Rosa26-Cas9 and H11-Cas9 constitutive lineages have different numbers of unique NHEJ indels.
Sanger sequencing of the Tyr4a-gRNA target exon amplified from tail tip genomic DNA using TyrHALF2 and TyrHARR2 primers as specified in Supplementary Table 3. Above: A single representative Sanger sequence trace of the bulk PCR product amplified from a Rosa26-Cas9, TyrCopyCat+ F2 animal (Rosa26 Family 1 in Extended Data Table 3) with either major or minor peaks called revealing two distinct alleles. Five Tyrchinchilla+ F3 offspring of this F2 individual each match one of the two alleles [marked 1 (insertion) and 2 (deletion)]. Below: A single representative sequence trace of the bulk PCR product amplified from an H11-Cas9, TyrCopyCat+ F2 animal (H11 Family 1 in Extended Data Table 3). Alternate alleles cannot be called because of the complexity of overlapping peaks. Five Tyrchinchilla+ F3 offspring each have one of four different alleles (marked 1, 2, 3, 4). Sequence trace data is representative of all 90 individuals total in five families of each constitutive strategy detailed in Extended Data Table 3.
Extended data table 1.
Coat color of F2 individuals that were constitutive Cas9+ and TyrCopyCat/chinchilla.
| Rosa26>Cas9 | H11>Cas9 | |
|---|---|---|
| White | 17 | 3 |
| Mosaic | 0 | 21 |
Extended data table 2.
Phenotypes and genotypes of all F3 progeny in Fig. 1c that are the offspring of a subset of F2 individuals listed in Extended Data Table 1.
| Chinchilla + F3 offspring |
||||||
|---|---|---|---|---|---|---|
| F2 parent color | Total F3 Chinchilla− |
Total F3 Chinchilla+ |
White (NHEJ mutation) |
Grey (no cut or functional repair) |
mChemy+ (HDR gene conversion) |
|
| Rosa26 Family 1 | white | 30 | 20 | 20 | 0 | 0 |
| Rosa26 Family 2 | white | 11 | 15 | 15 | 0 | 0 |
| Rosa26 Family 3* | white | 28* | 16 | 16 | 0 | 0 |
| Rosa26 Family 4 | white | 22 | 22 | 22 | 0 | 0 |
| Rosa26 Family 5 | white | 16 | 16 | 16 | 0 | 0 |
| H11 Family 1 | prim. white mosak | 14 | 15 | 15 | 0 | 0 |
| H11 Family 2 | print. grey mosak | 23 | 28 | 25 | 3 | 0 |
| H11 Family 3 | prim. white mosaic | 13 | 9 | 9 | 0 | 0 |
| H11 Family 4 | prim. white mosak | 12 | 15 | 15 | 0 | 0 |
| H11 Family 5 | mosaic | 23 | 24 | 18 | 6 | 0 |
indicates a family with offspring that were Tyrchinchilla negative and mCherry negative, suggesting a large deletion in the recipient chromosome may encompass the Tyrchinchilla SNP
“prim.” = “primarily” and refers to the coat color that covers the greatest total area when there was obviously not an equal proportion of white and grey
Extended data table 3.
Allelic complexity of the constitutive Rosa26- and H11-Cas9 families.
| Number of distinct NHEJ alleles in ‘n’ Tyrchinchilla+ offspring |
|
|---|---|
| Rosa26 Family 1 | 2 (n=9) |
| Rosa26 Family 2 | 3 (n=10) |
| Rosa26 family 3* | 2 (n=9) |
| Rosa26 Family 4 | 2 (n=9) |
| Rosa26 Family 5 | 3 (n=13) |
| H11 Family 1 | 4 (n=10) |
| H11 Family 2 | 6 (n=9) |
| H11 Family 3 | 4 (n=6) |
| H11 Family 4 | 2 (n=8) |
| H11 Family 5 | 7 (n=9) |
indicates a family with offspring that were Tyrchinchilla negative and mCherry negative, suggesting a large deletion in the recipient chromosome may encompass the Tyrchinchilla SNP. This was counted as one of the two unique NHEJ indels.
Extended data table 4.
Phenotypes and genotypes of all F4 progeny that are the offspring of a subset of F3 individuals listed in Supplementary Table 4.
 |
Supplementary Material
Acknowledgments:
We thank Kaisa Hanley for the DNA extraction protocol and Angela Green and An-Chih Chen for genotyping assistance. We thank Mai Tran for laser capture microdissection in an effort to genotype spermatogonia and Dr. Prashant Jain for assistance with fibroblast transfection. We also thank Dr. Heidi Cook-Andersen and Dr. Miles Wilkinson for conversations about mouse germline development and Dr. Luis Montoliu for discussion of the Tyrosinase locus. We thank Dr. Mark Tsuzynski for plasmids and for early support of the project. This work was funded by a Searle Scholar Award from the Kinship Foundation, a Pew Biomedical Scholar Award from the Pew Charitable Trusts, a Packard Fellowship in Science and Engineering from the David and Lucile Packard Foundation, and NIH grant R21GM129448 awarded to K.L.C. E.B. was supported by NIH grant R01GM117321, a Paul G. Allen Frontiers Group Distinguished Investigators Award, and a generous gift from the Tata Trusts in India to TIGS-UCSD and TIGS-India. H.A.G. was supported by a Ruth Stern Graduate Fellowship and by the NIH Cell and Molecular Genetics training grant T32GM724039; V.M.G. was supported by the NIH DP5OD023098.
Footnotes
Statistics and reproducibility: The five families in each of the constitutive crosses (Extended Data Table 2) are considered five independent experiments with each F3 offspring representing an early embryonic DSB repair event in the F2 parent. Each F4 offspring in Extended Data Table 4 represents a germline DSB repair event, an independent data point, and each family is considered an independent trial. No statistical method was used to predetermine sample size. Given breeding limitations, we assessed as many offspring as possible in each family and aimed to assess up to five families for each strategy. To detect gene conversion events, we used genetic linkage rather than a statistical test of inheritance greater than 50% expected by Mendelian segregation. Specifically, the receiver chromosome was marked with a SNP (Tyrch) located ~ 9.1 kb from the target site for gene conversion. Since the average genetic distance for mouse chromosome 7 is 0.52 cM/Mb30, the probability of a naturally occurring recombination event that would unite these ultra-tightly linked loci onto the same chromosome is 4.7×10−5.
Data availability: All genotyping data for F3 offspring of constitutive crosses and F4 offspring of germline conditional crosses is available at Zenodo.org with the identifier 10.5281/zenodo.2003087. Annotated sequence data for the TyrCopyCat transgene is available in Genbank with the accession number MK160997.
References:
- 1.Gantz VM & Bier E The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gantz VM et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. PNAS 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond A et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyrou K et al. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nature Biotechnology (2018). doi:10.1038/nbt.4245 [DOI] [PMC free article] [PubMed]
- 5.Gantz VM & Bier E The dawn of active genetics. Bioessays 38, 50–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould F Broadening the application of evolutionarily based genetic pest management. Evolution 62, 500–510 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Esvelt KM, Smidler AL, Catteruccia F & Church GM Emerging Technology: Concerning RNA-guided gene drives for the alteration of wild populations. eLife Sciences 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao Z, Bozzella M, Seluanov A & Gorbunova V Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyaoka Y et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Scientific Reports 6, 23549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X-RS, Gantz VM, Siomava N & Bier E CRISPR/Cas9 and active genetics-based trans-species replacement of the endogenous Drosophila kni-L2 CRM reveals unexpected complexity. eLife Sciences 6, e30281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama T et al. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res 18, 7293–7298 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen S-T et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol 393, 3–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyagishi M & Taira K U6 promoter-driven siRNAs with four uridine 3’ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol 20, 497–500 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Boshart M et al. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41, 521–530 (1985). [DOI] [PubMed] [Google Scholar]
- 15.Platt RJ et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiou S-H et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 29, 1576–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beermann F et al. Rescue of the albino phenotype by introduction of a functional tyrosinase gene into mice. EMBO J 9, 2819–2826 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keeney S, Giroux CN & Kleckner N Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Goedecke W, Eijpe M, Offenberg HH, Aalderen M van & Heyting, C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23, 194–198 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Gallardo T, Shirley L, John GB & Castrillon DH Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadate-Ngatchou PI, Payne CJ, Dearth AT & Braun RE Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Rooij DG & Grootegoed JA Spermatogonial stem cells. Curr. Opin. Cell Biol 10, 694–701 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Pepling ME From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 44, 622–632 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Burt A Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci 270, 921–928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deredec A, Burt A & Godfray HCJ The Population Genetics of Using Homing Endonuclease Genes in Vector and Pest Management. Genetics 179, 2013–2026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unckless RL, Clark AG & Messer PW Evolution of Resistance Against CRISPR/Cas9 Gene Drive. Genetics 205, 827–841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noble C, Olejarz J, Esvelt KM, Church GM & Nowak MA Evolutionary dynamics of CRISPR gene drives. Sci Adv 3, e1601964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall JM, Buchman A, Sánchez C HM & Akbari OS Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep 7, 3776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble C, Adlam B, Church GM, Esvelt KM & Nowak MA Current CRISPR gene drive systems are likely to be highly invasive in wild populations. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen-Seaman MI et al. Comparative Recombination Rates in the Rat, Mouse, and Human Genomes. Genome Res 14, 528–538 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.