Abstract
Renal transplant recipients have an increased risk of non-melanoma skin cancer (NMSC) compared to in the general population. Here, we show polygenic risk scores (PRS) calculated from genomewide association studies (GWAS) of NMSC in general, non-transplant setting, can predict risk of, and time to post-transplant skin cancer. Genetic variants, reaching pre-defined p-value thresholds were chosen from published squamous cell carcinoma (SCC) and basal cell carcinoma (BCC) non-transplant GWAS. Using these genome-wide association studies, BCC and SCC PRS were calculated for each sample across three European-ancestry renal-transplant cohorts (n=889) and tested as predictors of case: control status and time to NMSC post-transplant. BCC PRS calculated at p-value threshold 1×10−5 was the most significant predictor of case: control status of NMSC post-transplant (OR=1.65; adjusted P=0.0008; AUC(full model adjusted for clinical predictors and PRS)=0.81). SCC PRS at p-value threshold 1×10−5 was the most significant predictor of time to post-transplant NMSC (adjusted P=8.15×10−7; HR=1.42, concordance (full model)=0.74). PRS of non-transplant NMSC is predictive of case: control status and time to NMSC post-transplant. These results are relevant to how genomics can risk stratify patients to help develop personalised treatment regimens.
1. INTRODUCTION
Renal transplantation is an effective therapy for end-stage kidney failure, with one-year graft survival rates of between 89 and 91% for deceased donor grafts and graft half-life of 8.8 years in the United States1,2. As a result of this success, malignancy is a common transplant complication, with non-melanoma skin cancers (NMSC) accounting for 95% of post-transplant malignancies3. Renal transplant recipients have an estimated prevalence of NMSC ranging from 2%−18%4 and an approximate 16-fold excess risk of developing basal cell carcinoma (BCC), and 65-fold increased risk of developing squamous cell carcinoma (SCC) relative to age-matched non-transplanted individuals5. A number of studies have evaluated clinical predictors of NMSC post-transplant 6–8 and robust predictors include age, sex and type of immunosuppressive treatment6,8,9.
The immunosuppressive regime chosen for a patient can have a dramatic effect on their risk of developing skin cancer, as immunosuppressants can impair the immune system’s ability to scan for potentially oncogenic cells. Certain classes of immunosuppressant drugs have additional carcinogenic properties beyond their effects on the immune system9. For example, it has been suggested that azathioprine exposure when combined with UV light induces DNA damage, which can lead to tumorigenesis9. Conversely, mammalian target of rapamycin (mTOR) inhibitors, which is also used as an immunosuppressant in solid organ transplants, were reported to reduce skin cancer incidence post-transplantation10.
Despite the increased risk of skin cancer, only one study has tested for germline genetic associations with skin cancer post-transplant on a genomic scale7. Studying SCC in a kidney and heart transplant population, the authors highlighted increased age and azathioprine exposure as risk factors for skin cancer, but no genome-wide significant genetic predictors were detected7. This study had limited power (n=388 cases) compared to GWAS of SCC and BCC in non-transplant populations (n=8058+) where robust genetic risk factors for NMSC were identified11–14.
The genetic architecture of NMSC in a transplant setting would appear to mirror that of complex human traits generally, where common genetic variants individually tend to have small effect sizes and limited clinical relevance, with some notable exceptions15,16. However, by combining effect sizes across variants one can create an overall ‘polygenic risk score’ (PRS)17 which might produce a more clinically relevant measure of genetic load for a given trait. For the purpose of this study, PRS was defined as the sum of all alleles associated with a trait weighted by the effect size of that allele as determined by a previous GWAS.
Here we set out to leverage large, robust published GWAS, to develop a PRS of NMSC and test for ability to predict in a transplant setting, case-control status and time to developing NMSC.
2. MATERIALS AND METHODS
Written, informed consent was obtained from each participant in the study. Approval for ethics study protocols was received from appropriate review committees.
2.1. Cohort
Data on post-kidney transplant recipients was collected from Tennessee (Vanderbilt University Medical Centre, n=275), Dublin (Beaumont Hospital, n=280), Glasgow (Western Infirmary Glasgow, n=64) and Belfast (Belfast City Hospital, n=270). For the Dublin and Belfast cohorts, NMSC status was taken from de-identified National Cancer Registry Records. For the Tennessee cohort, NMSC status was retrieved from de-identified electronic medical records7. For the Glasgow cohort, cases were retrieved from hospital records. Due to the small case numbers in Glasgow (n-cases=4), these samples were merged with the Dublin cohort for further analyses.
Analysis was carried out on three phenotypes in our post-transplant cohorts: SCC, BCC and any NMSC. A number of individuals reported NMSC but did not have a specific sub-type recorded. Such individuals were included in the any NMSC analysis but not the BCC or SCC analyses. For the Cox analysis (see statistical analyses section below), cases were defined as those who developed NMSC post-transplantation; controls are those who have no record of NMSC post-transplantation. Controls were censored at last known follow-up, date of renal allograft failure or patient death. For the logistic regression (see below), cases were defined as those who developed NMSC within ten years’ post-transplantation. Controls had to be at least ten years’ cancer free post-transplantation. This cut-off was selected as it incorporated previously reported peak incidence rates of NMSC posttransplant whilst still allowing for a sufficient control group in the three cohorts 5. If individuals developed both SCC and BCC post-transplant, for the any NMSC Cox analysis – time to the first incidence of either was taken as the time of event. For the subtype analysis, time to the specific BCC or SCC event was taken as the time of event. Controls in the subtype (BCC and SCC) analysis had to be free of any NMSC post-transplant.
Exclusion criteria were as follows: non-European individuals as determined via principal components analysis (PCA, see below), related individuals (one from each pair of individuals with PIHAT score >0.1 removed) and individuals who had any incidence of any type of NMSC prior to transplant. Azathioprine exposure, era of transplantation (pre-2000 vs. post-2000), age at transplantation and sex of recipient were tested as predictors of post-transplant NMSC status. Azathioprine exposure data was unavailable for the Belfast cohort. Individuals in the Tennessee cohort were on an oral immunosuppressive regimen of calcineurin inhibitor (predominantly tacrolimus) with 38 on azathioprine in addition to tacrolimus7. In the Belfast cohort, 199 individuals received cyclosporin A and six individuals received tacrolimus (73 individuals had missing immunosuppression data). In the Dublin/Glasgow cohort, 330 individuals received cyclosporin A and eleven individuals received tacrolimus with 278 exposed to azathioprine. The majority of the Tennessee cohort were transplanted post-2000 (n=220), with 47 being transplanted between 1990–2000 and eight patients transplanted pre-1990, with the year of transplant ranging from 1970 to 2011. In the Belfast cohort, the year of transplant ranged from 1986 to 2004 with 54 patients transplanted post-2000, 153 patients between 1990–2000 and 63 individuals being transplanted pre-1990. In the Dublin/Glasgow cohort, 13 individuals were transplanted pre-1990, 266 individuals were transplanted between 1990–2000 and 65 individuals were transplanted post 2000 with the year of transplant ranging from 1981 to 2006.
2.2. Genotyping, imputation and data quality control
Belfast and Glasgow samples were genotyped using the Illumina® 660K array as part of the UK and Ireland Renal Transplant Consortium (ukirtc.org) and the Wellcome Trust Case Control Consortium 3 Study into Renal Transplant Dysfunction18. Genotyping details for the Dublin and Tennessee cohorts were described previously7,19.
Tennessee, Dublin and Glasgow datasets were imputed using Impute2 and the 1000 Genomes Phase 3 (1KG3) and Genomes of the Netherlands version 5 reference panels as previously described20–24. The Belfast dataset was phased and imputed via the Sanger Imputation Service using Eagle, PBWT, and 1KG3-reference panel25–28. SNP2HLA and the Type 1 Diabetes Genetic Consortium were used to impute HLA alleles for each cohort29,30.
Imputed genotype probabilities were converted to hard-called genotypes using PLINK (Chang et al., 2015). Calls with uncertainty greater than 0.05 were excluded using the hard-call threshold function. Variants with missingness >5%, info scores <0.95, minor allele frequency <0.05 and Hardy Weinberg p<1×10−6 were removed. PCA was performed using GCTA to identify population outliers31 and to control for population structure. PC (principal component) 1 and 2 were plotted using ggplot2 in R and outliers were identified32. To identify non-European descent individuals, each of the datasets was merged with the Human Genome Diversity Project (HGDP) dataset33 and PCs were calculated and plotted. Individuals that did not plot to the same region as the HGDP European groups were removed.
2.3. Polygenic risk scores
PRS was defined as the sum of all alleles associated with a trait weighted by the effect size of that allele as determined by a previous GWAS17. SNPs and associated effect sizes (in this case, log OR) that reached pre-defined p-value thresholds (pT; 0.0001, 1×10−5, 1×10−6, 1×10−7, 1×10−8, 1×10−9) were extracted from the discovery GWAS results data from two previously published GWAS papers, one of SCC13 and one of BCC, both in a non-transplant population14. The full set of discovery SNPs and associated effect sizes needed for calculating the PRS can be accessed via 23andme. For more details on accessing this data see 13 and 14. Effect sizes for the SNPs that reached the given pT, along with the effect allele and associated rsID were extracted from these results. These SNP sets (one for each pT) were then extracted from the transplanted cohorts’ genotype data using PLINK’s extract function. The genotype data was then pruned to remove SNPs in high linkage disequilibrium using PLINK with a 200kb sliding window, 10kb step size and r2 threshold of 0.2534. SCC PRS and BCC PRS were then calculated for each individual in our transplant cohorts using PLINK’s score function. In each cohort, PRSs were normalized to mean zero and standard deviation of one.
2.4. Statistical analyses
To evaluate the PRSs’ predictive value, they were tested as predictors of case: control status using logistic regression, and time to developing post-transplant NMSC using Cox proportional hazards models. Covariates included age at time of transplantation, era of transplant, site and the first eight PCs. Site was just included as a covariate for the Dublin-Glasgow analyses to account for the differences in obtaining NMSC status between the hospitals. Logistic regression was carried out using the glm function in R and Cox analysis was completed using the cox.ph function of the R survival package35,36.
To correct for multiple testing, we applied an approximate adjusted correction technique37. As the PRSs include subsets of the same overall group of SNPs, they are correlated. Bonferroni correction assumes tests are independent and therefore would be overly conservative. In this context, the following equation was applied37:
Padj = 1-(1-p(k))g(k)
Where g(k)=M1-r(.k)
Letpadj=adjusted p-value
M=number of PRSs tested × number of phenotypes tested
p(k)=unadjusted p-value for the given PRS score
r(.k)=mean correlation among the outcomes other than outcome k
After the p-values were adjusted for the number of PRSs tested using the above technique, further correction was applied to account for the three phenotypes being tested (NMSC, SCC and BCC) using Bonferroni correction.
Beta coefficients were meta-analysed using the metafor R package38. The percentage of variation across the results from the different sites in the meta-analysis was accessed for heterogeneity using the I2 statistic39. Receiver operator curves were calculated using the pROC package in R40.
We performed decile analysis using the most significant PRS from the NMSC logistic regression (BCC pT1×10−5) and Cox analysis (SCC pT1×10−5). We merged the four datasets and divided samples into decile groups with the lowest polygenic load in the first decile and the highest PRS in the tenth decile. Samples in each decile group were then compared to patients the first decile group and tested for association with NMSC using the previously described Cox and logistic regression analysis.
3. RESULTS
Polygenic risk analysis was performed on 889 transplant recipients (567 males and 322 females), 239 of whom developed any NMSC with 106 developing BCC and 150 developing SCC (see table 1) screened for European ancestry. Average time to developing NMSC was 2,837 days with a range from 91 to 7,343 days. In the total dataset, patients had a mean age of 47.3 years at time of transplantation and had an average follow-up (censoring at first skin cancer event, failure or death) of 9.0 years ranging from one to 10,398 days. During this follow-up period, there were 243 graft failures with an average of 6.7 years to graft failure. Patients demonstrated a one, five and ten-year graft survival of 95.4%, 85.8% and 65.3% respectively.
Table 1.
Summary statistics of total dataset
| NMSC | SCC | BCC | |
|---|---|---|---|
| Total N with skin cancer data | 889 | 593 | 549 |
| Number of events | 239 | 150 | 106 |
| Average days to event (stdev) | 2837(1920) | 2897(1927) | 3314(2121) |
| Average days of follow-up in controls (stdev) | 3456(1881) | 3387(1854) | 3387(1854) |
| No. (%) male - Total | 567(64%) | 371(63%) | 343(62%) |
| No. (%) male - Cases | 165(69%) | 105(70%) | 77(73%) |
| No. (%) male - Controls | 402(62%) | 266(60%) | 266(60%) |
| Average age (years) at transplant (stdev) - Total |
47.27 (14.37) | 48.72 (14.44) | 47.99 (14.39) |
| Average age (years) at transplant (stdev) - Cases |
52.26 (12.79) | 54.19 (12.56) | 52.69 (12.58) |
| Average age (years) at transplant (stdev) - Controls |
45.44 (14.49) | 46.86 (14.58) | 47.01 (14.69) |
Stdev = standard deviation, No. (%) male – Total/Cases/Controls = Number (and percentage) of males in total cohort, cases only and controls only, NMSC=non-melanoma skin cancer, SCC = squamous cell carcinoma, BCC= basal cell carcinoma, Total N with skin cancer data = number of individuals with skin cancer data available for given skin cancer type
In order to facilitate a comprehensive assessment of PRSs on post-transplant NMSC we first set out to identify the clinical variables influencing NMSC status in a post-transplant setting. In this context, we analysed age at time of transplantation, sex, era of transplant and azathioprine exposure using a logistic regression and Cox regression approach (see methods section). These were tested against NMSC, as well as the two main subtypes of NMSC: BCC and SCC to evaluate subtype specific differences. Age alone was found to be a significant predictor of NMSC, BCC and SCC in both the Cox and logistic regression analyses (see table 2) and was brought forward as a covariate in our analyses. Although era of transplantation was not a significant predictor of skin cancer, we felt it was appropriate to include it as a covariate as to account for differences in immunosuppression regimens which are known to influence the onset of NMSC post-transplant41.
Table 2.
Clinical meta-analyses results
| Logistic | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMSC | SCC | BCC | |||||||||||||
| Clinical Variable | OR |
Lower CI |
Upper CI |
Pbon | OR |
Lower CI |
Upper CI |
Pun | Pbon | OR |
Lower CI |
Upper CI |
Pun | Pbon | |
| Age | 1.02 | 1.016 | 2.56X10- 1.024 12 |
3.07X10- 11 |
1.02 | 1.01 | 1.03 | 3.27X10−13 | 3.92X10- 12 |
1.02 | 1.016 | 1.024 | 3.53X10−14 | 4.24X10- 13 |
|
| Era | 1.34 | 1.03 | 1.74 0.03 | 0.37 | 1.46 | 1.00 | 2.14 | 0.0521 | 0.63 | 1.50 | 0.89 | 2.51 | 0.1257 | 1 | |
| Sex | 0.85 | 0.76 | 0.96 0.007 | 0.08 | 0.84 | 0.75 | 0.95 | 0.005 | 0.06 | 0.90 | 0.79 | 1.02 | 0.09 | 1 | |
| Azathioprine | 1.13 | 0.95 | 1.34 0.18 | 1.00 | 1.11 | 0.93 | 1.32 | 0.24 | 1 | 1.11 | 0.91 | 1.34 | 0.32 | 1 | |
| Cox | |||||||||||||||
| NMSC | SCC | BCC | |||||||||||||
| Clinical Variable | HR |
Lower CI |
Upper CI |
Pun | Pbon | HR |
Lower CI |
Upper CI |
Pun | Pbon | HR |
Lower CI |
Upper P CI un |
Pbon | |
| Age | 1.06 | 1.04 | 1.08 | 3.35X10- 24 |
4.02X10- 23 |
1.07 | 1.05 | 1.09 | 3.85X10−18 | 4.62X10- 17 |
1.07 | 1.05 | 1.09 5.46X10−13 | 6.55X10- 12 |
|
| Era | 0.92 | 0.54 | 1.54 | 0.74 | 1.00 | 0.93 | 0.55 | 1.55 | 0.77 | 1 | 1.03 | 0.32 | 3.33 0.95 | 1 | |
| Sex | 0.65 | 0.47 | 0.91 | 0.01 | 0.12 | 0.64 | 0.45 | 0.91 | 0.01 | 0.12 | 1.57 | 0.72 | 3.43 0.31 | 1 | |
| Azathioprine | 2.23 | 1.04 | 4.78 | 0.04 | 0.48 | 3.06 | 0.84 | 11.17 | 0.09 | 1 | 0.76 | 0.44 | 1.32 0.33 | 1 | |
NMSC=non-melanoma skin cancer, SCC = squamous cell carCInoma, BCC= basal cell carCInoma, OR = odds ratio, HR= hazards ratio, Lower/UpperCI = Lower/Upper 95% confidence interval, Pun=uncorrected p value, Pbon=Bonferroni corrected p value (corrected for 12 tests). Estimate for sex is assoCIated with females, OR/HR for age represents change in odds per year of age, era = those who had transplant pre-2000 = 1 and those who had transplant from 2000 onwards = 2
To investigate the impact of PRS on time to NMSC, we carried out a Cox proportional hazards analysis adjusting for age, era, eight PCs and clinical centre for transplantation (site) where appropriate (see methods section). The Cox analysis was carried out separately for each site and then meta-analysed across sites. Analysing the impact of SCC and BCC PRS on case: control status of NMSC, we observed a number of PRSs significantly associated with time-to-event and case: control status of NMSC, BCC and SCC post-transplant (see table 3).
Table 3.
PRS logistic and Cox regression top results
| Logistic | ||||||||
|---|---|---|---|---|---|---|---|---|
|
Cancer Type |
Top PRS | OR |
Lower CI |
Upper CI |
Padj | Pbon |
N cases |
N |
| NMSC | BCC pT1×10−5 | 1.61 | 1.22 | 2.12 | 0.0022 | 0.03 | 165 | 438 |
| SCC | SCC pT1×10−9 | 1.97 | 1.32 | 2.93 | 0.0023 | 0.03 | 100 | 283 |
| BCC | SCC pT1×10−6 | 3.03 | 1.78 | 5.16 | 0.0001 | 0.0016 | 60 | 243 |
| Cox | ||||||||
|
Cancer Type |
Top PRS | HR |
Lower CI |
Upper CI |
Padj | Pbon |
N events |
N |
| NMSC | SCC_pT1×10−5 | 1.41 | 1.24 | 1.61 | 9.39E- 07 |
1.29E-05 | 239 | 889 |
| SCC | SCC_pT1×10−6 | 1.48 | 1.25 | 1.74 | 9.35E- 06 |
0.0001 | 150 | 593 |
| BCC | SCC_pT1×10−6 | 1.67 | 1.35 | 2.06 | 5.5E-06 | 7.02E-05 | 106 | 549 |
Meta-analysis results for logistic regression and Cox regression. Models were adjusted for age at time of transplant, era of transplant (pre-2000 vs 2000 onwards), recruitment site and the first 8 principal components. (NMSC=non-melanoma skin cancer, SCC = squamous cell carcinoma, BCC= basal cell carcinoma, Top PRS = most significant polygenic risk score associated with given skin cancer in given model. OR = odds ratio, HR= hazards ratio, Lower/Upper CI = lower/upper 95% confidence interval, OR and HR based on 1 standard deviation increase in normalized PRS, Padj=approximate adjusted p value, Pbon=Bonferroni corrected p value (corrected for 36 tests); N = number of individuals tested
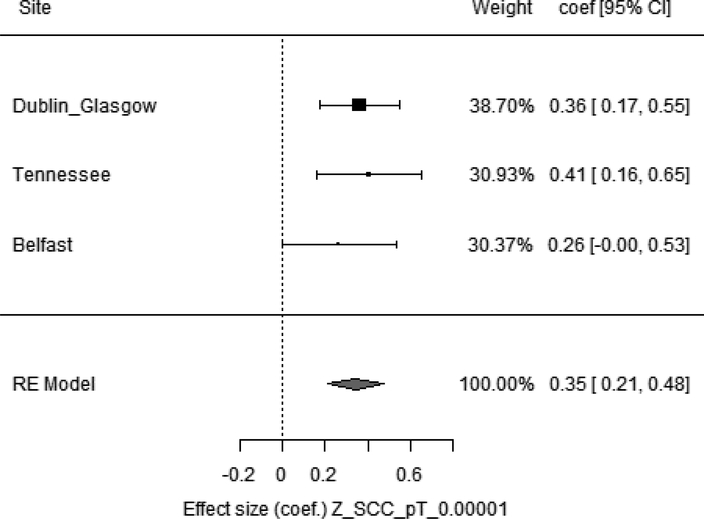
SCC pT1×10−5 PRS was the most significant predictor of time to NMSC post-transplant and BCC pT1×10−5 PRS was the most significant predictor of NMSC post-transplant case: control status, with a number of other SCC and BCC PRSs at different p-value thresholds also reaching significance. There was a large degree of overlap in the SNPs that reached pT1×10−5 in BCC and SCC GWASs13,14, with 69% of the SNPs in the SCC PRS found in the BCC PRS at this threshold (see Table S1 and S2 for SNPs used for these scores). This overlap is consistent with our finding that both SCC and BCC PRSs were significant of time to and case:control status of NMSC. Figure 1 illustrates how an increase in SCC pT1×10−5 PRS leads to a faster time to developing NMSC. This effect was consistent across cohorts and the same trend was seen in the BCC and SCC sub analyses (see table 3).
Figure 1. SCC pT1×10−5 as a predictor of time to developing NMSC post-transplant.
Weight= proportion of data the given site contributed to overall model; coef [95% CI] = effect size with lower and upper 95% confidence intervals. Z_SCC_pT_0.00001 = normalized PRS of SCC at p value threshold 0.00001, RE model = Random effects model, models were adjusted for age, recruitment site, 8 PCs and era of transplantation (pre-2000 vs post-2000).
Increased BCC pT1×10−5 PRS correlated with an increased risk of developing NMSC within the first ten years’ post-transplant (see table 3). The afore-mentioned most significant results all had an I2 of 0% indicating there was low heterogeneity across the variation of results at the different sites.
For the most significant PRS in the Cox and logistic NMSC analysis, we performed a decile analysis whereby samples in each decile group were compared to patients the first (lowest) decile group and tested for association with NMSC (see table 4). For this and for the proceeding receiver operator curve and concordance analyses we merged the three cohorts prior to analysing association with NMSC. We found that for each decile group, the lower PRS subset associated with lower risk of NMSC post-transplant and reduced time to NMSC post-transplant, with the highest polygenic load (10th decile) group having a hazard ratio of 3.11 when compared to the first decile group.
Table 4.
PRS logistic and Cox regression top results split by decile
| Logistic Top PRS BCC pT0.00001 |
||||||
|---|---|---|---|---|---|---|
| PRS by decile | OR | Lower CI |
Upper CI |
P | N | N Cases |
| 2nd Decile | 2.48 | 0.68 | 9.02 | 0.17 | 92 | 24 |
| 3rd Decile | 1.61 | 0.49 | 5.28 | 0.43 | 89 | 24 |
| 4th Decile | 2.20 | 0.76 | 6.35 | 0.15 | 94 | 31 |
| 5nd Decile | 3.17 | 0.91 | 11.04 | 0.07 | 101 | 26 |
| 6nd Decile | 1.32 | 0.40 | 4.38 | 0.65 | 91 | 27 |
| 7nd Decile | 4.10 | 1.20 | 13.98 | 0.024 | 91 | 27 |
| 8nd Decile | 6.06 | 1.67 | 21.92 | 0.006 | 96 | 34 |
| 9nd Decile | 4.35 | 1.35 | 13.99 | 0.014 | 96 | 31 |
| 10th Decile (highest) | 5.22 | 1.65 | 16.57 | 0.005 | 89 | 36 |
| Cox Top PRS SCC pT0.00001 |
||||||
| PRS by decile | HR |
Lower CI |
Upper CI |
P | N | N events |
| 2nd Decile | 1.24 | 0.58 | 2.67 | 0.58 | 178 | 34 |
| 3rd Decile | 1.49 | 0.75 | 2.97 | 0.25 | 178 | 39 |
| 4th Decile | 2.09 | 1.00 | 4.35 | 0.049 | 178 | 41 |
| 5th Decile | 2.00 | 0.98 | 4.08 | 0.06 | 177 | 37 |
| 6th Decile | 1.90 | 0.93 | 3.87 | 0.08 | 178 | 42 |
| 7th Decile | 0.99 | 0.47 | 2.12 | 0.99 | 178 | 37 |
| 8th Decile | 1.83 | 0.93 | 3.61 | 0.08 | 178 | 43 |
| 9th Decile | 3.37 | 1.82 | 6.22 | 0.0001 | 178 | 55 |
| 10th Decile (highest) | 3.11 | 1.60 | 6.06 | 0.0008 | 177 | 47 |
Samples in each decile group were compared to those in the first decile group (lowest PRS group). Analyses were adjusted for age, era of transplantation, geographic site and eight PCs. SCC = squamous cell carcinoma, BCC= basal cell carcinoma, Top PRS = most significant polygenic risk score associated with given skin cancer in given model, pT = p-value threshold, OR = odds ratio, HR= hazards ratio, Lower/Upper CI = lower/upper 95% confidence interval, P= p value, N = number of individuals tested, N events = number of skin cancer events observed, N cases = number of individuals with non-melanoma skin cancer.
To access the potential clinical applicability of the significant PRSs previously described, we performed a comparative receiver operator curve analysis that determined the area under the curves for the different predictive models (see figure 2). The most predictive model was the full model, which included age, site, era, SCC pT1×10−5 PRS and PCs. The area under the curve for the clinical model (which included ‘age’, ‘era’ and ‘site’) was 0.786 compared with 0.813 for the full model (which included ‘age’, ‘era’, ‘site’, ‘PRS’ and ‘PCs’) indicating that the genetic factors contribute additional predictive value. The area under the curve when PCs were excluded from the full model, leaving just clinical covariates and PRS, was 0.799. The PRS alone, without additional covariates, had an area under the curve of 0.60. To analyse the predictive value of the PRS in our Cox analysis of time to developing NMSC post-transplant we compared the concordance statistic (c-stat) of the full model (which included ‘age’, ‘era’, ‘site’, ‘PRS’ and ‘PCs’) to the clinical model (‘age’, ‘era’ and ‘site’). The c-stat measures the proportion of pairs of individuals in which the individual with the higher-risk predictor developed NMSC before the individual with the lower-risk predictor. We found that found the genetic component added additional predictive value when predicting time to NMSC post-transplant with a larger c-stat for the full model (0.74) compared to the clinical model (0.71) with the PRS contributing 0.02 and the PCs contributing 0.01.
Figure 2. Comparison of area under the curve in the logistic regression analysis.
Genetic model = Polygenic Risk Score (PRS) and first eight principal components (8 PCs); clinical model = age, era and site; Full model = PRS, first 8 PCs, age, era and site
4. DISCUSSION
Our findings show a significant association between polygenic effects of variants associated with NMSC in the general population and post-transplant NMSC. We found that individuals with increased polygenic risk for BCC and SCC as determined from a previous non-transplant GWAS had both increased risk of NMSC and faster time to NMSC post-transplant respectively. This effect was consistent across three different cohorts in over 800 patients. We found the most significant PRS in our NMSC Cox analysis was that at pT1×10−5 indicating that signals that are below the genome-wide significance threshold (5×10−8) are contributing to the speed at which individuals develop NMSC after transplantation.
To analyse the potential clinical application of this work, we compared the area under the curve of the full model, which included PRS to that with only the clinical variables for predicting case: control status of NMSC. The full model had a larger area under the curve than the model with clinical variables alone indicating the PRS is adding some positive predictive value to the model. Similarly, for the Cox analysis we compared the concordance statistics of the model with and without the PRS. Again, the PRS had additional predictive value beyond that explained by clinical variables. Although clinical variables have a large impact on the development of NMSC post-transplant, there is unexplained risk, and with the addition of the PRS we do get increased predictive value (albeit modest, 0.02 of the concordance explained by PRS).
Knowledge of a patient’s genetic risk of certain conditions could help clinicians to best determine what drugs would best suit what patients. Polygenic risk scores are reasonably cheap and easy to implement, with GWAS genotyping now costing less than €100 per individual. A recent study demonstrated the potential clinical utility of PRS in patients with bipolar affective disorder to stratify those who were at higher risk of non-response to lithium, finding that those with a high PRS for schizophrenia had a greater increased risk of non-response to lithium42. In transplant patients, clinicians have the option to tailor patients’ immunosuppression regimens and select from a number of different immunosuppressant treatments43. Incorporating PRSs for multiple outcomes such as post-transplant diabetes, cardiovascular risk as well as other forms of malignancy could allow for more accurate prediction of the risks for an individual patient on a specific treatment regime. For example, switching patients to sirolimus has been demonstrated to reduce risk of post-transplant skin cancer44. The knowledge of a patient’s polygenic risk of skin cancer post-transplant could also inform less drastic interventions such as frequency of checks with dermatologists and may also increase a patient’s likelihood to take on board lifestyle interventions such as wearing sunscreen and carrying out regular skin checks. This being said, although highly significant, the effect size of the PRS found in this study is relatively small and therefore only those in the top deciles of the distribution of polygenic burden (see table 4) could realistically benefit from interventions based on their PRS. However, this study is the first of its kind to look at GWAS based PRS in a transplant setting and is proof of concept of the potential clinical utility of these kinds of scores in transplant populations. The integration of genetic knowledge in the clinic could allow for more personalised post-transplant treatment regimens in the future and even get us one step closer to tolerant transplantations.
PRSs have the potential to inform clinical care through the identification of patients at high genetic risk for a certain condition. This could inform not only therapeutic interventions but also patient’s lifestyle choices and the frequency/need for disease screening45. However, PRSs have a number of limitations in this setting. Firstly, there is a higher level of uncertainty around the levels of risk predicted by a PRS compared to risk estimates based on familial genetic variant screening (such as screening for BRCA1/2 alleles)45. Also, PRS have yet to fully incorporate rare variants, which may change an individual’s PRS. Communication of polygenic risk is also a major obstacle in their application for clinical care. It is vital that patients and clinicians are fully aware of what a risk determined via a PRS really means for the patient’s prognosis45.
Our study had a number of limitations. Firstly, a number of clinical variables were unavailable including sun exposure or full immunosuppressant treatment data (such as dosage or changes in immunosuppression over time). Therefore, some of the unexplained variance from our models may have been characterised if there was more extensive clinical data available. This may have also lead to some confounding between the different cohorts as the Tennessee cohort was predominantly receiving tacrolimus whereas the Dublin/Glasgow and Belfast groups were predominantly treated with cyclosporine. However, we did attempt to control for this effect by including era of transplantation as a covariate in our analyses as different eras would have a predominant immunosuppression treatment regime. Previous studies have shown that NMSC risk is greater in those treated with cyclosporine compared to tacrolimus treated patients46. However, we did not see a large amount of heterogeneity between the sites in the I2 analysis.
Secondly, we cannot tell if the effect of PRS on skin cancer detected here is different from that expected within a non-transplant population. It is possible the predictive value we see in this transplant population is identifying those individuals at high risk of skin cancer regardless of setting. In this context, the study was limited by the lack of a non-screened population to test for difference in the effect size of PRS, across transplant and non-transplant populations. For this reason, we cannot say whether this effect is specific to post-transplant NMSC or applicable to NMSC in general nor can we determine if the effect is immunosuppression related. Further work will be required to answer this important question.
Finally, this study was done a homogenous European population and so further work is required to assess the predictive ability of this risk score in other more diverse, non-European populations. The alleles used to calculate the PRS were taken from two European GWA studies. For accurate interpretation of polygenic risk of NMSC in non-European populations a GWAS ethnically matched to the population being studied should be used to select alleles47.
In conclusion, we have illustrated that PRS can be used to predict case: control status of, and time to NMSC from the time of transplantation. We found that the PRS provides limited, but significant predictive value beyond what clinical variables alone can explain. As transplant populations are exposed to immunosuppression, surgery and, in most cases, long term chronic disease common genetic variants that may have small effects in the general population may have increased effects in these screened populations. Therefore, the utility of PRS in transplant settings could be a useful tool both clinically and for discovery of new biological mechanisms involved in transplant outcomes.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the patients who contributed their DNA and phenotype data, without whom these analyses would not have been possible.
We would like to thank the research participants and employees of 23andMe for data which allowed us to create the PRS.
We would also like to acknowledge Dr. Mark McCormack and Dr. Andrés Ingason for their scientific guidance.
C.P.S is supported by the Irish Research Council and Punchestown Kidney Research Fund (grant number EPSPG2015). P.J.P. is supported by an NRS Career Research Fellowship. Further thanks to funding support from Northern Ireland Kidney Research Fund and SFI-DfE (15/IA/3152).
ABBREVIATIONS
- GWAS
genome-wide association study
- NMSC
non-melanoma skin cancer
- SCC
squamous cell carcinoma
- BCC
basal cell carcinoma
- PRS
polygenic risk score
- pT
p-value threshold
- PCA
principal components analysis
- 1KG3
1000 Genomes Phase 3
- CI
confidence interval
- RE model
Random effects model
- OR
odds ratio
- HR
hazards ratio
- SE
standard error
- Padj
approximate adjusted p value
- Pbon
Bonferroni corrected p value
Footnotes
DISCLOSURES
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- 1.Gondos A, Dohler B, Brenner H, Opelz G. Kidney graft survival in Europe and the United States: strikingly different long-term outcomes. Transplantation. 2013;95(2):267–274. [DOI] [PubMed] [Google Scholar]
- 2.Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11(3):450462. [DOI] [PubMed] [Google Scholar]
- 3.Ulrich C, Kanitakis J, Stockfleth E, Euvrard S. Skin cancer in organ transplant recipients-where do we stand today? Am J Transplant. 2008;8(11):2192–2198. [DOI] [PubMed] [Google Scholar]
- 4.Bannon FJ, McCaughan JA, Traynor C, et al. Surveillance of nonmelanoma skin cancer incidence rates in kidney transplant recipients in Ireland. Transplantation. 2014;98(6):646652. [DOI] [PubMed] [Google Scholar]
- 5.Moloney FJ, Comber H, O’Lorcain P, O’Kelly P, Conlon PJ, Murphy GM. A population-based study of skin cancer incidence and prevalence in renal transplant recipients. The British journal of dermatology. 2006;154(3):498–504. [DOI] [PubMed] [Google Scholar]
- 6.Garrett GL, Blanc PD, Boscardin J, et al. Incidence of and Risk Factors for Skin Cancer in Organ Transplant Recipients in the United States. JAMA Dermatol. 2017;153(3):296–303. [DOI] [PubMed] [Google Scholar]
- 7.Sanders ML, Karnes JH, Denny JC, Roden DM, Ikizler TA, Birdwell KA. Clinical and Genetic Factors Associated with Cutaneous Squamous Cell Carcinoma in Kidney and Heart Transplant Recipients. Transplantation direct. 2015;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imao T, Ichimaru N, Takahara S, et al. Risk factors for malignancy in Japanese renal transplant recipients. Cancer. 2007;109(10):2109–2115. [DOI] [PubMed] [Google Scholar]
- 9.O’Donovan P, Perrett CM, Zhang X, et al. Azathioprine and UVA light generate mutagenic oxidative DNA damage. Science. 2005;309(5742):1871–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Euvrard S, Morelon E, Rostaing L, et al. Sirolimus and secondary skin-cancer prevention in kidney transplantation. The New England journal of medicine. 2012;367(4):329–339. [DOI] [PubMed] [Google Scholar]
- 11.Nan H, Xu M, Kraft P, et al. Genome-wide association study identifies novel alleles associated with risk of cutaneous basal cell carcinoma and squamous cell carcinoma. Human molecular genetics. 2011;20(18):3718–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey SN, Sulem P, Gudbjartsson DF, et al. Germline sequence variants in TGM3 and RGS22 confer risk of basal cell carcinoma. Human molecular genetics. 2014;23(11):3045–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chahal HS, Lin Y, Ransohoff KJ, et al. Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nature communications. 2016;7:12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chahal HS, Wu W, Ransohoff KJ, et al. Genome-wide association study identifies 14 novel risk alleles associated with basal cell carcinoma. Nature communications. 2016;7:12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack M, Alfirevic A, Bourgeois S, et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364(12):1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P, Lin JJ, Lu CS, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. The New England journal of medicine. 2011;364(12):1126–1133. [DOI] [PubMed] [Google Scholar]
- 17.International Schizophrenia C, Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. Am J Transplant. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Brien RP, Phelan PJ, Conroy J, et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin Transplant. 2013;27:379–387. [DOI] [PubMed] [Google Scholar]
- 20.International G, Translational Research in Transplantation N. Design and Implementation of the International Genetics and Translational Research in Transplantation Network. Transplantation. 2015;99(11):2401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nature genetics. 2014;46(8):818–825. [DOI] [PubMed] [Google Scholar]
- 22.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature genetics. 2012;44(8):955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3. 2011;1(6):457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin R Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics. 2014;30(9):1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh PR, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nature genetics. 2016;48(11):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rich SS, Concannon P, Erlich H, et al. The Type 1 Diabetes Genetics Consortium. Annals of the New York Academy of Sciences. 2006;1079:1–8. [DOI] [PubMed] [Google Scholar]
- 30.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PloS one. 2013;8(6):e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American journal of human genetics. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham H ggplot2 Elegant Graphics for Data Analysis Introduction. Use R. 2009:1-+. [Google Scholar]
- 33.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. [DOI] [PubMed] [Google Scholar]
- 34.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Therneau T A Package for Survival Analysis in S. . 2015.
- 36.R: A language and environment for statistical computing. [computer program]. Vienna, Austria: : R Foundation for Statistical Computing; 2017. [Google Scholar]
- 37.Sankoh AJ, Huque MF, Dubey SD. Some comments on frequently used multiple endpoint adjustment methods in clinical trials. Statistics in medicine. 1997;16(22):2529–2542. [DOI] [PubMed] [Google Scholar]
- 38.Viechtbauer W Conducting meta-analyses in R with the metafor package. Journal of Statistical Software. 2010;36(3):1–48. [Google Scholar]
- 39.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 40.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tremblay F, Fernandes M, Habbab F, de BEMD, Loertscher R,Meterissian S Malignancy after renal transplantation: incidence and role of type of immunosuppression. Ann Surg Oncol. 2002;9(8):785–788. [DOI] [PubMed] [Google Scholar]
- 42.International Consortium on Lithium G, Amare AT, Schubert KO, et al. Association of Polygenic Score for Schizophrenia and HLA Antigen and Inflammation Genes With Response to Lithium in Bipolar Affective Disorder: A Genome-Wide Association Study. JAMA Psychiatry. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalluri HV, Hardinger KL. Current state of renal transplant immunosuppression: Present and future. World J Transplant. 2012;2(4):51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alter M, Satzger I, Schrem H, Kaltenborn A, Kapp A, Gutzmer R. Non-melanoma skin cancer is reduced after switch of immunosuppression to mTOR-inhibitors in organ transplant recipients. Journal der Deutschen Dermatologischen Gesellschaft = Journal of the German Society of Dermatology : JDDG. 2014;12(6):480–488. [DOI] [PubMed] [Google Scholar]
- 45.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018. [DOI] [PubMed] [Google Scholar]
- 46.Wimmer CD, Angele MK, Schwarz B, et al. Impact of cyclosporine versus tacrolimus on the incidence of de novo malignancy following liver transplantation: a single center experience with 609 patients. Transpl Int. 2013;26(10):999–1006. [DOI] [PubMed] [Google Scholar]
- 47.Martin AR, Gignoux CR, Walters RK, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. American journal of human genetics. 2017;100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.