Abstract
Many proteins must translocate through the protein-conducting Sec61 channel in the eukaryotic endoplasmic reticulum membrane or the SecY channel in the prokaryotic plasma membrane1,2. Proteins with hydrophobic signal sequences are first recognized by the signal recognition particle (SRP)3,4 and then moved co-translationally through the Sec61/SecY channel by the associated translating ribosome. Substrates with less hydrophobic signal sequences bypass SRP and are moved through the channel post-translationally5,6. In eukaryotic cells, post-translational translocation is mediated by the association of the Sec61 channel with another membrane protein complex, the Sec62/Sec63 complex7–9, and substrates are moved through the channel by the luminal BiP ATPase9. How the Sec62/63 complex activates the Sec61 channel for post-translational translocation is unclear. Here, we report the electron cryo-microscopy (cryo-EM) structure of the Sec complex from S. cerevisiae, consisting of the Sec61 channel and the Sec62, Sec63, Sec71, and Sec72 proteins. Sec63 causes wide opening of the lateral gate of the Sec61 channel, priming it for the passage of low-hydrophobicity signal sequences into the lipid phase, without displacing the channel’s plug domain. Lateral channel opening is triggered by Sec63 interacting with both cytosolic loops in the C-terminal half of Sec61 and trans-membrane (TM) segments in the N-terminal half of the Sec61 channel. The cytosolic Brl domain of Sec63 blocks ribosome binding to the channel and recruits Sec71 and Sec72, positioning them for the capture of polypeptides associated with cytosolic Hsp70 (ref.10). Our structure shows how the Sec61 channel is activated for post-translational protein translocation.
The Sec61 channel is formed from the multi-spanning Sec61 protein and two single-spanning proteins (called Sbh1 and Sss1 in S. cerevisiae)7,8. Sec61 and its prokaryotic homolog SecY consist of two halves that form an hourglass-shaped pore with a constriction in the middle of the membrane, a plug domain in the luminal/extracellular cavity, and a lateral gate, which opens to the surrounding lipid11–16. The idle, closed Sec61 channel is first primed for protein translocation by the binding of a channel partner, the ribosome or the Sec62/63 complex1,2. Subsequently, the translocating polypeptide inserts as a loop into the channel, with the hydrophobic part of the signal sequence moving through the lateral gate into the lipid phase, and the following segment of the polypeptide chain remaining in the channel pore13,16.
In S. cerevisiae, the Sec complex contains the trimeric Sec61 complex and the tetrameric Sec62/Sec63 complex, consisting of Sec62, Sec63, Sec71 (Sec66), and Sec72 (refs.7,8). Sec62 and Sec63 are essential for cell viability and are predicted to have two and three TMs, respectively17,18. Sec63 contains a luminal J-domain that activates the BiP ATPase to bind to the incoming substrate, preventing its back-sliding into the cytosol9. Sec71 and Sec72 are not essential19 and do not exist in higher organisms. Sec71 is a single-spanning protein that anchors the Hsp70-interacting Sec72 protein to the ER membrane20. In humans, Sec62 and Sec63 are frequently mutated or overexpressed in various cancers and Sec63 mutants can cause polycystic kidney disease21.
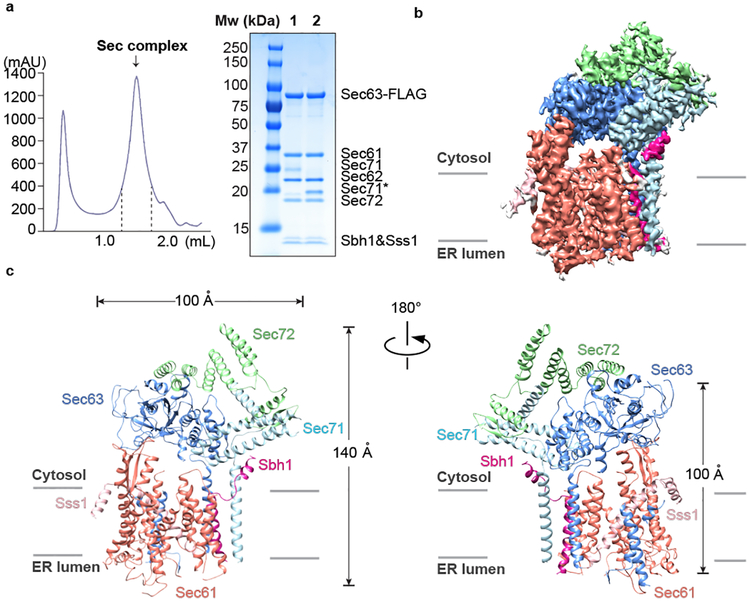
For structure determination, the Sec complex was purified in digitonin via the FLAG-tagged Sec63 subunit, followed by gel filtration (Fig. 1a). It contained all seven components, i.e. the Sec61 channel (Sec61, Sbh1, Sss1) and the Sec62, Sec63, Sec71, and Sec72 proteins in approximately stoichiometric amounts (Fig. 1a). The purified Sec complex was analyzed by single-particle cryo-EM. To reduce particle aggregation and preferred orientation on the grids, the complex was modified at surface-exposed lysines with low-molecular weight polyethylenglycol (PEG).
Fig. 1 |. Overall structure of the Sec complex.
a, The Sec complex was purified by utilizing a FLAG-tag on Sec63, followed by gel filtration (left panel). The indicated fractions were combined and subjected to SDS-PAGE and Coomassie blue staining (lane 1). Endoglycosidase H treatment (lane 2) cleaves the glycan from Sec71 (Sec71*). The experiment was independently repeated five times. b, Side view of the density map with regions of the Sec complex compoents in different colors. The map is shown at contour level 0.028, with the exception of the Sec71 part, which is shown at level 0.02. c, Side views of the models for the Sec complex components in a ribbon diagram.
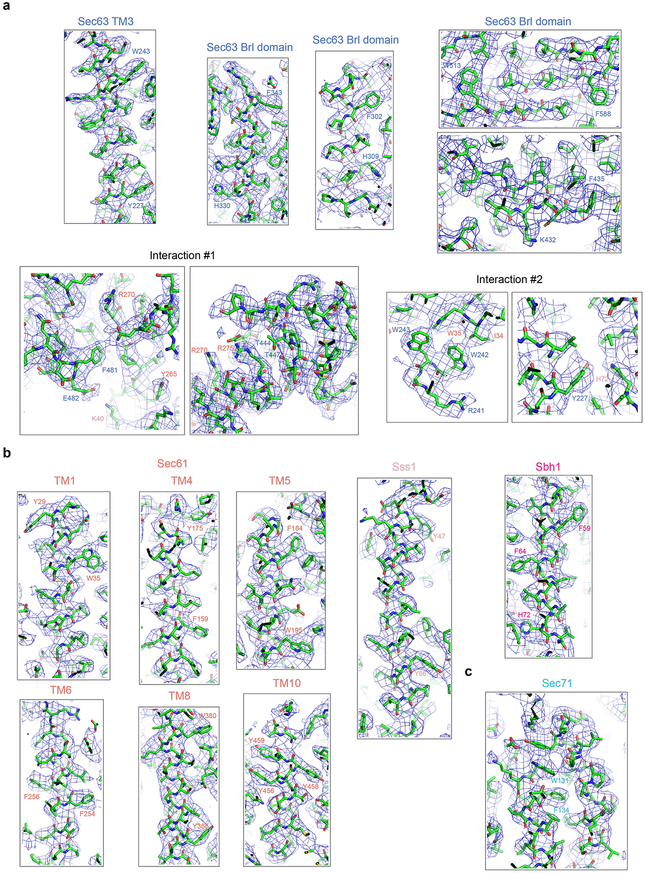
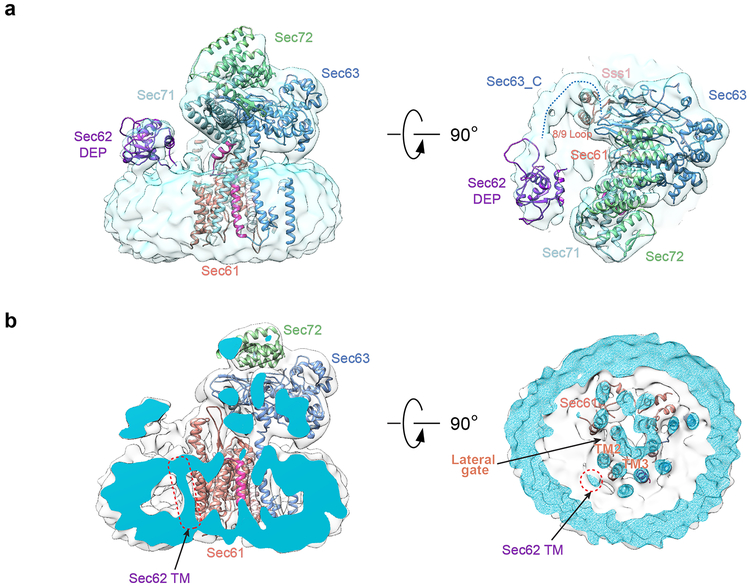
Following initial 2D classification, 3D classification and refinement of the cryo-EM particle images yielded a final electron density map at an overall resolution of 4.1 Å (Fig. 1b; Extended Data Fig. 1; Extended Data Table). Atomic models were built into the map for Sec61, Sbh1, Sss1, Sec63, and most parts of Sec71 and Sec72 (Fig. 1c; examples of the fit into the map are shown in Extended Data Fig. 2). The luminal J-domain of Sec63 is invisible and thus likely flexible. Density for Sec62 was weak, but sufficient to dock a homology model of its cytosolic DEP-like domain (Extended Data Fig. 3a) and the identification of one of its TMs (Extended Data Fig. 3b).
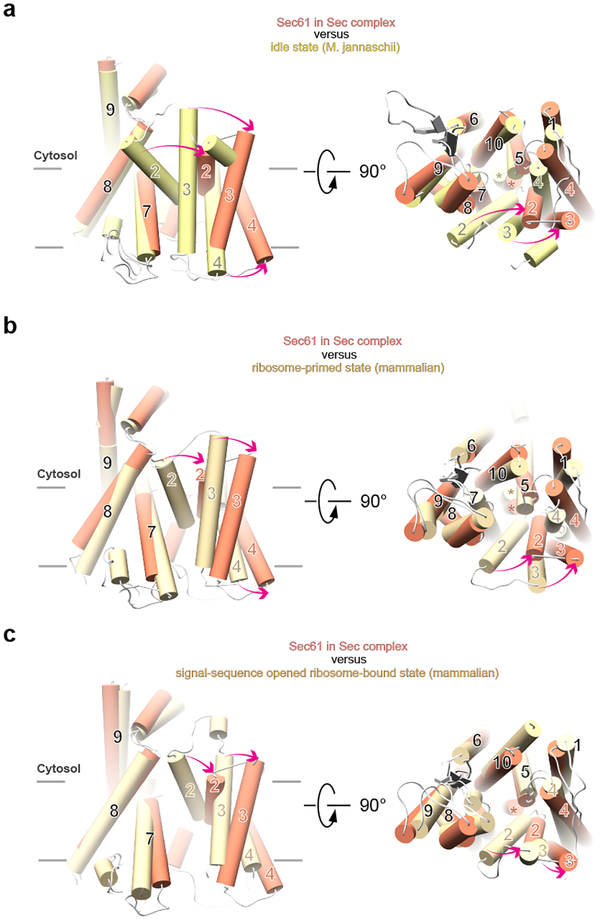
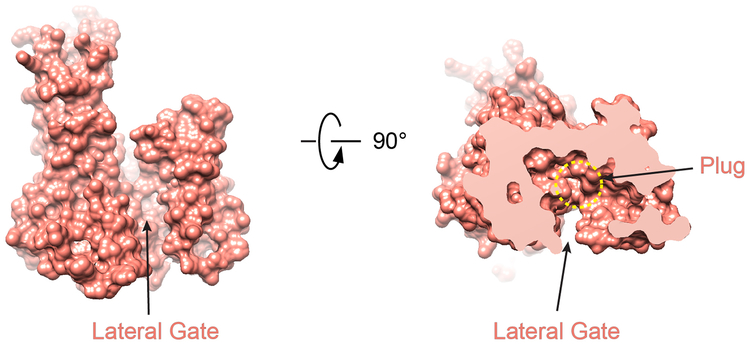
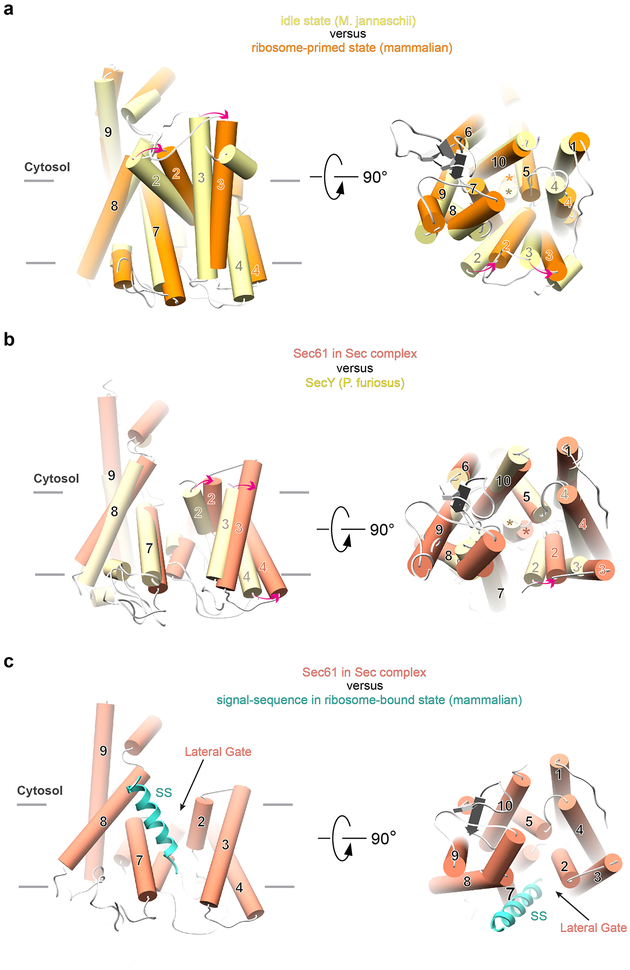
In the structure of the Sec complex, the Sec61 channel undergoes major conformational changes compared to its idle state, exemplified by a structure of the SecY channel from Methanococcus jannaschii (Fig. 2a)11. In the idle state, the channel is closed at its lateral gate, formed between TMs 7 and 8 on one side and TMs 2 and 3 on the other. In contrast, in the Sec complex, the lateral gate is wide open. Lateral gate opening is caused by TMs 2 and 3 moving outwards and by TM 4 tilting on the luminal side (Fig. 2a). The remaining TMs undergo little change. As in the idle state, the plug domain of Sec61 is located at the center of the channel (Fig. 2a; right panel), preventing ions and other small molecules from permeating (Extended Data Fig. 4). Thus, opening of the channel laterally and across the membrane are distinct events, with the latter requiring the insertion of a translocating polypeptide and displacement of the plug.
Fig. 2 |. Lateral gate changes of the Sec61 channel in the Sec complex.
a, The structure of Sec61 in the Sec complex is superimposed on that of the idle channel from M. jannaschii (PDB code 1RH5). Shown are the TMs as cylinders, viewed from the side (left panel) and from the cytosol (right panel). Movements of TMs are indicated by red arrows. The plug domains are indicated by stars in the right panel. b, As in a, but comparison with the ribosome-primed Sec61 complex (PDB code 3J7Q). c, As in a, but comparison with the signal-sequence opened ribosome-bound Sec61 channel (PDB code 3JC2).
The binding of the Sec62/63 complex to the Sec61 channel opens the lateral gate to an unprecedented extent. Ribosome binding opens the lateral gate much less (Fig. 2b; Extended Data Fig. 5a), and even after insertion of a nascent chain, the ribosome-associated Sec61 channel is not as open as after Sec62/63 binding (Fig. 2c)16. The width of the gate in the Sec complex even exceeds that in a crystal structure of the Pyrococcus furiosus SecY channel, in which opening was caused artificially by interaction with a neighboring SecY molecule (Extended Data Fig. 5b)14.
In the ribosome-primed state of the Sec61 channel, the lateral gate is only slightly open compared with the idle state (Extended Data Fig. 5a), so that it has to widen significantly more by thermal fluctuation to allow signal sequences to exit into lipid. This gate-opening energy must be compensated by energy gained from partitioning of a hydrophobic signal sequence into the hydrophobic lipid phase. In the post-translational Sec complex, the lateral channel gate does not require further opening to allow an α-helical signal sequence to exit into lipid (Extended Data Fig. 5c). Thus, low-hydrophobicity signal sequences, which gain less free energy from lipid partitioning, can still function in post-translational translocation. In agreement with this model, mutations in a polar cluster at the lateral gate permit translocation of low-hydrophobicity signal sequences22, likely by favoring the open-gate conformation. In bacteria, the association of the SecA ATPase with the SecY channel, which primes the channel for post-translational translocation, also opens the lateral gate significantly more than ribosome binding, although in this case, TMs 7 and 8 move, while TMs of the N-terminal half of the channel remain unchanged12,13. Thus, the lateral gates are generally more open in Sec61/SecY channels primed for post-translational translocation and may allow sequences of lower hydrophobicity to be functional.
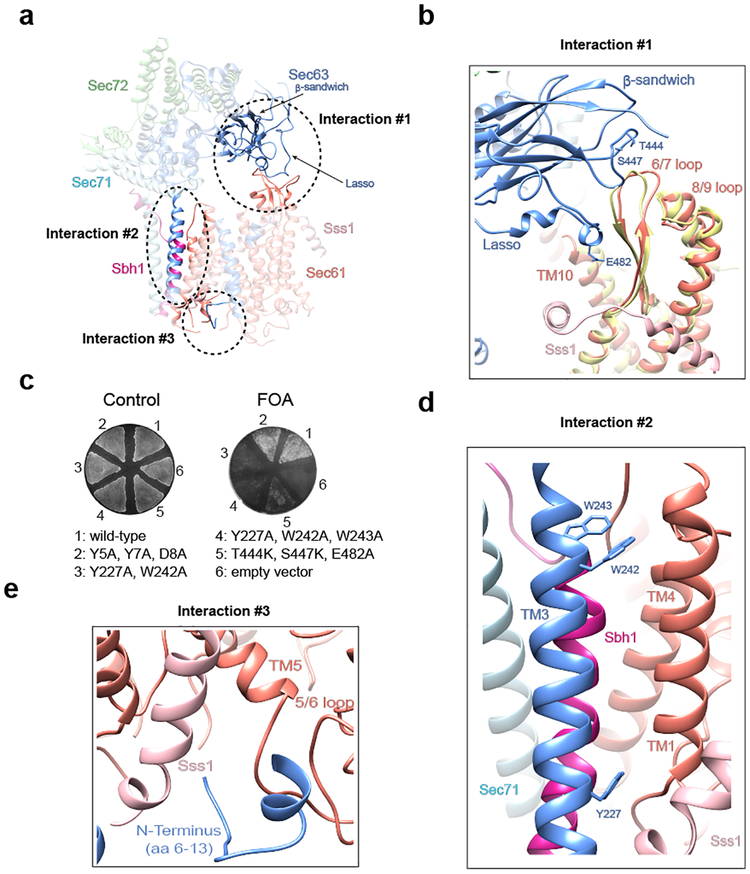
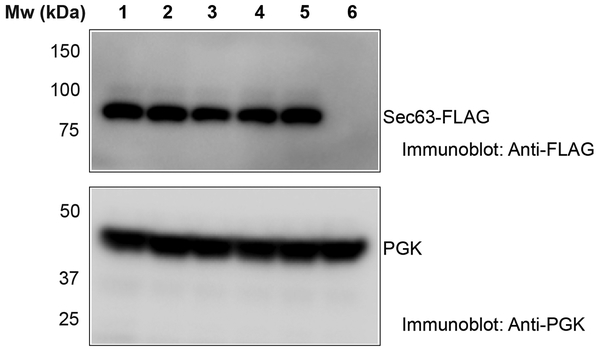
Opening of the lateral channel gate in the Sec complex is caused by Sec63. As predicted, Sec63 has three TMs and a large cytosolic Brl domain18,23. The Brl domain of Sec63 binds to the cytosolic 6/7 and 8/9 loops of Sec61 (Fig. 3a; interaction #1). Several conserved amino acid residues in the β-sandwich and lasso segments of the Brl domain face these loops, and the simultaneous mutation of these residues causes a strong defect in Sec63 function, as shown by compromised cell growth (Fig. 3b, c; sequence alignment and immunoblots for Sec63 expression are shown in Extended Data Figs. 6 and 7, respectively). A second interaction site is formed by TM3 of Sec63 binding to the other side of the channel, between TM1 of Sec61 and the TMs of Sbh1 and Sss1 (Fig. 3a; interaction #2; Fig. 3d). Several conserved aromatic residues of TM3 (Trp242, Trp243, Tyr227) are involved in the interaction with TM1 of Sec61, and their combined mutation drastically reduces Sec63 function (Fig. 3c). Finally, the N-terminus of Sec63 wedges on the luminal side of the channel between the TM 5/6 loop and Sss1 (Fig. 3a; interaction #3; Fig. 3e). This interface may be less important, as mutations in the N-terminus of Sec63 do not affect its function (Fig. 3c). Because the conformations of the 6/7 and 8/9 loops of Sec61 are essentially the same in the ribosome- and Sec62/63- bound channels (Fig. 3b), interaction #1 with the C-terminal half of Sec61 can be considered a static anchor point for Sec63. The lateral gate is pried open by Sec63’s additional interaction with the N-terminal half of Sec61 (interaction #2). Thus, Sec63 serves as a scaffold that imposes on Sec61 a gate-opened conformation. Since Sec63’s interactions #1 and #2 are required both for gate opening and cell viability (Fig. 3c), it is unlikely that the wide-open lateral gate is an artefact of sample preparation.
Fig. 3 |. Interactions of Sec63 with the Sec61 channel.
a, The interactions sites are indicated by dashed ovals and numbered. b, Magnified view of interaction #1. The Brl domain of Sec63 binds through the tip of a β-sandwich and a lasso loop to the 6/7 and 8/9 loops of Sec61. Mutated conserved Sec63 residues at the interfaces are shown as sticks. For comparison, the same Sec61 region is also shown for the ribosome-bound channel (yellow). c, Wild-type Sec63 was expressed in sec63Δ S. cerevisiae cells from a URA plasmid, together with wild-type or mutant Sec63. The cells were plated on FOA to select for cells that lost the URA plasmid. Controls were done without FOA. The sections on the plate are numbered as follows: 1) wild-type Sec63; 2) mutations in interface #3; 3) and 4) mutations in interface #2; 5) mutations in interface #1; 6) empty vector. The experiment was independently repeated twice. d, Magnified view of interaction #2. TM3 of Sec63 interacts with TM1 of Sec61 and the TMs of Sbh1 and Sss1. Trp and Tyr residues involved in the interaction are shown as sticks. e, Magnified view of interaction #3. The N-terminus of Sec63 wedges between Sss1 and the TM5/6 loop of Sec61.
Sec63 not only activates the Sec61 channel for post-translational translocation, but its Brl domain also obstructs ribosome binding to Sec61 (Fig. 1c). Thus, a nascent polypeptide chain cannot be transferred directly from the translating ribosome into the Sec61 channel. However, many proteins with low-hydrophobicity signal sequences begin their translocation through the Sec complex when the ribosome is still synthesizing the C-terminal part of the polypeptide chain24,25. The existence of free pools of Sec61 and Sec62/63 sub-complexes in the ER8 suggests that the Sec complex can dissociate, allowing the Sec61 channel to switch between the ribosome and Sec62/63 partners.
The Sec71/Sec72 sub-complex20 sits on top of Sec63’s Brl domain and engages in multiple interactions with Sec complex components (Fig. 1c). Sec62 binds through its cytosolic DEP domain to the acidic C-terminus of Sec63 (Extended Data Fig. 3a), consistent with previous data26, but the role of the DEP domain remains unclear, as it is dispensable (ref.26). The hydrophobic part of a channel-inserted signal sequence can be photo-crosslinked simultaneously to the lateral gate of Sec61 and either Sec62 or Sec71, which could not be distinguished in SDS-PAGE because of their similar size27. The structure now shows that the TM of Sec71 is far away from the lateral gate, while a TM of Sec62 is close to it (Extended Data Fig. 3b) and thus likely the crosslinking partner. This TM might thus interact with a signal sequence inside the membrane and facilitate its insertion.
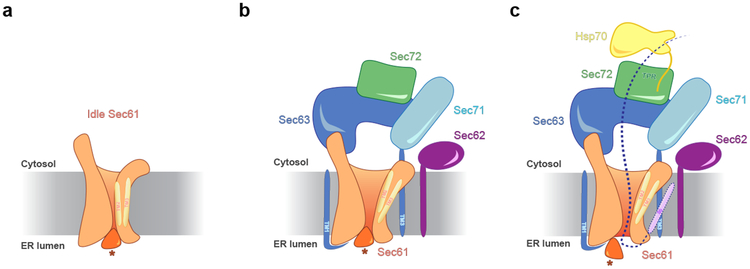
The structure leads to a model for post-translational protein translocation (Fig. 4). First, the Sec62/63 complex binds to the channel, priming it for translocation. Sec63 opens the lateral gate by serving as a scaffold for the Sec61 channel. When a post-translational polypeptide substrate arrives, it is initially associated with cytosolic chaperones28, including Hsp70, which cycle on and off. Then, the acidic C-terminal tail of Hsp70 (Ssa1–4 in S. cerevisiae) binds to the TPR domain of Sec72 (ref.20), positioning the bound substrate for subsequent transfer into the membrane channel. In higher organisms lacking Sec71 and Sec72, substrates might be targeted by calmodulin29. Next, the signal sequence moves through the lateral gate and docks into a groove on its outside13,16, while the following polypeptide segment is located in the actual pore. At this point, the polypeptide can no longer be associated with chaperones, as there is not enough space for them between the Brl domain of Sec63 and the Sec61 channel (Fig. 1c). The chaperone-stripped region of the polypeptide chain must comprise at least 40 residues following the signal sequence, as the distance from the top of the Brl domain to the luminal end of the Sec61 channel is ~100 Å (Fig. 1c). Indeed, all chaperones dissociate upon substrate insertion into the Sec complex28. The structure of the Sec complex explains why post-translational substrates are either short and devoid of bound chaperones29,30, or loosely folded polypeptides associated with Hsp70 (ref.10,28).
Fig. 4 |. Scheme for insertion of a post-translational substrate into the Sec complex.
a, Scheme of the idle Sec61 channel. The lateral and luminal gates are closed. The plug domain is indicated by a star. b, Scheme of the Sec62/63-primed channel. The lateral gate is opened, caused by Sec63 serving as a scaffold. The channel is closed across the membrane by the plug. c, A post-translational substrate binds through associated Hsp70 to Sec72; the C-terminus of Hsp70 binds to the TPR domain of Sec72. The polypeptide inserts as a loop into the Sec61 channel, with the signal sequence (SS) exiting the open lateral gate and binding to a groove on the outside. The plug is displaced. A TM segment of Sec62 might stabilize the inserted signal sequence.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Yeast strains and Plasmids
For purification of endogenous Sec complex, a FLAG tag was inserted at the C-terminus of Sec63 at its genomic locus in the wild-type strain BY4741. The strain used for 5-fluoroorotic acid (FOA) selection of Sec63 mutants was kindly provided by Reid Gilmore. The strain lacks the endogenous sec63, URA, and Leu genes and expresses instead wild-type Sec63 under its endogenous promoter from a plasmid containing the URA marker. This strain was transformed with pRS315 plasmids containing the Leu marker, which express either wild-type Sec63 or different mutants under the endogenous promoter, all with a FLAG tag at the C-terminus of Sec63.
Protein purification
The BY4741 strain with C-terminally FLAG tagged endogenous Sec63 was first grown in YPD containing 0.2 μg/ml geneticin in a shaker at 230 rpm overnight at 30°C. A large culture was inoculated by diluting the starter culture 1:80 into YPD containing 0.2 μg/ml geneticin and then incubated at 30°C for 20 hrs. The cells were pelleted and frozen until use.
Purification of the Sec complex was carried out as follows. 100 g of cell pellet were resuspended in 150 ml of buffer A (50 mM HEPES pH 7.4, 150 mM NaCl) supplemented with a home-made protease inhibitor cocktail. The cells were lysed in a BioSpec Beadbeater for 35 min with 20 s/60 s on/off cycles in a water-ice bath. After lysis, cell debris were pelleted by centrifugation at 8,000 g for 10 min. The supernatant was subjected to centrifugation in a Ti45 rotor (Beckman) at 43,000 rpm for 1 hr at 4°C. The pelleted membranes were resuspended with a Dounce homogenizer in buffer A and pelleted again by centrifugation. This washed membrane fraction was flash-frozen in liquid nitrogen and kept at −80 °C. For protein purification, the membranes were thawed and resuspended with a Dounce homogenizer and solubilized by stirring at 4°C for 2 hrs in buffer B (50 mM HEPES pH 7.5, 0.4 M NaCl, 10 % glycerol, 5 mM MgCl2, 5 mM EDTA, 1 % digitonin) and protease inhibitors. Insoluble material was then removed by centrifugation at 43,000 rpm for 30 min. The supernatant was incubated with 1 ml of anti-FLAG M2 resin (Sigma) for 3 hrs. The beads were washed with 10 ml of buffer C (50 mM HEPES pH 7.5, 0.4 M NaCl, 10 % glycerol, 0.05% digitonin), and the complex was eluted with 4 ml of buffer B containing 0.2 mg/ml of 3xFLAG peptide (Sigma). The complex was concentrated and further purified by size-exclusion chromatography on a Superose 6 3.2/300 Increase column, equilibrated with buffer D (25 mM HEPES pH 7.4, 150 mM NaCl, 0.05 % digitonin). Peak fractions were pooled and concentrated to 6 mg/ml for cryo-EM analysis.
Cryo-EM Sample Preparation and Data Acquisition
The concentrated sample was incubated with MS(PEG)12 methyl-PEG-NHS-ester (Thermo Fisher) at a 1:40 molar ratio for 2 hrs on ice to reduce preferred orientation of particles on the grids. The PEGylated sample was applied to a glow-discharged quantifoil grid (1.2/1.3, 400 mesh). The grids were blotted for 2.5 s at ~90 % humidity and plunge-frozen in liquid ethane using a Cryoplunge 3 System (Gatan).
Cryo-EM data were collected on a Titan Krios electron microscope (FEI) operated at 300 kV and equipped with a K2 Summit direct electron detector (Gatan) at HHMI Janelia Farm. A Gatan Imaging filter with a slit width of 20 eV was used to remove inelastically scattered electrons. All cryo-EM movies were recorded in super-resolution counting mode using SerialEM. The nominal magnification of 81,000× corresponds to a calibrated physical pixel size of 1.35 Å and 0.675 Å in the super-resolution mode. The dose rate was 5.48 electrons/Å2 × s. The total exposure time was 10 s, resulting a total dose of 54.8 electrons/Å2 fractionated into 50 frames (200 ms per frame). The defocus range for the sample was between 1.0 and 2.8 μm.
Image Processing
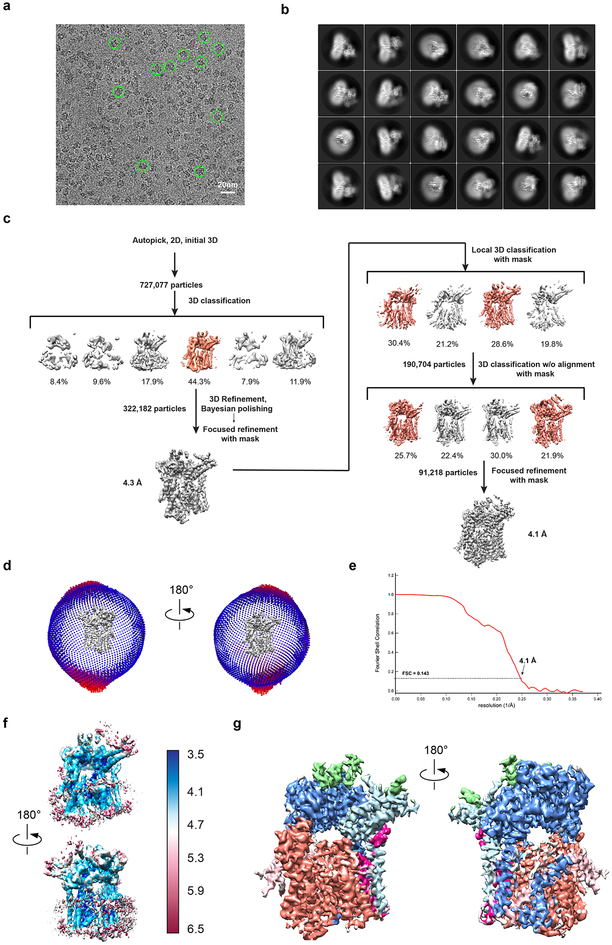
A total of 7504 dose-fractionated super-resolution movies were subjected to motion correction using the program MotionCor2 (ref.31) with a 2× binning, yielding a pixel size of 1.35 Å. A sum of all frames of each image stack (50 total) was calculated following a dose-weighting scheme and used for all image-processing steps except for defocus determination. The program Gctf32 was used to estimate defocus values of the summed images from all movie frames without dose weighting. Particles were autopicked by Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/). After manual inspection and sorting to discard poor images, classifications were done in Relion 3.0 (ref.33). A total of 2,280,162 particles were extracted and subjected to one round of reference-free 2D classification to remove false picks and obvious junk classes. To speed up 3D classification during data collection, data were analyzed in 4 batches in the order of their collection, and each batch was subjected to 3D classification, using as reference an initial model obtained from a previous small dataset collected on a Talos microscope. Only one class, containing 727,077 particles, showed protein features and particles from this class were combined for further classification. Another round of global 3D classification with 6 classes was carried out and one class with the complete Sec complex and good secondary structure features was selected (322,182 particles). Auto-refinement was done on this particle set using the reconstruction from previous 3D classification as initial model and a soft mask surrounding the protein and detergent micelle. After this round of refinement, particles were subjected to Bayesian polishing, followed by another round of auto-refinement and focused refinement using a mask encompassing Sec61/Sec63/Sec71/Sec72. The refinement at this step yielded a 4.3 Å map. Using the angle assignments obtained after the focused refinement, a 1.8 degree local 3D classification (2 sigma and T20) with an adaptive mask for Sec61/Sec63/Sec71/Sec72 was used to further classify the particles. 190,704 particles were selected and subjected to another round of auto-refinement. 3D classification (T30) without alignment, but with a mask, was used to further improve the quality of the map. After selection of 91,218 particles, a final round of auto-refinement followed by focused refinement using the adaptive mask yielded a map at 4.1 Å. Local resolution was calculated by Resmap34 and map sharpening was performed in Relion 3.0. All reported resolutions are based on gold-standard refinement procedures and the FSC=0.143 criterion. To generate a map filtered to local resolution, the map refined in Relion3.0 was imported into cryoSPARC2 (ref.35). A B-factor of −180 was applied. All software is supported by SBGrid36.
Model Building
All model building was done in Coot. For building of the Sec63 model, three TM helices could be easily identified and were initially built as poly-Ala. An homology model was generated for the large cytosolic Brl (Brr2-like) domain of Sec63 using RaptorX37, based on its similarity with domains found in RNA helicases, including the Brr2 protein involved in splicing23. The model was then docked into the density map, Cα-backbone atoms were adjusted, and the registry checked and modified, using secondary structure prediction and residues with bulky side-chains (Trp, Tyr, Phe, His, and Arg) as guidance. For building of models for Sec61, Sss1, and Sbh1, homology models for each individual component were first generated with RaptorX. These models were modified as for Sec63. For Sec71 and Sec72, homology models were generated using RaptorX, based on previous crystal structures of a thermophilic species20. Atomic models were built for these proteins, except for Sec72 residues 1–100. The manually built models were then refined using Phenix38. For the figures, the Sec72 segment (residues 1–100) was included and docked as a rigid body into the map with slight modifications. A homology model of the DEP-like domain of Sec62, predicted by RaptorX37 and other servers, was fit as a rigid body into the map.
Mutagenesis experiments
Constructs carrying wild-type Sec63 or mutants were transformed into the strain described above. Transformed yeast cells were plated on Leu/Ura double drop-out plates and allowed to grow for three days. Multiple colonies were mixed and streaked out again on Leu/Ura double drop-out plates. For each construct, the same number of cells were diluted in water and then grown on Leu drop-out plates containing FOA or on Leu/Ura double drop-out plates as a control. Plates were incubated at 30°C for 2–3 days before imaging.
Extended Data
Extended Data Fig. 1 |. Cryo-EM analysis of the Sec complex.
a, The image shows a representative cryo-EM image of Sec complex particles. Some particles are highlighted with green circles. Four EM grids were screened and had a similar particle distribution. b, Representative 2D class averages of picked particles collected from images of three grids. c, Image processing workflow for 3D classification and refinement. Shown are views of 3D reconstructions parallel to the membrane, with percentages of the particles in each class indicated. Classes in color were used for subsequent analysis. d, Euler angle distribution in two different views. e, Fourier shell correlation (FSC) curve with indicated resolution at FSC = 0.143. f, Local resolution was calculated from the unfiltered half-map and colored according to the scale on the side. Two different views are shown. g, Map filtered to local resolution using cryoSPARC2. Regions corresponding to the different proteins are colored as in Fig. 1b.
Extended Data Fig. 2 |. Examples of the fit of the models into the density map.
a, Density map and model for the indicated segments of Sec63. Interactions #1 and #2 show the interfaces between Sec63 and Sec61 complex components. Residues in salmon and pink belong to Sec61 and Sss1, respectively. b, As in a, but for segments of the Sec61 complex. c, As in a, but for the loop of Sec71 interacting with Sec63 and Sec61.
Extended Data Fig. 3 |. Localization of Sec62 in the Sec complex.
a, Shown is the unsharpened map (grey), low-pass filtered to 8Å, with models for the components of the Sec complex in ribbon diagram. A homology model for the N-terminal DEP domain of Sec62 (purple) was docked into the map. The acidic C-terminal tail of Sec63 (dotted line on the right panel) wraps around the 8/9 loop of Sec61 and interacts with the cytosolic DEP domain of Sec62. b, Cuts through the middle of the unsharpened map are shown in views from the side and from the cytosol. Density for a TM of Sec62 (highlighted by red, dotted ovals) is close to the lateral gate (arrow in right panel).
Extended Data Fig. 4 |. The Sec61 channel in the Sec complex has open lateral and closed luminal gates.
Space-filling model of the Sec61 channel in the Sec complex. The left panel shows a side view with the lateral gate in the front. The right panel shows a cut through the space-filling model viewed from the cytosol. Note that the plug domain keeps the luminal gate closed.
Extended Data Fig. 5 |. Lateral gate changes of the Sec61 channel.
a, The structure of Sec61 in the ribosome-primed state (PDB code 3J7Q) is superimposed on that of the idle channel from M. jannaschii (PDB code 1RH5). Shown are the TMs as cylinders, viewed from the side (left panel) and from the cytosol (right panel). Movements of TMs are indicated by red arrows. The plug domains are indicated by stars in the right panel. b, As in a, but comparison of the Sec61 channel in the Sec complex with the SecY channel from P. furiosus (PDB code 3MP7). c, The Sec61 channel in the Sec complex is shown together with the signal sequence (SS; in cyan) from a structure of the signal-sequence opened ribosome-bound Sec61 channel (PDB code 3JC2). The alignment of the Sec61 molecules was done as in Fig. 2c. Note that the lateral gate in the Sec complex in open enough to allow a helix to move through.
Extended Data Fig. 6 |. Alignment of Sec63 sequences from different species.
TM segments, and the Brl- and J- domains are shown as black bars above the sequences. The β-sandwich and lasso regions in the Brl domain are indicated as blue lines. Conserved residues are highlighted. Mutated residues involved in the interaction with the Sec61 channel are shown as colored circles and arrows: interaction #3 at the N-terminus in blue; interaction #2 in TM3 in red; interaction #1 in the β-sandwich and lasso segments of the Brl domain in green.
Extended Data Fig. 7 |. Sec63 expression in wild-type and mutant S. cerevisiae cells.
The strains used in Fig. 3c were analyzed for expression of FLAG-tagged Sec63. FLAG-tagged wild-type or mutant Sec63 was expressed in sec63Δ S. cerevisiae cells. Equal number of cells were analyzed by SDS-PAGE, followed by immunoblotting with FLAG antibodies. To control for equal loading, the samples were also analyzed with antibodies to phosphoglycerate kinase (PGK). The lanes correspond to expression of the following Sec63 proteins: 1) wild-type Sec63; 2) mutations in interface #3 of Sec63 (Y5A, Y7A, D8A); 3) mutations in interface #2 (Y227A, W242A); 4) mutations in interface #2 (Y227A, W242A, W243A); 5) mutations in interface #1 (T444K, S447K, E482A); 6) empty vector. The experiment was repeated twice.
Extended Data Table 1 |.
Cryo-EM data collection, refinement, and validation statistics.
| Sec Complex (EMDB-0440) (PDB 6ND1) | |
|---|---|
| Data collection and processing | |
| Magnification | 81,000 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 54.8 |
| Defocus range (μm) | −1.0 to −2.8 |
| Pixel size (Å) | 1.35 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 2,280,162 |
| Final particle images (no.) | 91,218 |
| Map resolution (Å) | 4.1 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 3.5 to 6.5 |
| Refinement | |
| Initial model used (PDB code) | |
| Model resolution (Å) | 4.1 |
| FSC threshold | 0.5 |
| Model resolution range (Å) | |
| Map sharpening B factor (Å2) | −180 |
| Model composition | |
| Non-hydrogen atoms | 9189 |
| Protein residues | 1179 |
| Ligands | |
| B factors (Å2) | |
| Protein | 72.61 |
| Ligand | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.254 |
| Validation | |
| MolProbity score | 1.68 |
| Clashscore | 7.55 |
| Poor rotamers (%) | 0.1 |
| Ramachandran plot | |
| Favored (%) | 96.16 |
| Allowed (%) | 3.76 |
| Disallowed (%) | 0.08 |
Acknowledgments
We thank Z. Yu, R. Huang, and H-T. Chou at the HHMI Janelia Cryo-EM Facility for help in microscope operation and data collection, Ed Twomey for help with data analysis, and Susan Shao and Marco Catipovic for comments on the manuscript. This work was supported by a Jane Coffin Child fellowship to X.W., and an NIGMS award (R01GM052586) to T.A.R. T.A.R. is a Howard Hughes Medical Institute Investigator.
Footnotes
Data availability
The coordinates of atomic models of the Sec complex without Sec62 were deposited in the Protein Data Bank with accession code 6ND1. The cryo-EM maps of the Sec complex before and after focused refinement with a mask were deposited with accession code EMDB-0440. All other data are available on reasonable request from the corresponding authors.
References
- 1.Voorhees RM & Hegde RS Toward a structural understanding of co-translational protein translocation. Curr. Opinion Cell Biol. 41, 91–99 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA, Li L & Park E Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol 33, 369–390 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Saraogi I & Shan S-O Co-translational protein targeting to the bacterial membrane. Biochim. Biophys. Acta 1843, 1433–1441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild K, Halic M, Sinning I & Beckmann R SRP meets the ribosome. Nature Struct. Mol. Biol 11, 1049–1053 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ng DT, Brown JD & Walter P Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269–278 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ast T, Cohen G & Schuldiner M A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Sanders SL, Feldheim DA & Schekman R Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Panzner S, Dreier L, Hartmann E, Kostka S & Rapoport TA Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Matlack KE, Misselwitz B, Plath K & Rapoport TA BiP acts as a molecular ratchet during posttranslational transport of prepro-afactor across the ER membrane. Cell 97, 553–564 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Deshaies RJ, Koch BD, Werner WM, Craig EA & Schekman R A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332, 800–805 (1988). [DOI] [PubMed] [Google Scholar]
- 11.van den Berg BVD et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Zimmer J, Nam Y & Rapoport TA Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L et al. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531, 395–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egea PF & Stroud RM Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Nat. Acad. Sci. USA 107, 17182–17187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voorhees RM, Fernandez IS, Scheres SH & Hegde RS Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 157, 1632–1643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voorhees RM & Hegde RS Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshaies RJ & Schekman R Structural and Functional Dissection of Sec62P, a Membrane-Bound Component of the Yeast Endoplasmic Reticulum Protein Import Machinery. Mol. Cell. Biol 10, 6024–6035 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldheim D, Rothblatt J & Schekman R Topology and Functional Domains of Sec63p, an Endoplasmic Reticulum Membrane Protein Required for Secretory Protein Translocation. Mol. Cell. Biol 12, 3288–3296 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldheim D, Yoshimura K, Admon A & Schekman R Structural and Functional Characterization of Sec66p - A New Subunit of the Polypeptide Translocation Apparatus in the Yeast Endoplasmic Reticulum. Mol. Biol. Cell 4, 931–939 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi A, Mandon EC, Gilmore R & Rapoport TA Two alternative binding mechanisms connect the protein translocation Sec71/Sec72 complex with heat shock proteins. J. Biol. Chem 292, 8007–8018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linxweiler M, Schick B & Zimmermann R Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct Target Ther 2, 17002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trueman SF, Mandon EC & Gilmore R A gating motif in the translocation channel sets the hydrophobicity threshold for signal sequence function. J. Cell Biol. 199, 907–918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponting CP Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem. J 351 Pt 2, 527–535 (2000). [PMC free article] [PubMed] [Google Scholar]
- 24.Jan CH, Williams CC & Weissman JS Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa EA, Subramanian K, Nunnari J & Weissman JS Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittke S, Dünnwald M & Johnsson N Sec62p, a component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the Sec-complex. Mol. Biol. Cell 11, 3859–3871 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plath K, Wilkinson BM, Stirling CJ & Rapoport TA Interactions between Sec complex and prepro-alpha-factor during posttranslational protein transport into the endoplasmic reticulum. Mol. Biol. Cell 15, 1–10 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plath K & Rapoport TA Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 151, 167–178 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao S & Hegde RS A calmodulin-dependent translocation pathway for small secretory proteins. Cell 147, 1576–1588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller G & Zimmermann R Import of honeybee prepromelittin into the endoplasmic reticulum: Energy requirements for membrane insertion. EMBO J. 7, 639–648 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng S, Palovcak E, Armache J-P, Cheng Y & Agard D MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Meth 14, 331–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zivanov J et al. RELION-3: new tools for automated high-resolution cryo-EM structure determination. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Meth 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Meth 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Morin A et al. Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Källberg M et al. Template-based protein structure modeling using the RaptorX web server. Nat. Prot 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biol. Cryst 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]