Abstract
Plant-parasitic nematodes (PPNs) cause severe damage to agricultural crops worldwide. As most chemical nematicides have negative environmental side effects, there is a pressing need for developing efficient biocontrol methods. Nematophagous microbes, the natural enemies of nematodes, are potential biocontrol agents against PPNs. These natural enemies include both bacteria and fungi and they use diverse methods to infect and kill nematodes. For instance, nematode-trapping fungi can sense host signals and produce special trapping devices to capture nematodes, whereas endo-parasitic fungi can kill nematodes by spore adhesion and invasive growth to break the nematode cuticle. By contrast, nematophagous bacteria can secrete virulence factors to kill nematodes. In addition, some bacteria can mobilize nematode-trapping fungi to kill nematodes. In response, nematodes can also sense and defend against the microbial pathogens using strategies such as producing anti-microbial peptides regulated by the innate immunity system. Recent progresses in our understanding of the signal pathways involved in microbe–nematode interactions are providing new insights in developing efficient biological control strategies against PPNs.
This article is part of the theme issue ‘Biotic signalling sheds light on smart pest management'.
Keywords: plant-parasitic nematode, biocontrol, nematophagous fungi, microbe–nematode interaction, signal transduction
1. Introduction
Plant-parasitic nematodes (PPNs), such as the root-knot nematode (RKN) and the cyst nematode, are causing an estimated global economic loss of more than US$157 billion per year in agriculture [1]. These parasitic nematodes not only cause direct damage to plant roots, but they can also facilitate infections by other phytopathogens, including those of fungi, bacteria and viruses. Traditional methods for controlling PPNs involves applying chemical nematicides; however, large amounts of chemical nematicides can cause severe environmental problems and harm human health [2,3]. As a result, most chemical nematicides have been banned or have restricted use in the world today. Because of their environmental-friendliness and safety for humans, biological control agents have attracted significant attention. As natural enemies of nematodes, the nematophagous fungi and bacteria are potential agents for the biological control of PPNs [4,5]. So far, the biocontrol of PPNs has mostly relied on direct applications of live microbes such as the nematophagous fungi Paecilomyces lilacinus and Pochonia chlamydosporia and nematophagous bacteria of the genus Bacillus. However, there are several challenges associated with biocontrol applications, including their relative low efficiency and high inconsistency in agricultural and forestry environments [5]. In this review, we focus on recent discoveries in the signalling pathways involved in the microbe–nematode interactions and discuss how such knowledge can facilitate the development of biocontrol applications.
2. The nematophagous fungi–nematode interaction
Nematophagous fungi are highly diverse. They capture nematodes using a variety of methods such as by specialized adhesive or mechanical trapping devices (nematode-trapping fungi), by infecting nematodes through conidial attachment and invasion from the cuticle (endo-parasitic fungi), by invading nematode eggs or females with their hyphal tips (egg parasitic fungi), or by secreting toxins to immobilize and kill nematodes (toxin-producing fungi) [6]. Here, we focus on the former two types of fungi as they have been more extensively studied.
(a). Signalling in trap formation of nematode-trapping fungi
Nematode-trapping fungi produce special hyphal structures called traps. Several types of trapping devices have been reported, including three-dimensional networks, constriction rings, adhesive hypha or knobs [7]. Among the nematode-trapping fungi, Arthrobotrys oligospora is among the most extensively studied [8]. Arthrobotrys oligospora produces adhesive three-dimensional networks to capture nematodes. The network production induced by nematodes is a signature of lifestyle transition from saprophytic to carnivorous lifestyles [9]. By analysing the genome and comparing the transcriptomes between the network and the vegetative mycelia, Yang et al. revealed that several components of the signalling pathways are upregulated during trap formation, indicating that the lifestyle switch is initiated by signalling cascades to alter its gene expression patterns in response to environmental cues [9].
(i). Signalling molecules in trap formation of nematode-trapping fungi
Identifying the signalling molecules involved in trap formations in nematode-trapping fungi is essential for understanding fungi–nematode interactions. Pramer & Stoll [10] found that the broth in which the nematode Neoaplectana glaseri had developed axenically can induce the mycelia of the predaceous fungus Arthrobotrys conoides to differentiate into traps. They named the active principal component from worm-free culture filtrates of predaceous fungi ‘nemin' [10]. Further study indicated that nemin is a peptide of relatively low molecular weight or possibly a single amino acid [11]. Partly inspired by nemin, Yang et al. found that the homogenate of the nematode Caenorhabditis elegans (called nematode extract, NE) can induce trap formation in A. oligospora [9].
Subsequent studies identified several specific compounds as potential inducers, including ascarosides and nitrogenous compounds such as NaNO3 and urea. Ascarosides are a family of pheromones secreted by nematodes that can regulate their development and behaviour, such as mate finding, aggregation, repulsion and developmental diapause [12–14]. Nematode-trapping fungi such as A. oligospora and related species can sense these small molecules to produce adhesive networks [15]. Our previous study reported that while (NH4)2SO4 and NH4Cl cannot induce trap formation, NaNO3, urea and yeast extract all can induce trap formation in A. oligospora [16]. The role of the nitrate assimilation pathway in trap formation in A. oligospora was confirmed by analysing strains with knockout of several key enzymes, including nitrate reductase (NiaD) and nitrite reductase (NiiA). The results showed that trap formation in this fungus could be induced by nitrate via a pathway distinctly different from those caused by nematodes. Interestingly, nitrate and nematodes can act synergistically to induce trap formation in A. oligospora [17].
(ii). AoMad1 helps the fungi in recognizing host signal molecules
Except ascarosides, other signal molecules found so far that are capable of inducing trap formation in A. oligospora or other nematode-trapping fungi are nitrogenous compounds. Interestingly, the preferred nitrogenous compound ammonia for fungal vegetative growth is a suppressor of trap formation. In addition, in the presence of other nitrogen sources at concentrations above a certain level, A. oligospora may fail to produce traps [17]. These results suggest that A. oligospora captures nematodes as a potential nitrogen source. In the paragraph below, we describe how nematode-trapping fungi distinguish signals from their prey and from other nitrogen sources.
In A. oligospora, there is a cell wall surface protein called AoMad1. This glycoprotein is the homologue of Mad1 from the entomopathogenic fungus, Metarhizium anisopliae. Mad1 is an adhesive protein located on the surface of conidia or blastospores that contribute to adhesion to insect cuticle [18]. When the AoMad1 protein coding gene is knocked out, the adhesive-like materials on the surface of traps, as well as the vegetative hyphae, disappears. Interestingly, the mutant strain can be induced to form traps by lower concentrations of certain nitrogenous compounds than the wild-type strain, suggesting that AoMad1 could prevent trap formation in the absence of nematodes, and keeps the fungus in the saprophytic lifestyle [16].
(iii). Signal cascades in trap formation
Many substances have been found capable of inducing trap formation in A. oligospora. Within fungal cells, these substances act as signals to affect morphological changes. Several signal transduction pathways are known to be involved in generating the morphological changes from saprophytic to predacious lifestyles in A. oligospora. These include the mitogen-activated protein kinase (MAPK) cascade, the G-protein-coupled receptors (GPCRs) or small GTPases. The MAPK Slt2 is a key molecule in regulating cell wall integrity in the budding yeast [19]. In the entomopathogenic fungus Beauveria bassiana, deletion of Slt2 resulted in a reduction of conidial production and cell viability, and increased sensitivity to cell wall stress substances, including Congo red and fungal cell wall–degrading enzymes [20,21]. The Slt2 orthologues in the nematode-trapping fungi A. oligospora and Monacrosporium haptotylum have been identified and characterized. Knockout of Slt2 in these two fungi caused defects in their growth, development and virulence against nematodes. Importantly, no trap was observed in the Slt2 mutants of both fungi, indicating that this MAPK plays a key role in trap formation [22].
Rab GTPases are a family of small, conserved proteins that act as switches in the signalling hub of molecular circuits and play a pivotal role in the secretion of proteins [23]. Two Rab GTPases, AoRab-7A and AoRab-2, have been characterized by gene knockout in A. oligospora. Between these two, the AoRab-7A mutant showed notable defects in mycelial growth and conidiation, and was unable to produce traps and capture nematodes. By contrast, the AoRab-2 mutant showed little defects [24].
(iv). Signals from secondary metabolites
Signalling between hyphae is an important cue in fungal morphological development such as hyphal fusion [25]. Recent studies showed that the morphology of nematode-trapping fungi can be regulated by secondary metabolites secreted by themselves. Several morphology-regulating arthrosporol metabolites were recently characterized from A. oligospora. These arthrosporol metabolites display significant autoregulatory effects on the formation of conidiophores and the transition of hypha to a three-dimensional trapping network in A. oligospora [26]. In a subsequent study on arthrosporol biosynthesis, disruption of a polyketide synthase gene ΔAOL_s00215g283 not only led to the total loss of the arthrosporol A but also resulted in a significant reduction in the production of secondary metabolites in the cultural broth of the mutant strain. Importantly, the mutant strain displayed a significant increase in trap formation and in nematicidal activity [27]. In another study, the metabolomes in A. oligospora were analysed and compared between those living on corn meal agar and potato dextrose agar media. These two media differ in their capacities to induce traps. A compound, maltol, was isolated and shown to be able to stimulate trap formation in A. oligospora [28]. Together, these studies identified many signal molecules and several signal transducers capable of inducing fungal trap formation. However, much remains unknown about the components of the pathways and how they interact with each other during trap development.
(b). Endo-parasitic fungi–nematode interaction
Endo-parasitic fungi are a diverse group of fungi that infect nematodes but are unable to develop trapping devices. The nematodes are attacked by spores of these fungi, which adhere to the cuticle of worms or are ingested by them [29]. The most commonly studied endo-parasitic fungi include Pa. lilacinus, Po. chlamydosporia, Clonostachys rosea and Drechmeria coniospora. In general, the infection starts with the adhesion of spores to the nematode cuticle. Under appropriate conditions, the spores germinate and invade the cuticle through both mechanical forces and cuticle-degrading enzymes, such as serine proteases, chitinases, etc. [30–33].
Drechmeria coniospora is an obligate endo-parasitic fungus of various species of nematodes. The matured conidia of D. coniospora have an adhesive knob at the distal end of the spore [34]. After adhesion to the nematode cuticle (usually close to the tubes of the sensory-organs), an appressorium forms and then penetrates the cuticle [35]. The invasive growth may also involve enzymatic actions, including those of acid phosphatases, similar to that in the nematode-trapping fungus A. oligospora [36]. A study by Jansson demonstrated that treatments of the nematode Panagrellus redivivus and the conidia of D. coniospora with proteases resulted in decreased adhesion of the conidia to the nematode [37]. This result suggests the signals involved in adhesion and infection of nematodes may be mediated by proteins on the adhesive bud of the conidia and in the proteinaceous matrix materials emanating from the sensilla pores of the nematode. The exact materials and chemotaxis mechanisms involved in conidial adhesion are still unclear.
(c). Signalling in immune responses of nematodes against endonematophagous fungi
In the model nematode C. elegans, the epidermis directly interacts with the environment and is expected to play a key role in defence. The collagen-rich cuticle that surrounds the nematode provides a permanent physical barrier to pathogens [38]. Although many bacterial pathogens, such as Microbacterium nematophilum, Xenorhabdus nematophila or Yersinia pestis, can adhere to the surface cuticle of worms, they cannot infect worms by penetrating the epidermis [39–41]. Unlike these bacteria, endonematophagous fungi can directly invade the cuticle of their hosts. In nematodes, the strategies to defend against fungal pathogens are different from those against bacterial pathogens. Microarray analysis showed that many antimicrobial peptides (AMPs)-coding genes are upregulated in C. elegans infected by D. coniospora [42]. Many AMPs act by disrupting microbial cell membranes [43]. Six nlp family AMPs with different structures were found located in a small gene cluster. A phylogenetic analysis indicated that the evolutionary diversification of this cluster, which might have evolved a broad repertoire of AMPs that enhance their defensive potential, is driven by natural selection [38]. One of the AMPs, NLP-31, showed strong activities against fungi, such as D. coniospora, Neurospora crassa and Aspergillus fumigatus [42]. The antifungal activity of NLP-31 is similar to that of the antifungal peptide drosomycin in Drosophila and to the antimicrobial peptide androctonin from scorpion blood [44]. Transgenic worms carrying supernumerary copies of the entire nlp-29 cluster showed increased resistance to fungal infections [38]. Like fungal infections, wounding in the epidermis of C. elegans or osmotic stress can also induce the expression of AMP gene nlp-29 [38,45]. Genetic inactivation of a Toll–interleukin 1 receptor (TIR) domain protein, tir-1 causes increased susceptibility to infection and downregulates the expression of nlp-29 and nlp-31. Meanwhile, the small GTPase Rab1 and the F subunit of ATP synthase interact physically with TIR-1 in vivo and synergistically regulate the expression of AMP genes [42]. The p38 MAPK pathway including MAP3 K NSY-1, MAP2 K SEK-1 and p38 MAPK PMK-1, acts downstream of the conserved adapter protein TIR-1 to regulate nlp-29 expression. NIPI-3, a kinase related to human Tribbles homologue 1, probably acts upstream of the MAPKK SEK-1 to participate in nlp-29 induction after D. coniospora infection [45]. A chaperone of the BiP/GRP78 family, HSP3, which was previously characterized as a regulator in unfolded protein response (UPR) and endoplasmic reticulum stress, is found to act upstream of NIPI-3 in a UPR-independent manner during D. coniospora infection [46]. As AMPs in C. elegans play important roles in the primary immunity against fungal infections, we hypothesize that there should be corresponding molecules in PPNs. At present, homologue genes of the nlp family have not been reported in the genome of Meloidogyne incognita [1].
3. The nematophagous bacteria–nematode interaction
Many bacteria can infect nematodes. Some of these bacteria are also human pathogens, including Pseudomonas aeruginosa, Staphylococcus aureus, etc. Others such as Bacillus. spp., Pasteurella punctata and many actinomycete species are not pathogenic to humans and do not cause human diseases. These non-human pathogenic bacteria are potential biocontrol agents against nematodes.
(a). The ‘Trojan-horse' mechanism by Bacillus nematocida
The nematophagous bacterium Bacillus nematocida infects nematodes using a ‘Trojan-horse' -like mechanism [47]. This bacterium can emit potent volatile organic compounds, such as 2-heptanone, which are much more attractive to nematodes than those from ordinary dietary bacteria. After swallowed by nematodes and the bacterium colonizes in the intestinal tract of nematodes and secretes virulence factors in the form of proteases to cause nematode death [48]. The 2-heptanone receptor is identified as a GPCR (STR-2) located in the AWC neurons of worms. Downstream signalling pathway elements in worms include two Gα subunits EGL-30 and GPA-3, the phospholipase C PLC-1and EGL-8, the calcium/calmodulin-dependent protein kinase CMK-1 and the calmodulin-like protein CAL-1 [49]. In B. nematocida, the ComP-ComA Quorum sensing system is involved in regulating the biosynthesis of attractants. Transcriptions of these two virulence proteases Bace16 and Bae16 are directly regulated by ComA [50]. Thus, the Quorum sensing system orchestras the ‘Trojan-horse'-like interaction between the bacterium and nematodes.
(b). Bacillus thuringiensis
Crystal (Cry) toxins produced by the invertebrate pathogen Bacillus thuringiensis (Bt) are a large family of pore-forming toxins (PFTs) that target the intestinal cells of insects and nematodes [51–53]. The high and specific toxicity makes B. thuringiensis a leading biocontrol agent [54]. The genes of Cry proteins are wildly used to generate transgenic insect-resistant plants [55]. Among the crystal toxins, Cry5B has especially high toxicity against PPNs Meloidogyne spp. [52,56], as well as the model nematode C. elegans [53]. The nematode-specific cadherin CDH-8 acts as a receptor for Cry5B toxin [57].
To study the defence of PFTs by host nematodes, Huffman et al. [58] investigated how C. elegans responded to Cry5B using microarrays. They found that two MAPK pathways were involved in response to the toxin. One is the p38 MAPK pathway composed of NSY-1, SEK-1 and PMK-1. Nematodes lacking either genes of the three kinases showed hypersensitivity to Cry5B. The other MAPK induced by Cry5B is the JNK-family MAPK genes, kgb-1 and kgb-2. kgb-1 mutant worms exhibited hypersensitivity to Cry5B. By contrast, knockdown of kgb-2 by RNAi did not enhance the susceptibility to Cry5B. Thus, the JNK MAPK pathway is required for protection against toxins. Moreover, two downstream targets of the p38 MAPK pathway, ttm-1 and ttm-2, were identified as required for defence against Cry5B [58]. Bischof et al. found that the UPR was activated upon exposure to PFTs in worms. Animals that lack either the ire-1-xbp-1 or the atf-6 arms of the UPR are more sensitive to PFT than wild-type animals. A subsequent study found that this pathway was located downstream of the P38 MAPK pathway [59].
4. Fungi–bacteria–nematode interaction
The above nematophagous bacteria can infect and kill nematodes. How do those bacteria without weapons like the ‘Trojan-horse' strategy or the pore PFTs maintain their populations when they encounter nematodes? An early study reported that diffusible compounds from cow dung could induce trap formation in A. oligospora, as soon as the conidia start germinating (called conidial traps) [60]. Wang et al. [61] isolated 126 bacteria strains from the cow dung. Of these bacteria, 55 strains induced trap formation in A. oligospora, and three Stenotrophomonas maltophilia isolates were the most efficient. These authors identified urea from the culture medium of S. maltophilia as the signal molecule to induce trap formation. A further study revealed that after entering fungi, urea is hydrolyzed into ammonia, which then induces trap formation in A. oligospora [61]. This study provides unique insights into the complex interplay among fungi, nematodes and bacteria. Recently, Li et al. [62] also found that 21 out of 47 fungus-associated bacteria isolated from A. oligospora GJ-1 mycelia are able to induce trap formation. The bacteria with induction activity are identified as Stenotrophomonas and Rhizobium species. Unlike the study from Wang et al., these bacteria do not secrete urea. Instead, the biofilms formed by these bacteria on the surface of A. oligospora hypha caused trap formation in this fungus. Four diketopiperazines are identified from Stenotrophomonas supernatant extract as inducers of trap formation in Arthrobotrys [62].
5. The sensing of attractive chemicals and its application in biocontrol of plant-parasitic nematodes
As mentioned above, nematodes can be attracted by molecules secreted by their pathogens through their olfactory neurons and receptors. Beside 2-heptanone secreted by B. nematocida, several nematophagous fungi have also been found to produce attractive materials. For example, the nematode-trapping fungus Monacrosporium rutgeriensis could produce three different compounds that attract the saprophytic nematode, P. redivivus [63]. Furthermore, the traps produced by several nematode-trapping fungi make these fungi more attractive to nematodes than those without traps [64]. By using gas-chromatographic mass-spectral analyses of A. oligospora-derived volatile metabolites, Hsueh et al. [65] identified several odours mimicking food cues that are attractive to nematodes. Interestingly, one compound, methyl 3-methyl-2-butenoate additionally triggered strong sex- and stage-specific attraction in several Caenorhabditis species [65]. Furthermore, the mycelia of the endo-parasitic fungus, Esteya vermicola as well as its infected pine seedling and dead blocks of infected pine trees were found to attract the pinewood nematode (PWN), Bursaphelenchus xylophilus [66–69]. In the pine wilt disease system, the ascarosides produced by PWNs, as well as its vector beetle, play a crucial role in mediating the symbiotic interspecific interaction between PWN, its vector beetle Monochamus alternatus and their associated fungi. The amplification effect of ascarosides on the invasive complex may be used to develop effective approaches for preventing the development of vector beetles and limiting the spread of PWN [70].
As a model species, C. elegans has been used to study the amphid chemosensory system of nematodes. The 12 pairs of neurons in C. elegans can detect a wide variety of volatile and water-soluble chemicals in the environment [71–73]. Among the 12 pairs of neurons, five pairs are responsible for olfaction, specialized in detecting volatile chemicals. Two of the pairs, AWA and AWC, respond to attractive odorants [74]. Olfactory receptors (especially GPCRs), G protein and cation channels are essential in the signal transduction of olfaction [75–77]. In C. elegans, CO2 is repulsive for adults but attractive for dauer larvae [78]. The URX neurons were identified to control CO2 response by coordinating the response to CO2 with the response to ambient O2 [79]. The olfactory circuit of C. elegans consists of a small number of highly interconnected neurons, allowing the nematode to sense and respond to at least 50 odorants [80] (see review in [81]). Together, these studies provide important clues to investigate the molecular mechanism of chemotaxes in PPNs.
The attraction phenomenon and mechanisms mentioned above can be used to develop biocontrol strategies against PPNs. It is reported that volatiles from nearby plants can modulate attraction of PPNs to host plants. For example, the crown daisy can produce lauric acid, which is attractive for PPNs at low concentrations but repulsive at high concentrations. Manipulating its concentration to disturb chemotaxes of M. incognita could potentially help protect crops like tomatoes [82]. However, traditional biocontrol agents made by nematophagous microbes generally have limited effects as direct contacts between these microbes and PPNs may be limited. Researchers in our laboratory have recently optimized a bionematicide by adding attractive substances for RKNs. This improved nematicide formulation has shown the same effect as the most widely used chemical nematicide, Avermectin (M.H. Mo, L.M. Liang, K.Q. Zhang 2018, unpublished data).
6. Conclusion and perspective
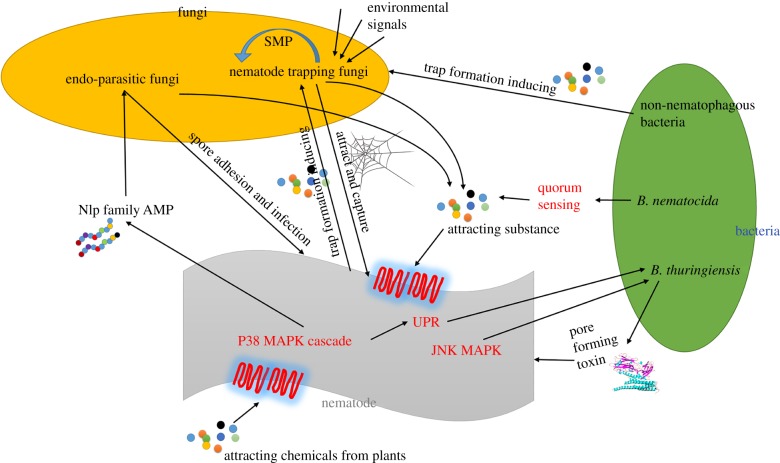
Complex interactions exist between nematodes and their pathogens (figure 1). Among these interactions, while several types have been elucidated, many issues remain unknown. Investigations on the nematophagous microbes started in the nineteenth century. Early studies focused on the isolations of microbial strains with high virulence, and on how their morphological features change during infections. Subsequent studies investigated the virulence factors of the microbial pathogens, such as cuticle-degrading enzymes [83–87]. In 2011, the first genome of a nematophagous fungus A. oligospora was sequenced [9]. The genome and transcriptome analysis revealed that several signal pathways are involved in fungal infection. Although many substances from the nematodes or from the environment are known to induce trap formation in nematode-trapping fungi, how the signals are transduced remains largely unknown. Similar to that in the bacterium B. nematocida, nematophagous fungi can also secrete chemicals to attract nematodes [64]. How these attracting chemicals are sensed by nematodes remains to be elucidated. Pratylenchus penetrans is a bacterial pathogen of many RKNs and it has been used broadly as a biocontrol agent. As an obligate parasite of RKNs, it has been difficult to obtain the pure culture of this bacterium, and thus efforts to sequence its whole genome has not been successful. Furthermore, limitations exist when translating the knowledge derived from the model nematodes in the laboratory to PPNs in agricultural and forest environments. Soil nematodes encounter countless soil microbes daily and thus have probably developed a sophisticated innate immune system to maintain their populations in nature. While many defence signalling pathways have been identified using the model nematode C. elegans, it is still unclear whether these pathways are also used by PPNs. As reported previously, M. incognita had fewer immune effectors in classes such as lysozymes, C-type lectins and chitinases than C. elegans, and lacked certain classes of immune effectors in C. elegans, including antibacterial genes such as abf and spp [88] and antifungal genes of several classes (nlp, cnc, fip, fipr) [1,89]. As the immune defence pathways may be used as molecular targets of biocontrol agents, an efficient strategy for genetic manipulation of Meloidogyne spp. and other PPNs needs to be developed to address these and other issues and to gain a comprehensive understanding on how these nemaotdes can only complete their life cycle in planta.
Figure 1.
The interactions between nematodes and their fungal and bacterial pathogens. When sensing the nematodes, the nematophagous fungi can develop various trapping devices to capture nematodes, or directly infect the nematodes using adhesive spores. The bacteria infect nematodes in different manners: mimic the food (‘Trojan-horse'-like mechanism), using pore-forming toxins, or with the help of nematode-trapping fungi. On the other hand, nematodes can develop elaborate defence strategies against microbial infections. SMP, secondary metabolic products; UPR, unfolded protein response; AMP, anti-microbial peptides. (Online version in colour.)
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (31572063, 31770064, 31770060 and 31860016), Yunnan Applied Basic Research Project (2016FB062) and grants from the Young Academic and Technical Leader Raising Foundation of Yunnan Province (to L.-M.L.).
References
- 1.Abad P, et al. 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. ( 10.1038/nbt.1482) [DOI] [PubMed] [Google Scholar]
- 2.Anastasiadis IA, Giannakou IO, Prophetouathanasiadou DA, Gowen SR. 2008. The combined effect of the application of a biocontrol agent Paecilomyces lilacinus, with various practices for the control of root-knot nematodes. Crop Protect. 27, 352–361. ( 10.1016/j.cropro.2007.06.008) [DOI] [Google Scholar]
- 3.Noling JW, Becker JO. 1994. The challenge of research and extension to define and implement alternatives to methyl bromide. J. Nematol. 26, 573–586. [PMC free article] [PubMed] [Google Scholar]
- 4.Morton CO, Hirsch PR, Kerry BR. 2004. Infection of plant-parasitic nematodes by nematophagous fungi: a review of the application of molecular biology to understand infection processes and to improve biological control. Nematology 6, 161–170. ( 10.1163/1568541041218004) [DOI] [Google Scholar]
- 5.Li J, Zou C, Xu J, Ji X, Niu X, Yang J, Huang X, Zhang KQ. 2015. Molecular mechanisms of nematode-nematophagous microbe interactions: basis for biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 53, 67–95. ( 10.1146/annurev-phyto-080614-120336) [DOI] [PubMed] [Google Scholar]
- 6.Zhang KQ, Hyde KD. 2014. Nematode-trapping fungi. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 7.Nordbring-Hertz B, Jansson HB, Tunlid A. 2006. Nematophagous fungi. In Encyclopedia of Life Sciences. Hoboken, NJ: Wiley-Blackwell ( 10.1038/npg.els.0004293 [DOI] [Google Scholar]
- 8.Niu XM, Zhang KQ. 2011. Arthrobotrys oligospora: a model organism for understanding the interaction between fungi and nematodes. Mycology 2, 59–78. ( 10.1080/21501203.2011.562559) [DOI] [Google Scholar]
- 9.Yang J, et al. 2011. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7, e1002179 ( 10.1371/journal.ppat.1002179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pramer D, Stoll NR. 1959. Nemin: a morphogenic substance causing trap formation by predaceous fungi. Science 129, 966–967. ( 10.1126/science.129.3354.966) [DOI] [PubMed] [Google Scholar]
- 11.Pramer D, Kuyama S. 1963. Symposium on biochemical bases of morphogenesis in fungi. II. Nemin and the nematode-trapping fungi. Bacteriol. Rev. 27, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan J, et al. 2008. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1119. ( 10.1038/nature07168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan J, et al. 2012. A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 10, e1001237 ( 10.1371/journal.pbio.1001237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher RA, Fujita M, Schroeder FC, Clardy J. 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3, 420–422. ( 10.1038/nchembio.2007.3) [DOI] [PubMed] [Google Scholar]
- 15.Hsueh YP, Mahanti P, Schroeder FC, Sternberg PW. 2013. Nematode-trapping fungi eavesdrop on nematode pheromones. Curr. Biol. 23, 83–86. ( 10.1016/j.cub.2012.11.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang L, Shen R, Mo Y, Yang J, Ji X, Zhang K-Q. 2015. A proposed adhesin AoMad1 helps nematode-trapping fungus Arthrobotrys oligospora recognizing host signals for life-style switching. Fungal Genet. Biol. 81, 172–181. ( 10.1016/j.fgb.2015.02.012) [DOI] [PubMed] [Google Scholar]
- 17.Liang L, Liu Z, Liu L, Li J, Gao H, Yang J, Zhang KQ. 2016. The nitrate assimilation pathway is involved in the trap formation of Arthrobotrys oligospora, a nematode-trapping fungus. Fungal Genet. Biol. 92, 33–39. ( 10.1016/j.fgb.2016.05.003) [DOI] [PubMed] [Google Scholar]
- 18.Wang C, St Leger RJ. 2007. The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot. Cell 6, 808–816. ( 10.1128/EC.00409-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32, 671–680. ( 10.1046/j.1365-2958.1999.01375.x) [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Zhu J, Ying S.-H, Feng M-G. 2014. Three mitogen-activated protein kinases required for cell wall integrity contribute greatly to biocontrol potential of a fungal entomopathogen. PLoS ONE 9, e87948 ( 10.1371/journal.pone.0087948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Keyhani NO, Yu X, He Z, Luo Z, Pei Y, Zhang Y. 2012. The MAP kinase Bbslt2 controls growth, conidiation, cell wall integrity, and virulence in the insect pathogenic fungus Beauveria bassiana. Fungal Genet. Biol. 49, 544–555. ( 10.1016/j.fgb.2012.05.002) [DOI] [PubMed] [Google Scholar]
- 22.Zhen Z, et al. 2018. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50. ( 10.1016/j.fgb.2018.04.011) [DOI] [PubMed] [Google Scholar]
- 23.Takai Y, Kaibuchi K, Kikuchi A, Kawata M. 2001. Small GTP-binding proteins. Trends Biochem. Sci. 14, 394–396. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, et al. 2018. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 102, 4601–4613. ( 10.1007/s00253-018-8929-1) [DOI] [PubMed] [Google Scholar]
- 25.Leeder AC, Palma-Guerrero J, Glass NL. 2011. The social network: deciphering fungal language. Nat. Rev. Microbiol. 9, 440–451. ( 10.1038/nrmicro2580) [DOI] [PubMed] [Google Scholar]
- 26.Zhang HX, et al. 2012. Morphology regulatory metabolites from Arthrobotrys oligospora. J. Nat. Prod. 75, 1419–1423. ( 10.1021/np300342w) [DOI] [PubMed] [Google Scholar]
- 27.Xu Z, Wang B, Sun H, Yan N, Zeng Z, Zhang K, Niu X. 2015. High trap formation and low metabolite production by disruption of the PKS gene involved in the biosynthesis of arthrosporols from nematode-trapping fungus Arthrobotrys oligospora. J. Agric. Food Chem. 63, 9076–9082. ( 10.1021/acs.jafc.5b04244) [DOI] [PubMed] [Google Scholar]
- 28.Wang BL, Chen YH, He JN, Xue HX, Yan N, Zeng ZJ, Bennett JW, Zhang KQ, Niu XM. 2018. Integrated metabolomics and morphogenesis reveals volatile signaling of the nematode-trapping fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. 84 e02749-17 ( 10.1128/AEM.02749-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barron GL. 1977. The nematode-destroying fungi. Guelph, Ontario, Canada: Canadian Biological Publications. [Google Scholar]
- 30.Yang J, Tian B, Liang L, Zhang K. 2007. Extracellular enzymes and the pathogenesis of nematophagous fungi. Appl. Microbiol. Biotechnol. 75, 21–31. ( 10.1007/s00253-007-0881-4) [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Liang L, Li J, Zhang K-Q. 2013. Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 97, 7081–7095. ( 10.1007/s00253-013-5045-0) [DOI] [PubMed] [Google Scholar]
- 32.Liang L, Liu S, Yang J, Meng Z, Lei L, Zhang K. 2011. Comparison of homology models and crystal structures of cuticle-degrading proteases from nematophagous fungi: structural basis of nematicidal activity. FASEB J. 25, 1894–1902. ( 10.1096/fj.10-175653) [DOI] [PubMed] [Google Scholar]
- 33.Liang L, et al. 2010. The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes. FASEB J. 24, 1391–1400. ( 10.1096/fj.09-136408) [DOI] [PubMed] [Google Scholar]
- 34.Jansson HB. 1994. Adhesion of conidia of Drechmeria coniospora to Caenorhabditis elegans wildtype and mutants. J. Nematol. 26, 430–435. [PMC free article] [PubMed] [Google Scholar]
- 35.Dijksterhuis J, Veenhuis M, Harder W. 1990. Ultrastructural study of adhesion and initial stages of infection of nematodes by conidia of Drechmeria coniospora. Mycol. Res. 94, 1–8. ( 10.1016/S0953-7562(09)81257-4) [DOI] [Google Scholar]
- 36.Veenhuis M, Nordbring-Hertz B, Harder W. 1985. An electron-microscopical analysis of capture and initial stages of penetration of nematodes by Arthrobotrys oligospora. Antonie Van Leeuwenhoek 51, 385–398. ( 10.1007/BF02275043) [DOI] [PubMed] [Google Scholar]
- 37.Jansson HB. 1993. Adhesion to nematodes of conidia from the nematophagous fungus Drechmeria coniospora. J. Gen. Microbiol. 139, 1899–1906. ( 10.1099/00221287-139-8-1899) [DOI] [Google Scholar]
- 38.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, Ewbank JJ. 2008. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 4, e1000105 ( 10.1371/journal.ppat.1000105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodgkin J, Kuwabara PE, Corneliussen B. 2000. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10, 1615–1618. ( 10.1016/S0960-9822(00)00867-8) [DOI] [PubMed] [Google Scholar]
- 40.Darby C, Hsu JW, Ghori N, Falkow S. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244. ( 10.1038/417243a) [DOI] [PubMed] [Google Scholar]
- 41.Couillault C, Ewbank JJ. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70, 4705–4707. ( 10.1128/IAI.70.8.4705-4707.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5, 488–494. ( 10.1038/ni1060) [DOI] [PubMed] [Google Scholar]
- 43.Shai Y. 2010. Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–248. ( 10.1002/bip.10260) [DOI] [PubMed] [Google Scholar]
- 44.Ehret-Sabatier L, Loew D, Goyffon M, Fehlbaum P, Hoffmann JA, Van DA, Bulet P.. 1996. Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. J. Biol. Chem. 271, 29 537–29 544. ( 10.1074/jbc.271.47.29537) [DOI] [PubMed] [Google Scholar]
- 45.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. 2008. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 18, 481–489. ( 10.1016/j.cub.2008.02.079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couillault C, Fourquet P, Pophillat M, Ewbank JJ. 2012. A UPR-independent infection-specific role for a BiP/GRP78 protein in the control of antimicrobial peptide expression in C. elegans epidermis. Virulence 3, 299–308. ( 10.4161/viru.20384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu Q, et al. 2010. A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl Acad. Sci. USA 107, 16 631–16 636. ( 10.1073/pnas.1007276107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niu Q, Tian Y, Zhang L, Xu X, Niu X, Xia Z, Lei L, Zhang KQ, Huang X. 2011. Overexpression of the key virulence proteases Bace16 and Bae16 in Bacillus nematocida B16 to improve its nematocidal activity. J. Mol. Microbiol. Biotechnol. 21, 130–137. ( 10.1159/000332805) [DOI] [PubMed] [Google Scholar]
- 49.Zhang C, Zhao N, Chen Y, Zhang D, Yan J, Zou W, Zhang K, Huang X. 2016. The signaling pathway of C. elegans mediates chemotaxis response to the attractant 2-heptanone in a ‘Trojan horse'- like pathogenesis. J. Biol. Chem. 291, 23 618–23 627. ( 10.1074/jbc.M116.741132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng XD, Tian YX, Niu QH, Xu XE, Shi H, Zhang HB, Liang LM, Zhang KQ, Huang XW. 2013. The ComP-ComA quorum system is essential for ‘Trojan horse’ like pathogenesis in Bacillus nematocida. PLoS ONE 8, e76920 ( 10.1371/journal.pone.0076920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnepf E, Crickmore N, Van RJ, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH.. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo S, et al. 2009. Crystal structure of Bacillus thuringiensis Cry8Ea1: an insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J. Struct. Biol. 168, 259–266. ( 10.1016/j.jsb.2009.07.004) [DOI] [PubMed] [Google Scholar]
- 53.Wei JZ, Hale K, Carta L, Platzer E, Wong C, Fang SC, Aroian RV. 2003. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl Acad. Sci. USA 100, 2760–2765. ( 10.1073/pnas.0538072100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosasgarcia NM. 2009. Biopesticide production from Bacillus thuringiensis: an environmentally friendly alternative. Recent Pat. Biotechnol. 3, 28–36. ( 10.2174/187220809787172632) [DOI] [PubMed] [Google Scholar]
- 55.Roh JY, Choi JY, Li MS, Jin BR, Je YH. 2007. Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J. Microbiol. Biotechnol. 17, 547–559. [PubMed] [Google Scholar]
- 56.Zhang F, Peng D, Ye X, Yu Z, Hu Z, Ruan L, Ming S. 2012. In vitro uptake of 140 kDa Bacillus thuringiensis mematicidal crystal proteins by the second stage juvenile of Meloidogyne hapla. PLoS ONE 7, e38534 ( 10.1371/journal.pone.0038534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng D, Wan D, Cheng C, Ye X, Sun M. 2018. Nematode-specific cadherin CDH-8 acts as a receptor for Cry5B toxin in Caenorhabditis elegans. Appl. Microbiol. Biol. 102, 3663–3673. ( 10.1007/s00253-018-8868-x) [DOI] [PubMed] [Google Scholar]
- 58.Huffman DL, Abrami L, Sasik R, Corbeil J, Goot FGVD, Aroian RV, Collier RJ. 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl Acad. Sci. USA 101, 10 995–11 000. ( 10.1073/pnas.0404073101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bischof LJ, Kao CY, Los FC. O., Gonzalez MR, Shen Z, Briggs SP, Goot FG. V. D, Aroian RV.. 2008. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog. 4, e1000176 ( 10.1371/journal.ppat.1000176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dackman C, Nordbring-Hertz B. 1992. Conidial traps: a new survival structure of the nematode. Mycol. Res. 96, 194–198. ( 10.1016/S0953-7562(09)80965-9) [DOI] [Google Scholar]
- 61.Wang X, et al. 2014. Bacteria can mobilize nematode-trapping fungi to kill nematodes. Nat. Commun. 5, 5776 ( 10.1038/ncomms6776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L, Yang M, Luo J, Qu Q, Chen Y, Liang L, Zhang K. 2016. Nematode-trapping fungi and fungus-associated bacteria interactions: the role of bacterial diketopiperazines and biofilms on Arthrobotrys oligospora surface in hyphal morphogenesis. Environ. Microbiol. 18, 3827–3839. ( 10.1111/1462-2920.13340) [DOI] [PubMed] [Google Scholar]
- 63.Balan J, Krizkova L, Nemec P, Kolozsvary A. 1976. A qualitative method for detection of nematode attracting substances and proof of production of three different attractants by the fungus Monacrosporium rutgeriensis. Nematologica 22, 306–311. ( 10.1163/187529276X00580) [DOI] [Google Scholar]
- 64.Field JJ, Webster J. 1977. Traps of predacious fungi attract nematodes. Trans. Brit. Mycol. Soc. 68, 467–469. ( 10.1016/S0007-1536(77)80210-6) [DOI] [Google Scholar]
- 65.Hsueh YP, Gronquist MR, Schwarz EM, Nath RD, Lee CH, Gharib S, Schroeder FC, Sternberg PW. 2017. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. Elife 6, e20023 ( 10.7554/eLife.20023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang CY, Wang Z, Mi LL, Li Z, Zhang DL, Liu L, Fang ZM. 2009. Attraction of pinewood nematode, Bursaphelenchus xylophilus, to the endoparasitic fungus Esteya vermicola. Afr. J. Microbiol. Res. 3, 637–640. [Google Scholar]
- 67.Liou JY, Shih JY, Tzean SS. 1999. Esteya, a new nematophagous genus from Taiwan, attacking the pinewood nematode (Bursaphelenchus xylophilus). Mycol. Res. 103, 242–248. ( 10.1017/S0953756298006984) [DOI] [Google Scholar]
- 68.Wang CY, Fang ZM, Sun BS, Gu LJ, Zhang KQ, Sung CK. 2008. High infectivity of an endoparasitic fungus strain, Esteya vermicola, against nematodes. J. Microbiol. 46, 380–389. ( 10.1007/s12275-007-0122-7) [DOI] [PubMed] [Google Scholar]
- 69.Wang CY, Fang ZM, Wang Z, Gu J, Sun BS, Zhang DL, Chang KS. 2009. High infection activities of two Esteya vermicola isolates against pinewood nematode. Afr. J. Microbiol. Res. 3, 581–584. [Google Scholar]
- 70.Zhao L, Ahmad F, Lu M, Zhang W, Wickham JD, Sun J. 2018. Ascarosides promote the prevalence of ophiostomatoid fungi and an invasive pathogenic nematode, Bursaphelenchus xylophilus. J. Chem. Ecol. 44, 701–710. ( 10.1007/s10886-018-0996-3) [DOI] [PubMed] [Google Scholar]
- 71.Bargmann CI. 1993. Genetic and cellular analysis of behavior in C. elegans. Annu. Rev. Neurosci. 16, 47–71. ( 10.1146/annurev.ne.16.030193.000403) [DOI] [PubMed] [Google Scholar]
- 72.Bargmann CI, Horvitz HR. 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742. ( 10.1016/0896-6273(91)90276-6) [DOI] [PubMed] [Google Scholar]
- 73.Mori I, Ohshima Y. 1997. Molecular neurogenetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. Bioessays 19, 1055–1064. ( 10.1002/bies.950191204) [DOI] [PubMed] [Google Scholar]
- 74.Zhang C, Yan J, Chen Y, Chen C, Zhang K, Huang X. 2013. The olfactory signal transduction for attractive odorants in Caenorhabditis elegans. Biotechnol. Adv. 32, 290–295. ( 10.1016/j.biotechadv.2013.10.010) [DOI] [PubMed] [Google Scholar]
- 75.Bargmann CI, Mori I. 1997. Chemotaxis and thermotaxis. In C. elegans II (eds DL Ridle, T Blumenthal, BJ Meyer, JR Priess), pp. 717–737. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 76.Mori I. 1999. Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. Annu. Rev. Genet. 33, 399–422. ( 10.1146/annurev.genet.33.1.399) [DOI] [PubMed] [Google Scholar]
- 77.Reed RR. 1990. G protein diversity and the regulation of signaling pathways. New Biol. 2, 957–960. [PubMed] [Google Scholar]
- 78.Hallem EA, Dillman AR, Hong AV, Zhang Y, Yano JM, Demarco SF, Sternberg PW. 2011. A sensory code for host seeking in parasitic nematodes. Curr. Biol. 21, 377–383. ( 10.1016/j.cub.2011.01.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carrillo MA, Guillermin ML, Rengarajan S, Okubo RP, Hallem EA. 2013. O2-sensing neurons control CO2 response in C. elegans. J. Neurosci. 33, 9675–9683. ( 10.1523/JNEUROSCI.4541-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hart AC, Chao MY. 2010. From odors to behaviors in Caenorhabditis elegans. In The neurobiology of olfaction (ed. A Menini), pp. 1–33. Boca Raton, FL: CRC Press/Taylor & Francis. [PubMed] [Google Scholar]
- 81.Rengarajan S, Hallem EA. 2016. Olfactory circuits and behaviors of nematodes. Curr. Opin. Neurobiol. 41, 136–148. ( 10.1016/j.conb.2016.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong L, Li X, Huang L, Gao Y, Zhong L, Zheng Y, Zuo Y. 2014. Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. JExB 65, 131–141. ( 10.1093/jxb/ert356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahman J, Ek B, Rask L, Tunlid A. 1996. Sequence analysis and regulation of a gene encoding a cuticle-degrading serine protease from the nematophagous fungus Arthrobotrys oligospora. Microbiology 142, 1605–1616. ( 10.1099/13500872-142-7-1605) [DOI] [PubMed] [Google Scholar]
- 84.Bonants P, Fitters P, Thijs H, den Belder E, Waalwijk C, Henfling J. 1995. A basic serine protease from Paecilomyces lilacinus with biological activity against Meloidogyne hapla eggs. Microbiology 141, 775–784. ( 10.1099/13500872-141-4-775) [DOI] [PubMed] [Google Scholar]
- 85.Khan A, Williams K, Molloy MP, Nevalainen H. 2003. Purification and characterization of a serine protease and chitinases from Paecilomyces lilacinus and detection of chitinase activity on 2D gels. Protein Expr. Purif. 32, 210–220. ( 10.1016/j.pep.2003.07.007) [DOI] [PubMed] [Google Scholar]
- 86.Liang L, Yang J, Li J, Mo Y, Li L, Zhao X, Zhang KQ. 2011. Cloning and homology modeling of a serine protease gene (PrC) from the nematophagous fungus Clonostachys rosea. Ann. Microbiol. 61, 511–516. ( 10.1007/s13213-010-0166-5) [DOI] [Google Scholar]
- 87.Morton CO, Hirsch PR, Peberdy JP, Kerry BR. 2003. Cloning of and genetic variation in protease VCP1 from the nematophagous fungus Pochonia chlamydosporia. Mycol. Res. 107, 38–46. ( 10.1017/S0953756202007050) [DOI] [PubMed] [Google Scholar]
- 88.Alegado RA, Tan MW. 2010. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol. 10, 1259–1273. ( 10.1111/j.1462-5822.2008.01124.x) [DOI] [PubMed] [Google Scholar]
- 89.Ewbank JJ. 2006. Signaling in the immune response. In Wormbook the online review of C elegans biology. See http://www.wormbook.org/citewb.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.