Abstract
Rice black-streaked dwarf virus (RBSDV) was known to be transmitted by the small brown planthopper (SBPH) in a persistent, circulative and propagative manner in nature. Here, we show that RBSDV major outer capsid protein (also known as P10) suppresses the protein kinase C (PKC) activity of SBPH through interacting with the receptor for activated protein kinase C 1 (LsRACK1). The N terminal of P10 (amino acids (aa) 1–270) and C terminal of LsRACK1 (aa 268–315) were mapped as crucial for the interaction. Confocal microscopy and subcellular fractionation showed that RBSDV P10 fused to enhanced green fluorescent protein formed vesicular structures associated with endoplasmic reticulum (ER) membranes in Spodoptera frugiperda nine cells. Our results also indicated that RBSDV P10 retargeted the initial subcellular localization of LsRACK1 from cytoplasm and cell membrane to ER and affected the function of LsRACKs to activate PKC. Inhibition of RACK1 by double stranded RNA-induced gene silencing significantly promoted the replication of RBSDV in SBPH. In addition, the PKC pathway participates in the antivirus innate immune response of SBPH. This study highlights that RACK1 negatively regulates the accumulation of RBSDV in SBPH through activating the PKC signalling pathway, and RBSDV P10 changes the subcellular localization of LsRACK1 and affects its function to activate PKC.
This article is part of the theme issue ‘Biotic signalling sheds light on smart pest management’.
Keywords: rice black-streaked dwarf virus, P10 protein, Laodelphax striatellus, receptor for activated protein C kinase 1, RACK1, protein kinase C
1. Introduction
Rice black-streaked dwarf virus (RBSDV) causes devastating diseases in rice and maize fields in China and Japan [1]. RBSDV belongs to the genus Fijivirus, family Reoviridae, and can be transmitted by Laodelphax striatellus (small brown planthopper, SBPH) in a persistent and circulative-propagative manner [1,2]. The RBSDV genome consists of 10 linear double-stranded RNAs (dsRNAs) and encodes thirteen proteins. Each dsRNA has one open reading frame (ORF) except RNA S5, S7 and S9 (two ORFs each) [3–5]. To date, the P6 protein has been shown to be an RNA silencing suppressor [6] and P7-1 was reported to form tubules in insect cells to facilitate virus spread from the midgut epithelium into the visceral circular muscle through the basal lamina, similar to that reported for southern rice black-streaked dwarf virus (SRBSDV) [7,8]. Protein P9-1 was shown to form viroplasm-like structures by itself in Arabidopsis protoplasts or in Spodoptera frugiperda 9 (Sf9) cells [9,10]. The P10 protein is known to be the viral major outer capsid protein (P10) with 558 amino acids and a molecular weight of about 63 kDa. Bioinformatic analyses revealed that RBSDV P10 has three predicted transmembrane domains (TM1, TM2 and TM3) [11]. It was reported that transient expression of RBSDV P10 fused to an enhanced green fluorescent protein (eGFP) in Nicotiana benthamiana epidermal cells or in rice protoplasts formed endoplasmic reticulum (ER)-associated vesicular-like structures [11]. More recent studies have indicated that the viral P10 is multifunctional and may function in almost every stage during virus infection in plants through specific interactions with other viral and/or host factors [12].
Insects have evolved effective antivirus innate immune responses, including humoral and cellular responses [13]. Humoral immune responses include synthesizing antimicrobial peptides, triggering enzymatic cascades that regulate coagulation, melanizing foreign materials in haemolymph, and producing reactive oxygen or nitrogen species [14]. Cellular-mediated immune responses include phagocytosis, nodulation, encapsulation and melanization [15]. To escape insect host immune responses, viruses have evolved strategies to avoid to be targeted by these immune responses [16].
The receptor for activated C kinase 1 (RACK1) is a 36 kDa protein with seven repeated tryptophan-aspartate (WD) domains and can form a propeller-like structure acting as a intracellular scaffold protein involved in many signalling events [17,18]. RACK1 was originally identified as a receptor for protein kinase C (PKC) members in mammals [19,20]. In animals, RACK1 is known as a multifunctional protein, important for its roles in regulating several cell surface receptors and intracellular protein kinases [21]. RACK1 was demonstrated to localize at the internal side of membranes and on cytoskeletons in cytosol [22,23]. RACK1 can bind to and stabilize PKCs to increase PKC-mediated phosphorylation. Sequences of RACK1 from yeast, plants and animals were highly conserved [24].
PKC is a multimember protein kinase family involved in regulating functions of many other proteins through phosphorylating the hydroxyl groups on serine and threonine amino acid residues. PKC itself can be activated by signalling molecules, such as increased concentrations of diacylglycerol or calcium ions (Ca2+) [25]. PKC enzymes are known to play important roles in several signal transduction cascades [26]. Previous reports have indicated that RACK1 can bind with PKCs and activate them to serve as an intracellular receptor, which anchors activated PKCs to cytoskeleton, and targets PKCs to membranes [22,23]. In addition to the above activities, PKCs were shown to regulate virus endocytic trafficking through phosphorylation [27]. Interaction of PKC with RACK1 is thought not only to target PKC to proper intracellular locations but also to hold PKC in an active conformation [28]. This model is based on the premise that the PKC isoforms that are capable of interacting with RACK1 all contain a pseudo-RACK1 binding site within its primary amino acid sequence [20]. PKC contains a pseudo-substrate sequence in the regulatory domain of PKC that binds to the substrate site to maintain the enzyme in the inactive form [20]. RACK1 can disrupt these interactions and stabilize the bound PKC in an open and active conformation [20,28].
Numerous viruses can be transmitted by insect vectors [29]. However, the molecular mechanism controlling virus transmission by insect vector was poorly understood [30]. To better understand the biological functions of viral capsid protein during virus transmission, we investigated the interaction between RBSDV P10 and its insect vector factors. The result of this study demonstrated that RBSDV P10 interacted with LsRACK1 of L. striatellus both in vivo and in vitro. Although previous reports had shown that RACK1 could bind with rice stripe virus (RSV) or beet western yellow virus (BWYV) particles, based on two-dimensional electrophoresis (2-DE), the nature of protein(s) on virus particles that interacted specifically with RACK1 were unclear [31,32]. In this study, we further investigated the roles of LsRACK1 and the PKC cascade during RBSDV transmission by L. striatellus. Our results demonstrated that LsRACK1 and the PKC cascade have negative effects on RBSDV accumulation in the insect vector. These new findings provide useful information on the molecular mechanism by which Fijivirus interacts with its insect vectors.
2. Material and methods
(a). Virus resource, host plant and insect care
RBSDV Kaifeng isolate was obtained from an infected rice plant in a field in the Henan Province, China. The virus was characterized through reverse transcription-polymerase chain reaction (RT-PCR) using specific primers and serological assays using an anti-RBSDV monoclonal antibody [33]. RBSDV in this plant was transmitted to new rice seedlings by L. striatellus Fallen and the infected plants were maintained in an insect-free greenhouse until further use. Laodelphax striatellus was maintained on rice seedlings in an insect culture room as described previously [31]. Briefly, non-viruliferous L. striatellus were allowed to grow on rice seedlings cv. Huainan No. 5 in large glass beakers covered with a layer of nylon mesh inside an insect growth room, set at 25°C and a 16 L : 8 D photoperiod cycle. Rice seedlings were changed once every 10–14 days.
(b). Vector construction and yeast two-hybrid assay
The RBSDV P10 gene was amplified from the total RNA extracted from a RBSDV-infected rice plant through RT-PCR and cloned into the pGBKT7 (BD) vector (Clontech, Mountain View, CA, USA) to generate a bait vector pGBKT7-P10 (BD-P10). Construction and screening of an L. striatellus cDNA library was conducted as instructed by the manufacturer (Clontech). Full-length sequence of the LsRACK1 gene of L. striatellus was obtained through searching the database at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/pubmed). The coding sequence of LsRACK1 was then PCR-amplified from an L. striatellus cDNA and inserted into the pGADT7 (AD) or BD vector. The coding sequences of RBSDV P10 deletion mutant (muP10) or LsRACK1 deletion mutant (muRACK1) was also RT-PCR amplified with specific primers and cloned individually into the vector AD or BD. The muP10 mutants were made based on the predictions of their transmembrane domains using online software (http://www.cbs.dtu.dk/services/TMHMM/). The muRACK1 mutants were made based on the prediction of its seven WD40 domains. Primers used in this study are listed in the electronic supplementary material, table S1 and the authenticity of each construct was confirmed through DNA sequencing.
Analysis of protein–protein interaction in yeast was performed as described previously [34]. Briefly, combinations of two different plasmids were transformed into Saccharomyces cerevisiae strain Gold. Saccharomyces cerevisiae transformed with pGADT7-T (AD-T, encoding simian virus 40 (SV40) large T antigen) and pGBKT7-p53 (BD-p53), AD-T and pGBKT7-Lamin (BD-Lam, encoding human lamin C protein), or BD and AD empty vectors were used as the positive, negative and blank negative controls, respectively. The transformed S. cerevisiae cells were grown at 30°C for 72 h on a triple-selective medium (SD-LTH/3-AT/Aba, lacking leucine, tryptophan and histidine, supplemented with 25 mM 3-amino-1,2,4-triazole (3-AT) and 70 ng ml−1 aureobasidin A (Aba)), or on a quadruple-selective medium (SD-LTHA/Aba, lacking leucine, tryptophan, histidine and adenine, supplemented with 70 ng ml−1 Aba).
(c). Protein analysis, co-immunoprecipitation assay and glutathione S-transferases pull-down assay
For protein expression analyses, total protein was extracted from L. striatellus tissues or Sf9 cells in a 2×sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (1 : 1, w/v, mg ml−1). After 10 min boiling, the total protein extracts were separated in SDS-PAGE gels through electrophoresis. The separated proteins were transferred to nitrocellulose membranes followed by detections of specific proteins using indicated antibodies. For co-immunoprecipitation (Co-IP) assay, total protein was extracted from Sf9 cells in ice-cold immunoprecipitation buffer (25 mM Tris, pH 7.5, containing 10% glycerol (v/v), 0.15% nonidet P-40 (v/v), 150 mM NaCl and 1 × protease inhibitor cocktail (Roche)). Protein extracts were incubated with a Myc antibody for 2 h at 4°C, and then incubated with protein A/G beads for 4 h at 4°C. The precipitated proteins were washed three times with ice-cold immunoprecipitation buffer at 4°C and analysed by Western blot using an anti-Myc or an anti-HA antibody (Abcam).
Based on the results from the protein–protein interaction assays in yeast, a full-length LsRACK1 gene was PCR-amplified and cloned into the pGEX4T-3 vector to express glutathione S-transferases (GST)-tagged LsRACK1 fusion protein in Escherichia coli strain BL21 (DE3). A 6X His-P10-HA fusion protein was expressed in Sf9 cells. The GST-LsRACK1 protein was purified using a GST-bind resin (Novagen) and the 6X His-P10-HA fusion protein was purified using a Ni-NTA His*bind resin (Novagen) as instructed by manufacturers. Approximately 1 mg of individual purified GST fusion protein or GST was incubated with 1 mg 6X His-P10-HA fusion protein at 4°C for 3 h in 1 ml buffer A (50 mM Tris–HCl, pH 7.5, containing 100 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.2% Triton X-100, 0.1% β-mercaptoethanol, 1 mM PMSF and protease inhibitor cocktail). The beads were washed three times with ice-cold buffer B (50 mM HEPES, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM PMSF and protease inhibitor cocktail) at 4°C. The washed beads were boiled for 10 min in 2x SDS-PAGE loading buffer and the proteins were separated in SDS-PAGE gels followed by Western blot using an anti-HA antibody.
(d). Baculovirus expression assay
DNA fragments representing full-length RDSDV P10, LsRACK1 or various deletion mutants were PCR-amplified. The resulting PCR products were purified and inserted individually into vector pFastBac (Invitrogen, Carlsbad, CA, USA). The recombinant baculovirus vectors were introduced individually into E. coli DH10Bac cells (Invitrogen) prior to transposition into the bacmid vector. The recombinant bacmid vectors were then individually used to transfect Sf9 cells in the presence of cellfectin. The transfected cells were incubated inside a 27°C humidified incubator for 72 h. The baculovirus expression assays were performed as instructed (Invitrogen). The cells expressing protein were examined using an inverted Zeiss LSM780 confocal laser-scanning microscope (Zeiss, Göttingen, Germany) and the images were processed using ZEN software (Zeiss).
(e). Immunofluorescence microscopy
Midguts, ovaries and testes were dissected from non-viruliferous and viruliferous L. striatellus in cold distilled water on a glass plate, and fixed in 4% paraformaldehyde for 2 h at room temperature (RT). After being permeabilized with osmotic buffer (0.01 M phosphate-buffered saline (PBS) containing 2% Triton X-100, pH 7.4) for 4 h, the tissues were blocked with 1% bovine serum albumin for 30 min at RT. The samples were probed with an anti-RBSDV P10 murine monoclonal antibody conjugated with fluorescein isothiocyanate (FITC), and an anti-LsRACK1 rabbit polyclonal antibody, overnight at 4°C. After washing with 0.01 M PBS containing 1% Tween-20 (pH 7.4), the tissues were incubated in a goat anti-rabbit IgG conjugated with Alexa Flour 555 (Sangon Biotech) for 2 h at room temperature. Nuclei in cells were stained blue using DAPI (Beyotime Biotechnology) in accordance with the manufacturer's instructions. The images were viewed under an inverted Zeiss LSM780 confocal laser-scanning microscope (Zeiss). Thirty copies of each treatment were tested.
(f). Subcellular fractionation through sucrose gradient centrifugation
Subcellular fractionation was conducted as previously described [35] with minor modifications. Briefly, at 48 h post-transfection, Sf9 cells were washed twice in ice-cold 0.01 M PBS, pelleted, resuspended in PBS containing 10 mM EDTA and incubated at 4°C for 5–10 min. The cells were pelleted, resuspended in ice-cold PBS and pelleted again. The resulting cells were resuspended in a hypotonic buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA) supplemented with complete mini protease inhibitor cocktail as instructed (1 tablet/10 ml buffer, Roche Applied Science, Mannheim, Germany), kept at −80°C for about 30 min, and then allowed the cells to swell at RT for 15–20 min. The cells were disrupted by sonication. The suspensions were added to NaCl to a final concentration of 150 mM, and centrifuged at 1000g for 10 min at 4°C to remove cell debris. SDS was added to the supernatant to reach a final concentration of 0.1%. The supernatant (300 μl) was mixed with 1.6 ml 80% (w/v) sucrose solution and placed underneath a layer of 65% sucrose (7.0 ml) inside a centrifuge tube. The top layer was 3.1 ml 10% sucrose. After centrifugation at 151 000g for 4 h at 4°C, the sample was fractionated nine equal parts (1.5 ml per fraction) starting from the top of the centrifuge tube. The membrane-associated vesicles were floated in the 65% and 10% sucrose gradients. Each fraction was subjected to Western blot analysis.
(g). Double stranded RNAs preparation and transfection of Spodoptera frugiperda 9 cells
DsRNAs were synthesized using the MEGAscript T7 Transcription Kit (catalogue no. AM1333, Ambion, Austin, TX, USA). Briefly, DNA fragments used for productions of dsRNAs were PCR-amplified using specific primers with a T7 RNA polymerase promoter sequence (electronic supplementary material, table S1). A 414 bp segment representing a partial sequence of GFP gene was also PCR-amplified and used as a control. The resulting DNA fragments were used (200 ng per reaction) to produce dsRNAs. The synthesized dsRNAs were concentrated via LiCl precipitation and then resuspended in nuclease-free water. Quality and size of the dsRNAs were visualized in 1% agarose gels after electrophoresis. Sf9 cells were transfected with dsRNAs (0.5 μg μl−1) in the presence of cellfectin as instructed and gene silence efficiency was determined at 72 h post transfection through real-time quantitative RT-PCR (qRT-PCR) and Western blot.
(h). Quantification of rice black-streaked dwarf virus proliferation in RNA interference planthoppers
Microinjection of dsRNA into L. striatellus was performed as previously reported [36]. Second-instar L. striatellus nymphs were randomly collected from the L. striatellus culture and anaesthetized with CO2 for 30 s. For LsRACK1 knockdown, approximately 25 ng dsRNA was injected into the thorax between the mesocoxa and the hind coxa using a FemtoJet microinjection system (Eppendorf, Hauppauge, NY, USA). A total of 200–250 nymphs were injected for each treatment. At 4 h post-injection, the nymphs were allowed to feed on RBSDV-infected rice plants for 3 d and then on healthy rice seedlings. To determine the efficiency of LsRACK1 silencing, total RNA was extracted from 20 individual nymphs at 2 d post microinjection and analysed through qRT-PCR. The remaining nymphs were allowed to continue to feed on the RBSDV-infected rice plants for one more day and then on healthy rice seedlings for 15 d. The resulting nymphs were used to determine the effect of LsRACK1 silencing on RDSDV P10 gene expression by qRT-PCR and Western blot.
Total RNA was isolated from L. striatellus using TRIzol reagent (Invitrogen). Rever Tra Ace® qPCR RT Master Mix with gDNA remover was used to conduct reverse transcription as instructed (Toyobo, Osaka, Japan). RT-PCR and qRT-PCR were performed as described previously [37]. All primers used in assays are described in the electronic supplementary material, table S1. Specific primers for qRT-PCR were designed using the Primer Premier 5 software.
(i). Measurement of protein kinase C activity
Microinjection of 10 µl 1 μM PKCs specific inhibitor Calphostin C (Sigma-Aldrich, Santa Clara, CA, USA) or 1 μM PKCs activator phorbol-12-myristate-13-acetate (PMA) (Sigma-Aldrich) into L. striatellus was performed to suppress or activate the PKC activity as previously reported [38]. Planthopper tissues or cell lysate were assayed for PKC activity using the PepTag® Non-Radioactive PKC Assay kit as instructed (Promega, Madison, WI, USA). Briefly, 0.1 g tissue or 5 × 106 to 1 × 107 cultured Sf9 cells were homogenized in 0.5 ml cold PKC extraction buffer (25 mM Tris–HCl, pH 7.4, 0.5 mM EDTA, 0.5 mM EGTA, 0.05% Triton X-100, 10 mM β-mercaptoethanol, 1 μg ml−1 leupeptin, 1 μg ml−1 aprotinin, 0.5 mM PMSF) using a cold homogenizer. The homogenates were centrifuged at 14 000g for 5 min at 4°C in a micro-centrifuge tube and the pellets were discarded. Reaction reagent was made by mixing 5 μl PepTag® PKC reaction 5X buffer, 5 μl PepTag® C1 peptide, 5 μl sonicated PKC activator 5X solution and 8 μl water in a 0.5 ml micro-centrifuge tube set on ice as instructed by the manufacturer. The tube was incubated in a 30°C water bath for 2 min followed by addition of 2 μl supernatant described above or PKC. The mixed samples were incubated again at 30°C for 30 min followed by boiling the sample for 2 min or by heating the sample in a 95°C heating block for 10 min to stop the reaction. For electrophoresis, 1 μl 80% glycerol was added to each sample prior to loading the samples into wells in 0.8% agarose gels to ensure all the samples remained in the wells. The activity of PKC in each sample was calculated based on the content of the phosphorylated substrate. Statistical significance between the two treatments was determined using Student's t-test and p < 0.05 was considered to be statistically significant.
3. Results
(a). Interaction between rice black-streaked dwarf virus P10 and LsRACK1
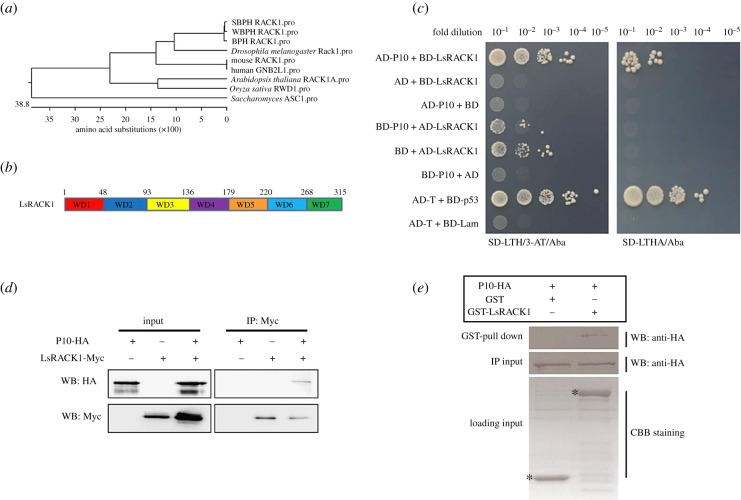
RBSDV P10 was used as a bait to screen a L. striatellus cDNA library by yeast two-hybrid (Y2H) assay. From this assay, a 687 bp DNA fragment showing 94% nucleotide sequence similarity with a L. striatellus gene, i.e. LsRACK1 (nucleotide position 262–948) was identified. Subsequently, a full-length coding sequence for LsRACK1 was cloned using specific primers. Sequence analysis revealed that the full-length LsRACK1 ORF contains 948 nucleotides (nt) (GenBank accession No. HQ385972.1) and encodes a protein with 315 amino acids (aa) with seven WD40 domains, which has high homology with that of Sogatella furcifera Horváth and Nilaparvata lugens (figure 1a,b).
Figure 1.
Interaction between RBSDV P10 and LsRACK1. (a) Phylogenetic analysis using RACK1 amino acid (aa) sequences from different sources. (b) Schematic of RACK1. WD40 domains are short structural motifs with about 40 aa and the numbers above the diagram represent the aa positions. (c) Results of yeast two-hybrid assays. RBSDV P10 and LsRACK1 were cloned individually into pGADT7 (AD) and pGBKT7 (BD) vectors. Serial dilutions of yeast cells co-transfected with two various combined vectors were plated on the SD-LTH/3-AT/Aba or SD-LTHA/Aba medium. Cells co-transfected with AD-T and BD-p53 and with AD-T and BD-Lam were, respectively, used as a positive and a negative control. Positive interactions are indicated by the growth of the cells. (d) Co-IP. LsRACK1-Myc and P10-HA were separately or co-expressed in Sf9 cells by transfection. LsRACK1-Myc expressed alone in cells served as a negative control. At 48 h post transfection, Sf9 cell lysates were immunoprecipitated with anti-Myc beads followed by Western blot (WB) using an anti-HA (upper panel) or an anti-Myc antibody (lower panel). (e) GST pull-down assay. GST or GST-LsRACK1 was incubated with 6X His-P10-HA and then mixed with glutathione-sepharose beads. The beads were washed and analysed by Western blot using an anti-HA antibody (upper panel). The middle panel shows the Western blot results of inputs of HA-tagged P10 proteins from pull-down assays. Equal volume of glutathione-sepharose beads carrying GST-LsRACK1 or GST were analysed by SDS-PAGE and stained with Coomassie blue. The position of GST-LsRACK1 or GST is indicated with asterisks.
RBSDV P10 and LsRACK1 fusion expression vectors with either a GAL4 activation domain (e.g. AD-LsRACK1) or a GAL4 DNA binding domain (e.g. BD-P10) were constructed. Saccharomyces cerevisiae cultures co-transfected with various combinations of two different vectors were plated on the SD-LTH/3-AT/Aba or SD-LTHA/Aba medium to determine the activation of reporter genes. Results showed that yeast harbouring both AD-P10 and BD-LsRACK1 or AD-T and BD-p53 vectors were able to grow on both SD-LTH/3-AT/Aba and SD-LTHA/Aba media (figure 1c). Yeast harbouring both BD-P10 and AD-LsRACK1 or AD-LsRACK1 and the empty GAL4 DNA binding domain vector (BD) were able to grow on the SD-LTH/3-AT/Aba medium but not the SD-LTHA/Aba medium (figure 1c). Yeast harbouring AD and BD-LsRACK1, AD-P10 and BD, BD-P10 and AD or AD-T and BD-Lam were unable to proliferate on both media (figure 1c), suggesting that RBSDV P10 did interact with LsRACK1 after been expressed in yeast.
To further confirm the above Y2H results, we further analysed the interaction between RBSDV P10 and LsRACK1 using a Co-IP assay. The HA-tagged P10 (P10-HA) and the Myc-tagged LsRACK1 (LsRACK1-Myc) were expressed individually or together in Sf9 cells. Cell lysates from different treatments were collected at 48 h post transfection and total protein extracts were analysed through Co-IP using anti-Myc antibody coupled agarose beads. The resulting precipitates were analysed by Western blot using an anti-HA antibody. The result indicated that RBSDV P10 was indeed co-immunoprecipitated with LsRACK1-Myc (figure 1d).
We then tested the interaction between RBSDV P10 and LsRACK1 in vitro by GST pull-down assay. The GST-tagged LsRACK1 (GST-LsRACK1) was expressed in E. coli and the double-tagged RBSDV P10 (6X His-P10-HA) was expressed in Sf9 cells. The 6X His-P10-HA was incubated with a glutathione-agarose purified GST-LsRACK1 or GST tag. The final pull-down products were separated in SDS-PAGE gels and then subjected to Western blot detection with an anti-HA antibody. The 6X His-P10-HA was detected only when both 6X His-P10-HA and GST-LsRACK1 were present in the reaction, further confirming that LsRACK1 interacted with RBSDV P10 (figure 1e).
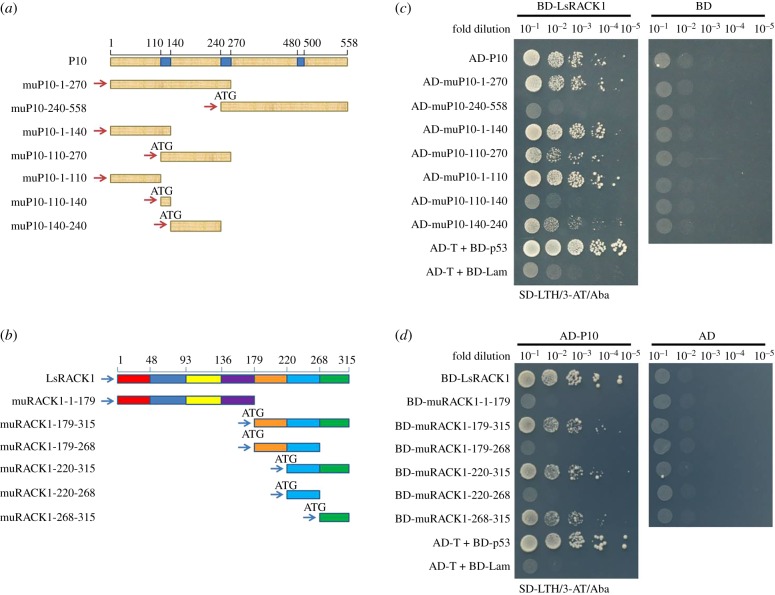
(b). Minimum domains required for rice black-streaked dwarf virus P10–LsRACK1 interaction
To determine which region(s) in the P10 or the LsRACK1 are responsible for their interaction, seven P10 deletion mutants (muP10-1-270, muP10-240-558, muP10-1-140 and muP10-110-270, muCP1-110, muCP110-140, muCP140-240; figure 2a) and six LsRACK1 deletion mutants (muRACK1-1-179, muRACK1-179-315, muRACK1-179-268, muRACK1-220-315, muRACK1-220-268 and muRACK1-268-315; figure 2b) were made and cloned into vector pGADT7 and pGBKT7, respectively. Interactions between various P10 and LsRACK1 constructs were investigated by Y2H assays. Results of the assays indicated that co-presence of muP10-1-110, muP10-1-140, or muP10-1-270 and LsRACK1 led to a strong positive reaction (figure 2c). This interaction was, however, not observed when muP10-1-270 was replaced with muP10-240-558 (figure 2c), demonstrating that the P10 N terminal region covering aa 1–270 was responsible for the strong interaction with LsRACK1. muP10-110-270 or muP10-140-240 had a weak interaction with LsRACK1 (figure 2c). We then tested various LsRACK1 mutants and determined that mutant muRACK1-1-179, muRACK1-179-268 and muRACK1-220-268 failed to interact with RBSDV P10, while mutant muRACK1-179-315, muRACK1-220-315 and muRACK1-268-315 all gave a strong interaction with RBSDV P10 (figure 2d), indicating that the seventh WD40 domain, covering LsRACK1 aa 268–315, is essential for the interaction with RBSDV P10. No positive interaction was observed when individual mutants were co-expressed with empty pGADT7 or pGBKT7 (figure 2c,d).
Figure 2.
Determination of domains needed for RBSDV P10 and LsRACK1 interaction. (a) Schematic of RBSDV P10 and its deletion mutants. The three blue boxes inside the diagram represent transmembrane domains in the RBSDV P10. The numbers above the diagram represent the positions of amino acids. The seven short fragments below the P10 diagram represent the seven deletion mutants and their respective positions. (b) Schematic of LsRACK1. Different colours shown in the diagram represent seven WD40 domains, respectively. The fragments below the diagram represent the LsRACK1 deletion mutants. (c) Yeast two-hybrid assay results showing the positive or negative interactions between AD-P10 and its deletion mutants with BD-LsRACK1. Co-expression of AD-P10 or its deletion mutants with the BD vector were used as negative controls. (d) Yeast two-hybrid assay results showing the positive or negative interactions between BD-LsRACK1 and its deletion mutants with AD-P10. The cell co-expressing AD-T and BD-p53 was used as a positive control, and the cell co-expressing AD-T and BD-Lam was used as a negative control.
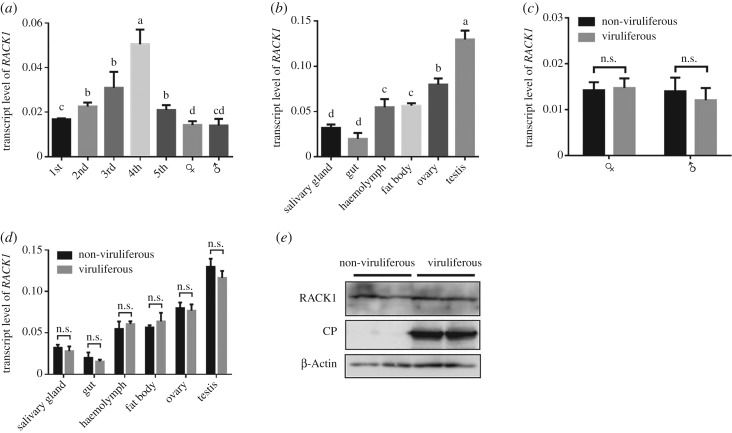
(c). Influence of rice black-streaked dwarf virus infection on LsRACK1 expression
We reasoned that the accumulation of RBSDV in viruliferous L. striatellus might interfere with the expression of LsRACK1. To address this concern, we analysed the temporal and spatial expression pattern of LsRACK1 in non-viruliferous and viruliferous L. striatellus using qRT-PCR. In the non-viruliferous L. striatellus, the highest LsRACK1 expression level was found in the four-instar nymphs while the lowest expression level was found in the adult (figure 3a). In adult L. striatellus, the highest LsRACK1 expression was found in testis followed by the ovary, and the lowest expression was found in salivary glands and the gut (figure 3b). The expression levels of LsRACK1 transcript and LsRACK1 protein in the non-viruliferous or viruliferous adult males or females were similar (figure 3c,e). In various organs, the expression levels of LsRACK1 were also similar between the non-viruliferous and viruliferous L. striatellus (figure 3d), indicating that the accumulation of RBSDV had no effect on LsRACK1 expression in L. striatellus.
Figure 3.
Expression pattern of LsRACK1 in L. striatellus. (a) Relative expression levels of LsRACK1 in non-viruliferous L. striatellus at various developmental stages. (b) Relative expression levels of LsRACK1 in various tissues of non-viruliferous L. striatellus. (c) Relative expression levels of LsRACK1 in viruliferous and non-viruliferous female or male L. striatellus. (d) Relative expression levels of LsRACK1 in various tissues of viruliferous and non-viruliferous L. striatellus. All the results of figures a–d were determined by qRT-PCR using LsRACK1-specific primers and were normalized against the expression level of L. striatellus β-Actin gene. Each bar represents the mean of three biological repeats. Different lowercase letters above the bars indicate a significant statistical difference between the treatments. n.s., no significant statistical difference determined by the Tukey's multiple comparison tests (p > 0.05). (e) Western blot assay showing LsRACK1 and RBSDV P10 accumulation in adult viruliferous and non-viruliferous L. striatellus. The proteins were probed with an anti-RACK1, an anti-RBSDV P10, or an anti β-Actin antibody, respectively. The expression of L. striatellus β-Actin was used as a control.
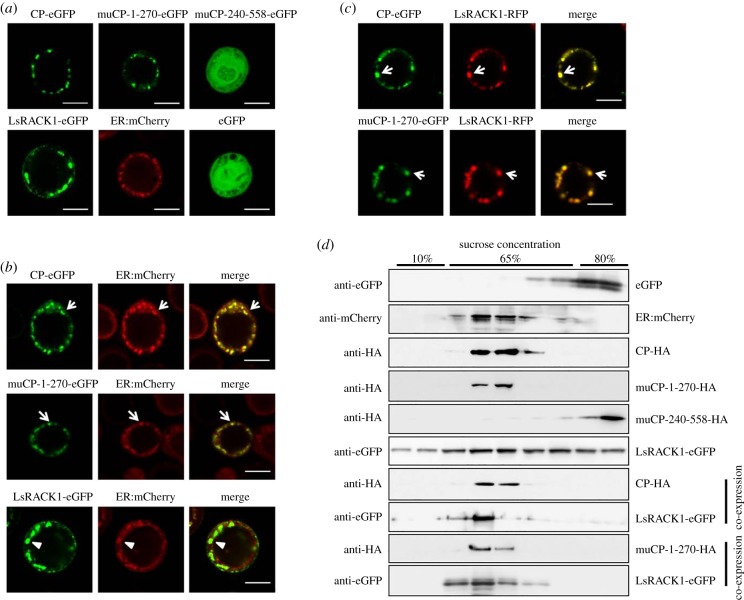
(d). Rice black-streaked dwarf virus P10 altered the subcellular localization pattern of LsRACK1
Subcellular localization patterns of RBSDV P10 and LsRACK1 were investigated using a baculovirus expression system. RBSDV P10, muP10-1-270 and muP10-240-558 were, respectively, tagged with the eGFP at their C-terminus, and LsRACK1 was tagged with a green or red fluorescent protein at its C-terminus. Under the confocal microscope, P10-eGFP, muP10-1-270-eGFP and ER:mCherry (a maker with an ER retention signal) were found to form granular-like structures in cytoplasm of Sf9 cells while muP10-240-558-eGFP was found to be free in the cells, similar to that shown by the free eGFP (figure 4a). Expressing of LsRACK1-eGFP in Sf9 cells also showed granular-like structures in cells (figure 4a).
Figure 4.
Expression of RBSDV P10 altered the subcellular localization pattern of LsRACK1. (a) Subcellular localization of P10-eGFP, muP10-1-270-eGFP, muP10-240-558-eGFP, ER:mCherry and LsRACK1-eGFP in Sf9 cells. The eGFP-expressed cell was used as a control. The transfected Sf9 cells were examined by confocal microscopy at 48 h post-transfection. Scale bars, 10 μm. (b) Sf9 cells were co-transfected with ER:mCherry and P10-eGFP, muP10-1-270-eGFP or LsRACK1-eGFP. The transfected cells were examined under a confocal microscope. The images were then processed using ZEISS ZEN confocal microscope software. White arrows indicate the granules of ER:mCherry, P10-eGFP or muP10-1-270-eGFP. The white arrows indicate the co-localized granules (yellow colour) in the superimposed images. Expressed LsRACK1 also formed granules (white arrowhead) in cells, which did not co-localize with ER:mCherry formed granules (white arrowhead). Scale bars, 10 μm. (c) Co-expression of P10-eGFP or muP10-1-270-eGFP with LsRACK1-RFP in Sf9 cells. Results of the assays indicated that the granules formed by P10-eGFP or muP10-1-270-eGFP co-localized with the granules formed by LsRACK1-RFP (white arrows). Scale bars, 10 μm. (d) Membrane flotation assays. After sucrose gradient centrifugation, proteins were separated in SDS-PAGE gels followed by a Western blot analysis using specific antibodies, namely anti-GFP, anti-mCherry and anti-HA antibody. Each experiment was repeated three times and similar results were obtained.
To determine whether these granular-like structures were associated with the ER network, P10-eGFP and ER:mCherry were co-expressed in Sf9 cells. Results of the study showed that co-expression of these two proteins in Sf9 cells yielded co-localized green and red granular-like structures, as indicated by the yellow fluorescent structures in the superimposed image (figure 4b, the top image of the right side column). Similar results were obtained when muP10-1-270-eGFP was co-expressed with ER:mCherry in Sf9 cells (figure 4b). By contrast, co-expression of LsRACK1-eGFP and ER:mCherry in Sf9 cells did not produce co-localized granular-like structures, in a manner of incomplete overlap, as indicated by the green fluorescent structures in the superimposed image (figure 4b, the bottom image of the right side column).
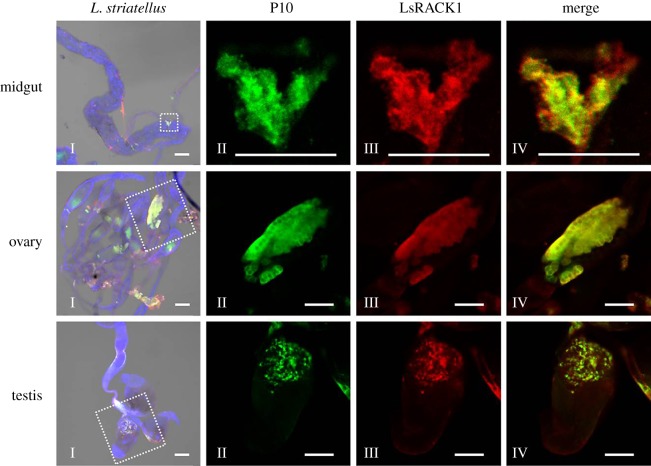
Because co-expression of P10-eGFP and LsRACK1-red fluorescent protein (RFP) in Sf9 cells also resulted in a co-localization pattern with green and red granular-like structures (figure 4c), we further analysed the midgut, ovary and testis from viruliferous L. striatellus by immunohistochemistry using a mixture of an anti-RBSDV P10 murine monoclonal antibody conjugated with FITC and an anti-LsRACK1 rabbit polyclonal antibody followed by a goat anti-rabbit IgG conjugated with Alexa Flour 555. Results of the assays indicated that the tissues which had accumulated RBSDV P10 also accumulated high-level LsRACK1, leading to a co-localized green and red florescence pattern (figure 5). We further analysed the ovary proteins from viruliferous or non-viruliferous L. striatellus by Western blot using an anti-RBSDV P10 monoclonal antibody, and the result proved further that P10 can be localized on the ovary of the viruliferous L. striatellus (electronic supplementary material, figure S1), which was consistent with the result of immunohistochemistry (figure 5).
Figure 5.
Immunocytochemical detection of RBSDV P10 and LsRACK1 in different organs of RBSDV-infected L. striatellus. Midguts, ovaries and testes were separated from viruliferous L. striatellus, and immunostained with a mixture of an anti-RBSDV P10 murine monoclonal antibody conjugated with FITC and an anti-LsRACK1 rabbit polyclonal antibody followed by a goat anti-rabbit IgG conjugated with Alexa Flour 555. The labelled tissues were examined under a confocal microscope. Nuclei in cells were stained blue using DAPI. Images in the left column (Is) were merged and low magnification images show the labelling signal in whole tissues. Images in the middle two columns (IIs and IIIs) are enlarged images from the boxed regions inside the I column images. Accumulation of RBSDV P10 was indicated by the green florescence from FITC and the accumulation of LsRACK1 was indicated by the red florescence from Alexa Fluor 555. Images in the right column (IVs) were the enlarged superimposed images. Images shown in this figure are representative images from the assay. Scale bars, 50 μm.
To further support our above results, a membrane flotation assay was conducted to show that RBSDV P10 could alter LsRACK1 distribution in cells. Sf9 cells were lysed in a hypotonic buffer at 48 h post-transfection and the lysates were separated by ultra-centrifugation and analysed by Western blot using specific antibodies. Results indicated that cell membrane-associated proteins were enriched in the 10% sucrose gradient fractions and the membrane vesicles, as well as RBSDV P10, were mainly present in the 65% sucrose gradient fractions close to the last 10% sucrose gradient fraction (figure 4d). To validate the separation efficiency, individual fractions were also analysed for the presence of eGFP and ER:mCherry. As expected, eGFP was remained predominantly in the 80% sucrose gradient fractions while ER:mCherry was mostly in the 65% sucrose gradient fractions, owing mainly to its association with the membrane through its histidine-aspartic acid-glutamate-leucine sequence (the two upmost panels in figure 4d). The enrichment of LsRACK1-eGFP in 65% sucrose gradient fractions, in the presence of P10-HA or muP10-1-270-HA, indicated that the presence of RBSDV P10 in Sf9 cells altered LsRACK1 migration pattern during gradient centrifugation. Together with the above subcellular localization results, we conclude that the ER membrane-associated RBSDV P10 captures cytosolic LsRACK1 in granular-like forms through protein–protein interaction.
(e). Silencing LsRACK1 expression in Laodelphax striatellus enhanced rice black-streaked dwarf virus accumulation
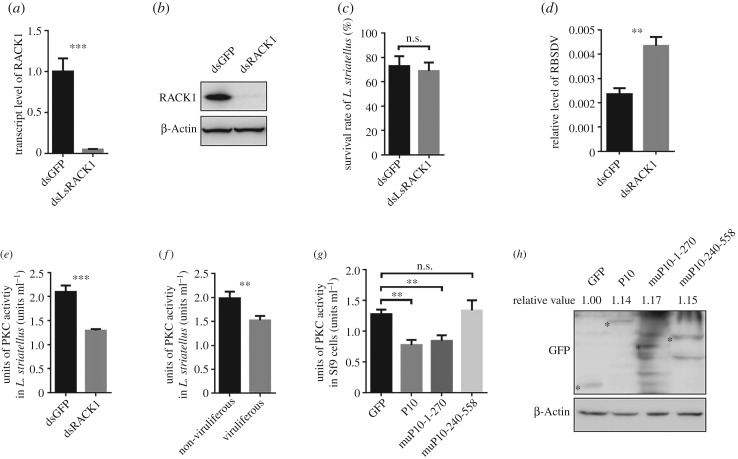
To investigate whether LsRACK1 is important for RBSDV accumulation in L. striatellus, we knocked down the expression level of LsRACK1 in L. striatellus through LsRACK1-dsRNA microinjection. The microinjected L. striatellus nymphs were analysed for LsRACK1 expression by qRT-PCR. Results showed that the expression level of LsRACK1 in LsRACK1-dsRNA-injected nymphs decreased by about 94.9% compared with the gfp-dsRNA-injected nymphs at 48 hpi (figure 6a). Results of Western blot analyses agreed with the results from qRT-PCR assays and showed that a very low amount of LsRACK1 had accumulated in the LsRACK1-silenced L. striatellus nymphs at 48 hpi (figure 6b). Because the survival rate of the LsRACK1-silenced L. striatellus nymphs was similar to that of the gfp-dsRNA-injected nymphs (control, figure 6c), we speculated that the expression of LsRACK1 might not have significant roles in L. striatellus growth and development. In a separate experiment, LsRACK1-silenced or gfp-dsRNA-injected L. striatellus nymphs were allowed to feed on RBSDV-infected rice plants for 3 d and then transferred to healthy rice seedlings for 15 d followed by qRT-PCR using RBSDV P10 specific primers. Results of the assay demonstrated that, compared with the gfp-dsRNA-injected L. striatellus nymphs, much higher RBSDV accumulation was found in the LsRACK1-silenced nymphs (figure 6d).
Figure 6.
LsRACK1 negatively regulated RBSDV accumulation in L. striatellus through suppressing the PKC signalling pathway. (a) Relative expression level of LsRACK1 in LsRACK1-dsRNA or gfp-dsRNA-microinjected L. striatellus at 48 h post-injection (hpi). Results were the means of 30 L. striatellus per treatment. (b) Western blot analysis of LsRACK1 in LsRACK1-dsRNA or gfp-dsRNA-microinjected L. striatellus at 48 hpi. The lower panel shows the accumulation of β-Actin and is used to show sample loadings. (c) Survival rates of LsRACK1-dsRNA or gfp-dsRNA-microinjected L. striatellus at 24 hpi. (d) Relative accumulation levels of RBSDV P10 in LsRACK1-dsRNA or gfp-dsRNA-microinjected L. striatellus at 15 d post virus acquisition. (e) Activities of PKC kinase in LsRACK1-dsRNA or gfp-dsRNA-microinjected L. striatellus. Thirty L. striatellus were used in each treatment. (f) Activities of PKC kinase in non-viruliferous and viruliferous L. striatellus. Each treatment had 30 L. striatellus. (g) PKC kinase activities in Sf9 cells expressing RBSDV P10, muP101-270, muP10-240-558 or eGFP. Bars in (a–g) are the means of three biological replicates ±s.d. *p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant difference (p > 0.05). (h) Western blot analyses of RBSDV P10-eGFP, muP10-1-270-eGFP, muP10-240-558-eGFP or eGFP expressed in Sf9 cells. The experiments presented in this figure were repeated three times. The relative expression levels of GFP, CP or CP mutants in each treatment were quantified using ImageJ software (https://imagej.nih.gov/ij/). Mean values of target were obtained after normalizing to β-Actin control and the protein accumulation of GFP was set as standard ‘1’.
(f). Rice black-streaked dwarf virus infection impaired protein kinase C activity in Laodelphax striatellus
Previous reports have indicated that virus infection could affect host PKC activities [39–41], leading to an increased or reduced phosphorylation of various substrates and then influence cellular functions. In this study, we first evaluated the impact of LsRACK1 on PKC activities. Second-instar L. striatellus nymphs were microinjected with LsRACK1-dsRNA or gfp-dsRNA, and then placed on healthy rice seedlings for 2 d. Analyses using total protein from L. striatellus demonstrated that the activities of PKCs in the LsRACK1-silenced L. striatellus was significantly reduced compared with that of gfp-injected L. striatellus (figure 6e). To further confirm this finding, we analysed PKC activity of the crude extracts from uninfected or RBSDV-infected L. striatellus. Results shown in figure 6f indicated that the activity of PKCs in the extract from RBSDV-infected L. striatellus was significantly decreased compared with that in the extract from the uninfected L. striatellus.
(g). Expression of rice black-streaked dwarf virus P10 in Spodoptera frugiperda 9 cell impaired the activities of PKCs
RACK1 is known as a receptor of PKCs and a multifunctional scaffold protein involved in many important signal transduction pathways. To further determine if interaction between RBSDV P10 and LsRACK1 affects the activities of PKCs in L. striatellus, we analysed the activities of PKCs in the P10-, muP10-1-270- or muP10-240-558-transfected Sf9 cells harvested at 2 dpi. Results indicated that the activities of PKCs in the P10, or muP10-1-270-transfected Sf9 cells were significantly reduced compared with the gfp-transfected control cells (figure 6g). By contrast, in the muP10-240-558-transfected Sf9 cells, the activities of PKCs remained unchanged (figure 6g). It is noteworthy that the expression levels of GFP, P10 or P10 mutants in each treatment were similar to that determined by Western blot (figure 6h).
(h). Protein kinase C activities negatively regulated rice black-streaked dwarf virus accumulation
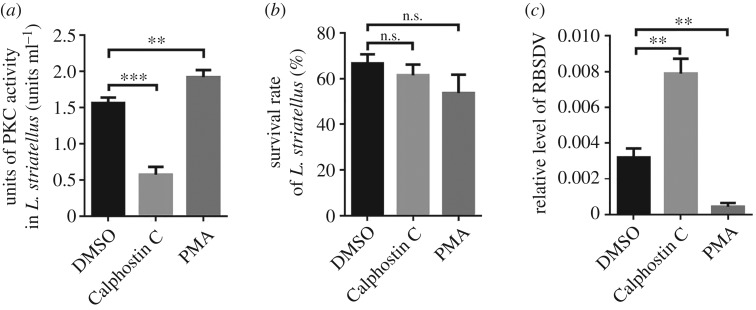
To further determine the role of PKCs during RBSDV replication, PKCs specific inhibitor (10 µl 1 μM Calphostin C) or PKCs activator (10 µl 1 μM PMA) was injected into L. striatellus. Laodelphax striatellus injected with 1% dimethyl sulfoxide (DMSO) was used as a control. The activities of PKCs in the Calphostin C-injected L. striatellus decreased approximately 64.2% compared with the control L. striatellus. By contrast, a 22.8% increase of PKCs activities was observed in the PMA-injected L. striatellus at 24 hpi (figure 7a). In this assay, the survival rate of injected L. striatellus was compared, and the result indicated that Calphostin C and PMA had no significant effect on L. striatellus viability (figure 7b). We then analysed the effect of PKCs activities on the accumulation of RBSDV P10 by qRT-PCR. The results demonstrated that the Calphostin C-injection treatment caused an increase of virus accumulation in L. striatellus while the PMA-injection treatment negatively affected the accumulation of RBSDV (figure 7c).
Figure 7.
Inhibition of PKC activity enhanced RBSDV accumulation in L. striatellus. (a) Activities of PKC kinase in L. striatellus treated with DMSO, Calphostin C or PMA at 24 hpi. The assay was conducted using the PepTag® non-radioactive PKC assay kit (Promega). Each treatment had 30 L. striatellus. (b) Survival rate of L. striatellus injected with Calphostin C, PMA or 1% DMSO (control) was measured by counting the survival insects at 24 hpi. Each bar represents the mean of 30 L. striatellus per treatment ±s.d. (c) Relative accumulation level of RBSDV RNA in Calphostin C-, PMA- or DMSO-injected L. striatellus at 15 d post-RBSDV acquisition. The accumulation was determined through qRT-PCR using a set of P10 specific primers. The bars represent the mean of three biological repetitions per treatment ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant (p > 0.05). All the experiments were repeated three times and similar results were obtained.
4. Discussion
It is well known that the capsid protein of viruses plays an important role in vector transmission of viruses. Deciphering the function and mechanism of RBSDV P10 involved in the process of RBSDV transmission by L. striatellus is important to uncover the epidemic tendency and establish a scientific system for prevention and control of RBSDV.
Insect transmission of plant viruses needs interactions between viral proteins and vector factors [8,29,42,43]. Consequently, characterization of viral proteins that interact with specific vector factors and understanding how viruses combat vector defence systems during their replication and movement in their insect vectors will provide important evidence and knowledge for developing new strategies for virus disease management. Rice stripe virus (RSV)-L. striatellus interaction is the most widely studied case for planthopper-borned viruses. Huo et al. [44] reported that RSV could use a vector transovarial transportation system for its transovarial transmission, and the vitellogenin (Vg) of L. striatellus played a critical role in this process [44]. In another report, RSV was reported to manipulate the c-Jun N-terminal kinase (JNK) signalling pathway in L. striatellus and the activation of JNK promoted RSV replication in its vector [38]. Further study revealed that the coat protein of RSV competitively bound with the G protein pathway suppressor 2 (GPS2) to release GPS2 from inhibiting the activation of the JNK signalling pathway [38].
RACK1, also known as guanine nucleotide-binding protein subunit beta-2-like 1 (GNB2L1) in humans, is a scaffold protein composed of seven WD40 domains and is involved in a variety of signalling transductions. The WD repeat sequence in RACK1 is highly conserved in a diverse range of species, including plants [45], and genetically malleable species such as Drosophila melanogaster [46]. The LsRACK1 identified in this study shears the highest amino acid sequence identity (81.6%) with that of D. melanogaster followed by human and mouse (figure 1a). We also amplified the RACK1 gene from the white-backed planthopper (Sogatella furcifera) and brown planthopper (Nilaparvata lugens) and found that their amino acid homology reached 99.7% and 99.4% with that of L. striatellus, respectively. RACK1 is known to be ubiquitously expressed in tissues of higher mammals and humans [47,48]. In this study, qRT-PCR analyses revealed that LsRACK1 expressed in all tested tissues of L. striatellus at different levels, suggest that it may have an important functional role in most, if not all, cells.
The RACK1 of aphid vector, M. persicae, can bind with wild-type BWYV virion, but much less efficiently with virions of two BWYV mutants during virus endocytosis/transcytosis, suggesting that RACK1 may play a key role in the process of virus endocytosis/transcytosis [32]. Although the interaction between RSV virion and LsRACK1 has been reported previously [31], which protein of RSV interacts with LsRACK1 was unknown. Results from this study demonstrated for the first time to our knowledge, that RBSDV P10 interacted directly with the LsRACK1 of L. striatellus. Further study showed that the seventh WD 40 domain at the C terminus (aa 268–315) of LsRACK1 and the N terminus (aa 1–270) of RBSDV P10 are critical for the interaction between RBSDV P10 and LsRACK1. Earlier reports have also indicated that human RACK1 was localized at the internal side of membranes and on cytoskeletons in cytosol [22,23]. It was also reported that the C terminus of RACK1 interacted with various signal transduction components and brought these components into stable complexes [49]. Particularly, the sixth WD 40 domain is responsible for binding with and activating PKC both in vitro and in vivo [20,23]. Moreover, the correct localization of RACK1 was important for its biological function [22,23]. ER is known to be the major site for protein synthesis and the accumulation of RBSDV P10 was reported to associate with ER in plant cells [11].
We found that LsRACK1 appeared in both cytoplasm and the cell membrane as small granules when expressed alone in Sf9 cells, while RBSDV P10 was co-expressed with LsRACK1 in cells, the interaction between the two proteins resulted in co-localized granules in or associated with ER, suggesting that RBSDV P10 interacted with LsRACK1 and brought it to the ER. We also found that the interaction between RBSDV P10 and the seventh WD 40 domain changed the subcellular localization pattern of LsRACK1. This new finding shades a new light on the special function of RBSDV P10 during virus transmission and infection in its vector.
DsRNA-induced gene silencing through microinjection has been widely used to down-regulate gene expression in planthoppers [36,50]. Our experimental results demonstrated that knockdown of LsRACK1 gene expression in L. striatellus through microinjection did not cause obvious morphological changes and death of the insect. In a different study, we determined that the accumulation of RBSDV was significantly increased after silencing LsRACK1 gene expression in L. striatellus. Thus, we consider that LsRACK1 is an antiviral gene in L. striatellus, and possibly in other insect vectors too, but has no significant role in L. striatellus growth and development. RACK1 is a receptor for activated C-kinase and responsible for binding with active forms of PKC family enzymes. We found that the PKC activity in the RBSDV-infected L. striatellus was much lower than that in the non-viruliferous L. striatellus. Moreover, the PKC activity in Sf9 cells expressing RBSDV P10 or a P10 N terminus deletion mutant, i.e. muP10-1-270 was significantly lower than that in the control Sf9 cells expressing GFP only. Those study results indicated that expression of RBSDV P10 could down-regulate activities of PKCs, a downstream signalling molecule of the RACK1 pathway. Further study found that inhibition of PKC activity using specific inhibitor Calphostin C significantly enhanced RBSDV accumulation in vectors, whereas activation of PKC using activator PMA dramatically inhibited RBSDV accumulation. Therefore, we think that the PKC pathway is important for the antivirus response in L. striatellus. Thus, we speculate that the decrease of PKC activity in viruliferous insects is a result caused by the interaction between RBSDV P10 and LsRACK1 that changes the subcellular localization and function of LsRACK1.
Our speculation is supported by a precious study showing that the ZEBRA protein of Epstein-Barr virus (EBV) could bind with RACK1 in vitro, and EBV infection altered the subcellular localization of RACK1 and impaired the activity and translocation of PKCα/β in infected monocytes [51]. Such disruption of PKC translocation/activation contributed to the ability of the EBV ZEBRA protein to interact with RACK1 [39]. Moreover, the HIV-1 Nef protein binds with RACK1 and interferes with PKCθ activity in T lymphocytes [51,52]. Thus, disruption of association between PKC and RACK1 can significantly affect PKC activity. Taken together, we propose that during virus infection, virus-encoded protein(s) interacts with RACK1 to suppress PKC activities leading to a weakened downstream antivirus defence response. Our detailed study showed that the N terminus 270 aa of RBSDV P10 is the key region for the interaction between the P10 and LsRACK1. Consequently, this region has the function to regulate the antiviral defence response of insect vectors, like phagocytosis.
Recently, RACK1 was identified as the first plant mitogen-activated protein kinase (MAPK) scaffold protein that connected heterotrimeric G protein with a MAPK cascade to form a unique signalling pathway in plant immunity [53]. Rice is known to contain two RACK1 genes: RACK1A and RACK1B. RACK1A has also been identified as an interactor with Rac1, a small GTPase, which is also known as Ras-related C3 botulinum toxin substrate 1, and involved in rice innate immunity [54]. The study also found that Rac1 regulated the expression of RACK1A at both transcriptional and post-transcriptional levels [54]. Transcription of RACK1A can be induced by a fungal elicitor or by abscisic acid, jasmonate and auxin, respectively [54]. RACK1A also plays a role in the production of reactive oxygen species and in disease resistance against rice blast infection [55]. Together, RACK1 is involved in blocking the release of the virus particle, activation of MAPK which controls antiviral responses, cell survival, proliferation and cell viability [53,56].
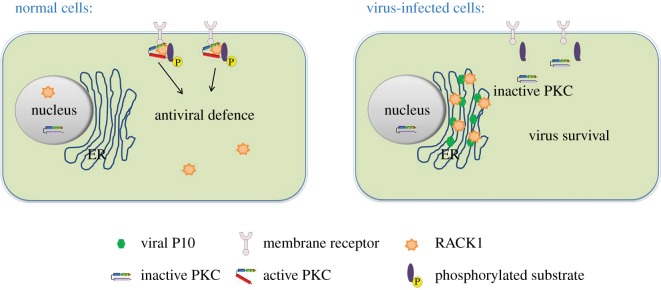
In summary, our findings presented here provide new evidence showing that the N-terminus region of RBSDV P10 is responsible for impairing the innate immunity in L. striatellus, and this function is achieved through altering the subcellular location pattern of LsRACK1 and modulating the structure and function of PKC in its vector. Therefore, we provide a model to summarize how RBSDV manipulates the PKC signalling pathway to increase its replication in L. striatellus (figure 8). We consider that LsRACK1 and the PKC pathway play critical roles in antiviral defence in L. striatellus and other insect vectors. This new finding sheds new light on the special function of RBSDV P10 during virus transmission and infection, and should contribute to our knowledge on the mechanisms controlling RBSDV transmission through L. striatellus.
Figure 8.
Model of RBSDV regulating PKC signalling pathway in L. striatellus to increase its replication. The PKC pathway participates in antiviral defence in L. striatellus. RBSDV P10 is synthesized on ER membranes and forms ER membrane-associated granules. RBSDV P10 brings LsRACK1, and makes it located on ER membranes through protein–protein interaction. The altered LsRACK1 conformation leads to an impaired PKC activity in RBSDV-infected L. striatellus cells.
Supplementary Material
Acknowledgements
We thank Dr Xinshun Ding, Plant Biology Division, the Samuel Roberts Noble Foundation, Oklahoma, USA, (retired) for his comments and language editing on the manuscript. The authors also thank Dr Yijun Zhou, the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, China, for providing L. striatellus populations.
Data accessibility
Additional data is supplied as the electronic supplementary material.
Authors' contribution
J.W. and X.Z. conceived and designed the study, secured funding and contributed to data analysis; L.L. and Q.W. conducted laboratory work and drafted the paper. D.H. and Q.X. performed assays and contributed to data analysis. All the authors contributed to manuscript revisions and gave their final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China (nos. 31571976; 31471768), the National Basic Research Program (973) of China (2014CB138400), the National Key Research and Development Project of China (no. 2016YFD0300706), the Earmarked Fund for Modern Agro-industry Technology Research System (nycytx-001) and Dabeinong Funds for Discipline Development and Talent Training in Zhejiang University.
References
- 1.Zhang P, Mar TT, Liu W, Li L, Wang X. 2013. Simultaneous detection and differentiation of rice black streaked dwarf virus (RBSDV) and southern rice black streaked dwarf virus (SRBSDV) by duplex real time RT-PCR. Virol. J. 10, 24 ( 10.1186/1743-422X-10-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin X, Zheng FQ, Tang W, Zhu QQ, Li XD, Zhang GM, Liu HT, Liu BS. 2013. Genetic structure of rice black-streaked dwarf virus populations in China. Arch. Virol. 158, 2505–2515. ( 10.1007/s00705-013-1766-8) [DOI] [PubMed] [Google Scholar]
- 3.Azuhata F, Uyeda I, Kimura I, Shikata E. 1993. Close similarity between genome structures of rice black-streaked dwarf and maize rough dwarf viruses. J. Gen. Virol. 74, 1227–1232. ( 10.1099/0022-1317-74-7-1227) [DOI] [PubMed] [Google Scholar]
- 4.Firth AE, Atkins JF. 2009. Analysis of the coding potential of the partially overlapping 3′ ORF in segment 5 of the plant fijiviruses . Virol. J. 6, 32 ( 10.1186/1743-422X-6-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isogai M, Uyeda I, Lee BC. 1998. Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 79, 1487–1494. ( 10.1099/0022-1317-79-6-1487) [DOI] [PubMed] [Google Scholar]
- 6.Zhang LD, Wang ZH., Wang XB, Zhang WH, Li DW, Han CG, Zhai YF, Yu JL. 2005. Two virus-encoded RNA silencing suppressors, P14 of beet necrotic yellow vein virus and S6 of rice black streak dwarf virus. Chin. Sci. Bull. 50, 305–310. ( 10.1007/BF02897570) [DOI] [Google Scholar]
- 7.Gallina A, Rossi F, Milanesi G. 2001. Rack1 binds HIV-1 Nef and can act as a Nef-protein kinase C adaptor. Virology 283, 7–18. ( 10.1006/viro.2001.0855) [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Jia D, Chen H, Chen Q, Xie L, Wu Z, Wei T. 2011. The P7-1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch. Virol. 156, 1729–1736. ( 10.1007/s00705-011-1041-9) [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Liu Y, Liu L, Lou Z, Zhang H, Miao H, Hu X, Pang Y, Qiu B. 2008. Rice black streaked dwarf virus P9-1, an α-helical protein, self-interacts and forms viroplasms in vivo. J. Gen. Virol. 89, 1770–1776. ( 10.1099/vir.0.2008/000109-0) [DOI] [PubMed] [Google Scholar]
- 10.Maroniche GA, Mongelli VC, Llauger G, Alfonso V, Taboga O, del Vas M. 2012. In vivo subcellular localization of Mal de Río Cuarto virus (MRCV) non-structural proteins in insect cells reveals their putative functions. Virology 430, 81–89. ( 10.1016/j.virol.2012.04.016) [DOI] [PubMed] [Google Scholar]
- 11.Sun Z, Yang D, Xie L, Sun L, Zhang S, Zhu Q, Li J, Wang X, Chen J. 2013. Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology 447, 131–139. ( 10.1016/j.virol.2013.09.001) [DOI] [PubMed] [Google Scholar]
- 12.Weber PH, Bujarski JJ. 2015. Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 196, 140–149. ( 10.1016/j.virusres.2014.11.014) [DOI] [PubMed] [Google Scholar]
- 13.Lavine MD, Strand MR. 2001. Surface characteristics of foreign targets that elicit an encapsulation response by the moth Pseudoplusia includens. J. Insect. Physiol. 47, 965–974. ( 10.1016/S0022-1910(01)00071-3) [DOI] [PubMed] [Google Scholar]
- 14.Lavine MD, Strand MR. 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32, 1295–1309. ( 10.1016/S0965-1748(02)00092-9) [DOI] [PubMed] [Google Scholar]
- 15.Marmaras VJ, Lampropoulou M. 2009. Regulators and signalling in insect haemocyte immunity. Cell. Signal. 21, 186–195. ( 10.1016/j.cellsig.2008.08.014) [DOI] [PubMed] [Google Scholar]
- 16.Sim S, Dimopoulos G. 2010. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS ONE 5, e10678 ( 10.1371/journal.pone.0010678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. 2002. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62, 1261–1273. ( 10.1124/mol.62.6.1261) [DOI] [PubMed] [Google Scholar]
- 18.Sklan EH, Podoly E, Soreq H. 2006. RACK1 has the nerve to act: structure meets function in the nervous system. Prog. Neurobiol. 78, 117–134. ( 10.1016/j.pneurobio.2005.12.002) [DOI] [PubMed] [Google Scholar]
- 19.Mochly-Rosen D, Khaner H, Lopez J. 1991. Identification of intracellular receptor proteins for activated protein kinase C. Proc. Natl Acad. Sci. USA 88, 3997–4000. ( 10.1073/pnas.88.9.3997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. 1994. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl Acad. Sci. USA 91, 839–843. ( 10.1073/pnas.91.3.839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi DS, Young H, McMahon T, Wang D, Messing RO. 2003. The mouse RACK1 gene is regulated by nuclear factor-kappa B and contributes to cell survival. Mol. Pharmacol. 64, 1541–1548. ( 10.1124/mol.64.6.1541) [DOI] [PubMed] [Google Scholar]
- 22.Ron D, Jiang Z, Yao L, Vagts A, Diamond I, Gordon A. 1999. Coordinated movement of RACK1 with activated betaIIPKC. J. Biol. Chem. 274, 27 039–27 046. ( 10.1074/jbc.274.38.27039) [DOI] [PubMed] [Google Scholar]
- 23.Ron D, Mochly-Rosen D. 1995. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc. Natl Acad. Sci. USA 92, 492–496. ( 10.1073/pnas.92.2.492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams DR, Ron D, Kiely PA. 2011. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 9, 22 ( 10.1186/1478-811X-9-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson CH, Ali ES, Scrimgeour N, Martin AM, Hua J, Tallis GA, Rychkov GY, Barritt GJ. 2015. Steatosis inhibits liver cell store-operated Ca2+ entry and reduces ER Ca2+ through a protein kinase C-dependent mechanism. Biochem. J. 466, 379–390. ( 10.1042/BJ20140881) [DOI] [PubMed] [Google Scholar]
- 26.Ali ES, Hua J, Wilson CH, Tallis GA, Zhou FH, Rychkov GY, Barritt GJ. 2016. The glucagon-like peptide-1 analogue exendin-4 reverses impaired intracellular Ca2+ signalling in steatotic hepatocytes. Biochim. Biophys. Acta 1863, 2135–2146. ( 10.1016/j.bbamcr.2016.05.006) [DOI] [PubMed] [Google Scholar]
- 27.Sieczkarski SB, Whittaker GR. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83, 1535–1545. ( 10.1099/0022-1317-83-7-1535) [DOI] [PubMed] [Google Scholar]
- 28.Dorn GW, Mochly-Rosen D. 2002. Intracellular transport mechanisms of signal transducers. Annu. Rev. Physiol. 64, 407–429. ( 10.1146/annurev.physiol.64.081501.155903) [DOI] [PubMed] [Google Scholar]
- 29.Hogenhout SA, Ammarel D, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359. ( 10.1146/annurev.phyto.022508.092135) [DOI] [PubMed] [Google Scholar]
- 30.Blanc S, Drucker M, Uzest M. 2014. Localizing viruses in their insect vectors. Annu. Rev. Phytopathol. 52, 403–425. ( 10.1146/annurev-phyto-102313-045920) [DOI] [PubMed] [Google Scholar]
- 31.Li S, Xiong R, Wang X, Zhou Y. 2011. Five proteins of Laodelphax striatellus are potentially involved in the interactions between rice stripe virus and vector. PLoS ONE 6, e26585 ( 10.1371/journal.pone.0026585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seddas P, Boissinot S, Strub JM, Van Dorsselaer A, Van Regenmortel MH, Pattus F.. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325, 399–412. ( 10.1016/j.virol.2004.05.014) [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Ni Y, Liu H, Rao L, Zhou Y, Zhou X. 2013. Development and use of three monoclonal antibodies for the detection of rice black-streaked dwarf virus in field plants and planthopper vectors. Virol. J. 10, 114 ( 10.1186/1743-422X-10-114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X. 2011. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus beta-satellite. Plant Physiol. 157, 1394–1406. ( 10.1104/pp.111.184648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentham M, Mazaleyrat S, Harris M. 2006. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J. Gen. Virol. 87, 563–571. ( 10.1099/vir.0.81200-0) [DOI] [PubMed] [Google Scholar]
- 36.Xu HJ, et al. 2015. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. ( 10.1038/nature14286) [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Xie Y, Zhou X. 2009. Efficient virus-induced gene silencing in plants using a modified geminivirus DNA1 component. Plant Biotechnol. J. 7, 254–265. ( 10.1111/j.1467-7652.2008.00395.x) [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Zhao W, Li J, Luo L, Kang L, Cui F. 2017. The c-Jun N-terminal kinase pathway of a vector insect is activated by virus capsid protein and promotes viral replication. Elife 6, e26591 ( 10.7554/eLife.26591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardif M, Savard M, Flamand L, Gosselin J. 2002. Impaired protein kinase C activation/translocation in Epstein-Barr virus-infected monocytes. J. Biol. Chem. 277, 24 148–24 154. ( 10.1074/jbc.M109036200) [DOI] [PubMed] [Google Scholar]
- 40.Park R and Baines JD. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80, 494–504. ( 10.1128/JVI.80.1.494-504.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartjen P, et al. 2013. The NTPase/helicase domain of hepatitis C virus nonstructural protein 3 inhibits protein kinase C independently of its NTPase activity. Cell. Mol. Biol. Lett. 18, 447–458. ( 10.2478/s11658-013-0099-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray S, Gildow FE. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41, 539–566. ( 10.1146/annurev.phyto.41.012203.105815) [DOI] [PubMed] [Google Scholar]
- 43.Ammar el-D, Tsai CW, Whitfield AE, Redinbaugh MG, Hogenhout SA. 2009. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54, 447–468. ( 10.1146/annurev.ento.54.110807.090454) [DOI] [PubMed] [Google Scholar]
- 44.Huo Y, et al. 2014. Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog. 10, e1003949 ( 10.1371/journal.ppat.1003949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwak JM, Kim SA, Lee SK, Oh SA, Byoun CH, Han JK, Nam HG. 1997. Insulin-induced maturation of Xenopus oocytes is inhibited by microinjection of a Brassica napus cDNA clone with high similarity to a mammalian receptor for activated protein kinase C. Planta 201, 245–251. ( 10.1007/s004250050063) [DOI] [PubMed] [Google Scholar]
- 46.Bini L, Heid H, Liberatori S, Geier G, Pallini V, Zwilling R. 1997. Two-dimensional gel electrophoresis of Caenorhabditis elegans homogenates and identification of protein spots by microsequencing. Electrophoresis 18, 557–562. ( 10.1002/elps.1150180337) [DOI] [PubMed] [Google Scholar]
- 47.Guillemot F, Billault A, Auffray C. 1989. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc. Natl Acad. Sci. USA 86, 4594–4598. ( 10.1073/pnas.86.12.4594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou YC, Chou CC, Chen YK, Tsai S, Hsieh FM, Liu HJ, Hseu TH. 1999. Structure and genomic organization of porcine RACK1 gene. Biochim. Biophys. Acta 1489, 315–322. ( 10.1016/S0167-4781(99)00213-4) [DOI] [PubMed] [Google Scholar]
- 49.Wang W, et al. 2002. Identification and characterization of AGTRAP, a human homolog of murine angiotensin II receptor-associated protein (Agtrap). Int. J. Biochem. Cell. Biol. 34, 93–102. ( 10.1016/S1357-2725(01)00094-2) [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Wu W, Chen H, Liu Q, Jia D, Mao Q, Chen Q, Wu Z, Wei T. 2013. An insect cell line derived from the small brown planthopper supports replication of rice stripe virus, a tenuivirus. J. Gen. Virol. 94, 1421–1425. ( 10.1099/vir.0.050104-0) [DOI] [PubMed] [Google Scholar]
- 51.Baumann M, Gires O, Kolch W, Mischak H, Zeidler R, Pich D, Hammerschmidt W. 2000. The PKC targeting protein RACK1 interacts with the Epstein-Barr virus activator protein BZLF1. Eur. J. Biochem. 267, 3891–3901. ( 10.1046/j.1432-1327.2000.01430.x) [DOI] [PubMed] [Google Scholar]
- 52.Smith BL, Krushelnycky BW, Mochly-Rosent D, Berg P. 1996. The HIV nef protein associates with protein kinase C theta. J. Biol. Chem. 271, 16 753–16 757. ( 10.1074/jbc.271.28.16753) [DOI] [PubMed] [Google Scholar]
- 53.Su J, Xu J, Zhang S. 2015. RACK1, scaffolding a heterotrimeric G protein and a MAPK cascade. Trends Plant Sci. 20, 405–407. ( 10.1016/j.tplants.2015.05.002) [DOI] [PubMed] [Google Scholar]
- 54.Nakashima A, et al. 2008. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 20, 2265–2279. ( 10.1105/tpc.107.054395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirasu K, Schulze-Lefert P. 2003. Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 8, 252–258. ( 10.1016/S1360-1385(03)00104-3) [DOI] [PubMed] [Google Scholar]
- 56.Daniels CC, Rovnak J, Quackenbush SL. 2008. Walleye dermal sarcoma virus Orf B functions through receptor for activated C kinase (RACK1) and protein kinase C . Virology 375, 550–560. ( 10.1016/j.virol.2008.01.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data is supplied as the electronic supplementary material.