Abstract
Nucleus pulposus (NP) cells reside in the hypoxic niche of the intervertebral disc. Studies have demonstrated that RNA-binding protein HuR modulates hypoxic signaling in several cancers, however, its function in the disc is unknown. HuR did not show cytoplasmic translocation in hypoxia and evidenced levels of Hif-1α and HIF-targets in NP cells. RNA-Sequencing data revealed important extracellular matrix-related genes including several collagens, MMPs, aggrecan, Tgf-β3 and Sdc4 were regulated by HuR. Analysis of HuR-silenced NP cells confirmed that HuR maintained expression of these matrix genes. We confirmed decreased levels of secreted collagen I and Sdc4 and increased pro-MMP13 in HuR-knockdown cells. In addition, messenger ribonucleoprotein immunoprecipitation demonstrated HuR binding to Tgf-β3 and Sdc4 mRNAs. Interestingly, while HuR bound to Hif-1α and Vegf mRNAs, it was clear that compensatory mechanisms sustained their expression when HuR was silenced. Noteworthy, despite the presence of multiple HuR-binding sites and reported interaction in other cell types, HuR showed no binding to Pgk1, Eno1, Pdk1 and Pfkfb3 in NP cells. Metabolic studies showed a significant decrease in the extracellular acidification rate (ECAR) and mitochondrial oxygen consumption rate (OCR) and acidic pH in HuR-silenced NP cells, without appreciable change in total OCR. These changes were likely due to decreased Ca12 expression in HuR silenced cells. Taken together, our study demonstrates for the first time that HuR regulates extracellular matrix (ECM) and pH homeostasis of NP cells and has important implications in the maintenance of intervertebral disc health.
Keywords: Human antigen R (HuR), HIF-1α, intervertebral disc, nucleus pulposus, extracellular matrix (ECM)
Introduction
Low back pain (LBP) is a common health problem and its lifetime prevalence has been reported to be as high as 84% [1, 2]. LBP is the leading cause of years lived with disability in both developed and developing countries [3–5] and results in an enormous economic burden [6, 7]. Back pain is strongly associated with degeneration of the intervertebral discs (IVD) [8], which is characterized by the degradation of extracellular matrix (ECM), including proteoglycans and collagens, along with upregulation of matrix degrading enzymes [9, 10]. The intervertebral disc comprises an outer fibrocartilaginous annulus fibrosus (AF) and inner proteoglycan-rich and notochord-derived nucleus pulposus (NP) [11–14], which resides in an avascular niche. HIF-1α, is a master transcription factor which plays a critical role in maintenance of NP cell matrix synthesis and regulation of glycolytic metabolism [15–17]. Recent studies have demonstrated that HIF-1α is required for postnatal NP cell survival [18] and to maintain intracellular pH homeostasis through regulation of carbonic anhydrases 9 and 12 [19].
Noteworthy, control of HIF-1α levels and activity in NP cells is unique and represents adaptation of these cells to their physiologically hypoxic microenvironment [16, 20–23]. Our previous studies have shown that prolyl hydroxylase (PHD) 2 and PHD3 play opposing roles in controlling HIF-1α protein stability and, therefore, activity in NP cells [20, 21, 24]. Thus, while regulation of HIF-1α has been studied at the protein level, far less is known about control of its transcript levels, mRNA stability, and translation. Regulatory steps between transcription of the pre-mRNA, cytoplasmic export and translation of the mature mRNA are regulated by cis elements within 5’ and 3’ untranslated regions (UTRs) and/or coding region of the mRNA, primarily through interaction with two classes of trans binding factors [25]. These comprise RNA-binding proteins (RBPs), and non-coding RNAs, including antisense RNAs and microRNAs (miRNAs) [26]. Concerning mRNA stability, a HIF antisense (aHIF) transcript, that forms a negative feedback loop with HIF-1α [27], and mRNA-destabilizing protein tristetraprolin (TTP) [28] has been shown to decrease the HIF-1α mRNA stability. On the other hand, miRNAs miR-17–92 and miR-199a have been shown to repress HIF-1α translation [29, 30], while the RBPs polypyrimidine tract-binding protein (PTB), cytoplasmic polyadenylation-element-binding protein (CPEB), and HuR enhance HIF-1α translation [31–33].
HuR (ELAVL1), a member of the embryonic lethal, abnormal vision, Drosophila-like protein family [34, 35], is a ubiquitously expressed RBP. Through its three RNA recognition motifs (RRM), HuR binds to many target mRNAs containing U- or AU-rich sequences, typically present in their UTRs [36–38]. Under basal conditions, HuR is predominantly localized to the nucleus and translocates to the cytoplasm under stressful conditions includinghypoxia and glucose deprivation to stabilize expression of specific, pro-survival mRNAs [39–41]. Noteworthy, increased HuR expression has been correlated to poor clinical outcomes in many cancers [42–44]. Recent studies have shown relationship between HuR and hypoxic signaling in several cancers, where HuR binds and controls levels of HIF-1α, VEGF and other hypoxia/HIF-1 responsive genes [33, 39, 44, 45]. HuR is proposed to control both cytoplasmic export and translation of HIF-1 [33, 46]. However, the role of HuR in NP cells, in their physiologically hypoxic environment, remains unknown. We therefore investigated whether HuR is a regulator of HIF-1α signaling pathway in these cells. In addition, we investigated other functions of HuR relevant to physiology and function of the intervertebral disc that have been not described before. Our studies showed for the first time that HuR does not play a major role in maintenance of HIF-1α signaling in NP cells but does control extracellular matrix (ECM) and pH homeostasis of NP cells, suggesting important implications in maintenance of intervertebral disc health.
Results
Hypoxia does not promote HuR nucleocytoplasmic shuttling or its expression in NP cells:
We first investigated the expression of HuR in the healthy mouse intervertebral disc. Immunohistochemistry showed robust expression of HuR in NP cells that was co-localized with DAPI (Fig. 1A), indicating prominent nuclear expression of HuR in the physiologically hypoxic NP niche. Weaker expression was also seen in the cytoplasm of NP cells. Previous study has shown HuR mRNA induction by hypoxia in other cell type [47]. However, there was no significant change in HuR mRNA expression in response to hypoxia in NP cells (Fig. 1B). Previously, we demonstrated in pancreatic cancer cells that HuR translocated from the nucleus to the cytoplasm upon hypoxic stress [39]. To test whether hypoxia promotes cytoplasmic translocation in NP cells, we cultured rat NP cells under hypoxia for up to 72 h and analyzed HuR expression by Western blot. Surprisingly, HuR protein was prominently expressed in both the nucleus and the cytoplasm of NP cells even under normoxia (Fig. 1C). Furthermore, the cellular distribution of HuR was unaltered by culturing under hypoxic conditions (1% pO2) (Fig. 1C–E). Similarly, immunofluorescent staining of HuR protein in NP cells showed no changes in nucleo-cytoplasmic distribution of HuR in response to hypoxia (Fig. 1F).
Figure 1.
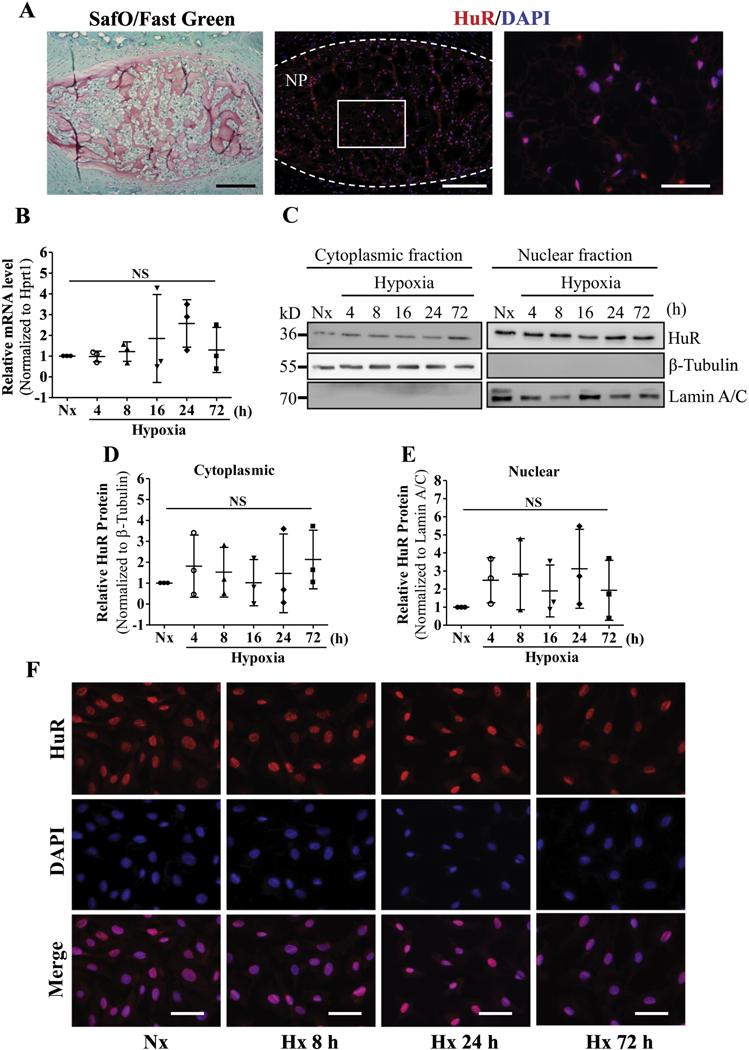
A) Safranin O/Fast Green staining of mouse intervertebral disc. Scale bar: 100 μm. A’, A’’) Immunofluorescence staining of HuR in NP. Scale bar: 100 μm (A’); 50 μm (A’’). B) HuR mRNA level in NP cells cultured under hypoxia up to 72 h. C) Cytoplasmic and nuclear levels of HuR protein in NP cells culture under hypoxia upto 72 h. D, E) Corresponding densitometric quantification of HuR cytoplasmic and nuclear protein fractions. F) Immunofluorescence staining of HuR in NP cells cultured in hypoxia. Western blot images shown in C are from one representative experiment that was performed independently at least thrice. Quantitative data are presented as mean ± SD of at least three independent experiments. NS, not significant; Nx, Normoxia; Hx, Hypoxia.
HuR silencing showed no effects on Hif-1α and select canonical HuR/HIF-1α target genes in NP cells:
In other cell types, HuR regulates the hypoxic response largely by stabilizing key transcription factor HIF-1α. To investigate the contribution of HuR in NP cells in controlling Hif-1α as well HIF-1α transcriptional targets that have been shown or predicted to directly bind HuR in other cells [33, 48, 49], we knocked down HuR by transducing rat NP cells with lentivirus expressing HuR-directed shRNA under either normoxic or hypoxic conditions. The level of HuR protein was robustly decreased in total (Fig. 2A and B), cytoplasmic, and nuclear (Fig. 2C) cellular protein fractions, confirming a successful knockdown. Proliferation rate of NP cells was not affected by HuR inhibition under hypoxia (Fig. 2D). Surprisingly, HuR knockdown had no impact on the mRNA level of Hif-1α (Fig. 2E). Moreover, regardless of the oxygen tension, select HIF-1α and/or HuR targets including Phd2, Phd3, Pdk1, Pgk1, Pfkfb3, Vegf, Eno1, and Ldhb were not affected by HuR knockdown and also did not alter hypoxic inducibility of select targets (Fig. 2 E and F). Likewise, Western blot and corresponding densitometric analysis showed no significant changes in protein levels of HIF-1α and selected HuR/HIF-1α targets in HuR silenced NP cells (Supplemental Fig. 1A–C).
Figure 2.
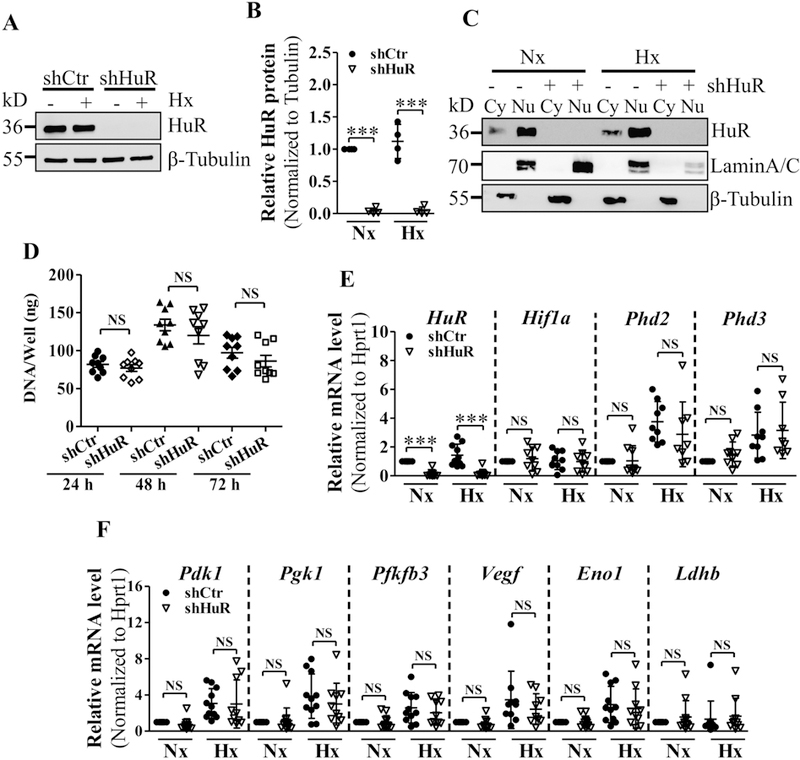
A, B) Measurement of HuR protein by Western blot (A) and corresponding densitometric analysis (B) confirmed successful knockdown of HuR after transduction with HuR-directed shRNA under normoxia and hypoxia. C) Both cytoplasmic levels of HuR significantly decreased following NP cell transduction with shHuR. D) Proliferation potential of HuR-silenced and control NP cells under hypoxia up to 72 h. E, F) HuR silencing exhibited no effects on select HuR/HIF-1α transcriptional targets - Hif-1α, Phd2, Phd3, Pdk1, Pgk1, Pfkfb3, Vegf, Eno1, Ldhb under both normoxia and hypoxia. Western blot images shown in A and C are from one representative experiment performed in ≥ 3 biological replicates. Quantitative data are represented as mean ± SD of at least three independent experiments. ***, p ≤ 0.001; NS, not significant; Cy, Cytoplasmic; Nu, Nuclear; Nx, Normoxia; Hx, Hypoxia.
RNA sequencing reveals a broader role of HuR in regulation of matrix homeostasis in NP cells:
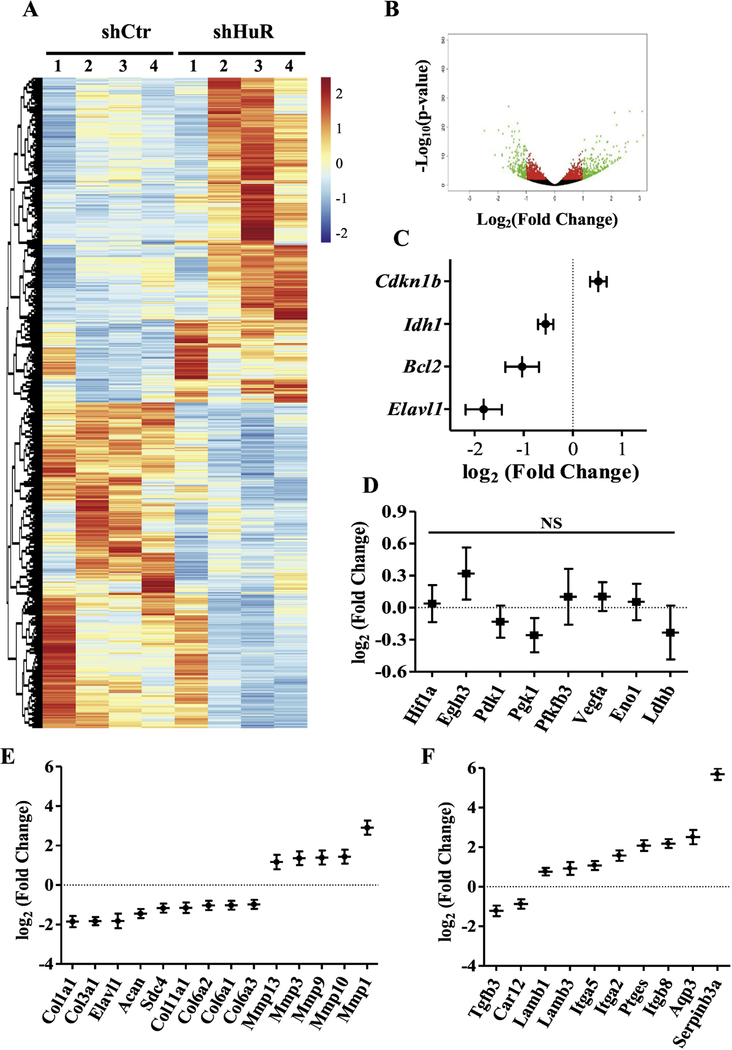
In order to understand the overall role of HuR in NP cells and identify previously unknown targets in an unbiased manner, we subjected HuR-knockdown cells to RNA sequencing. Our analysis identified 2,115 transcripts that were differentially expressed between HuR knockdown and control NP cells (adjusted p-value < 0.05) (Fig. 3 A and B). We first validated our RNA-Seq data by examining levels of known HuR target genes in knockdown cells. As expected, expression of HuR along with Bcl2 and Idh1 [41] were decreased while expression of CDKN1B was increased in HuR knockdown cells (Fig. 3C). Again, Hif-1α and select HIF-1α/HuR targets including Egln3, Pdk1, Pgk1, Pfkfb3, Vegfa, Eno1, and Ldhb showed no statistically significant differences between knockdown and control cells (Fig. 3D). Surprisingly, expression of several key extracellular matrix genes and genes involved in matrix homeostasis were altered by HuR knockdown (Fig. 3E and F). Ingenuity Pathway Analysis (IPA) was used to identify the top pathways, functions and networks associated with the list of differentially expressed genes (Supplemental Tables 1, 2 and 3). Many genes associated with Connective Tissue Development and Function, Skeletal and Muscular System Development, which are critical in context of disc matrix metabolism and degeneration, were identified by the IPA analysis.
Figure 3.
A, B) Heat map and volcano plot depicting differentially expressed genes between control and HuR knockdown NP cells. C) RNA sequencing of HuR-silenced NP cells confirmed decreased expression of HuR (Elavl1), along with significant changes in positively regulated (Bcl2, Idh1) and negatively regulated (Cdkn1b) known HuR target genes. Log2(fold change) values are shown. D) RNA sequencing of HuR-silenced NP cells showed no significant changes of select HuR/HIF-1α transcriptional targets - Hif-1α, Phd3, Pdk1, Pgk1, Pfkfb3, Vegf, Eno1, Ldhb. E, F) Log2(fold change) values for matrix-related transcripts that were differentially expressed between control and HuR knockdown NP cells. NS, not significant.
Distinct role of HuR in regulation of various genes involved in matrix homeostasis in NP cells:
We next used qRT-PCR to measure the expression levels of select matrix-related genes that were identified by the RNA-Sequencing analysis. In agreement with RNA-Seq results, expression levels of Acan, Sdc4, and Tgfb3, were positively regulated by HuR, irrespective of the oxygen tension (Fig. 4 A). There was no change in expression levels of Col2a1 with HuR silencing (Fig. 4B). Expression levels of several collagens (Fig. 4C), laminins (Fig. 4D), integrins (Fig. 4E), and MMPs (Fig. 4F) were also responsive to HuR knockdown. Noteworthy, some genes were regulated by HuR knockdown under both normoxia and hypoxia while others were controlled by HuR only in hypoxic conditions. HuR negatively regulated levels of Ptges, Aqp3, and Serpinb3a under both normoxic and hypoxic conditions (Fig. 4G).
Figure 4.
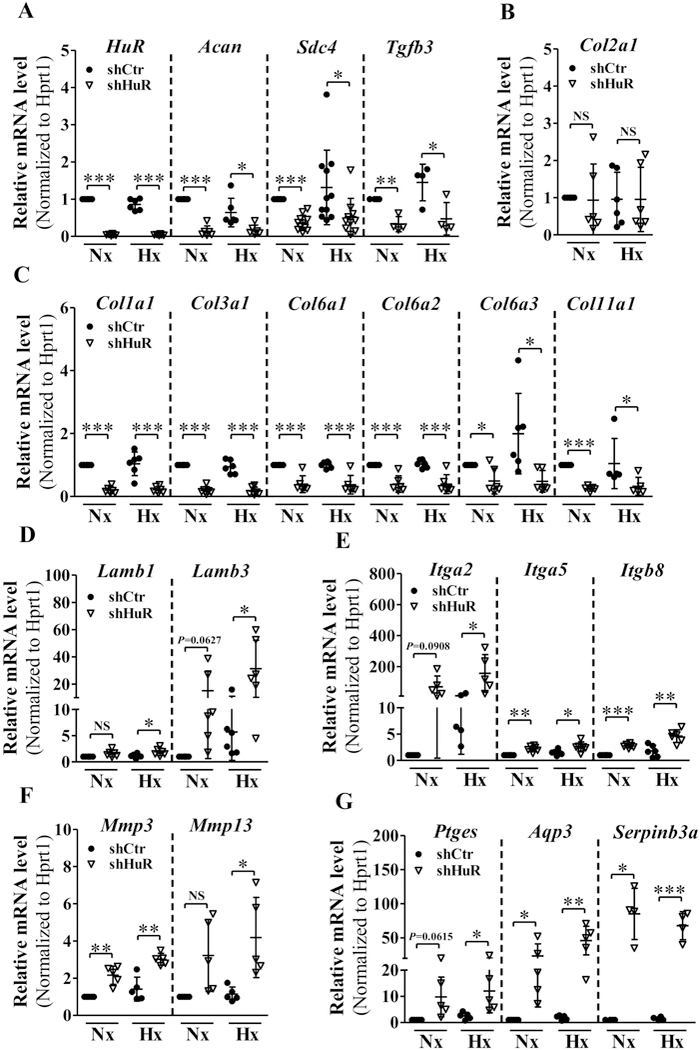
A-C) qRT-PCR analysis of mRNA levels of Acan, Sdc4, Tgfb3 (A) and C Col1a1, Col3a1, Col6a1, Col6a2, Col6a3, Col11a1 (C) were significantly decreased under both normoxia and hypoxia following HuR silencing. Col2a1 (B) mRNA level did not change with HuR silenced NP cells. D-G) Levels of Lamb1, Lamb3 (D), Itga2 (E), Mmp13 (F) and Ptges (G) mRNA levels increased only under hypoxia in HuR silenced cells. Whereas, Itga5, Itgb8, Mmp3 (F), Aqp3 and Serpinb3a (G) mRNA levels increased under both normoxia and hypoxia following HuR knockdown. PCR data is represented as Mean ± SD of at least three independent experiments performed in duplicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; NS, not significant; Nx, Normoxia; Hx, Hypoxia.
HuR silencing reduces protein levels of collagen I and aggrecan in NP cells:
To extend our finding that HuR regulates mRNA levels of Col I and Acan in NP cells, we performed immunofluorescence staining for their protein products. We found a significant decrease in collagen I expression level and a small reduction in aggrecan expression level in HuR knockdown NP cells (Fig. 5A). In contrast, there was no change in collagen II protein level (Fig. 5A). To investigate whether the production and secretion of collagen I was affected, we precipitated collagens from the cell media. As indicated in Figure 5B, the production of collagen I diminished in the HuR-silenced group under both normoxic and hypoxic conditions (Fig. 5B). In line with mRNA expression results, SDC4 protein level was also reduced in the HuR stable knockdown group in both normoxia and hypoxia (Fig. 5 C and D). Interestingly, in HuR knockdown cells, while levels of secreted pro-MMP13 (60 kDa) increased, there was a small decrease in the active form of the enzyme which has a molecular weight of 48 kDa (Fig. 5E, G). There was also no appreciable difference in MMP3 levels between control and HuR knockdown cells (Fig. 5F, G). While MMP2 (gelatinase A) activity remained unaffected, a small decrease in MMP9 (gelatinase B) enzymatic activity was seen in HuR knockdown cells under hypoxia as measured by gelatin zymography (Fig. 5H).
Figure 5.
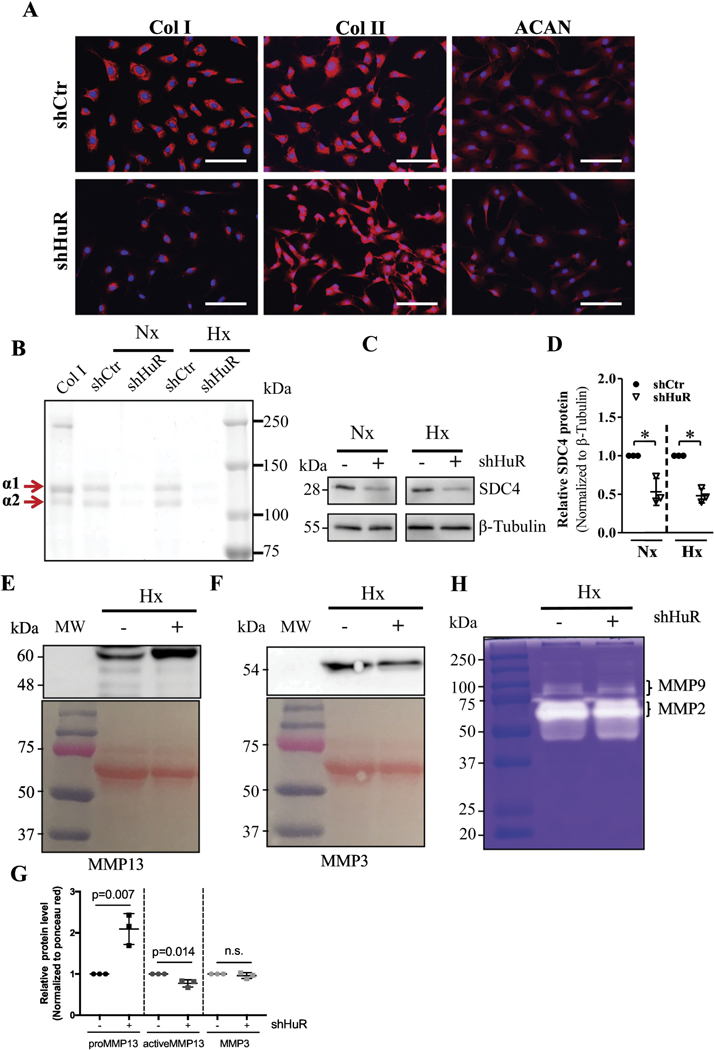
A) Immunofluorescence staining of NP cells demonstrated a dramatic decrease in Collagen I and a slight reduction in Aggrecan expression after HuR knockdown, while no change was observed in Collagen II levels. B) Electrophoresis of precipitated collagens from NP cell conditioned medium and Coomassie blue staining of precipitated fraction showed both α1 and α2 chains of Collagen I were diminished in HuR-silenced cells. C, D) Western blot and corresponding densitometric quantification showed decreased SDC4 protein levels after stable HuR knockdown under both normoxia and hypoxia. E) Western blot showed increased in levels of pro-MMP13 (60 kDa) but a slight decrease in active form (48 kDa) after stable HuR knockdown under hypoxia. F) Levels of MMP3 were not appreciably different between shcontrol and shHuR groups. G) Densitometry of Western blots from 3 independent experiments in E and F. H) Gelatin zymography showed decreased enzyme activity of MMP9 after HuR knockdown under hypoxia but no changes in MMP2 activity were seen. Images shown are from a representative experiment, which was performed at least at three independent times. *, p ≤ 0.05; **, p ≤ 0.01; NS, not significant; Nx, Normoxia; Hx, Hypoxia.
HuR silencing reduces ECAR and extracellular proton production rate possibly through targeting Ca12 in NP cells:
RNA-Seq data showed that carbonic anhydrase 12 (Ca12) was positively regulated by HuR under hypoxia; qPCR and Western blot analysis confirmed CA12 mRNA and protein levels were attenuated after HuR silencing (Fig. 6 A–C). We therefore investigated whether HuR affects NP cell metabolism by measuring the extracellular acidification rate (ECAR) and oxygen consumption rates (OCR) using a Seahorse XF Analyzer. Interestingly, culturing HuR-silenced NP cells under hypoxia for 24 h resulted in a significant decrease in the ECAR (Fig. 6, D and E). Whereas, a small reduction in Mitochondrial OCR, without any change in total OCR, was also observed in HuR silenced NP cells (Fig. 6F–H). We next explored the source of extracellular proton production that contributed to decreased ECAR: H+ produced via anaerobic glycolysis or CO2 hydration catalyzed by CA12. By measuring the extracellular lactate concentrations, we found that H+ generated from glycolysis remained unaffected following HuR knockdown (Fig. 6J). Noteworthy, a significant drop of CO2 hydration component of proton production rate (PPR) was detected after silencing HuR, which may result in decreased ECAR in HuR-silenced NP cells (Fig. 6I).
Figure 6.
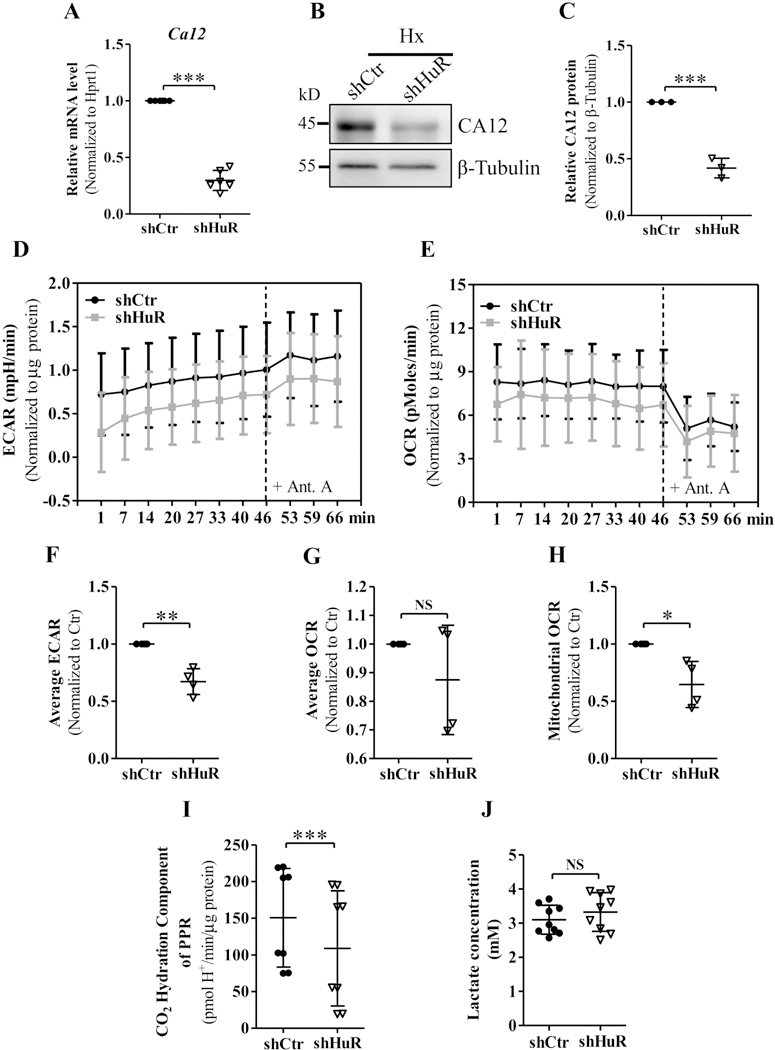
A-C) qRT-PCR and Western blot analysis showed carbonic anhydrase 12 (Ca12) mRNA and protein levels were attenuated after silencing HuR in hypoxia. D, E) Extracellular acidification rate (ECAR) of control and HuR-silenced NP cells subjected to hypoxia for 24 h, measured by Seahorse XF Analyzer, exhibited a significant decrease in the ECAR of HuR-silenced NP cells. F, G and H) HuR silencing did not affect total Oxygen consumption rate (OCR) but resulted in a slight reduction in Mitochondrial OCR of NP cells. I) CO2 hydration component of proton production rate (PPR) was reduced after HuR silencing. J) Extracellular lactate concentrations were unaffected in HuR silenced NP cells. Western blot images shown in (B) are from one representative experiment, which was performed at three independent times. Quantitative data are represented as mean ± SD of at least three independent experiments. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; NS, not significant; Ant. A, Antimycin A; Hx, Hypoxia.
Analysis of HuR binding to target mRNAs in NP cells:
To determine whether HuR directly binds HIF-1α and its target mRNAs in NP cells, messenger ribonucleoprotein immunoprecipitation (mRNP-IP) assays were performed using control and HuR-silenced cells (Fig. 7A and B). As expected, HuR protein directly bound to HuR mRNA but not to Gapdh mRNA [50] (Fig. 7C), which served as a negative control. In control cells, HuR protein interacted with Tgfβ3 and Sdc4 mRNAs, as was expected based on earlier results, and these interactions were greatly diminished in HuR-knockdown cells (Fig. 7D). Interestingly, we also detected some HuR binding to Hif-1α and Vegf mRNAs (Fig. 7E), despite the fact that no significant changes were found in expression levels of Hif-1α protein or mRNA or Vegf mRNA after stable knockdown of HuR. This may be due to compensatory regulation of Hif-1α and Vegf in HuR-silenced NP cells. Noteworthy, interaction between HuR and COL1A1 mRNA was detected in control NP cells (Fig. 7E), although there were no predicted potential HuR binding sites in Col1a1 mRNA (Supplemental Table 4). No interactions were detected between HuR protein and other select HIF-1α target mRNAs including Pgk1, Pdk1, Eno1, Pfkfb3, Acan and Ca12 (Fig. 7F).
Figure 7.
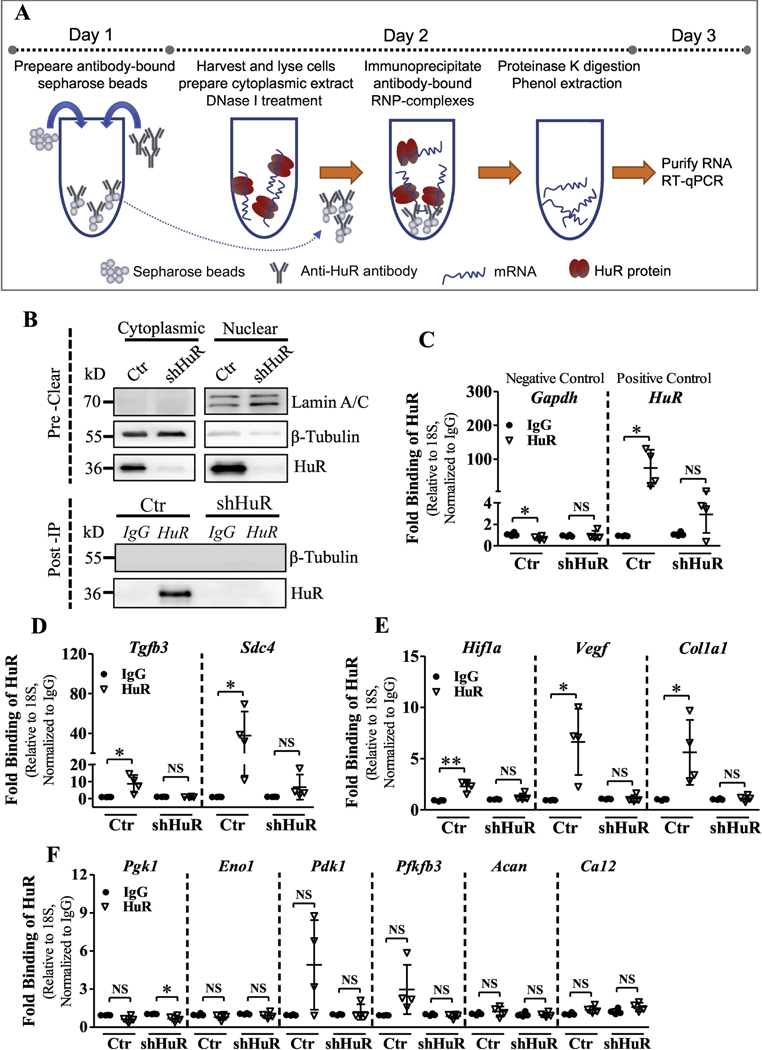
A) Schematic showing the workflow of the Messenger ribonucleoprotein immunoprecipitation (mRNP-IP or RIP) assay. B) Validation of cell lysates and post-IP samples from HuR RIP assay. HuR was immunoprecipitated (IP) using a rabbit polyclonal antibody, and IP was validated by a mouse monoclonal antibody by Western blot analysis. β-Tubulin was used as a loading control for the input and a negative control for the post-IP samples. Lamin A/C was used as a control to detect nuclear contamination in the input. C-E) The relative abundance of Tgfb3, Sdc4, Hif-1α, Vegf, Col1a1, Pgk1, Pdk1, Eno1, Pfkfb3, Acan and Ca12 mRNA bound to HuR, normalized to IgG isotype controls, was determined by qRT-PCR using 18S rRNA as a loading control. Gapdh was used as a negative control and HuR as a positive control. Western blot images shown in B are from one representative experiment out of 4 independent experiments. Quantitative data are represented as Mean ± SD. *, p ≤ 0.05; **, p ≤ 0.01; NS, not significant.
Discussion
HuR (ELAVL1), an RBP, is ubiquitously expressed and is primarily localized to the nucleus under basal conditions [36–38]. Previous studies have shown that various stresses, including hypoxia, induce shuttling of HuR to the cytoplasm, where it stabilizes expression of target mRNAs [39]. Cytoplasmic translocation of HuR has been correlated to poor clinical prognosis in many cancers and promotes resistance to hypoxia in tumor cells [42–44, 47]. Importantly, HuR is a known regulator of HIF-1α [33, 46]. However, the role of HuR in NP cells, which exist in a physiologically hypoxic environment and exhibit novel modes of HIF-1α regulation [16, 20–23] remains unexplored. In the current study, we discovered that hypoxia failed to induce HuR nucleocytoplasmic shuttling in NP cells, which may represent an adaptation to their physiologically hypoxic conditions.
It has been previously reported that direct binding of HuR increases HIF and VEGF mRNA stability and promotes their translation [33, 48]. Surprisingly, we found that loss of HuR function did not decrease levels of Hif-1α or levels of select HIF/HuR-target genes, including Phd2, Phd3, Pdk1, Pgk1, Pfkfb3, Vegf, Eno1 and Ldhb in NP cells. Interestingly, while binding of HuR to Hif-1α and Vegf mRNAs was observed, it was evident that compensatory mechanisms sustained their expression when HuR was silenced. Noteworthy, despite the presence of multiple HuR-binding sites, Pgk1, Eno1, Pdk1 and Pfkfb3 did not bind HuR in NP cells. HuR has also been shown to promote proliferation of cancer cells through multiple mechanisms, including regulation of cell cycle genes [51–53]. However, HuR did not contribute to regulation of NP cell proliferation, reflecting post-mitotic and limited proliferative capacity of this tissue compartment in vivo. These results were supported by RNA sequencing that showed no effect of HuR silencing on expression of major cell cycle regulatory genes; cyclins D1, E1, A2, and B1, p21, and C-myc in NP cells. These results indicated that, unlike in cancer cells, the primary function of HuR in NP cells was not to act on traditional genes concerned with hypoxia- or proliferation-response.
Intervertebral disc degeneration is driven by the imbalance between anabolism and catabolism of ECM secreted by NP and AF cells and degraded by matrix degrading enzymes such as matrix metalloproteinases (MMPs). Our RNA sequencing revealed that HuR acts as an important regulator of many key ECM-related genes including several collagens (Col1a1, Col3a1, Col6a1, Col6a2, Col6a3 and Col11a1), aggrecan, TGF-β3 and SDC4, which was confirmed by qPCR. Of note, in fibroblasts, HuR has been shown to regulate TGF-β-mediated induction of Col1a and Col3a1 [54]. We also observed that HuR knockdown resulted in decreased levels of secreted collagen I, aggrecan, and SDC4 in NP cells. In line with previously reported bioinformatics predictions [49], we found that HuR directly binds to Tgf-β3 and Sdc4 mRNAs by using messenger ribonucleoprotein immunoprecipitation (mRNP-IP) assay. HuR knockdown significantly elevated levels of Mmp13, mRNA and secreted pro-protein which is consistent with a previous study in human articular chondrocytes [55]. Surprisingly, secreted MMP3 showed no difference in expression levels, while, MMP9 showed a trend of decreased activity following HuR knockdown, implying presence of differential mechanisms in regulating mRNA and protein expression and/ or activity of these MMPs. Again, expression of Mmp9 has been shown previously to be sensitive to HuR in mesangial cells of kidney [56]. Additionally, we observed HuR regulation of Itga2, Itga5, Itgb8, Lamb1 and Lamb3 expression; these molecules modulate cell-cell and cell-matrix signaling in the NP [57]. All of these findings suggested that HuR may play a crucial role in maintaining ECM homeostasis in NP cells, a function of HuR that has yet to receive much attention and may have important implications for cancer cell metastasis and tissue fibrosis. Recent data suggest that HuR binds to TonEBP, an osmo-sensitive transcription factor TonEBP (NFAT5) that is one of the important regulators of ECM homeostasis in the NP niche [58–62]. Considering that HuR does not stabilize Hif-1α in NP cells, it is plausible that HuR’s actions on ECM could occur through regulation of TonEBP activity or other key transcription factors.
In addition to its regulatory role on important ECM molecules, we found that HuR positively regulated CA12 expression in NP cells. We have recently demonstrated that CA12 is a critical regulator of NP cell pH, metabolism, and survival [19]. Metabolic studies showed that there was a significant decrease in the ECAR and mitochondrial OCR in HuR-silenced NP cells, without any change in total OCR. By using analytical methods proposed by Mookerjee, et al. [63], we found that the decreased ECAR in HuR silenced NP cells mainly resulted from the significant drop in CO2 hydration component of proton production rate (PPR). As expected, extracellular lactate concentration was unaffected by HuR silencing. Thus, decreased CA12 expression may serve as one of the possible mechanisms for the observed changes in metabolic parameters in HuR silenced cells.
Taken together, our study demonstrates for the first time that HuR regulates extracellular matrix (ECM) production and pH homeostasis of NP cells and could have important implications in maintenance of intervertebral disc health. Our striking observation that HuR does not regulate Hif-1α or its canonical target genes in NP cells is further evidence for the unique adaptation of these cells to their hypoxic microenvironment. It is plausible that the cell-type specific posttranslational modification of HuR such as phosphorylation, methylation, ubiquitination, NEDDylation and/or cleavage products may impact HuR activity or ability to bind certain mRNA species in NP cells [64]. Noteworthy, we have identified HuR as a critical mediator of several ECM genes tightly linked to disc degeneration as well as other fibrotic disorders. This observation has further implications in the context of cancer metastasis where ECM degradation is required for cell movement and could have implications for HuR-directed therapeutics.
Materials and Methods
Isolation of NP cells and hypoxic culture:
Rat NP cells were isolated using a method previously reported by Risbud et al [15]. After isolation, cells were maintained in normoxia in Dulbecco’s Modified Eagles Medium (DMEM) (Corning, 10–013-CV) with 10% FBS (Sigma-Aldrich, F6178) supplemented with antibiotics. Cells from passage 2–4 were used for experiments. To mimic the in vivo hypoxic environment, cells were cultured in a Hypoxia Work Station (Invivo2 400, Baker Ruskinn, Bridgend, UK) with a mixture of 1% O2, 5% CO2, and 94% N2 in medium pre-equilibrated at 1% O2 for up to 72 hours.
Plasmids and reagents:
Lentiviral ShHuR (TRCN0000276126) and control ShRNA pLKO.1 plasmids were purchased from Sigma, and psPAX2 (plasmid 12260) and pMD2.G (plasmid 12259) developed by Dr. Didier Trono were obtained from Addgene.
Lentiviral particle production and transduction:
HEK-293T cells (ATCC, CRL-3216) were plated in 10-cm plates at a seeding density of 5 × 106 cells/plate in DMEM with 10% heat-inactivated FBS one day before transfection. Cells were transfected with 9 μg of either ShCtr or ShHuR plasmids, plus 6 μg psPAX2 and 3 μg pMD2.G using Calcium Phosphate Transfection Kit (Clontech, 631312). After 16 h, transfection medium was replaced with DMEM with 10% heat-inactivated FBS and antibiotics. Lentiviral particles were harvested at 48 and 60 h after transfection, and mixed with 7% PEG 6000 (Sigma Aldrich, 81253) solution and incubated overnight at 4 oC to precipitate virus particles. PEG solution was removed from virus medium before transduction by centrifugation at 1,500 × g for 30 min to pellet virus particles. NP cells were transduced with DMEM with 10% heat-inactivated FBS containing viral particles along with 8 μg/ml polybrene (Sigma Aldrich, H9268). After 24 h transduction, the medium was replaced with fresh DMEM with 10% FBS. Cells were collected 5 days after transduction to ensure maximum knockdown efficiency without affecting cell viability.
RNA Sequencing:
Illumina TruSeq Stranded Total RNA Sample Prep with Ribo-Zero was used to prepare the library. Libraries were chemically denatured and applied to an Illumina HiSeq v4 single read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR Cluster Kit v4-cBot. After transfer of the flow cell to an Illumina HiSeq 2500, a 50-cycle single-read sequence run was performed (HiSeq SBS Kit v4). For data analysis, Rn5 Ensembl annotations (Build 75) were downloaded and converted to genePred format. Reads were aligned to the transcriptome reference index using NovoAlign (v2.08.01), allowing up to 50 alignments for each read. Read counts were generated using the USeq Defined Region Differential Seq application and used in DESeq2 to measure the differential expression between each condition, controlling for sample preparation batch. Data has been submitted to the GEO Database (Accession # GSE110804). For Ingenuity Pathway Analysis (IPA), differentially expressed gene lists were used as input to identify related pathways, diseases, and networks.
Real-time Quantitative RT-PCR:
Total RNA was extracted from rat NP cells using RNAeasy mini columns (Qiagen) and treated with RNase-free DNase I (Qiagen) before elution from the column. The purified, DNA-free RNA was reverse transcribed to cDNA using EcoDryTM premix (Clontech). Template cDNA and gene-specific primers were added to the SYBR Green master mixture (Applied Biosystems), and mRNA expression was quantified using the StepOnePlus real time PCR System (Applied Biosystems). Hypoxanthine-guanine phosphoribosyltransferase 1 (Hprt1) was used to normalize gene expression. Melting curves were analyzed to verify the specificity of the RT-qPCR and the absence of primer dimer formation. Each sample was analyzed in duplicate and included a template-free control. All primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Protein extraction and Western blotting:
Conditioned medium was collected and cells were placed on ice immediately after treatment and washed with ice-cold PBS. Wash buffer and lysis buffer contained 1X-protease inhibitor cocktail (Roche), NaF (4mM) and Na3VO4 (20mM), NaCl (150mM), β-glycerophosphate (50mM), and DTT (0.2mM). Nuclear and cytoplasmic proteins were isolated using the NE-PER™ Nuclear and Cytoplasmic Extraction kit (Thermo Fisher Scientific, 78833). Equal amounts of total or fractionated cell proteins were resolved on 8–10 % SDS-polyacrylamide gels and transferred to PVDF membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat dry milk in TBST and incubated overnight at 4 oC in blocking buffer with anti-HuR (1:400, Santa Cruz, sc-5261); anti-HIF-1α (1:500, R&D Systems, MAB1536); anti-PHD2 (1:1000, Cell Signaling Technology, 4835); anti-PHD3 (1:1000, Novus Biologicals, NB100–139); anti-PDK1 (1:1000, Novus Biologicals, NBP1–85955); anti-PGK1 (1:1000, Abcam, ab137575); anti-PFKFB3 (1:1000, Abcam, ab181861); anti-ENO1 (1:1000, Novus Biologicals, NB100–65252); anti-SDC4 (1:1000, EMD Millipore, ABT157); anti-MMP3 (1:2000, Abcam, ab52915);anti-MMP13 (1:3000, Abcam, ab39012); anti-CA12 (1:1000, Cell Signaling Technology, 5865); anti-lamin A/C (1:1000, Cell Signaling Technology, 2032); or anti-β-tubulin (1:3000, DSHB, E-7). Immunolabeling was detected with ECL reagent (Amersham Biosciences). Chemiluminescence was detected using Digital Imaging System ImageQuant LAS-4000 (GE Healthcare). Densitometric analysis included blots from at least 3 independent experiments and was performed using ImageQuant software. Protein levels were first normalized to the levels of housekeeping protein (β-tubulin) and then represented as relative change to the control group.
Gelatin zymography:
Conditioned medium from NP cells transduced with ShCtr and ShHuR and cultured in serum free DMEM in hypoxia for 24 h was collected. After concentration of conditioned medium using 10 kDa vivaspin 6 columns (Sartorius, Gloucestershire, UK), protein was quantified using BCA method. Equal amount of protein was loaded on to the polyacrylamide gel containing 0.1% gelatin into resolving gel. Electrophoresis was carried out under SDS non-reducing condition at constant voltage. The gel was incubated with 2.5% Triton X-100, 50mM Tris-HCl (pH 7.4), 5mM CaCl2 and 1 μM ZnCl2 at room temperature for 1–2 h to remove SDS. The gel was washed 3–4 times with 1% Triton X-100, 50mM Tris-HCl (pH 7.4), 5mM CaCl2, 1 μM ZnCl2 and incubated in the same buffer for overnight at 37 oC. Gel was stained with Coomassie Brilliant Blue R250 for 1h, destained and the metalloproteinases activity was visualized using EPSON Expression 1640XL gel scanner.
Immunohistochemistry:
Tail motion segments of 17 week-old mice were sectioned in the coronal plane (7 μm thickness). For antigen retrieval, deparaffinized sections were incubated in boiled citrate-based unmasking solution (Vector Laboratories, H-3301). Next, the sections were incubated for 1 hour in blocking solution and subsequently incubated overnight at 4°C with anti-HuR primary antibody (1:25, Santa Cruz Biotechnology, sc-5261). After washing with PBS, the tissues were incubated for 10 minutes with biotinylated probe. Excess probe was washed off with PBS, and the sections were incubated 1 hour with Alexa Fluor®−594 Streptavidin secondary antibody (1:700, Jackson ImmunoResearch Lab, Inc.). The sections were washed with PBS before mounting with ProLong® Gold Antifade Mountant containing DAPI (Thermo Fisher Scientific, P36934), and imaged by fluorescence microscopy (Axio Imager 2, Carl Zeiss). The mouse-on-mouse M.O.M.™ Immunodetection Kit (Vector Laboratories, BMK-2202) was used for blocking and incubation solutions.
Collagen precipitation and Coomassie blue staining:
HuR-silenced and control NP cells were plated in 6-well plate at 300,000 cells/well seeding density. The next day, cell media was changed with serum-free DMEM containing 40 μg/ml L-ascorbic acid 2-phosphate (Sigma Aldrich, A8960) and cultured in hypoxia for 24 h. After treatment, cell media was collected and mixed with 25% PEG 8000 stock solution to obtain the final concentration of 5% PEG (Sigma Aldrich, P5413). The mixture was incubated overnight at 4 oC to precipitate collagen. Subsequently, collagen pellet was collected by centrifugation at 16,000 × g for 30 min. To remove procollagen propeptides, precipitated samples were digested with the mixture of Trypsin and Chymotrypsin. Next, collagen chains were resolved in 6 % SDS-polyacrylamide gels and visualized with Bio-Safe Coomassie Stain solution (Bio-rad, 161–0786).
Immunofluorescence staining:
HuR-silenced and control NP cells were plated on glass coverslips and cultured in hypoxia for 72 h. After treatment, cells were fixed with 4% paraformaldehyde at room temperature for 15 minutes, washed with PBS and then blocked with 5% normal goat serum in PBS with 0.3% Triton X-100 (Sigma Aldrich, T8787) for 1 h at room temperature. Cells were then incubated with anti-Collagen I antibody (1:500, Abcam, ab34710), anti-Collagen II antibody (1:100, Fitzgerald, 70R-CR008), or anti-Aggrecan antibody (1:100, EMD Millipore, AB1031) in blocking buffer at 4°C overnight. Cells were washed with PBS and incubated with Alexa-fluor-594-conjugated anti-rabbit secondary antibody (1:800, Jackson ImmunoResearch Lab, 711–586-152) for 1 h at room temperature. After washing with PBS, cells were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, P36934) and visualized using a Zeiss AxioImager A2 (Carl Zeiss, Germany).
Messenger ribonucleoprotein immunoprecipitation (mRNP-IP or RIP) assay:
HuR-silenced and control NP cells were cultured in hypoxia for 24 h. After treatment, cells were collected and cytoplasmic extracts were isolated using the NE-PER™ Nuclear and Cytoplasmic Extraction kit (Thermo Fisher Scientific, 78833). The endogenous RNA-protein complexes were immunoprecipitated (IP) from cytoplasmic extracts using Protein A-Sepharose beads (Sigma, P9424) that were precoated with 30 μg of either rabbit IgG (Santa Cruz, sc-2027), or anti-HuR antibodies (Medical and Biological Laboratories, RN004P). After IP, the RNA was isolated from the IP materials via acid: phenol method, was purified, and reverse transcribed to cDNA. qRT-PCR detection of HuR-bound mRNAs was carried out as previously described [65–68].
Seahorse XF Analyzer Respiratory Assay:
HuR-silenced and control NP cells were plated in a 24-well XF Analyzer plate (Seahorse Bioscience, 100777–004) at a density of 20,000 cells/well in 1 g/L glucose DMEM with 10% FBS and cultured in hypoxia for 24 h. After treatment, the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using a Seahorse XF24 Analyzer (Seahorse Bioscience, North Billerica, MA), as described by Silagi et al. [19]. Basal levels of ECAR and OCR were created by first eight readings before the injection of Antimycin A (Sigma Aldrich). Mitochondrial OCR was calculated by subtracting the final OCR value after Antimycin A treatment from the average of the three OCR values before Antimycin A treatment. All measurements were normalized to total protein in our experiments.
Extracellular Lactate Measurement:
HuR-silenced and control NP cells were plated in a 6-well plate at a seeding density of 200,000 cells/well. After culturing the cells in hypoxia for 24 h, lactate concentrations were measured in each sample using a clinical lactate measurement kit from Trinity Biotech (#735–10) per the manufacturer’s instruction.
DNA measurement:
The Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, P7589) was used to quantify cell proliferation. HuR-silenced and control NP cells were seeded in 6-well plate at a density of 250,000 cells/well and incubated in hypoxia for 24 h, 48 h and 72h. After treatment, the media was removed and the cell layers processed for DNA measurement according to the manufacturer’s protocol. The fluorescence intensity was measured using an Infinite M1000 Pro microplate reader (Tecan, Switzerland). Quantification of DNA content was assessed using a dsDNA standard curve.
Statistical analysis:
All experiments are performed at least three independent times. Data is presented as the Mean ± SD. Differences between groups were analyzed by ANOVA and Student’s T-test using GraphPad Prism Software. P < 0.05 was considered statistically significant.
Supplementary Material
Figure 8.
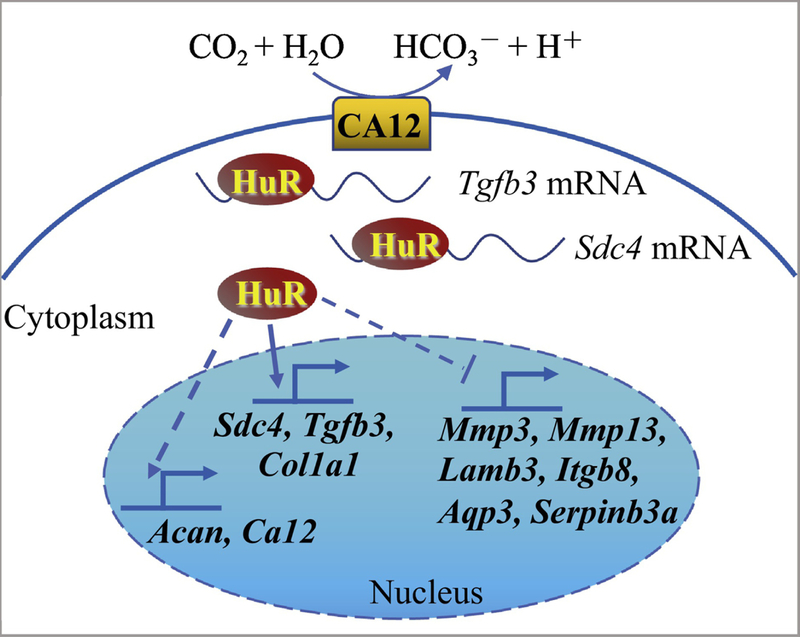
Schematic of proposed HuR regulation of extracellular matrix (ECM) and pH homeostasis in NP cells. HuR positively regulates Tgfb3, Sdc4, Col1a1, Acan and Ca12 expression through direct binding of Tgfb3, Sdc4 and Col1a1 mRNAs, and negatively regulates levels of Mmp3, Mmp13, Lamb3, Itgb8, Ptges, and Serpinb3a.
Highlights.
HuR regulates expression levels of major extracellular matrix molecules and MMPs in NP cells.
HuR does not control expression levels of HIF-1α and HIF-1 targets in NP cells
HuR does not bind to several known HuR mRNA targets in NP cells.
HuR may control pH homeostasis of NP cells through maintenance of Ca12 expression.
Acknowledgments:
This study is supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) AR055655 and AR064733 to MVR. The authors would like to thank Steven Tessier for helping with immunohistochemistry staining, Andrzej Steplewski for helping with collagen precipitation, Erin L. Seifert and Elizabeth S. Silagi for assistance with Seahorse experiments. In addition, we would like to thank the China Scholarship Council for supporting Hehai Pan (fellowship award #201506380080).
Abbreviations
- HuR
human antigen R
- HIF
hypoxia inducible factor
- IVD
intervertebral disc
- NP
nucleus pulposus
- AF
annulus fibrosus
- ECM
extracellular matrix
- LBP
low back pain
- SDC4
syndecan 4
- TGF-β3
transforming growth factor beta 3
- PHD2
prolyl hydroxylase 2
- PHD3
prolyl hydroxylase 3
- PGK1
phosphoglycerate kinase 1
- ENO1
enolase 1
- PDK1
pyruvate dehydrogenase kinase 1
- PFKFB3 6
phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- VEGF
vascular endothelial growth factor
- LDHB
lactate dehydrogenase B
- CDKN1B
cyclin dependent kinase inhibitor 1B
- IDH1
isocitrate dehydrogenase 1
- BCL2 B
Cell CLL/Lymphoma 2
- ACAN
aggrecan
- COL
collagen
- LAM
laminin
- ITG
integrin
- MMP
matrix metallopeptidase
- PTGES
prostaglandin E synthase
- AQP3
aquaporin 3
- SERPINB3A
serpin family B member 3a
- ECAR
extracellular acidification rate
- PPR
proton production rate
- OCR
oxygen consumption rate
- shRNA
short-hairpin RNA
- CA12
carbonic anhydrase 12
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors all declare no conflicts of interest.
References:
- [1].Hoy D, March L, Brooks P, Blyth F, Woolf A, Bain C, Williams G, Smith E, Vos T, Barendregt J, Murray C, Burstein R, Buchbinder R, The global burden of low back pain: estimates from the Global Burden of Disease 2010 study, Ann. Rheum. Dis 73 (2014) 968–974. [DOI] [PubMed] [Google Scholar]
- [2].Balague F, Mannion AF, Pellise F, Cedraschi C, Non-specific low back pain, Lancet 379 (2012) 482–91. [DOI] [PubMed] [Google Scholar]
- [3].Murray CJ, Atkinson C, Bhalla K, Birbeck G, Burstein R, Chou D, Dellavalle R, Danaei G, Ezzati M, Fahimi A, et al. , The state of US health, 1990–2010: burden of diseases, injuries, and risk factors, JAMA 310 (2013) 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang G, Wang Y, Zeng Y, Gao GF, Liang X, Zhou M, Wan X, Yu S, Jiang Y, Naghavi M, Vos T, Wang H, Lopez AD, Murray CJ, Rapid health transition in China, 1990–2010: findings from the Global Burden of Disease Study 2010, Lancet 381 (2013) 1987–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Bolliger I, Charlson F, Davis A, Degenhardt L, Dicker D, et al. , Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet 386 (2015) 743–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Battie MC, Videman T, Lumbar disc degeneration: epidemiology and genetics, J. Bone Joint Surg. Am 88 Suppl 2 (2006) 3–9. [DOI] [PubMed] [Google Scholar]
- [7].Steenstra IA, Prognostic factors for duration of sick leave in patients sick listed with acute low back pain: a systematic review of the literature, Occup. Environ. Med 62 (2005) 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Risbud MV, Shapiro IM, Role of cytokines in intervertebral disc degeneration: pain and disc content, Nat. Rev. Rheumatol 10 (2014) 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Roughley PJ, Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix, Spine (Phila Pa 1976) 29 (2004) 2691–9. [DOI] [PubMed] [Google Scholar]
- [10].Le Maitre CL, Freemont AJ, Hoyland JA, Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc, J. Pathol 204 (2004) 47–54. [DOI] [PubMed] [Google Scholar]
- [11].Silagi ES, Shapiro IM, Risbud MV, Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation and the pathogenesis of disc degeneration, Matrix Biol (2018). doi: 10.1016/j.matbio.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Choi H, Tessier S, Silagi ES, Kyada R, Yousefi F, Pleshko N, Shapiro IM, Risbud MV, A novel mouse model of intervertebral disc degeneration shows altered cell fate and matrix homeostasis, Matrix Biol (2018). doi: 10.1016/j.matbio.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tian Y, Yuan W, Li J, Wang H, Hunt MG, Liu C, Shapiro IM, Risbud MV, TGFbeta regulates Galectin-3 expression through canonical Smad3 signaling pathway in nucleus pulposus cells: implications in intervertebral disc degeneration, Matrix Biol 50 (2016) 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Binch A, Shapiro IM, Risbud MV, Syndecan-4 in intervertebral disc and cartilage: Saint or synner? Matrix Biol 52–54 (2016) 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, Albert TJ, Shapiro IM, Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment, J. Cell. Biochem 98 (2006) 152–9. [DOI] [PubMed] [Google Scholar]
- [16].Agrawal A, Guttapalli A, Narayan S, Albert TJ, Shapiro IM, Risbud MV, Normoxic stabilization of HIF-1alpha drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk, Am J Physiol Cell Physiol 293 (2007) C621–31. [DOI] [PubMed] [Google Scholar]
- [17].Agrawal A, Gajghate S, Smith H, Anderson DG, Albert TJ, Shapiro IM, Risbud MV, Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc, Arthritis Rheum 58 (2008) 3798–808. [DOI] [PubMed] [Google Scholar]
- [18].Merceron C, Mangiavini L, Robling A, Wilson TL, Giaccia AJ, Shapiro IM, Schipani E, Risbud MV, Loss of HIF-1alpha in the notochord results in cell death and complete disappearance of the nucleus pulposus, PLoS One 9 (2014) e110768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Silagi ES, Schoepflin ZR, Seifert EL, Merceron C, Schipani E, Shapiro IM, Risbud MV, Bicarbonate Recycling by HIF-1-Dependent Carbonic Anhydrase Isoforms 9 and 12 Is Critical in Maintaining Intracellular pH and Viability of Nucleus Pulposus Cells, J. Bone Miner. Res 33 (2018) 338–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fujita N, Chiba K, Shapiro IM, Risbud MV, HIF-1alpha and HIF-2alpha degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc, J. Bone Miner. Res 27 (2012) 401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fujita N, Markova D, Anderson DG, Chiba K, Toyama Y, Shapiro IM, Risbud MV, Expression of prolyl hydroxylases (PHDs) is selectively controlled by HIF-1 and HIF-2 proteins in nucleus pulposus cells of the intervertebral disc: distinct roles of PHD2 and PHD3 proteins in controlling HIF-1alpha activity in hypoxia, J. Biol. Chem 287 (2012) 16975–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hirose Y, Johnson ZI, Schoepflin ZR, Markova DZ, Chiba K, Toyama Y, Shapiro IM, Risbud MV, FIH-1-Mint3 axis does not control HIF-1 transcriptional activity in nucleus pulposus cells, J. Biol. Chem 289 (2014) 20594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Suyama K, Silagi ES, Choi H, Sakabe K, Mochida J, Shapiro IM, Risbud MV, Circadian factors BMAL1 and RORalpha control HIF-1alpha transcriptional activity in nucleus pulposus cells: implications in maintenance of intervertebral disc health, Oncotarget 7 (2016) 23056–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schoepflin ZR, Silagi ES, Shapiro IM, Risbud MV, PHD3 is a transcriptional coactivator of HIF-1alpha in nucleus pulposus cells independent of the PKM2-JMJD5 axis, FASEB J 31 (2017) 3831–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Keene JD, RNA regulons: coordination of post-transcriptional events, Nat. Rev. Genet 8 (2007) 533–43. [DOI] [PubMed] [Google Scholar]
- [26].Galban S, Gorospe M, Factors interacting with HIF-1alpha mRNA: novel therapeutic targets, Curr Pharm Des 15 (2009) 3853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C, Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha, J. Biol. Chem 279 (2004) 14871–8. [DOI] [PubMed] [Google Scholar]
- [28].Chamboredon S, Ciais D, Desroches-Castan A, Savi P, Bono F, Feige JJ, Cherradi N, Hypoxia-inducible factor-1alpha mRNA: a new target for destabilization by tristetraprolin in endothelial cells, Mol. Biol. Cell 22 (2011) 3366–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T, Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster, Cancer Res 68 (2008) 5540–5. [DOI] [PubMed] [Google Scholar]
- [30].Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M, Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes, Circ. Res 104 (2009) 879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S, The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia, Nucleic Acids Res 33 (2005) 6884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hagele S, Kuhn U, Boning M, Katschinski DM, Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1alpha mRNA 3’-UTR and modulate HIF-1alpha protein expression, Biochem. J 417 (2009) 235–46. [DOI] [PubMed] [Google Scholar]
- [33].Galban S, Kuwano Y, Pullmann RJ, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M, RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha, Mol. Cell. Biol 28 (2008) 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H, Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein, J. Biol. Chem 271 (1996) 8144–51. [DOI] [PubMed] [Google Scholar]
- [35].Hinman MN, Lou H, Diverse molecular functions of Hu proteins, Cell. Mol. Life Sci 65 (2008) 3168–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lopez DSI, Zhan M, Lal A, Yang X, Gorospe M, Identification of a target RNA motif for RNA-binding protein HuR, Proc Natl Acad Sci U S A 101 (2004) 2987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Brennan CM, Steitz JA, HuR and mRNA stability, Cell. Mol. Life Sci 58 (2001) 266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Srikantan S, Gorospe M, HuR function in disease, Front Biosci (Landmark Ed) 17 (2012) 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner-Kober N, Londin E, Rigoutsos I, Sawicki JA, Risbud MV, Witkiewicz AK, McCue PA, Jiang W, Rui H, Yeo CJ, Petricoin E, Winter JM, Brody JR, The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells, Oncogene 35 (2016) 2529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, Cozzitorto JA, Rigoutsos I, Yeo CJ, Brody JR, Winter JM, HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer, RNA Biol 10 (2013) 1312–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K, Mambelli-Lisboa NC, Moffat C, Blanco FF, Chand SN, Jimbo M, Cozzitorto JA, Jiang W, Yeo CJ, Londin ER, Seifert EL, Metallo CM, Brody JR, Winter JM, Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells, Cancer Res 77 (2017) 4460–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM, Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells, J. Clin. Invest 108 (2001) 1657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, Sakai H, High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer, PLoS One 8 (2013) e59095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nabors LB, Gillespie GY, Harkins L, King PH, HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs, Cancer Res 61 (2001) 2154–61. [PubMed] [Google Scholar]
- [45].Sakuma T, Nakagawa T, Ido K, Takeuchi H, Sato K, Kubota T, Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human meningiomas, J Neurooncol 88 (2008) 143–55. [DOI] [PubMed] [Google Scholar]
- [46].Sheflin LG, Zou AP, Spaulding SW, Androgens regulate the binding of endogenous HuR to the AU-rich 3’UTRs of HIF-1alpha and EGF mRNA, Biochem Biophys Res Commun 322 (2004) 644–51. [DOI] [PubMed] [Google Scholar]
- [47].Gauchotte G, Hergalant S, Vigouroux C, Casse JM, Houlgatte R, Kaoma T, Helle D,Brochin L, Rech F, Peyre M, Labrousse F, Vallar L, Gueant JL, Vignaud JM, Battaglia-Hsu SF, Cytoplasmic overexpression of RNA-binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia, J. Pathol 242 (2017) 421–434. [DOI] [PubMed] [Google Scholar]
- [48].Levy NS, Chung S, Furneaux H, Levy AP, Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR, J. Biol. Chem 273 (1998) 6417–23. [DOI] [PubMed] [Google Scholar]
- [49].Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano MJ, Tuschl T, Ohler U, Keene JD, Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability, Mol. Cell 43 (2011) 327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yi J, Chang N, Liu X, Guo G, Xue L, Tong T, Gorospe M, Wang W, Reduced nuclear export of HuR mRNA by HuR is linked to the loss of HuR in replicative senescence, Nucleic Acids Res 38 (2010) 1547–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, Chen A, Zhao YD, Herman T, Ramesh R, Munshi A, HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells, Sci Rep 7 (2017) 9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Guo X, Hartley RS, HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells, Cancer Res 66 (2006) 7948–56. [DOI] [PubMed] [Google Scholar]
- [53].He Y, Zhang X, Zeng X, Huang Y, Wei JA, Han L, Li CX, Zhang GW, HuR-mediated posttranscriptional regulation of p21 is involved in the effect of Glycyrrhiza uralensis licorice aqueous extract on polyamine-depleted intestinal crypt cells proliferation, J. Nutr. Biochem 23 (2012) 1285–93. [DOI] [PubMed] [Google Scholar]
- [54].Bai D, Gao Q, Li C, Ge L, Gao Y, Wang H, A conserved TGFbeta1/HuR feedback circuit regulates the fibrogenic response in fibroblasts, Cell. Signal 24 (2012) 1426–32. [DOI] [PubMed] [Google Scholar]
- [55].McDermott BT, Ellis S, Bou-Gharios G, Clegg PD, Tew SR, RNA binding proteins regulate anabolic and catabolic gene expression in chondrocytes, Osteoarthritis Cartilage 24 (2016) 1263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Huwiler A, Akool E, Aschrafi A, Hamada FM, Pfeilschifter J, Eberhardt W, ATP potentiates interleukin-1 beta-induced MMP-9 expression in mesangial cells via recruitment of the ELAV protein HuR, J. Biol. Chem 278 (2003) 51758–69. [DOI] [PubMed] [Google Scholar]
- [57].Bridgen DT, Gilchrist CL, Richardson WJ, Isaacs RE, Brown CR, Yang KL, Chen J, Setton LA, Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc, J. Orthop. Res 31 (2013) 1661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].DuMond JF, Ramkissoon K, Zhang X, Izumi Y, Wang X, Eguchi K, Gao S, Mukoyama M, Burg MB, Ferraris JD, Peptide affinity analysis of proteins that bind to an unstructured NH2-terminal region of the osmoprotective transcription factor NFAT5, Physiol. Genomics 48 (2016) 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tsai TT, Danielson KG, Guttapalli A, Oguz E, Albert TJ, Shapiro IM, Risbud MV, TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc, J. Biol. Chem 281 (2006) 25416–24. [DOI] [PubMed] [Google Scholar]
- [60].Tsai TT, Guttapalli A, Agrawal A, Albert TJ, Shapiro IM, Risbud MV, MEK/ERK signaling controls osmoregulation of nucleus pulposus cells of the intervertebral disc by transactivation of TonEBP/OREBP, J. Bone Miner. Res 22 (2007) 965–74. [DOI] [PubMed] [Google Scholar]
- [61].Hiyama A, Gajghate S, Sakai D, Mochida J, Shapiro IM, Risbud MV, Activation of TonEBP by calcium controls {beta}1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc, J. Biol. Chem 284 (2009) 9824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Johnson ZI, Shapiro IM, Risbud MV, RNA Sequencing Reveals a Role of TonEBP Transcription Factor in Regulation of Pro-inflammatory Genes in Response to Hyperosmolarity in Healthy Nucleus Pulposus Cells: A HOMEOSTATIC RESPONSE? J. Biol. Chem 291 (2016) 26686–26697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mookerjee SA, Goncalves RL, Gerencser AA, Nicholls DG, Brand MD, The contributions of respiration and glycolysis to extracellular acid production, Biochim Biophys Acta 1847 (2015) 171–81. [DOI] [PubMed] [Google Scholar]
- [64].Grammatikakis I, Abdelmohsen K, Gorospe M, Posttranslational control of HuR function, Wiley Interdiscip Rev RNA 8 (2017). doi: 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M, Brody JR, The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase, Cancer Res 69 (2009) 4567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S, Cozzitorto JA, Nevler A, Scolaro L, Londin E, Jiang W, Meisner-Kober N, Pishvaian MJ, Knudsen KE, Yeo CJ, Pascal JM, Winter JM, Brody JR, Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors, Cancer Res 77 (2017) 5011–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kuwano Y, Kim HH, Abdelmohsen K, Pullmann RJ, Martindale JL, Yang X, Gorospe M, MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90, Mol. Cell. Biol 28 (2008) 4562–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cozzitorto JA, Jimbo M, Chand S, Blanco F, Lal S, Gilbert M, Winter JM, Gorospe M, Brody JR, Studying RNA-binding protein interactions with target mRNAs in eukaryotic cells: native ribonucleoprotein immunoprecipitation (RIP) assays, Methods Mol Biol 1262 (2015) 239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.