Abstract
Eustigmatophytes, a class of stramenopile algae (ochrophytes), include not only the extensively studied biotechnologically important genus Nannochloropsis but also a rapidly expanding diversity of lineages with much less well characterized biology. Recent discoveries have led to exciting additions to our knowledge about eustigmatophytes. Some proved to harbor bacterial endosymbionts representing a novel genus, Candidatus Phycorickettsia, and an operon of unclear function (ebo) obtained by horizontal gene transfer from the endosymbiont lineage was found in the plastid genomes of still other eustigmatophytes. To shed more light on the latter event, as well as to generally improve our understanding of the eustigmatophyte evolutionary history, we sequenced plastid genomes of seven phylogenetically diverse representatives (including new isolates representing undescribed taxa). A phylogenomic analysis of plastid genome-encoded proteins resolved the phylogenetic relationships among the main eustigmatophyte lineages and provided a framework for the interpretation of plastid gene gains and losses in the group. The ebo operon gain was inferred to have probably occurred within the order Eustigmatales, after the divergence of the two basalmost lineages (a newly discovered hitherto undescribed strain and the Pseudellipsoidion group). When looking for nuclear genes potentially compensating for plastid gene losses, we noticed a gene for a plastid-targeted acyl carrier protein that was apparently acquired by horizontal gene transfer from Phycorickettsia. The presence of this gene in all eustigmatophytes studied, including representatives of both principal clades (Eustigmatales and Goniochloridales), is a genetic footprint indicating that the eustigmatophyte–Phycorickettsia partnership started no later than in the last eustigmatophyte common ancestor.
Keywords: acyl carrier protein, Eustigmatophyceae, horizontal gene transfer, Ochrophyta, Phycorickettsia, plastid genome
Introduction
Eustigmatophytes are an interesting group of unicellular, primarily freshwater algae. These organisms generally have a simple coccoid morphology, yet they exhibit a suite of cytological and biochemical features making them unlike any other algal taxon (Eliáš et al. 2017). Phylogenetically they constitute a well-delineated lineage, typically classified as an independent class of ochrophytes, a prominent algal phylum that emerged within the stramenopiles by acquisition of a plastid (pt) through a higher-order endosymbiotic event (Dorrell et al. 2017; Dorrell and Bowler 2017). Although recognized as a separate phylogenetic unit nearly five decades ago, eustigmatophytes were long considered a relatively insignificant taxon with limited species diversity and minor ecological importance (Graham et al. 2009). Hence, until recently, research on fundamental aspects of eustigmatophyte biology was essentially limited to taxonomic studies that slowly expanded the number of eustigmatophyte taxa, combined with basic characterization of eustigmatophyte morphology, ultrastructure, and composition of photosynthetic pigments (reviewed in Eliáš et al. [2017]).
More recently, a renewed interest in algae as resources for biotechnologies has sparked an extensive research program on eustigmatophytes, but attention has been given almost exclusively to a single eustigmatophyte lineage traditionally classified as the genus Nannochloropsis (e.g., Ma et al. 2016). These studies include not only physiological, biochemical, and cell biological investigations but also application of genomic approaches yielding complete or draft genome sequences for nearly all species of the genus (Pan et al. 2011; Radakovits et al. 2012; Vieler et al. 2012; Corteggiani Carpinelli et al. 2014; Wang et al. 2014). For the sake of taxonomic accuracy, it should be stressed that two Nannochloropsis species have recently been segregated into a separate (yet related) genus Microchloropsis (Fawley et al. 2015), so the whole group will hereafter be referred to as the Nanno-/Microchloropsis clade.
In this article, we are primarily concerned with pt genomes. The first eustigmatophyte pt genomes were sequenced as part of the genome project for Microchloropsis (then Nannochloropsis) gaditana (Radakovits et al. 2012), but more detailed analyses came only with two subsequent studies that collectively provided pt genome sequences from nearly all known species of the Nanno-/Microchloropsis group (Wei et al. 2013; Starkenburg et al. 2014). These investigations yielded a number of important findings concerning the gene repertoire of pt genomes in eustigmatophytes and indicated the degree of variation in their pt genome architecture and coding capacity across different species of the single eustigmatophyte lineage. However, a question remained as to what extent the findings from the Nanno-/Microchloropsis clade are representative of eustigmatophytes across their actual phylogenetic breadth.
Eustigmatophytes are indeed much more than Nannochloropsis. Presently, the class comprises ∼30 species classified in 15 different genera, but molecular markers obtained from a number of unidentified isolates revealed a much higher taxonomic diversity that yet needs to be properly studied (Fawley et al. 2014; Eliáš et al. 2017). Phylogenetically, the class is deeply divided into two major monophyletic clades, one of them being the order Eustigmatales and the other presently referred to as the clade Goniochloridales. This clade was defined according to the PhyloCode but (for technical taxonomic reasons) so far lacks a standing in the traditional Linnaean taxonomy governed by the International Code of Nomenclature for algae, fungi, and plants (Fawley et al. 2014). However, for the purpose of this study, we will consider the group having the same rank as the order Eustigmatales and will refer to it simply to as Goniochloridales (not italicized).
The order Eustigmatales includes all eustigmatophytes defined by the pioneering work of Hibberd (1981), whereas Goniochloridales comprise more recently recognized eustigmatophytes, including taxa transferred to eustigmatophytes from xanthophytes based on molecular and other supporting evidence (e.g., Přibyl et al. 2012; Fawley and Fawley 2017) or described anew (Nakayama et al. 2015). However, the majority of known Goniochloridales representatives are unidentified algal isolates, many of which probably represent species and genera yet to be described (Fawley et al. 2014). Phylogenetic analyses based on 18S rRNA and rbcL gene sequences led to the recognition of four main (generally well supported) Goniochloridales lineages, specifically the genus Pseudostaurastrum and clades IIa, IIb, and IIc, but their relationships remain unresolved (Fawley et al. 2014; Fawley and Fawley 2017). Eustigmatales include three robustly supported main clades. One of them corresponds to the family Monodopsidaceae (which includes the Nanno-/Microchloropsis group and other taxa), whereas the other two lineages are not yet recognized in the formal taxonomy of the group and are informally referred to as the Eustigmataceae group and the Pseudellipsoidion group (Fawley et al. 2014). 18S rDNA phylogenies suggest that the Pseudellipsoidion group is basal to the other two lineages, but this branching order is only moderately supported (Eliáš et al. 2017; Fawley and Fawley 2017; supplementary fig. S1, Supplementary Material online), whereas the rbcL marker suggests the basal position of Monodopsidaceae (supplementary fig. S2, Supplementary Material online). Phylogenetic analyses based on more markers are clearly needed to resolve the relationships within both Eustigmatales and Goniochloridales.
The broad phylogenetic diversity of eustigmatophytes and the availability of many representative cultures provide opportunities for genomic explorations beyond the well-studied Nanno-/Microchloropsis group. As part of a such an endeavor, we recently reported on both pt and mitochondrial (mt) genomes of three eustigmatophytes representing three lineages progressively more distantly related to the Nanno-/Microchloropsis group, namely the genus Monodopsis, the Eustigmataceae group, and the Goniochloridales clade IIa (Ševčíková et al. 2015, 2016; Yurchenko et al. 2016). These data enabled us to obtain various additional interesting insights into the evolution of the eustigmatophyte organellar genomes as such, as well as of eustigmatophytes themselves. For example, by carrying out a phylogenomic analysis utilizing pt genome data from three eustigmatophytes representing both principal clades of the group, that is, Eustigmatales (Nannochloropsis oceanica and Microchloropsis gaditana) and Goniochloridales (Trachydiscus minutus), we could solidify the phylogenetic relationship of eustigmatophytes to other ochrophytes, supporting their classification within the group Limnista together with chrysophytes (Ševčíková et al. 2015).
The most striking discovery came, however, with sequencing the pt genomes of Vischeria sp. CAUP Q 202 and Monodopsis sp. MarTras21. Both turned out to harbor a unique six-gene cluster never seen before in any pt genome, yet homologous to a novel operon, denoted ebo. The operon occurs in diverse bacteria and encodes enzymes of an uncharacterized biochemical pathway that was hypothesized to produce a specific secondary metabolite (Yurchenko et al. 2016). Phylogenetic analyses suggested that the ebo operon was introduced via a single horizontal transfer event into the pt genome of a common ancestor of Vischeria and Monodopsis, implying its secondary loss from the Nanno-/Microchloropsis lineage (Monodopsis is more closely related to this lineage than to Vischeria). Furthermore, the putative donor of the ebo operon was suggested to be a bacterium belonging to the phylum Bacteroidetes (Yurchenko et al. 2016).
An interesting new twist in this story came with our recent discovery of a novel bacterial endosymbiont of T. minutus and characterization of its genomes (Yurchenko et al. 2018). The endosymbiont, described as Candidatus Phycorickettsia trachydisci (P. trachydisci for short), represents a new genus-level lineage of the family Rickettsiaceae, a group of obligate intracellular endosymbionts, primarily parasites, of various eukaryotes. Close relatives of P. trachydisci were detected in several other eustigmatophytes, representing both Goniochloridales and Eustigmatales, suggesting a widespread and possibly specific association of the Phycorickettsia lineage with eustigmatophytes. Most strikingly, the P. trachydisci genome also harbors a copy of the ebo operon (uniquely among the whole order Rickettsiales) and phylogenetic analyses demonstrated that the ebo operons from P. trachydisci and eustigmatophyte pt genomes are specifically related. We proposed that eustigmatophytes gained the ebo operon from their rickettsiacean endosymbiont, which was the actual recipient of the operon from a Bacteroidetes-related donor. Given the presumed antiquity of the transfer event from Phycorickettsia to the pt genome of the Vischeria and Monodopsis ancestor, we hypothesized that Phycorickettsia and eustigmatophytes have been engaged in a long-term evolutionary partnership (Yurchenko et al. 2018).
Further studies are obviously required to test this hypothesis. One of the key questions is how widespread the ebo operon is in eustigmatophyte pt genomes. Knowing this is critical for pinpointing the operon transfer event in eustigmatophyte phylogeny and may also give insights into the functional significance of the presence of the ebo operon. To answer this question, pt genome data from a wider array of phylogenetically diverse eustigmatophyte lineages are needed. With this in mind, we used the Illumina technology to generate large volumes of DNA sequence data from several phylogenetically diverse eustigmatophytes, which allowed us to extract and assemble complete sequences of their pt genomes. We exploited the new sequences to build a robust “backbone” phylogeny of the group and to address salient questions of pt genome evolution, including but not restricted to the origin and distribution of the ebo operon. In addition, analyses of partial nuclear genome data generated alongside the pt genome sequences enabled us to shed light on specific aspects of coevolution of the pt and nuclear genomes in eustigmatophytes, providing an unexpected new insight into the history of Phycorickettsia–eustigmatophyte interaction.
Materials and Methods
Algal Cultures
Six algal cultures were grown for the purpose of this study (supplementary table S1, Supplementary Material online). Pseudellipsoidion edaphicum strain CAUP Q 401 was obtained from the Culture Collection of Algae of Charles University in Prague (https://botany.natur.cuni.cz/algo/caup.html; last accessed January 22, 2019), spread out onto three agar plates (2% agar in Z-medium) of 90 mm diameter, sealed with Parafilm, and grown at the incident light intensity of 50 µmol m−2 s−1 and at room temperature for 3 weeks. The algal culture ACOI 456 came from the Coimbra Collection of Algae (http://acoi.ci.uc.pt/spec_detail.php? cult_id=520; last accessed January 22, 2019) and cultivated up to a volume of 200 ml in liquid Desmideacean Medium (Schlösser 1994) pH 6.4–6.6, at 23 °C, a 16:8 h photoperiod and under light intensity of 50 µmol photons m−2 s−1 provided by cool white fluorescent lamps. The previously reported unidentified eustigmatophyte strain Chic 10/23 P-6w (Fawley et al. 2014) and three newly characterized eustigmatophyte strains Bat 8/9-7w, NDem 8/9 T-3m6.8, and Mont 10/10-1w (for their origin see supplementary table S1, Supplementary Material online) were grown in liquid WH+ medium (Fawley et al. 2013) at 20 °C with a 14 h:10 h light:dark cycle using cool white fluorescent lights. Light intensity used was 50 µmol m−2 s−1 for strains Chic 10/23 P-6w, Bat 8/9-7w and NDem 8/9 T-3m6.8, whereas Mont 10/10-1w was grown at ∼20 µmol m−2 s−1. To confirm the phylogenetic position of the three previously uncharacterized strains, complete 18S rRNA gene sequences were identified in the respective genome assemblies (see below) and added to a manually curated alignment of all available eustigmatophyte 18S rRNA gene sequences. The ML tree was inferred from the alignment using RAxML v8.2.10 (Stamatakis 2014) and the GTR + Γ + I substitution model, and branch support was assessed by the rapid bootstrapping algorithm implemented in the program.
DNA Isolation
Cells of the ACOI 456 culture were collected by centrifugation and disrupted using glass beads and a mixer mill (MM200, Retsch, Haan, Germany) for 5 min. Genomic DNA was extracted from the homogenate using Invisorb Spin Plant Mini Kit (Stratec). DNA from the strains Chic 10/23 P-6w, Bat 8/9-7w, NDem 8/9 T-3m6.8, and Mont 10/10-1w was isolated using the procedure of Fawley and Fawley (2004) with minor modifications. Briefly, cells from rapidly growing cultures were harvested by centrifugation in 1.5 ml screw-top tubes and washed with 200 µl DNA extraction buffer (1 M NaCl, 70 mM Tris, 30 mM disodium salt of ethylenediaminetetraacetic acid, pH 8.6). Strain Mont 10/10-1w, which grows in large clumps, was agitated without glass beads for about 2 s with a MiniBeadBeater (BioSpec Products, Bartlesville, OK) before proceeding. Samples were then centrifuged and the extraction buffer was removed. Two hundred microliters of extraction buffer, 25 µl of 10% DTAB, glass beadsl and 200 µl of fresh molecular biology grade chloroform were added to each sample. Samples were shaken using a MiniBeadBeater for 30 sec at highest cycle rate. After centrifugation for 4 min at 5,000 rpm, the aqueous phase was removed and the DNA prepared using GenCleanTurbo Cartridges (MP Biomedicals North America, Solon, OH) following the manufacturers protocol using the genomic salt solution. DNA was quantified with the Qubit HE system (Thermo Fisher Scientific, USA).
DNA Sequencing and Assembly
DNA from P. edaphicum CAUP Q 401 was sequenced using the HiSeq 2500 technology by the Genomics Core Facility, EMBL, Germany (insert length 300 bp, read length 100 bp). Reads were processed using Trimmomatic v0.32 (Bolger et al. 2014), adjusted using FLASH v1.2.9 (Magoč and Salzberg 2011), and assembled using SOAPdenovo-127mer v2.04 from the SOAP package (Luo et al. 2012) with the Kmer size set to 77. The assembly was further improved using GapCloser v1.12 from the SOAP package. The remaining five DNA samples were sequenced using the HiSeq 2500 technology by Macrogen Inc. (Seoul, South Korea), with additional deep sequencing of the ACOI 456 culture performed using HiSeq X technology also at Macrogen. Reads were processed by Trimmomatic v0.36, and assembled using Spades v3.11.1 (Bankevich et al. 2012). The resulting assemblies were searched using Blast (Altschul et al. 1997) with conserved pt genes as queries, and scaffolds or contigs corresponding to pt genomes were identified. In the case of the ACOI 456 culture, pt scaffolds of two very different read coverage values were assigned to the two different species, Characiopsis acuta (the only algal component of the culture observed in a light microscope) and an unidentified Vischeria sp. (based on the high sequence similarity to the previously sequenced pt genome from Vischeria sp. CAUP Q 202), although this alga was not seen in the culture and apparently represents a very minor fraction of the cells. Full assemblies of seven pt genome sequences were obtained manually with the aid of iterative read mapping with Bowtie2 v2.3.4.1 (Langmead and Salzberg 2012) onto intermediate and final assemblies. The presence of two copies of inverted repeats was considered, supported by the coverage of the respective scaffolds being two times higher than scaffolds representing single-copy regions of the same pt genome. Finally, all assemblies were validated by visual inspection of mapped reads in Tablet 1.14.04.10 (Milne et al. 2013).
Annotation of pt Genome Sequences
Initial annotation of the pt genomes was obtained using MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl; last accessed January 22, 2019). Identification and delimitation of the genes provided by the program was checked manually by building multiple alignments of the newly annotated genes with homologs from previously sequenced eustigmatophyte and other ochrophyte organellar genomes. Modifications to the initial annotation, particularly concerning the definition of the initiation codon, were introduced when supported by sequence conservation in homologous sequences. Possible remote homologies of proteins encoded by eustigmatophyte-specific organellar open-reading frames (ORFs) were searched using HHpred (https://toolkit.tuebingen.mpg.de/#/tools/hhpred; last accessed January 22, 2019; Zimmermann et al. 2018). The presence of transmembrane helices in proteins was assessed using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/; last accessed January 22, 2019; Krogh et al. 2001). Short conserved protein-coding genes (e.g., secG or psaM), the conserved intron-containing trnL(uaa) gene, and the ssrA gene specifying tmRNA, missed by MFannot, were identified and manually integrated into the genome annotation. Putative intergenic regions were further systematically checked by BlastN and BlastX to detect possible additional genes that escaped automated annotation. MFannot does not distinguish genes specifying initiator fMet-tRNA, elongator Met-tRNA, and Ile-tRNA cognate to the AUA codon, which all exhibit the anticodon CAU (with the cytosine post-transcriptionally modified to lysidine in case of Ile-tRNACAU). The proper annotation of these genes was achieved by careful comparison to homologs in previously annotated organellar genomes. All seven newly sequenced and annotated pt genome sequences were deposited at GenBank with accession numbers listed in supplementary table S2, Supplementary Material online.
Plastid Phylogenomic Analysis
Based on preliminary analyses, 68 pt protein-coding genes were selected to construct a supermatrix for a phylogenomic analysis. Specifically, for each conserved pt gene, the respective protein sequences were collected from the newly sequenced species, previously reported eustigmatophyte genomes (selecting only a single representative for each Nannochloropsis and Microchloropsis species), a selection of other ochrophytes (maximizing the representation of different classes), and a selection of nonochrophyte taxa (cryptophytes and haptophytes) and the outgroup. The protein sequences were aligned using MAFFT v7 (Katoh and Standley 2013), the alignments were trimmed using TrimAl v. 1.3 (Capella-Gutierrez et al. 2009), with the “Gappyout” setting to remove poorly conserved regions, and the trimmed alignments were concatenated using FASconCAT v1.04 (Kück and Meusemann 2010). All alignments are available upon request. Phylogenetic trees were inferred from the supermatrices using the maximum likelihood (ML) method implemented in IQ-TREE v1.5 (Nguyen et al. 2015) and Bayesian inference implemented in PhyloBayes-MPI v1.7 (Lartillot et al. 2013). The ML tree was inferred using the most complex approach available for modeling the substitution process, that is, employing the posterior mean site frequency (PMSF) model (Wang et al. 2018), specifically the cpREV + C20 + F + Γ-PMSF model, and ultrafast bootstrapping (based on 1,000 replicates) was used for assessing branch support. Two chains of PhyloBayes were run for 4,002 and 4,042 generations, respectively, reaching convergence (maxdiff of 0.0). Eight hundred generations were removed as burn-in, and the consensus tree was calculated by summing every tenth generation from the rest. Trees were visualized using iTOL (http://itol.embl.de/upload.cgi; last accessed January 22, 2019) and adjusted in a graphical editor.
Analyses of Nuclear Genes for pt-Targeted Proteins
Using BlastP or TBlastN, nuclear homologs of selected pt genes were searched in available eustigmatophyte nuclear genome data, including the previously published genome assemblies from Nanno-/Microchloropsis species, partial nuclear genome sequences present in assemblies of Illumina sequencing data obtained for this study, and incomplete nuclear genome and transcriptome data generated in frame of our on-going sequencing projects for T. minutus, Vischeria sp. CAUP Q 202, and Monodopsis sp. MarTras21 (which also yielded the previously published organellar genomes from these species; see Ševčíková et al. 2015, 2016; Yurchenko et al. 2016). Significantly similar hits (e-value ≤0.01) were evaluated and coding sequences (exon-intron structures) were manually validated or deduced de novo based on transcriptome data (if available) or sequence conservation among homologs. For phylogenetic analyses of the FtrB, Rpl26 and acyl carrier protein (ACP) families, a selection of homologs was gathered by searching the NCBI nonredundant protein sequence database and, in order to sample ochrophyte classes not represented in the NCBI database, transcriptome assemblies generated by the MMETSP project (Keeling et al. 2014). Sequences were aligned using MAFFT, the alignments were trimmed manually to remove poorly conserved regions, and trees were inferred using IQ-TREE, with the most appropriate substitution models selected by the program. N-terminal presequences in nucleus-encoded putatively pt-targeted proteins were evaluated using ASAFind (Gruber et al. 2015) for proteins from ochrophytes, haptophytes, and chrompodellids; a combination of PredSL (Petsalaki et al. 2006) and ChloroP 1.1 (Emanuelsson et al. 1999) for proteins from other algae with secondary plastids; and ChloroP for proteins from eukaryotes with the primary plastid. The presence of mt transit peptides in the putative mitochondrion-targeted ACP proteins was confirmed using TargetP 1.1 (Emanuelsson et al. 2000). Sequences of relevant nuclear genes extracted from our unpublic genome and transcriptome assemblies were deposited at GenBank with accession numbers provided in supplementary tables S3–S5, Supplementary Material online. The present working assemblies are available upon request, whereas full publication of these data is planned after critical improvements (including removal of contaminations) and more systematic analyses have been carried out.
Results and Discussion
Eustigmatophyte Phylogeny Resolved by pt Phylogenomic Analyses
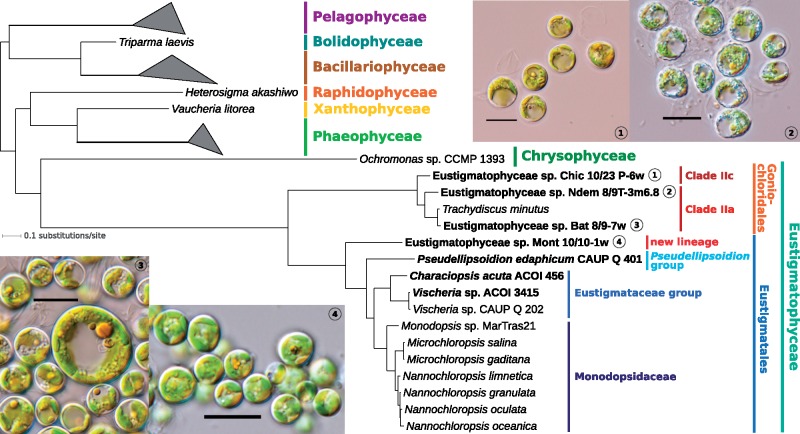
In total, we obtained complete pt genome sequences from seven eustigmatophytes. The sequencing targets were selected based on their phylogenetic position inferred from the 18S rRNA gene sequences (see supplementary fig. S1, Supplementary Material online) to sample across the eustigmatophyte phylogenetic diversity. Thus, we report on the first organellar genomes from the clades IIc in Goniochloridales (strain Chic 10/23 P-6w) and from the Pseudellipsoidion group in Eustigmatales (P. edaphicum CAUP Q 401). Particularly notable is the inclusion of the newly characterized strain Mont 10/10-1w in the analysis. This strain proved to represent a novel eustigmatophyte lineage sister to the known Eustigmatales (fig. 1). Sequencing the pt genomes of C.acuta ACOI 456 improved sampling of the highly diverse Eustigmataceae group, whereas inclusion of the strains Bat 8/9-7w and NDem 8/9 T-3m6.8 allowed us to evaluate the pt genome variation within the Goniochloridales clade IIa.
Fig. 1.
—Eustigmatophyte phylogeny inferred from pt genome data. The tree shown was inferred using PhyloBayes-MPI v1.7 and the site-heterogeneous substitution model CAT + GTR from an alignment of 18,378 amino acid positions (derived from 68 conserved pt genome-encoded proteins). All branches received maximal support (posterior probability of 1.0). For simplicity, only the ochrophyte subtree is shown (omitting thus the outgroup comprising cryptophytes and haptophytes) and classes (other than eustigmatophytes) with multiple representatives are collapsed as triangles. Species (strains) with pt genomes sequenced in this study are highlighted in bold. Supplementary figure S10, Supplementary Material online, shows a full version of the essentially identical ML tree inferred from the same supermatrix using IQ-TREE v1.6.5. GenBank accession numbers of pt genomes employed in the phylogenomic analysis are listed in supplementary table S2 and in the legend to supplementary figure S10, Supplementary Material online. Light microphotographs are provided for the four sequenced strains whose appearance has been previously documented in the literature; scale bar: 10 μm.
The seventh pt genome was sequenced accidentally as a result of microscopically unnoticed minor component of the ACOI 456 culture. Based on sequences of the 18S rRNA gene (supplementary fig. S1, Supplementary Material online), rbcL gene (supplementary fig. S2, Supplementary Material online) and the pt genome (fig. 1), it was identified as a representative of the genus Vischeria, hereafter referred to as of Vischeria sp. ACOI 3415. This was a welcome addition to our sampling, as it enabled us to assess the conservation of unusual features of previously sequenced pt genome of Vischeria sp. CAUP Q 202, particularly the presence of the ebo operon, among the relatives of this alga. The degree of sequence divergence between the two Vischeria strains (fig. 1) is comparable to that exhibited by different formally recognized species of the genera Nannochloropsis and Microchloropsis, so for the purpose of this study we consider the two Vischeria strains as representatives of two different species. Whether they can be identified as some of the formally described Vischeria species or whether any of them represent a new species remains to be resolved, together with the very much needed critical taxonomic revision of the genus as a whole (see Kryvenda et al. 2018).
All sequenced pt genomes are conventional circular-mapping molecules (supplementary figs. S3–S9, Supplementary Material online). The size and the gene content of the newly sequenced genomes are comparable to the previously sequenced ones (table 1 and supplementary table S2, Supplementary Material online). All seven genomes include the typical inverted repeat regions separated by short and long single-copy regions, with the number of genes in the inverted repeat slightly varying between taxa (table 1).
Table 1.
Basic Characteristics of the Newly Sequenced Plastid Genomes
| Eustigmatales |
Goniochloridales |
||||||
|---|---|---|---|---|---|---|---|
| Eustigmataceae Group |
Pseudellipsoidion Group |
New Lineage |
Clade IIa |
Clade IIc |
|||
| Vischeria sp. ACOI 3415 | Characiopsis acuta ACOI 456 | Pseudellipsoidion edaphicum CAUP Q 401 | Strain Mont 10/10-1w | Strain Ndem 8/9T-3m6.8 | Strain Bat 8/9-7w | Strain Chic 10/23 P-6w | |
| Size (bp) | 126,352 | 121,124 | 123,658 | 117,466 | 120,715 | 120,042 | 125,946 |
| inverted repeat (bp) | 9,783 | 10,379 | 9,777 | 9,323 | 9,434 | 9,446 | 10,072 |
| LSC region (bp) | 62,223 | 55,382 | 55,318 | 54,099 | 56,274 | 55,928 | 55,586 |
| SSC region (bp) | 44,563 | 44,984 | 48,786 | 44,721 | 45,573 | 45,232 | 50,216 |
| Total GC content (%) | 32.43 | 33.51 | 32.18 | 32.35 | 33.83 | 33.66 | 34.59 |
| Gene content (total) | 166 | 161 | 166 | 157 | 164 | 162 | 170 |
| Common conserved plastid protein-coding genes | 127 | 127 | 128 | 125 | 130 | 128 | 130 |
| Conserved group-specific genes (ycf95, orf1_gon, orf1_eust) | 2 | 2 | 1 | 1 | 2 | 2 | 2 |
| ebo operon genes | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ORFs without homologs | 0 | 0 | 5 | 0 | 0 | 0 | 6 |
| rRNA genes | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| tRNA genes | 27 | 28 | 28 | 27 | 28 | 28 | 28 |
| Other noncoding RNA genes (only ssrA) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Number of genes in inverted repeat | 12 | 12 | 12 | 11 | 12 | 12 | 13 |
Note.—Genes present in inverted repeats are counted only once for the gene counts provided in the table. Abbreviations used: LSC, long single-copy region; SSC, short single-copy region; ORFs, open-reading frames.
Plastid genomes have proven to be a particularly useful source of data to reconstruct the phylogenetic relationships of plants and algae at various phylogenetic scales (e.g., Fučíková et al. 2016; Leliaert et al. 2016; Jackson et al. 2018; Gitzendanner et al. 2018), and also provided insights into the phylogeny of ochrophytes (Ševčíková et al. 2015) and their specific sublineages, especially diatoms (Yu et al. 2018). Hence, we exploited the newly obtained sequences to improve our understanding of the phylogeny of eustigmatophytes. To this end, we built a supermatrix of 68 conserved proteins encoded by the pt genomes of eustigmatophytes, other ochrophytes, and selected other algal lineages (comprising 18,378 reliably aligned amino acid positions), and inferred trees using two different methods (Bayesian inference and ML) and different substitution models (CAT + GTR and cpREV + C20 + F + Γ-PMSF, respectively). Both analyses arrived at essentially the same topology (fig. 1 and supplementary fig. S10, Supplementary Material online), with the only difference concerning a position of the diatom Thalassiosira pseudonana (sister to all other diatoms in the PhyloBayes tree or to Trieres sinensis in the ML tree). All branches in the PhyloBayes tree and most branches in the ML tree (including the stem and internal branches of the eustigmatophyte subtree), received maximal support. The inferred relationships among the main ochrophyte lineages are congruent with previous analyses of pt genome data (Ševčíková et al. 2015; TajiMa et al. 2016). Eustigmatophytes are resolved sister to the lineage represented by the chrysophyte Ochromonas sp. CCMP 1393 (and presumably including other ochrophyte taxa so far lacking pt genome data, such as synchromophytes; see Yang et al. 2012).
The topology of the eustigmatophyte subtree is generally in accord with the analysis of the 18S rRNA gene (see supplementary fig. S1, Supplementary Material online). The added value of the pt genome analysis is primarily complete resolution of the internal topology of the Eustigmatales. Thus, the strain Mont 10/10-1w is resolved with maximal support as the new sister branch of Eustigmatales as currently circumscribed. This confirms that Mont 10/10-1w represents a novel lineage that is best classified as a separate family of Eustigmatales. The full support in the pt phylogenomic analysis for P. edaphicum (representing the Pseudellipsodion group) as a sister group of Eustigmataceae group plus Monodopsidaceae, and for the strain NDem 8/9 T-3m6.8 as being sister to the other clade IIa representatives, resolves conflicting positions of these taxa in the 18S rRNA-based tree (supplementary fig. S1, Supplementary Material online) and the rbcL-based tree (supplementary fig. S2, Supplementary Material online).
Defining the Ancestral pt Gene Set of Eustigmatophytes
The highly improved sampling of eustigmatophyte pt genomes and the robustly resolved phylogeny of the group enabled us to reconstruct the evolutionary history of the changes of the pt gene content in the eustigmatophyte lineage in an unprecedented detail. We start with a description of the inference of the ancestral eustigmatophyte pt gene complement. A gene was considered ancestral if it was present in at least some representatives of both principal eustigmatophyte groups (Eustigmatales and Goniochloridales) or if it was shared by pt genomes of at least some eustigmatophytes and another algal group.
This reasoning yielded at least 167 genes, including 135 protein-coding genes (counting the split clpC gene as two separate genes, see below) and 32 genes specifying noncoding RNAs evidently present in the pt genome of the last eustigmatophyte common ancestor (supplementary table S2, Supplementary Material online). The vast majority of them are typical conserved genes with readily identifiable orthologs in other taxa, whereas establishing orthology of a few genes required a more careful analysis. Specifically, the expanded sampling now enabled us to recognize that the eustigmatophyte gene previously referred to as orf60 (Yurchenko et al. 2016) is in fact an ortholog of secG. The assignment is supported by an HHpred search and a position of the gene just downstream of petM that is shared with many other ochrophyte pt genomes. Considering synteny as a clue we were able to identify a previously missed (unannotated) secG gene in the pt genomes of pelagophytes, Heterosigma akashiwo, and Ochromonas sp. CCMP 1393 (supplementary fig. S11, Supplementary Material online). The secG gene thus belongs to the core of ubiquitous ochrophyte pt genes (supplementary table S2, Supplementary Material online).
Another update concerns the eustigmatophyte gene previously referred to as orf403/457 (Yurchenko et al. 2016). The gene is rapidly evolving, as evident from the fact that simple BlastP searches are not sensitive enough to reveal homology of the respective sequences from distantly related eustigmatophytes, and no candidate homologs outside eustigmatophytes could previously be identified even when using more sensitive methods. However, the substantially expanded sampling of orf403/457 sequences improved the sensitivity of an HHpred search, which suggested homology of the C-terminal half of the Orf403/457 protein to the Pfam TIC20 family (PF16166) with the probability value >90. The TIC20 family includes not only the key component of the TIC translocon at the inner plastidial membrane but also the Ycf60 protein encoded by pt genomes of several algal groups including rhodophytes, haptophytes, and, notably, some ochrophytes, namely phaeophytes and xanthophytes (Le Corguillé et al. 2009), but arguably not eustigmatophytes (Yurchenko et al. 2016). To better understand the distribution of the ycf60 gene in ochrophytes, we employed the known Ycf60 sequence from the brown alga Ectocarpus siliculosus as a query for a PSI-Blast search, which led to the identification of previously unrecognized ycf60 genes in pt genomes of pelagophytes and the bolidophyte Triparma laevis. Interestingly, the ycf60 gene in the T. laevis pt genome is positioned (in the head-to-head orientation) next to the secA gene, exactly as the “orf403/457” gene in strain Mont 10/10-1w and P. edaphicum CAP Q 401; the relative position of secA and “orf403/457” in other eustigmatophytes differs only by an insertion of an extra gene (orf1_eust and orf1_gon, see below) between them (supplementary fig. S12A, Supplementary Material online). In addition, the pt genome from Ochromonas sp. CCMP 1393 includes an unannotated gene (orf215) occupying the same position with respect to secA as the ycf60 gene in T. laevis and eustigmatophytes (supplementary fig. S12, Supplementary Material online) yet lacking any significant hits when investigated by BlastP. We aligned it with homologs identified in chrysophyte transcriptome assemblies and carried out an HHpred search, which indicated homology to the TIC20 family with the probability value >94. The respective eustigmatophyte and chrysophyte proteins all include multiple predicted transmembrane helices, similar to known Ycf60 proteins (supplementary fig. S12, Supplementary Material online). The most parsimonious interpretation of these observations is that the eustigmatophyte “orf403/457” and the chrysophyte “orf215” genes are highly divergent forms of the ancestrally present ycf60 gene; hence, we reannotated them accordingly. The functional significance of the N-terminal extension acquired by the eustigmatophyte Ycf60 proteins (see supplementary fig. S12, Supplementary Material online) remains unknown.
We previously reported that the eustigmatophytes are the only ochrophyte lineage that has retained the ycf49 gene in their pt genome (Ševčíková et al. 2015). Here, we not only document that this still holds true despite the substantially improved sampling of the ochrophyte pt genomes, but we unveil one more such gene. The trnL(caa) gene, so far known only from the T. minutus pt genome (Yurchenko et al. 2016), has orthologs in other Goniochloridales as well as two of the newly sequenced Eustigmatales species (P. edaphicum and C. acuta), so we can infer that it was present in the ancestral eustigmatophyte pt genome (supplementary table S2, Supplementary Material online). Furthermore, despite being absent from all other ochrophyte pt genomes sequenced so far, it has homologs in pt genomes from some other algal groups, including red algae and cryptophytes. The similarly patchy distribution of ycf49 led Dorrell and Bowler (2017) to propose that the gene was introduced into the eustigmatophyte lineage by a horizontal gene transfer (HGT) event, so the same explanation is analogously applicable to the trnL(caa) gene. However, as previously discussed for ycf49 (Ševčíková et al. 2015) and as pointed to here for trnL(caa), both genes in eustigmatophyte pt genomes occupy positions that are syntenic with the location of the genes in pt genomes of other algae. Specifically, ycf49 is flanked by petL and ycf4 in both eustigmatophytes and cyanidiophytes, and trnL(caa) is upstream of the sufB gene in both eustigmatophytes and cryptophytes. In our view, this favors the scenario invoking vertical inheritance of these genes in the eustigmatophyte lineage (and multiple losses in other ochrophytes) over the alternative explanation employing HGT.
Nevertheless, the eustigmatophyte pt genomes do include genes that seem to have been created or acquired more recently. We previously documented stepwise additions to the pt gene number in the Limnista lineage by fragmentation of the ancestral clpC gene, initiated by separation of regions encoding the N- and C-terminal parts of the ClpC protein before the divergence of chrysophytes and eustigmatophytes, followed by separation of the region encoding the very N-terminal domain of the ClpC protein in the eustigmatophyte stem lineage (Ševčíková et al. 2015). Our expanded sampling documents that the state with the three separate genes encoding different parts of the ClpC protein is common to all eustigmatophytes (supplementary table S2, Supplementary Material online).
Further potential novelties of the eustigmatophyte pt genome are found among some of the remaining unidentified ORFs. One encodes a protein of ∼120 amino acids conserved in all eustigmatophyte pt genomes sequenced (supplementary fig. S13, Supplementary Material online), but we could not identify any candidate homologs outside of this group even when using HHpred or when considering its position relative to other genes as a clue. Following the convention for naming conserved pt genes of unknown function (Hallick and Bairoch 1994) and considering ycf94 being the most recently described gene in this category (Song et al. 2018), we designate the respective ORF as ycf95. It is possible that the encoded Ycf95 mediates some eustigmatophyte-specific pt function, but we cannot rule out the possibility that ycf95 is a highly divergent ortholog of some of the standard pt genes apparently missing from eustigmatophytes (such as some of the broadly conserved ycf genes; supplementary table S2, Supplementary Material online).
All eustigmatophytes, except for P. edaphicum and the strain Mont 10/10-1w, additionally exhibit an unidentified ORF located at the syntenic position upstream of the ycf60 gene (supplementary figs. S3–S9, Supplementary Material online). Comparison of the encoded protein sequences supports homology of the ORFs within Goniochloridales and within Eustigmatales, but there is no indication (other than synteny) that the proteins would be homologous across eustigmatophytes as a whole. Hence, it remains unclear whether this is an ancestral eustigmatophyte gene that has followed very different evolutionary trajectories in the two main eustigmatophyte lineages, or whether the ORFs emerged independently in Goniochloridales and Eustigmatales. Given this uncertainty, these genes are provisionally annotated as orf1_gon and orf1_eust, depending on which undeniable group of homologs they belong to.
Pinpointing the Acquisition of the ebo Operon in Eustigmatophyte Evolution
In addition to contributing to the ancestral pt gene set of eustigmatophytes as a whole, gene acquisition has been one of the sources of variation of pt genomes among different eustigmatophytes (fig. 2). Two different eustigmatophyte lineages have their pt gene complements potentially expanded by ORFs lacking discernible orthologs in other eustigmatophytes or other organisms, including Phycorickettsia. Six such ORFs (in two clusters comprising two and four ORFs, respectively) occur in the pt genome of the Chic 10/23 P-6w strain, and five ORFs (in three unique genomic regions including one, two, and two ORFs, respectively) are found in the P. edaphicum pt genome (supplementary table S2 and supplementary figs. S3 and S7, Supplementary Material online). Whether these ORFs are translated to produce functional proteins or not, what might be the function of the encoded proteins, and what is the origin of the respective DNA regions remains presently unknown.
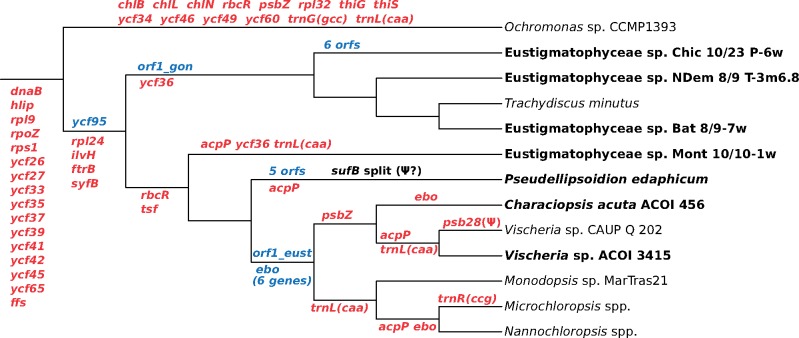
Fig. 2.
—Major events in the evolution of the pt genomes in eustigmatophytes. The scheme shows inferred events of gene loss and gain along the eustigmatophyte phylogeny. The pt genome of the chrysophyte Ochromonas sp. CCMP 1393 (a representative of the presumed sister lineage of eustigmatophytes) is included to provide a broader phylogenetic context. Genes lost are in red, genes gained are in blue (“ebo” refers to gain or loss of the whole operon). Losses mapped before divergence of Ochromonas sp. and eustigmatophytes may have happened at various deep branches of the ochrophyte phylogeny. Possible homology of orf1_gon and orf1_eust genes is discussed and detail concerning the split sufB gene (or pseudogene?) are provided in the main text. Note labels of some genes (e.g., acpP) at multiple branches in the tree, indicating multiple independent losses during the eustigmatophyte evolution. Taxa printed in bold are those for which the pt genome sequence is reported in this study.
More obvious is the origin and functional significance of the acquisition of the ebo operon. As mentioned in Introduction, we previously identified this novel six-gene cluster in pt genomes of two eustigmatophytes, Vischeria sp. CAUP Q 202 and Monodopsis sp. MarTras21 (Yurchenko et al. 2016). Of the seven newly sequenced pt genomes, only one—the one from the second Vischeria representative—proved to contain a copy of the ebo operon (supplementary fig. S9, Supplementary Material online), whereas all six ebo genes are missing from the remaining six pt genomes. Considering the occurrence of the operon and phylogenetic relationships in eustigmatophytes, the most parsimonious scenario implies gain of the operon, most likely from a bacterial endosymbiont representing the Phycorickettsia lineage (Yurchenko et al. 2018), in an exclusive ancestor of the Eustigmataceae group and Monodopsidaceae (fig. 2). The lack of the operon from pt genomes of C. acuta (in the Eustigmataceae group) and the Nanno-/Microchloropsis clade (in Monodopsidaceae) must then reflect at least two independent losses of the operon. An alternative scenario would be independent acquisition of the operon in the Vischeria and Monodopsis lineages. However, gene gain by HGT seem to be a priori a less likely event than gene loss, and the syntenic position of the ebo operon in the pt genomes of both Vischeria strains and Monodopsis sp. MarTras21 (see supplementary fig. S9, Supplementary Material online, and Yurchenko et al. 2016) strongly favors a single gain event.
Answering the question as to why the ebo operon has been retained by some eustigmatophytes but lost by others awaits identification and functional characterization of the metabolite putatively synthesized by the suite of enzymes encoded by the ebo operon. Very recently, a genetic study of the ebo operon in the cyanobacterium Nostoc punctiforme indicated that the product of the Ebo proteins-mediated pathway is critical for the export of a precursor of the natural sunscreen scytonemin from the cytoplasm into the periplasm (Klicki et al. 2018). However, the identity of the product and the mechanism of its action have not been clarified, and it is presently unclear how the findings from the cyanobacterium relate to the role of the ebo operon in other taxa, including eustigmatophytes, which do not make scytonemin.
Gene Loss in Eustigmatophyte pt Genome Evolution
Like organellar genomes in general, the dominant process that has sculpted the content of eustigmatophyte pt genomes is gene loss. Twenty conserved pt genes present in various ochrophyte lineages are missing from all eustigmatophyte pt genomes sequenced (supplementary table S2, Supplementary Material online). Of these, the most intriguing is the absence of the gene rpl24, which is retained in all other ochrophyte pt genomes sequenced so far. All other conserved pt genes universally missing from eustigmatophytes are also absent from at least some other ochrophytes. Three of them, ilvH, ftrB, and syfB, are present in the pt genome of the chrysophyte Ochromonas sp. CCMP 1393 representing the putative sister lineage of eustigmatophytes, suggesting that they were lost in the eustigmatophyte stem lineage independently of various other ochrophytes (fig. 2). The absence of 16 genes (dnaB, hlip, rpl9, rpoZ, rps1, ycf26, ycf27, ycf33, ycf35, ycf37, ycf39, ycf41, ycf42, ycf45, ycf65, and ffs) is shared with the pt genome of Ochromonas sp. CCMP 1393 and usually some other ochrophytes (supplementary table S2, Supplementary Material online), so these might have been dispensed with prior to the radiation of the Limnista clade (fig. 2).
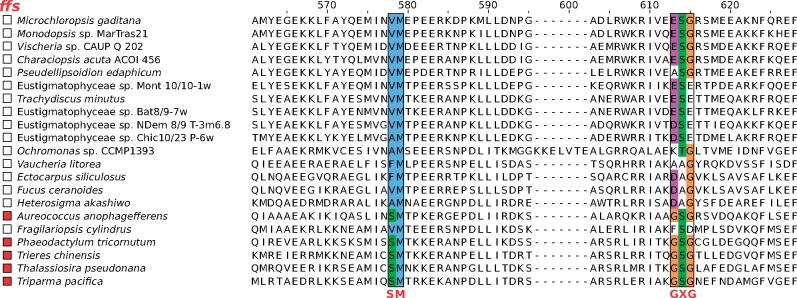
It remains theoretically possible that some of the apparent gene absences are due to their extreme divergence obscuring their identity rather than their genuine loss. This was particularly pertinent in the case of the ffs gene specifying the RNA component (4.5S RNA) of the plastidial signal recognition particle (cpSRP), because genes for noncoding RNAs are often poorly conserved and difficult to recognize. However, differential occurrence of 4.5S RNA in ochrophyte plastids (with eustigmatophytes not considered) was noticed previously and the absences were linked to specific mutations in the protein SRP component, the protein cpSRP54, that presumably abolish the ability of the protein to bind the RNA partner (Träger et al. 2012). Here, we report that eustigmatophytes and representatives of some other previously unstudied ochrophyte groups (the chrysophyte Ochromonas sp. CCMP 1393, the xanthophyte Vaucheria litorea, and the raphidophyte Heterosigma akashiwo) all exhibit mutations in the two motifs of their cpSRP54 critical for 4.5S RNA binding (fig. 3), and in perfect correlation with these changes they all lack discernible ffs gene in their pt genomes (supplementary table S2, Supplementary Material online). This supports the notion that the ffs gene is truly absent from eustigmatophytes and many other ochrophytes rather than being difficult to detect.
Fig. 3.
—Recurrent loss of 4.5S RNA (the RNA component of the plastidial SRP specified by the ffs gene) in ochrophytes. The figure shows a segment of a multiple sequence alignment of cpSRP54 (the protein component of the plastidial SRP) including two motifs critical for binding of the 4.5S RNA molecule. The presence of a discernible ffs gene in the pt genome of the respective species is indicated on the left by a red square. Note the perfect correlation between the conservation of the 4.5S RNA-binding motif in cpSRP54 (shown in red beneath the alignment) and the presence of the ffs gene.
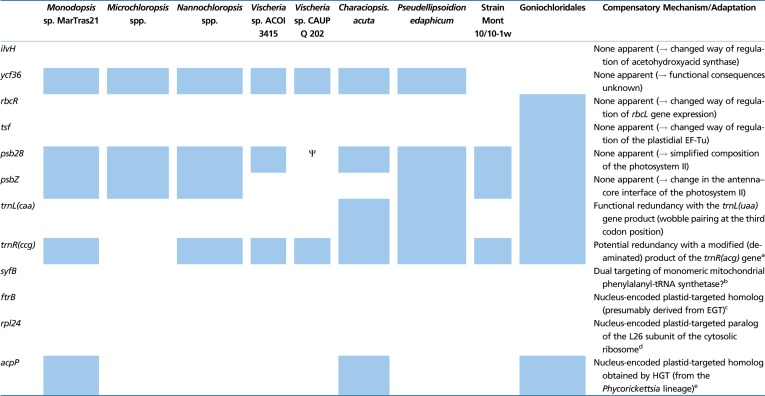
Further losses have differentially affected individual eustigmatophyte lineages, thus contributing to the variation in the pt genome content of different species (table 2 and supplementary table S2, Supplementary Material online). Two genes, rbcR and tsf, have been retained by Goniochloridales but lost in all Eustigmatales (incl. strain Mont 10/10-1w), whereas ycf36 is missing from all Goniochloridales and from the strain Mont 10/10-1w, indicating two independent loss events. Some Eustigmatales lack the trnL(caa) gene as a result of at least three independent loss events, and the genus Microchloropsis has further lost the trnR(ccg) gene. The acpP gene exhibits sporadic distribution in Eustigmatales due to four inferred independent losses (fig. 2). The previously reported absence of the psbZ gene in Vischeria sp. CAUP Q 202 (Yurchenko et al. 2016) seems to be shared by not only the second Vischeria species sequenced but also C. acuta ACOI 456, so it may be a specific feature of the whole Eustigmataceae group.
Table 2.
Plastid Gene Loss and Compensatory Mechanisms or Adaptations in Eustigmatophytes
|
Note.—The table lists all conserved plastid genes inferred to have been lost at any point of the eustigmatophyte phylogeny (including the stem branch). Presence/absence of the genes in individual taxa is indicated by blue and white squares, respectively (for simplicity, related species or strains with the same gene occurrence pattern were grouped into broader taxa). The Greek letter Ψ indicates that the psb28 gene in Vischeria sp. CAUP Q 202 is likely a pseudogene. For each gene, a mechanism or adaptation that seems to compensate for the respective gene loss in the taxa concerned is indicated; if none is apparent, putative functional consequences of the gene loss are specified.
See Gile et al. (2015) for an analogous situation is some other ochrophytes.
See supplementary figure S15 and supplementary table S3, Supplementary Material online.
See figure 4 and supplementary table S4, Supplementary Material online.
See figure 5 and supplementary table S5, Supplementary Material online.
Some eustigmatophyte pt genomes record an intermediate stage of gene loss. We previously observed apparent pseudogenization of the psb28 (psbW) gene in Vischeria sp. CAUP Q 202 (Yurchenko et al. 2016). Apparently, this is a recent event, not shared by the newly sequenced Vischeria sp. ACOI 3415. The sufB gene in the pt genome of P. edaphicum may represent another case of ongoing pseudogenization. The sufB coding sequence has been interrupted by a frame-shift mutation due to a single-nucleotide insertion (supplementary fig. S14, Supplementary Material online). The insertion is not an artifact of the sequence assembly, as it is unambiguously supported by visual inspection of mapped raw reads and is present also in a contig found in the transcriptome assembly and representing a partial transcript derived from the P. edaphicum sufB gene. The apparent sufB disruption does not seem to be compensated for by the existence of a nuclear copy of the gene (based on our searches of genome and transcriptome data from P. edaphicum), which is surprising, because the SufB protein is an essential component of the plastidial SUF system for the assembly of Fe–S clusters (Lu 2018). However, as detailed in supplementary figure S14, Supplementary Material online, and the associated legend, it is possible that the gene is still functional, producing an N-terminally truncated SufB protein alone or together with a separately translated short protein representing the α-helical N-terminus of the full-length SufB. Sequencing of pt genomes of additional members of the Pseudellipsoidion group will help to clarify the functionality of the sufB gene in P. edaphicum.
Different Modes of Functional Compensation of pt Gene Loss in Eustigmatophyte Evolution
Gene loss from organellar genomes is a pervasive phenomenon, but each individual loss raises a question whether and how it is functionally compensated. Some losses may manifest a degree of inherent redundancy of the system. For example, the loss of the trnL(caa) gene from several Eustigmatales probably reflects the ability of the product of the trnL(uaa) gene (common in eustigmatophyte pt genomes) to decode the UUG codon thanks to wobble pairing of the first anticodon position with the third codon position. Other losses of pt genes perhaps signify true loss of the respective functionality, entailing simplification of metabolic or regulatory functions in the organelle. Finally, specific adaptations providing direct or alternative functional complementation are associated with still other organellar gene losses. These are typically ensured by targeting into the organelle specific proteins encoded by nuclear genes, often homologous to the missing pt genes. We exploited available nuclear genomic data from diverse representatives of the group (both published genome assemblies from Nanno-/Microchloropsis species and the partial data obtained by us as part of the pt genome sequencing) to illuminate the functional significance of the gene losses that have impacted eustigmatophyte pt genomes. For the sake of focus, we ignored losses that seem to predate the divergence of eustigmatophytes from other ochrophyte classes. We searched for homologs of the missing pt genes and evaluated the presence of the characteristic N-terminal bipartite presequence that would indicate plastid targeting. The results are summarized in table 2 and the most interesting cases are discussed below.
In most cases, no functional compensation is readily apparent in the eustigmatophyte genomic data. One example is the ilvH gene inferred to have been lost in the eustigmatophyte stem lineage (fig. 2). The IlvH protein is a regulator of acetohydroxyacid synthase, an enzyme coded for by the ilvB gene (present in eustigmatophyte pt genomes) and catalysing the committed step of the branched amino acid biosynthesis. It was previously noted that the ilvH gene is likewise absent from the Nannochloropsis nuclear (and mt) genomes (Starkenburg et al. 2014), and we now extend this observation to eustigmatophytes as a whole. Similarly, neither of the losses specific for different eustigmatophyte subgroups, except for acpP (see the next section), seem to have been compensated by emergence of nucleus-encoded pt-targeted versions of the respective proteins, suggesting genuine lineage-specific simplification of the plastidial functioning. For example, the loss of the psbZ gene in the Eustigmataceae group indicates a change in the antenna–core interface of the photosystem II (see Wei et al. 2016). Another case of photosystem II simplification likely occurred due to apparent pseudogenization of the psb28 gene in Vischeria sp. CAUP Q 202 and the lack of a homologous gene in the nuclear genome of this organism (see Weisz et al. 2017). This is in contrast to some diatoms, which have been reported to exhibit a nuclear gene encoding a pt-targeted Psb28 homolog, in addition to the standard Psb28 encoded by the pt genome (Jiroutová et al. 2010).
The syfB gene, another gene probably lost in the eustigmatophyte stem lineage, encodes the beta subunit of a dimeric plastidial phenylalanyl-tRNA synthetase, that is, a protein mediating a function essential for plastidial translation. However, it has been retained in only some ochrophytes, together with the alpha subunit encoded by a nuclear cyanobacteria-derived syfH gene (Ruck et al. 2014; Gile et al. 2015). A possible transfer of the syfB gene into the eustigmatophyte nuclear genome was ruled out by our searches of available genome and transcriptome data, which also demonstrated the lack of the nuclear syfH gene in this group. We thus predict that like in plants or syfB/H-lacking diatoms (Gile et al. 2015), the production of phe-tRNA in the eustigmatophyte plastid is ensured by dual targeting of the monomeric mitochondrial phenylalanyl-tRNA synthetase (table 2).
In contrast to the missing pt genes discussed so far, several others do have equivalents in eustigmatophyte nuclear genomes that encode proteins evidently dedicated to function in the pt as likely compensatory devices. Thus, all eustigmatophytes investigated complement the missing pt gene for the catalytic (beta) subunit of ferredoxin-thioredoxin reductase (ftrB) by a nucleus-encoded isoform exhibiting a clear N-terminal bipartite pt-targeting signal (supplementary table S3, Supplementary Material online). To understand the origin of this nuclear gene, we inferred a tree of the FtrB family using a broad sample of sequences from eukaryotes and their most similar bacterial homologs (supplementary fig. S15, Supplementary Material online). FtrB sequences are short (providing only 94 reliably aligned amino acid positions), so the inferred phylogeny of the family needs to be interpreted with caution. However, the tree suggests that the nuclear FtrB genes in ochrophytes are likely derived from two different sources. One group, including genes from diatoms, bolidophytes, pelagophytes, and dictyochophytes, that is, Diatomista (see Derelle et al. 2016), is contained in a well-supported clade together with nuclear FtrB genes from a variety of algal (and plant) lineages with both primary and secondary plastids. This suggests that the pt ftrB gene missing from all Diatomista studied (supplementary tables S2 and S3, Supplementary Material online) was replaced by a nuclear version gained by HGT from another algal lineage (although its exact identity cannot be resolved at present). The second ochrophyte nuclear ftrB gene group is restricted to eustigmatophytes, but it seems to be specifically related to nuclear genes from the “chrompodellid” algae Chromera velia and Vitrella brassicaformis (supplementary fig. S15, Supplementary Material online). This is curious in light of a previous hypothesis that these algae obtained their plastid via a higher-order endosymbiosis of an ochrophyte alga (Ševčíková et al. 2015), possibly a specific relative of eustigmatophytes (Sobotka et al. 2017). The origin of the nuclear eustigmatophyte (and secondarily chrompodellid) ftrB gene cannot be decided with confidence from the data available, but its branching next to the pt ftrB from Phaeomonas parva (a pinguiophyte) suggests that it may simply result from endosymbiotic gene transfer (EGT) of the pt ftrB into the nucleus in the eustigmatophyte lineage.
An interesting situation was encountered when we investigated the case of the rpl24 gene, which was lost by eustigmatophytes uniquely among all ochrophytes studied so far. This gene encodes the L24 protein, a conserved component of the large ribosomal subunit likely important for the functionality of the plastidial ribosome. However, searches of the eustigmatophyte nuclear genomic data returned only an L24 homolog that carries an apparent mitochondrion-localization signal (data not shown), ruling out the possibility that the rpl24 absence is compensated for by a nuclear copy of the gene. We instead noticed that all eustigmatophytes investigated encode in their nuclear genome two different paralogs of the ribosomal L26 protein that is a part of the cytosolic large ribosomal subunit (fig. 4). Significantly, one of the paralogs carries an N-terminal extension with characteristics of the typical ochrophyte bipartite pt-targeting signal and hence is likely localized in the eustigmatophyte plastid (supplementary table S4, Supplementary Material online). The eukaryotic L26 protein is in fact an evolutionary and functional equivalent of the eubacterial (and hence plastidial and mt) L24 protein, as they both represent the same ortholog family (called uL24 according to the unified nomenclature of ribosomal proteins; Ban et al. 2014). We thus propose that in eustigmatophytes the original plastid-encoded eubacterial L24 protein was functionally replaced by a newly evolved nucleus-encoded copy of the eukaryotic L26 protein. Interestingly, a similar replacement was recently observed in the plastid of euglenophytes (Záhonová et al. 2018), suggesting that there are no serious mechanical obstacles to accommodate the eukaryotic L26 protein into the context of the eubacterial ribosome.
Fig. 4.
—A novel pt-targeted paralog of the ribosomal protein L26 in eustigmatophytes. The ML tree was inferred from an alignment of 115 amino acid position using IQ-TREE v1.6.5 and the optimal substitution model selected by the program (LG + I + Γ4). Bootstrap values <75 are omitted from the figure. The tree displayed unveils the existence of a separate clade of L26-related sequences encoded by nuclear genomes of all eustigmatophytes investigated and possessing an N-terminal extension fitting the structure of a bipartite pt-targeting presequence (supplementary table S4, Supplementary Material online). This clade is clearly separated from conventional L26 proteins lacking the N-terminal targeting sequence and expected to function as components of the cytosolic ribosomes, as well as from the presumably independently evolved group of pt-targeted L26 proteins from euglenophytes. The tree was arbitrarily rooted between sequences from Archaea and eukaryotes.
A plastid-Targeted Protein Encoded by a Phycorickettsia-Derived Gene in Eustigmatophytes
Based on the survey of eustigmatophyte pt genomes reported in this study, the current best estimate of the timing of the ebo operon gain by the eustigmatophyte pt genome lineage is an exclusive common ancestor of the Eustigmataceae group and Monodopsidaceae (see above). This would mean that bacteria of the Phycorickettsia lineage might have started to infect eustigmatophytes only after the emergence and initial diversification of the host algal group. However, a new exciting finding suggests a different scenario. We came across it when trying to explain what allowed most Eustigmatales to lose the acpP gene, which encodes ACP essential for plastidial fatty acid biosynthesis. It turned out that not only the species devoid of the pt acpP gene but all eustigmatophytes possess a nucleus-encoded ACP homolog with an obvious N-terminal bipartite pt-targeting signal (supplementary table S5, Supplementary Material online). This suggests functional redundancy conducive to the loss of the pt-encoded copy in eustigmatophytes (which happened at least four times independently; fig. 2).
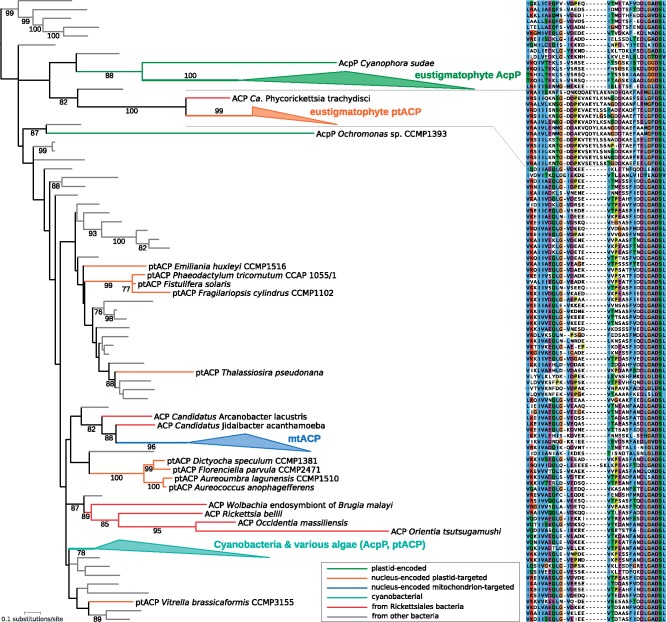
Strikingly, when used as queries against the nr protein sequence database at NCBI, the eustigmatophyte nuclear plastid-targeted ACP (ptACP) proteins gave as their best hit an ACP protein from P. trachydisci. We therefore carried out a phylogenetic analysis of a broad selection of ACP sequences, which showed that the eustigmatophyte ptACP and the Phycorickettsia ACP indeed form a clade well separated from other ACP sequences (fig. 5; supplementary fig. S16, Supplementary Material online). Specific relationship between the eustigmatophyte ptACP and the Phycorickettsia ACP is further supported by a shared unique insertion absent from any other ACP homologs (fig. 5), making it unlikely that the eustigmatophyte ptACP and the Phycorickettsia ACP branch together due to an artifact caused by their admittedly divergent nature. None of the ACP proteins encoded by nuclear genomes of other ochrophyte classes are related to the eustigmatophyte/Phycorickettsia clade. Some of them belong to a group represented in many other eukaryotes and comprising ACP presumably targeted to the mitochondrion, whereas others also bear predicted plastid-targeting presequences (supplementary table S5, Supplementary Material online) but correspond to apparently independent acquisitions from different bacterial groups or (in the case of phaeophytes) are likely derived by EGT from the original pt acpP gene (fig. 5; supplementary fig. S16, Supplementary Material online).
Fig. 5.
—Phylogenetic analysis of ACP sequences. The ML tree was inferred from an alignment of 70 amino acid position using IQ-TREE v1.6.5 and the optimal substitution model selected by the program (LG + I + Γ4). Bootstrap values <75 are omitted from the figure. Four ACP types are distinguished by different abbreviations: ACP, bacterial; AcpP, plastid genome encoded; ptACP, nucleus-encoded plastid targeted; mtACP, nucleus-encoded mitochondrion targeted. For simplicity, several clades were collapsed and their composition is shown on the right. Tip labels of sequences from bacteria, except those from Rickettsiales, are omitted for simplicity, the respective branches are rendered in gray. Note the strongly supported relationship between ACP from P. trachydisci and the eustigmatophyte ptACP. Full version of the tree is provided as supplementary figure S16, Supplementary Material online. The right part of the figure shows a segment of a multiple alignment of the ACP sequences included into the phylogenetic analysis (listed in the same vertical order as they appear in the tree) that includes a characteristic insertion shared by ACP from P. trachydisci and ptACP from eustigmatophytes.
Thus, it is highly likely that the ptACP gene in the eustigmatophyte nucleus and the ACP gene in Phycorickettsia share a specific ancestor, implying an HGT event between the two organismal lineages. Three observations lead us to favor the transfer being from the Phycorickettsia lineage to eustigmatophytes. First, the eustigmatophyte nuclear ACP genes possess introns, so invoking eustigmatophytes as the donor lineage necessitate to postulate that the transfer into the Phycorickettsia lineage must have happened before the acquisition of introns or that the transfer was mediated by a retrotranscribed intron-less copy of the eustigmatophyte gene. Second, the Phycorickettsia lineage must have always possessed an ACP gene (encoding an essential component of the fatty acid biosynthesis machinery), whereas the eustigmatophyte nuclear ptACP gene, whatever its origin, is an evolutionary innovation initially redundant with the pt acpP gene. Third, the genomic context of the Phycorickettsia ACP gene is conserved with other Rickettsiales. Specifically, the ACP gene in these organisms is flanked by two genes encoding enzymes also involved in fatty acid biosynthesis, FabB (3-oxoacyl-[acyl-carrier-protein] synthase II) upstream and FabG (3-oxoacyl-[acyl-carrier protein] reductase) downstream. The Phycorickettsia FabB and FabG genes are undeniably of rickettsiacean origin, so despite its considerable divergence, the ACP gene in between was also most likely vertically inherited by Phycorickettsia from its bacterial ancestors rather than acquired horizontally from an external source. It is worth noting that the eustigmatophyte FabB and FabG proteins are encoded by genes unlinked to the ptACP gene and not specifically related to the Phycorickettsia homologs (data not shown).
Altogether, there is sufficient evidence to propose that the eustigmatophyte ptACP gene has been acquired from a relative of extant Phycorickettsia. The presence of the ptACP gene in all eustigmatophytes examined, including representatives of both principal eustigmatophyte clades (Eustigmatales and Goniochloridales) further implies that the transfer event must have happened before the radiation of extant eustigmatophytes. The apparent absence of this gene from noneustigmatophyte ochrophytes (judging admittedly from an incomplete sampling of the ochrophyte diversity) further suggests that the HGT event occurred only after the eustigmatophyte lineage had diverged from other ochrophytes, that is, that the recipient was a stem eustigmatophyte. This means that the affair between eustigmatophytes and Phycorickettsia must have started earlier than evidenced so far, and that the whole crown eustigmatophyte group is evolutionarily influenced by the interaction with the Phycorickettsia endosymbiont.
Conclusions
The seven new plastid genome sequences reported in this study make eustigmatophytes one of the best sampled algal classes in terms of pt genomics. This is remarkable, given the fact that eustigmatophytes were for long considered an insignificant taxon and the knowledge of their biology still suffers from fundamental gaps. The wealth of the pt genome data enabled us to resolve phylogenetic relationships among main eustigmatophyte lineages, providing a useful framework for interpretation of the evolution of different eustigmatophyte traits. The improved sampling also enabled us to recognize some of the previously enigmatic eustigmatophyte genes as divergent homologs of known genes conserved in other taxa. The detailed reconstruction of the evolution of the eustigmatophyte pt gene complement provided interesting examples illustrating different events accompanying the organellar genome evolution: sequence divergence (nearly) beyond recognition, gene splitting, pseudogenization, complete gene loss, gene gain from external sources, and possibly de novo formation of truly novel genes. We also discovered examples of different configurations associated with loss of particular genes from the eustigmatophyte pt genomes—apparent loss of the respective functionality as well as functional compensation by employing nuclear genes. The latter may have various origins, including EGT, duplication of pre-existing nuclear genes and HGT from external sources. The latter case, concerning the nuclear ACP gene, is particularly interesting, as it points to a very early beginning of coevolution of eustigmatophytes and bacteria of the Phycorickettsia lineage. A systematic survey of eustigmatophyte nuclear genomes and of diverse Phycorickettsia representatives is now in order to understand the actual magnitude and significance of gene traffic between these two organismal groups.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Pavel Přibyl (Institute of Botany, Czech Academy of Sciences) for growing the culture of Pseudellipsoidion edaphicum CAUP Q 401 for DNA isolation. This work was supported by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 642575; ERD Funds, project OPVVV CZ.02.1.01/0.0/0.0/16_019/0000759 (Centre for research of pathogenicity and virulence of parasites); Czech Science Foundation grant 18-13458S; the infrastructure grant “Přístroje IET” (CZ.1.05/2.1.00/19.0388); the project LO1208 of the National Feasibility Programme I of the Czech Republic; the National Science Foundation (grant no. DEB1145414); and the Arkansas INBRE program through a grant (P20 GM103429) from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnote
Data deposition: All newly reported nucleotide sequences were deposited at GenBank under the accessions MK281411–MK281458.
Literature Cited
- Alkatib S, et al. 2012. The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet. 8(11):e1003076.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corteggiani Carpinelli E, et al. 2014. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol Plant 7(2):323–335. [DOI] [PubMed] [Google Scholar]
- Derelle R, López-García P, Timpano H, Moreira D.. 2016. A phylogenomic framework to study the diversity and evolution of Stramenopiles (=Heterokonts). Mol Biol Evol. 33(11):2890–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell RG, Bowler C.. 2017. Secondary plastids of stramenopiles. Adv Bot Res. 84:57–103. [Google Scholar]
- Dorrell RG, et al. 2017. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. Elife 6:e23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliáš M, et al. 2017. Eustigmatophyceae In: Archibald JM, Simpson AG, Slamovits CH, editors. Handbook of the protists. New York: Springer International Publishing; p. 367–406. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G.. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300(4):1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G.. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8(5):978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawley KP, Eliáš M, Fawley MW.. 2014. The diversity and phylogeny of the commercially important algal class Eustigmatophyceae including the new clade Goniochloridales. J Appl Phycol. 26(4):1773–1782. [Google Scholar]
- Fawley MW, Fawley KP.. 2017. Rediscovery of Tetraëdriella subglobosa Pascher a member of the Eustigmatophyceae. Fottea 17(1):96–102. [Google Scholar]
- Fawley MW, Fawley KP. 2004. A simple and rapid technique for the isolation of DNA from microalgae. J Phycol. 40(1):223–225 [Google Scholar]
- Fawley MW, Fawley KP., Hegewald E. 2013. Desmodesmus baconii (Chlorophyta), a new species with double rows of arcuate spines. Phycologia 52(6):565–572. [Google Scholar]
- Fawley MW, Jameson I, Fawley KP.. 2015. The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae) with descriptions of N. australis sp. nov. and Microchloropsis gen. nov. Phycologia 54(5):545–552. [Google Scholar]
- Fučíková K, Lewis PO, Lewis LA.. 2016. Chloroplast phylogenomic data from the green algal order Sphaeropleales (Chlorophyceae, Chlorophyta) reveal complex patterns of sequence evolution. Mol Phylogenet Evol. 98:176–183. [DOI] [PubMed] [Google Scholar]
- Gile GH, Moog D, Slamovits CH, Maier UG, Archibald JM.. 2015. Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol Evol. 7(6):1728–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS, Wong GK, Ruhfel BR, Soltis DE.. 2018. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am J Bot. 105(3):291–301. [DOI] [PubMed] [Google Scholar]
- Graham LE, Graham JE, Wilcox LW.. 2009. Algae. 2nd ed San Francisco (CA: ): Benjamin Cummings. [Google Scholar]
- Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T.. 2015. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 81(3):519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick RB, Bairoch A.. 1994. Proposals for the naming of chloroplast genes III. Nomenclature for open reading frames encoded in chloroplast genomes. Plant Mol Biol Rep. 12(2):S29–S30. [Google Scholar]
- Hibberd DJ. 1981. Notes on the taxonomy and nomenclature of the algal classes Eustigmatophyceae and Tribophyceae (synonym Xanthophyceae). Bot J Linn Soc. 82(2):93–119. [Google Scholar]
- Jackson C, Knoll AH, Chan CX, Verbruggen H.. 2018. Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci Rep. 8(1):1523.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiroutová K, Kořený L, Bowler C, Oborník M.. 2010. A gene in the process of endosymbiotic transfer. PLoS One 5(10):e13234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12(6):e1001889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicki K, et al. 2018. The widely conserved ebo cluster is involved in precursor transport to the periplasm during scytonemin synthesis in Nostoc punctiforme. MBio 9:e02266–e02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kryvenda A, Rybalka N, Wolf M, Friedl T.. 2018. Species distinctions among closely related strains of Eustigmatophyceae (Stramenopiles) emphasizing ITS2 sequence-structure data: eustigmatos and Vischeria. Eur J Phycol. 53(4):471–491. [Google Scholar]
- Kück P, Meusemann K.. 2010. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol. 56(3):1115–1118. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615. [DOI] [PubMed] [Google Scholar]
- Le Corguillé G, et al. 2009. Plastid genomes of two brown algae Ectocarpus siliculosus and Fucus vesiculosus: further insights on the evolution of red-algal derived plastids. BMC Evol Biol. 9:253.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, et al. 2016. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci Rep. 6:25367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. 2018. Assembly and transfer of iron-sulfur clusters in the plastid. Front Plant Sci. 9:336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XN, Chen TP, Yang B, Liu J, Chen F.. 2016. Lipid production from Nannochloropsis. Mar Drugs 14(4):61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, et al. 2013. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinformatics 14(2):193–202. [DOI] [PubMed] [Google Scholar]
- Nakayama T, et al. 2015. Taxonomic study of a new eustigmatophycean alga, Vacuoliviride crystalliferum gen. et sp. nov. J Plant Res. 128(2):249–257. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K, et al. 2011. Nuclear monoploidy and asexual propagation of Nannochloropsis oceanica (Eustigmatophyceae) as revealed by its genome sequence. J Phycol. 47(6):1425–1432. [DOI] [PubMed] [Google Scholar]
- Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ.. 2006. PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics 4(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzinger H, Weil JH, Pillay DT, Guillemaut P.. 1990. Codon recognition mechanisms in plant chloroplasts. Plant Mol Biol. 14(5):805–814. [DOI] [PubMed] [Google Scholar]
- Přibyl P, Eliáš M, Cepák V, Lukavský J, Kaštánek P.. 2012. Zoosporogenesis morphology ultrastructure pigment composition and phylogenetic position of Trachydiscus minutus (Eustigmatophyceae Heterokontophyta). J Phycol. 48(1):231–242. [DOI] [PubMed] [Google Scholar]
- Radakovits R, et al. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 3:686.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck EC, Nakov T, Jansen RK, Theriot EC, Alverson AJ.. 2014. Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol Evol. 6(3):644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlösser UG. 1994. SAG—Sammlung von Algenkulturen at the University of Göttingen Catalogue of strains. Bot Acta 107(3):113–186. [Google Scholar]
- Ševčíková T, et al. 2015. Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci Rep. 5:10134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ševčíková T, et al. 2016. A comparative analysis of mitochondrial genomes in eustigmatophyte algae. Genome Biol Evol. 8(3):705–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotka R, et al. 2017. Extensive gain and loss of photosystem I subunits in chromerid algae photosynthetic relatives of apicomplexans. Sci Rep. 7(1):13214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, et al. 2018. A novel chloroplast gene reported for flagellate plants. Am J Bot. 105(1):117–121. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg SR, et al. 2014. A pangenomic analysis of the Nannochloropsis organellar genomes reveals novel genetic variations in key metabolic genes. BMC Genomics. 15:212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N, et al. 2016. Sequencing and analysis of the complete organellar genomes of Parmales, a closely related group to Bacillariophyta (diatoms). Curr Genet. 62(4):887–896. [DOI] [PubMed] [Google Scholar]
- Träger C, et al. 2012. Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: the signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell 24(12):4819–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A, et al. 2012. Genome functional gene annotation and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8:e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. 2014. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10(1):e1004094.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Minh BQ, Susko E, Roger AJ.. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol. 67(2):216–235. [DOI] [PubMed] [Google Scholar]
- Wei L, et al. 2013. Nannochloropsis plastid and mitochondrial phylogenomes reveal organelle diversification mechanism and intragenus phylotyping strategy in microalgae. BMC Genomics. 14:534.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, et al. 2016. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534(7605):69–74. [DOI] [PubMed] [Google Scholar]
- Weisz DA, et al. 2017. Mass spectrometry-based cross-linking study shows that the Psb28 protein binds to cytochrome b559 in Photosystem II. Proc Natl Acad Sci U S A. 114(9):2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EC, et al. 2012. Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163(2):217–231. [DOI] [PubMed] [Google Scholar]
- Yu M, et al. 2018. Evolution of the plastid genomes in diatoms. Adv Bot Res. 85:129–155. [Google Scholar]
- Yurchenko T, et al. 2018. A gene transfer event suggests a long-term partnership between eustigmatophyte algae and a novel lineage of endosymbiotic bacteria. ISME J. 12(9):2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko T, Ševčíková T, Strnad H, Butenko A, Eliáš M.. 2016. The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite. Open Biol. 6(11):160249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Záhonová K, et al. 2018. Peculiar features of the plastids of the colourless alga Euglena longa and photosynthetic euglenophytes unveiled by transcriptome analyses. Sci Rep. 8(1):17012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, et al. 2018. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J Mol Biol. 430(15):2237–2243. [DOI] [PubMed] [Google Scholar]
- Dorrell RG, et al. 2017. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. Elife 6:e23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawley MW, Fawley KP. 2004. A simple and rapid technique for the isolation of DNA from microalgae. J Phycol. 40(1):223–225. [Google Scholar]
- Fawley MW, Fawley KP, Hegewald E. 2013. Desmodesmus baconii (Chlorophyta), a new species with double rows of arcuate spines. Phycologia. 52(6):565–572. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.