Abstract
Baculoviridae is a family of large DNA viruses that infect insects. They have been extensively used as safe and efficient biological agents for the control of insect pests. As a result of coevolution with their hosts, baculoviruses developed unique life cycles characterized by the production of two distinctive virion phenotypes, occlusion-derived virus and budded virus, which are responsible for mediating primary infection in insect midgut epithelia and spreading systemic infection within infected insects, respectively. In this article, advances associated with virus–host interactions during the baculovirus life cycle are reviewed. We mainly focus on how baculoviruses exploit versatile strategies to overcome diverse host barriers and establish successful infections. For example, in the midgut, baculoviruses encode enzymes to degrade peritrophic membranes and use a series of per os infectivity factors to initiate primary infection. A viral fibroblast growth factor is expressed to attract tracheoblasts that spread the virus for systemic infection. Baculoviruses use different strategies to suppress host defence systems, including apoptosis, melanization and RNA interference. Additionally, baculoviruses can manipulate host physiology and induce ‘tree-top disease’ for optimal virus replication and dispersal. These advances in our understanding of baculoviruses will greatly inform the development of more effective baculoviral pesticides.
This article is part of the theme issue ‘Biotic signalling sheds light on smart pest management’.
Keywords: baculovirus, insect, primary infection, systemic infection, immune system, behaviour
1. Introduction
Baculoviridae is a diverse family of large DNA viruses that infect insects, and over 600 host species have been described. Based on phylogenetic analyses, Baculoviridae is divided into four genera, including Alphabaculovirus and Betabaculovirus isolated from Lepidoptera, Gammabaculovirus from Hymenoptera and Deltabaculovirus from Diptera [1]. In nature, baculoviruses play a vital role in controlling insect populations and they have been recommended by the Food and Agriculture Organization/World Health Organization for pest control since the 1970s. To date, over 50 baculoviral products have been applied as bioinsecticides worldwide. Baculoviruses are environmentally friendly and highly specific to their hosts. However, problems such as narrow host range, slow killing rates and low efficacy towards late instar larvae have limited the application of baculoviral pesticides. Therefore, in-depth investigations of virus–host interactions are crucial for the genetic improvement of baculoviral pesticides.
During millions of years of coevolution with insect hosts, baculoviruses developed unique bi-phasic life cycles characterized by the production of two morphologically distinct virion phenotypes: the budded virus (BV) and the occlusion-derived virus (ODV). ODVs are embedded within a protective proteinaceous matrix to form occlusion bodies (OBs). In nature, ingested OBs are dissolved under the alkaline conditions in insect midguts to release ODVs, which initiate primary infection (or per os infection) of midgut columnar epithelial cells (from the apical side). BVs are then produced and released from the basolateral side of the infected midgut epithelial cells to infect other tissues, resulting in systemic infection within the larval body [2]. BVs and ODVs share identical genomes but differ with regard to envelope protein composition, which determines the tissue-tropisms of these two virion phenotypes [3].
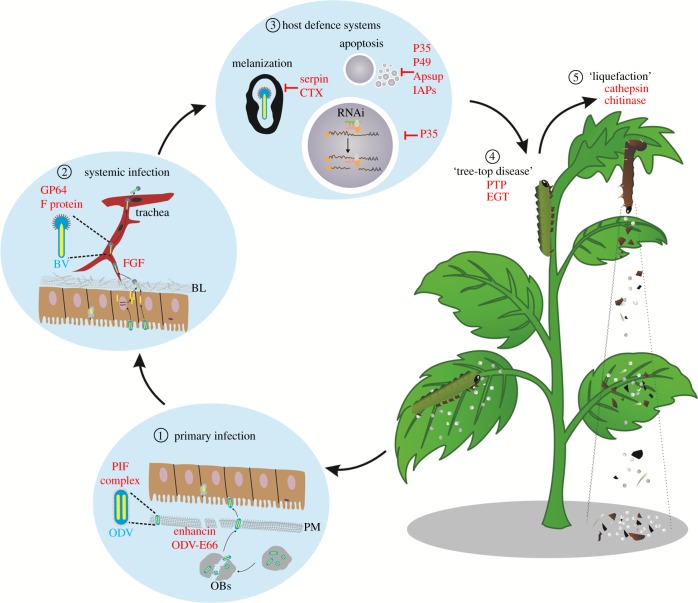
To establish a successful infection, baculoviruses must overcome several host defence barriers within insects. For example, the midgut is protected by a layer of dense extracellular matrix called the peritrophic membrane (PM) and the haemocoel is separated from the midgut by basal laminae (BL). In addition, the powerful innate immune system of insects poses another barrier to baculovirus infection. Correspondingly, baculoviruses have evolved versatile strategies to overcome these barriers as well as finely regulated host physiology and behaviours to favour optimal virus replication and transmission (figure 1). In this review, we summarize the advances of baculovirus–host interactions, particularly how baculoviruses overcome hosts defence systems to achieve successful infection.
Figure 1.
Model of how baculoviruses overcome different host barriers to establish successful infections. After being ingested by susceptible larvae, OBs are dissolved under the alkaline conditions of the insect midgut to release ODVs. Two enzymes (enhancin and ODV-E66) can degrade the PM to allow the access of ODV to the midgut epithelia. Primary infection is then initiated by a group of ODV-specific envelope proteins (PIFs) (①). To egress from the BL barrier at the basal side of the midgut epithelial cells, baculoviruses use vFGF for the chemotaxis of tracheoblasts to facilitate BV passage, and they use BV-specific envelope proteins (GP64 or F proteins) to spread systemic infection (②). Baculoviruses exploit different strategies to suppress host defence systems, including melanization, apoptosis and RNAi for efficient virus replication (③). Baculoviruses can also regulate host physiology and behaviour, such as inducing a ‘tree-top disease’ via PTP and EGT (④) and ‘liquefaction of infected larval bodies' by chitinase and cathepsin (⑤) for optimal virus dispersal. BL: basal laminae; BV: budded virus; ODV: occlusion-derived virus; OB: occlusion body; PM: petritrophic membrane. Viral proteins are in red.
2. Primary infection
(a). Cross the peritrophic membrane barrier
After being ingested by susceptible host insects, ODVs released from OB must first pass through the PM before they gain access to midgut cells (figure 1, ①). The PM is a thin extracellular mucinous matrix comprised of chitin, glycoproteins and proteoglycans, which regulates the passage of nutrients and serves as a defensive barrier against ingested pathogens, including viruses. The PM pore sizes of lepidopteran insects are much smaller than ODV sizes, thus preventing the direct passage of ODVs. Accordingly, baculoviruses have developed their own approaches to overcome the PM barrier, as evidenced by the degradation of PM proteins and increase in PM permeability in the infected larvae. To date, two kinds of OB/ODV-specific proteins are thought to play pivotal roles in PM penetration by baculoviruses.
Enhancin (also called the ‘synergistic factor’), first found in betabaculoviruses, is a virus-encoded metalloproteinase that can enhance infection potency by degrading the PM. Enhancin appears to proteolyse insect intestinal mucin, a major mucinous protein constituent of the PM, and increase the permeability of the PM to facilitate baculovirus infection [4]. Enhancin is identified in approximately 30% of lepidopteran baculoviral genomes, and several baculoviruses contain multiple copies of enhancin genes. In contrast to betabaculoviruses, of which enhancins are enriched in the OB matrix, the counterpart in an alphabaculovirus (Lymantria dispar nucleopolyhedrovirus, LdMNPV) was associated with the ODV envelope. The OB matrix or ODV envelope residence might allow enhancin to immediately interact with PM after ODV release. Since enhancins can increase the infectivity of heterologous baculoviruses, they are potentially adjuvants for baculoviral insecticides.
ODV-E66 is one of the major ODV-specific envelope proteins and is conserved in all lepidopteran baculoviruses. The deletion of odv-e66 resulted in a significant decrease in Autographa californica multiple nucleopolyhedrovirus (AcMNPV) oral infectivity [5]. ODV-E66 proteins of AcMNPV and Bombyx mori nucleopolyhedrovirus (BmNPV) were identified as substrate-specific endo-chondroitin lyses [6,7], and the crystal structure of AcMNPV ODV-E66 resembles that of a polysaccharide lyase [8]. In vitro biochemistry analyses showed that BmNPV ODV-E66 could digest chondroitin sulfates from silkworm PM, suggesting that ODV-E66 functions by destroying the PM of the host's midgut to facilitate ODV infection [7].
(b). Infection of midgut epithelia
After crossing the PM barrier, baculovirus ODVs are able to establish primary infection within the midgut epithelia of host insects. This process is largely dependent on a group of ODV-specific envelope proteins called per os infectivity factors (PIFs). To date, nine PIFs (P74 (also called PIF0) and PIF1–8) have been identified as conserved in all baculoviruses sequenced thus far [9,10]. These PIFs vary considerably in their molecular weights (ranging from approximately 7 kDa of PIF7 to approximately 95 kDa of PIF8) and protein sequences [9]. However, deletion of any one of the individual PIFs leads to substantial impairment or complete loss of oral infectivity.
ODV enters the midgut epithelial cells via direct membrane fusion between the ODV envelope and the host cell membrane. To date, the detailed role of each PIF during per os infection remains largely enigmatic, and no specific insect receptors have been identified. Among PIFs, P74, PIF1 and PIF2 may be involved in the specific binding of ODV to midgut cells. P74 undergo proteolysis during ODV entry, and cleavage is probably critical for its optimal function [11,12]. It is known that many viral envelope fusion proteins (EFPs) require proteolytic cleavage for the activation of their fusogenic activities, and P74 seems to comply with this feature. However, whether it functions as an actual ODV fusion protein requires further investigation.
Similar to those in other large DNA viruses, such as poxviruses and herpesviruses, which also encode multiprotein entry-fusion complexes, PIFs interact with each other and might act in concert during the entry of ODVs into midgut epithelial cells. A PIF complex of approximately 480 kDa was identified, and it contains at least eight PIFs, including PIF0–4, PIF6–8, while PIF5 was excluded from the complex [10,13–15]. The formation of an intact PIF complex may provide individual PIFs with resistance to proteolytic degradation under the alkaline conditions of the insect midgut [16].
Interestingly, homologues of certain PIFs are also present in other inveterate large DNA viruses, including white spot syndrome virus, salivary gland hypertrophy virus, Apis mellifera filamentous virus, bracovirus and nudivirus [9]. The widespread occurrence of PIFs suggests that PIF-mediated per os infection is an ancient and evolutionarily conserved entry mechanism shared by these invertebrate DNA viruses.
3. Systemic infection
(a). Central role of tracheae in spreading systemic infection
After infection of midgut epithelial cells, BV particles bud from the basolateral side to spread the infection systemically within infected larval bodies. Insect haemocytes were previously mistaken as factors that mediate the systemic dissemination of baculovirus [17]. However, the results of subsequent studies suggested that the host tracheal system serves as a conduit for the spread of the virus to other tissues [18] (figure 1, ②).
A layer of tight, semi-permeable fibrous extracellular matrix called the BL occurs on the basal side of the midgut epithelium. The BL comprises various glycoproteins secreted by midgut epithelial cells. Similar to the PM, the BL provides a protective barrier that prevents the escape of pathogens from the midgut into haemocoelic tissues. To establish a successful systemic infection, baculoviruses must bypass the BL barrier. Considering the small size of the pore in the BL (less than 15 nm in diameter), direct penetration by the virus is unlikely to be an efficient route. Insect tracheal cells have long extensions that penetrate the midgut-associated BL, and this could provide a channel for virus particles to circumvent the BL barrier. Using a lacZ-expressing recombinant AcMNPV, Engelhard et al. [18] showed that secondary infections always originated from tracheoblasts and proceeded along tracheal branches, followed by infections in haemocytes and other tissues [18]. Similarly, in BmNPV-infected Bombyx mori larvae, infections were only observed in tissues that were in contact with tracheae, but not in those without associated tracheae [19]. These results suggest a major role of the trachea in spreading systemic infection.
Viral fibroblast growth factor (vFGF) is a baculovirus-encoded protein that facilitates virus dissemination from infected midgut epithelial cells to other tissues. FGFs are a large family of growth factors that play diverse roles in regulating cell proliferation, migration and differentiation. FGFs are widespread in vertebrates and invertebrates, but their viral homologs (vFGFs) have only been identified in baculoviruses. Deletion of vfgfs from baculoviruses significantly delayed the speed of host insect killing [20,21]. vFGFs probably function via a two-step mechanism: (i) attract tracheoblast migration towards infected midgut cells; and (ii) stimulate a signalling cascade involving the activation of matrix metalloproteases and effector caspases, resulting in the degradation of the BL barrier and acceleration of the establishment of systemic infection [22]. Host factor Breathless, a receptor for FGF in Drosophila, was found to be required for vFGF-induced cell migration activity in vitro and suggested to be a receptor for vFGF [23]. It has been proposed that vfgfs might be captured by baculoviruses from insect hosts. They are found in most alphabaculoviruses and betabaculoviruses, but not in gammabaculoviruses and deltabaculoviruses which are more ancient, and midgut-restricted baculoviruses. The acquisition of vfgfs by alphabaculoviruses and betabaculoviruses might be an evolutionary advantage that contributed to virus dissemination and pathogenesis as well as host range expansion.
(b). Budded virus-specific envelope fusion proteins
The entry of BV into host cells is mediated by specific EFPs, namely GP64 or F proteins. These two types of EFPs play similar essential roles during virus infection processes such as virus-cell receptor binding, low pH-dependent membrane fusion and efficient budding. However, they are quite different in their distributions, protein sequences/structures and modes of action.
GP64 homologues are closely related EFPs (greater than 74% amino acid identity), and they only occur in Group I alphabaculoviruses. Crystal structure analysis revealed that GP64 belongs to class III viral fusion proteins [24]. By contrast, F protein homologues are much less conserved (20–40% amino acid identity), but they are more widely spread throughout the Baculoviridae (Group II alphabaculoviruses, betabaculoviruses and deltabaculoviruses). F proteins share common features of class I viral fusion proteins, and they require the proteolytic cleavage by a furin-like protease to activate fusogenicity [25]. Interestingly, apart from GP64, Group I alphabaculoviruses also encode non-fusogenic F proteins, designated as F-like proteins. Although their precise functions remain still unclear, F-like proteins appear to have an auxiliary function for virus infection, as evidenced by enhancing viral infectivity both in vivo and in vitro [26,27].
Evolutionary hypotheses suggested that F proteins might be ancestral baculovirus fusion proteins, while gp64 seemed to be a recent acquisition by Group I alphabaculoviruses. The acquisition of gp64 led to the inactivation of fusogenic F proteins to F-like protein. This hypothesis was supported by the experimental reconstruction of a possible major step in the evolution of baculoviruses [28] and by the recent discovery of a betabaculovirus containing both functional F protein and GP64 [29]. It was speculated that gp64 entered ancestral baculoviruses via recombination with arboviruses, specifically thogotoviruses, which also employ GP64 homologs (GP75) as their EFPs [30]. The functional analogy between GP64 and F proteins is not reciprocal, because F proteins could readily replace the function of GP64 [31,32]. However, GP64 is unable to efficiently substitute Group II alphabaculovirus F proteins by itself [33].
Numerous efforts have been made towards the identification of baculovirus cellular receptors. Current data suggest that negatively charged host cell surface molecules, such as phospholipids, are involved in baculovirus entry [34]. A recent report found that a host membrane protein, BmREEPa, could interact with GP64 and facilitate the entry of BmNPV into silkworm cells [35]. Regarding F proteins, it seems that they exploit a distinct cellular receptor from GP64 to gain entry into cells [36]. However, the specific cellular receptors for both GP64 and F proteins remain to be discovered.
4. Modulation of host defence systems
Insect hosts possess powerful innate immune systems that combat viral infections. These include RNA interference (RNAi), apoptosis, autophagy, melanization and a series of conserved immune signalling pathways (Jak/STAT, NF-κB mediated Toll and Imd pathways), etc. [37]. To date, a humoral immune response, melanization and two cellular mechanisms (RNAi and apoptosis) have been implicated as host defences against baculoviruses. Correspondingly, baculoviruses developed multiple strategies to antagonize these defence systems (figure 1, ③). A recent review also summarizes research progress on the interactions between baculoviruses and host immune systems [38].
(a). Melanization
Melanization is a unique innate defence mechanism in invertebrates, which encapsulates and kills invading pathogens and parasites. Melanization pathways consist of a cascade of serine proteases that convert prophenoloxidase (PPO) into active phenoloxidase (PO), further catalysing the formation of melanin. PPO is mainly present in the haemolymph, and it probably makes up the first line of the insect defence system. Melanization has been demonstrated to play an important role in the protection of insects against baculovirus infection. In vitro studies showed that melanized insect haemolymph could completely inactivate baculovirus infectivity [39]. In AcMNPV-infected resistant larvae, significant melanization and encapsulation were found in the tracheal epidermis, which limited the spread of infection to haemocytes and other tissues [40]. However, this phenomenon was not found in virus-infected permissive larvae, suggesting that baculoviruses developed effective strategies to antagonize melanization responses during the coevolution of viruses and their hosts. A recent report suggested that Helicoverpa armigera nucleopolyhedrovirus (HearNPV) inhibited host melanization pathways via two mechanisms: (i) global downregulation of expression of genes involved in the melanization pathway; and (ii) simultaneous upregulation of specific negative regulators (serine protease inhibitors, serpins) associated with this pathway [39]. Interestingly, a serpin orthologue has been found in the genome of a baculovirus, which exhibited inhibitory effects to a subset of host serine proteases, including those possibly involved in the melanization pathway [41]. A viral conotoxin-like gene encodes a small cysteine-rich polypeptide abrogates insect haemolymph melanization with unknown mechanisms [42].
(b). Apoptosis
Apoptosis is programmed cell death, which is important for the development and immunity of multicellular organisms. Apoptosis is executed by a cascade activation of initiator and effector caspases, and the activation is regulated by cellular inhibitors of apoptosis proteins (IAPs). During baculovirus infection, viral DNA replication triggers host DNA damage responses, which result in the depletion of cellular IAPs and the initiation of apoptosis [43,44]. Moreover, baculoviruses encode several suppressors to inhibit apoptosis. The first identified baculovirus suppressor of apoptosis was P35 [45], which inhibits apoptosis via the inactivation of a wide range of effector caspases. The P35 protein family also contains a highly divergent homologue, P49. Unlike P35, P49 inhibits both initiator and effector caspases. Another baculovirus apoptosis suppressor, Apsup, inhibits initiator caspase [46]. Apart from P35 family proteins, baculovirus also encoded another type of apoptosis suppressor, IAPs. In fact, IAPs were first identified in baculoviruses before their counterparts were identified in cells [47]. Unlike P35, P49 and Apsup which are present in only a few baculoviruses, IAPs exist in most baculoviruses. Baculovirus-encoded IAPs are phylogenetically classified into five lineages (iap1–5). Among these, IAP3 is the most widely spread and its anti-apoptotic activity has been confirmed in many baculoviruses. The model of action of IAP3 appears to be interacting with and stabilizing cellular IAPs, thereby suppressing apoptosis [48]. Some IAPs do not appear to block apoptosis and they even trigger apoptosis in certain cases [49].
(c). RNA interference
The RNAi-based antiviral response plays an important role in the infection of insects by many RNA viruses and some DNA viruses. Deep sequencing of baculovirus-infected larvae resulted in the discovery of a large quantity of viral short-interfering RNAs (v-siRNAs). Moreover, the knock-down of Dicer-2, which is responsible for siRNA processing, significantly enhanced viral DNA replication, suggesting that RNAi acts as a host defence mechanism against baculovirus infection [50]. Subsequent experiments indicated that AcMNPV infection could inhibit RNAi response of host cells, and viral protein P35 is a broadly active viral suppressor of RNAi. The mechanism that allows P35 to inhibit the RNAi response is not clear at the moment, but it is downstream in the RNAi pathway and independent of the P35 anti-apoptotic activity [51]. In addition, baculoviruses also encode microRNA, another type of RNAi response, to regulate a wide variety of viral or cellular genes for optimal replication [52].
5. Manipulation of host physiology and behaviour
(a). Prevention of host insect moulting
Insects stop feeding and sometimes even fail to survive during the moulting stage, and this is unfavourable for the optimal replication of baculoviruses. Virus-encoded ecdysteroid (UDP)-glucosyltransferase (EGT) is able to catalyse the sugar conjugation of ecdysteroids, resulting in the inactivation of moulting hormones [53]. Consequently, normal insect moulting is blocked in virus-infected larvae, leading to prolonged insect feeding times and increased yield of viral progeny. When egt is deleted, the efficacy of baculoviral pesticides could be significantly improved, as evidenced by considerably reduced feeding and earlier mortality compared to wild-type viruses [54]. Considering the widespread presence of egt homologues in diverse insects, baculoviruses probably captured this gene from their host insects to achieve maximum proliferation.
(b). Enhanced locomotory activity and virus dissemination
Baculoviruses have long been known to induce enhanced locomotory activity (ELA) of infected host insects. Infected larvae exhibit ‘hyperactivity’ and ‘tree-top disease’ (climb to the top or edge of leaves before dying), and these behaviour changes are thought to increase the dispersal of OBs after the death of the infected larvae. Two viral proteins have been implicated in inducing ELA of hosts (figure 1, ④).
A baculovirus-encoded protein tyrosine phosphatase (PTP), is responsible for the enhanced ‘wandering-like’ behaviour. Silkworms infected with BmNPV showed increased ELA at the late stage of virus infection, with the highest intensity occurring approximately 12–24 h before death. Via the systematic screening of a gene-knockout library of BmNPV, a ptp-deletion mutant was found to have almost completely eliminated host ELA, and the effect of ELA could be enhanced by light [55]. The ptp-induced host ELA is probably an evolutionarily conserved mechanism associated with a subset of baculoviruses [56]. However, its mode of action may differ among various baculoviruses. In BmNPV, the phosphate activity of PTP is dispensable for ELA induction [57], while in AcMNPV, which contains a PTP with 97% amino acid identity with the BmNPV homologue, the catalytic activity of PTP is required for the induction of hyperactive behaviour [56]. Homologues of ptp are only present in Group I alphabaculoviruses, and ptp is probably another example of a captured gene from insect hosts. A host-derived PTP homologue could partially rescue the ELA of a ptp-deleted BmNPV [55]. Furthermore, PTP may be involved in a variety of signalling pathways that regulate diverse physiological processes, but how this enzyme induces ELA by influencing these pathways remains unclear.
Tree-top disease, also coined as ‘Wipfelkrankheit,’ was first reported in Germany in the late nineteenth century. Until 2011, the egt gene of a Group II alphabaculovirus (LdMNPV; lacks a ptp gene) was discovered to be involved in this behaviour. The egt-deletion LdMNPV-infected Lymantria dispar larvae died at low positions, while the wild-type virus-infected larvae died at elevated positions [58]. Similar effects of egt on tree-top disease were also observed in Spodoptera exigua nucleopolyhedrovirus (SeMNPV)-infected Spodoptera exigua larvae [59]. However, in AcMNPV-infected Trichoplusia ni and S. exigua, the virus-induced tree-top disease was not affected by the egt gene [60]. Instead, the climbing behaviour appeared to be moulting-related. Using RNAi and exogenous hormone treatment, a recent report showed that 20-hydroxyecdysone (20E), a substrate of EGT, inhibited virus-induced tree-top disease in HearNPV-infected larvae [61]. One possible explanation is that the higher 20E hormone titre may trigger apolysis, leading to an immobilized state of larvae and death at lower positions. These results suggested that EGT-induced tree-top disease might, to some extent, be an indirect effect of 20E-mediated moulting.
ELA is thought to promote the transmission of baculoviruses because insects climbing to the top of the tree enhance visibility to predators. Furthermore, when they die, foliage or soils are contaminated with OBs. After death, larval tissues undergo liquefaction at the final stage of infection, a process mainly mediated by two additional virus-encoded enzymes, chitinase and cathepsin [62,63] (figure 1, ⑤). A typical baculovirus-infected larval corpse may contain more than 100 million progeny OBs after liquefaction, and they are released to the environment, waiting to be indigested by susceptible hosts to initiate another round of the infection cycle.
6. Conclusion and future perspectives
Baculoviruses have co-evolved with their insect hosts for over 300 million years. They encode an excessively large number of genes and have adapted dedicated mechanisms to precisely regulate the physiology, immunity and behaviours of insects to favour their own replication and dispersal. As summarized in figure 1, to gain access to the midgut, baculoviruses encode two enzymes (enhancin and ODV-E66) to degrade the PM, and then a group of ODV-specific envelope proteins (PIFs) is used to initiate primary infection (figure 1, ①). To overcome the BL barrier, baculoviruses express vFGF to attract tracheoblasts as the conduit for BV passage, and they use BV-specific envelope proteins (GP64 or F proteins) to mediate systemic infection (figure 1, ②). Baculoviruses exploit diverse strategies to suppress host defence systems, including melanization, apoptosis and RNAi for efficient virus replication (figure 1, ③). In addition, baculovirus can also manipulate host physiology and behaviour, such as inducing a ‘tree-top disease’ (PTP and EGT) (figure 1, ④) and the ‘liquefaction of infected larval bodies' (chitinase and cathepsin) (figure 1, ⑤) for optimal virus transmission. Interestingly, some of these mechanisms seem to be ‘learned’ from their hosts or other symbiotic organisms.
Over the past few decades, remarkable achievements have been made regarding the different aspects of baculovirus–host interactions, but these insights have raised a series of really interesting questions. For example, what are the structures and functions of the PIF complex? What are the cellular receptors for ODV and BV entry? Are there more virus factors involved in the modulation of antiviral immunity and the manipulation of host behaviours? With the development of more advanced technologies, such as modern genomic/metabolic/proteomic approaches, Cryo-EM, Crispr-Cas9, etc., these issues are expected to be addressed in the near future. The answers will undoubtedly provide new insights into baculovirus infection mechanisms, and they will also shed light on the improvement of baculoviral pesticides.
Data accessibility
This article has no additional data.
Authors' contributions
M.W. and Z.H. wrote the manuscript. M.W. drew the figure.
Competing interests
We have no competing interests.
Funding
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB11030400 to Z.H.), the Key Research Program of Frontier Sciences of Chinese Academy of Sciences (grant no. QYZDJ-SSW-SMC021 to Z.H. and M.W.), the National Natural Science Foundation of China (grant no. 31621061 to Z.H.) and the Virology Key Frontier Science Program of State Key Laboratory of Virology (grant no. klv-2016-03 to Z.H.).
References
- 1.Harrison RL, et al. 2018. ICTV virus taxonomy profile: Baculoviridae. J. Gen. Virol. 99, 1185–1186. ( 10.1099/jgv.0.001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohrmann GF. 2013. Baculovirus molecular biology (3rd edn). Bethesda, MD: National Center for Biotechnology Information (US). [PubMed] [Google Scholar]
- 3.Hou D, et al. 2013. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J. Virol. 87, 829–839. ( 10.1128/JVI.02329-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P, Granados RR. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl Acad. Sci. USA 94, 6977–6982. ( 10.1073/pnas.94.13.6977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang X, Chen L, Hu X, Yu S, Yang R, Wu X. 2011. Autographa californica multiple nucleopolyhedrovirus ODV-E66 is an essential gene required for oral infectivity. Virus Res. 158, 72–78. ( 10.1016/j.virusres.2011.03.012) [DOI] [PubMed] [Google Scholar]
- 6.Sugiura N, Setoyama Y, Chiba M, Kimata K, Watanabe H. 2011. Baculovirus envelope protein ODV-E66 is a novel chondroitinase with distinct substrate specificity. J. Biol. Chem. 286, 29 026–29 034. ( 10.1074/jbc.M111.251157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugiura N, Ikeda M, Shioiri T, Yoshimura M, Kobayashi M, Watanabe H. 2013. Chondroitinase from baculovirus Bombyx mori nucleopolyhedrovirus and chondroitin sulfate from silkworm Bombyx mori. Glycobiology 23, 1520–1530. ( 10.1093/glycob/cwt082) [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi Y, Sugiura N, Kimata K, Kimura M, Kakuta Y. 2013. The crystal structure of novel chondroitin lyase ODV-E66, a baculovirus envelope protein. FEBS Lett. 587, 3943–3948. ( 10.1016/j.febslet.2013.10.021) [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Liu X, Makalliwa GA, Li J, Wang H, Hu Z, Wang M. 2017. Per os infectivity factors: a complicated and evolutionarily conserved entry machinery of baculovirus. Sci. China Life Sci. 60, 806–815. ( 10.1007/s11427-017-9127-1) [DOI] [PubMed] [Google Scholar]
- 10.Boogaard B, van Oers MM, van Lent JW. M.. 2018. An advanced view on baculovirus per os infectivity factors. Insects 9, 84 ( 10.3390/insects9030084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slack JM, Lawrence SD, Krell PJ, Arif BM. 2008. Trypsin cleavage of the baculovirus occlusion-derived virus attachment protein P74 is prerequisite in per os infection. J. Gen. Virol. 89, 2388–2397. ( 10.1099/vir.0.2008/002543-0) [DOI] [PubMed] [Google Scholar]
- 12.Peng K, van Lent JW, Vlak JM, Hu Z, van Oers MM.. 2011. In situ cleavage of baculovirus occlusion-derived virus receptor binding protein P74 in the peroral infectivity complex. J. Virol. 85, 10 710–10 718. ( 10.1128/JVI.05110-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng K, van Lent JW, Boeren S, Fang M, Theilmann DA, Erlandson MA, Vlak JM, van Oers MM.. 2012. Characterization of novel components of the baculovirus per os infectivity factor complex. J. Virol. 86, 4981–4988. ( 10.1128/JVI.06801-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng K, van Oers MM, Hu Z, van Lent JW, Vlak JM.. 2010. Baculovirus per os infectivity factors form a complex on the surface of occlusion-derived virus. J. Virol. 84, 9497–9504. ( 10.1128/JVI.00812-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javed MA, et al. 2017. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J. Virol. 91, e02115–16 ( 10.1128/JVI.02115-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boogaard B, van Lent JWM, Theilmann DA, Erlandson MA, van Oers MM.. 2017. Baculoviruses require an intact ODV entry-complex to resist proteolytic degradation of per os infectivity factors by co-occluded proteases from the larval host. J. Gen. Virol. 98, 3101–3110. ( 10.1099/jgv.0.000974) [DOI] [PubMed] [Google Scholar]
- 17.Keddie BA, Aponte GW, Volkman LE. 1989. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243, 1728–1730. ( 10.1126/science.2648574) [DOI] [PubMed] [Google Scholar]
- 18.Engelhard EK, Kam-Morgan LN, Washburn JO, Volkman LE. 1994. The insect tracheal system: a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc. Natl Acad. Sci. USA 91, 3224–3227. ( 10.1073/pnas.91.8.3224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman MM, Gopinathan KP. 2004. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 101, 109–118. ( 10.1016/j.virusres.2003.12.027) [DOI] [PubMed] [Google Scholar]
- 20.Katsuma S, Horie S, Daimon T, Iwanaga M, Shimada T. 2006. In vivo and in vitro analyses of a Bombyx mori nucleopolyhedrovirus mutant lacking functional vfgf. Virology 355, 62–70. ( 10.1016/j.virol.2006.07.008) [DOI] [PubMed] [Google Scholar]
- 21.Detvisitsakun C, Cain EL, Passarelli AL. 2007. The Autographa californica M nucleopolyhedrovirus fibroblast growth factor accelerates host mortality. Virology 365, 70–78. ( 10.1016/j.virol.2007.03.027) [DOI] [PubMed] [Google Scholar]
- 22.Means JC, Passarelli AL. 2010. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl Acad. Sci. USA 107, 9825–9830. ( 10.1073/pnas.0913582107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katsuma S, Daimon T, Mita K, Shimada T. 2006. Lepidopteran ortholog of Drosophila breathless is a receptor for the baculovirus fibroblast growth factor. J. Virol. 80, 5474–5481. ( 10.1128/JVI.00248-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15, 1024–1030. ( 10.1038/nsmb.1484) [DOI] [PubMed] [Google Scholar]
- 25.Westenberg M, Wang H, Goldbach RW, Vlak JM, Zuidema D. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76, 178–184. ( 10.1128/JVI.76.1.178-184.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lung OY, Cruz-Alvarez M, Blissard GW. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77, 328–339. ( 10.1128/JVI.77.1.328-339.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Tan Y, Yin F, Deng F, Vlak JM, Hu Z, Wang H. 2008. The F-like protein Ac23 enhances the infectivity of the budded virus of gp64-null Autographa californica multinucleocapsid nucleopolyhedrovirus pseudotyped with baculovirus envelope fusion protein F. J. Virol. 82, 9800–9804. ( 10.1128/JVI.00759-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, et al. 2014. Unraveling the entry mechanism of baculoviruses and its evolutionary implications. J. Virol. 88, 2301–2311. ( 10.1128/JVI.03204-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardisson-Araujo DM, Melo FL, Clem RJ, Wolff JL, Ribeiro BM. 2016. A betabaculovirus-encoded gp64 homolog codes for a functional envelope fusion protein. J. Virol. 90, 1668–1672. ( 10.1128/JVI.02491-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson MN, Rohrmann GF. 2002. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 76, 5301–5304. ( 10.1128/JVI.76.11.5301-5304.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76, 5729–5736. ( 10.1128/JVI.76.11.5729-5736.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin F, Wang M, Tan Y, Deng F, Vlak JM, Hu Z, Wang H. 2013. Betabaculovirus F proteins showed different efficiencies when rescuing the infectivity of gp64-null Autographa californica nucleopolyhedrovirus. Virology 436, 59–66. ( 10.1016/j.virol.2012.10.017) [DOI] [PubMed] [Google Scholar]
- 33.Wang M, Yin F, Shen S, Tan Y, Deng F, Vlak JM, Hu Z, Wang H. 2010. Partial functional rescue of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus infectivity by replacement of F protein with GP64 from Autographa californica multicapsid nucleopolyhedrovirus. J. Virol. 84, 11 505–11 514. ( 10.1128/JVI.00862-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tani H, Nishijima M, Ushijima H, Miyamura T, Matsuura Y. 2001. Characterization of cell-surface determinants important for baculovirus infection. Virology 279, 343–353. ( 10.1006/viro.2000.0699) [DOI] [PubMed] [Google Scholar]
- 35.Dong XL, Liu TH, Wang W, Pan CX, Wu YF, Du GY, Chen P, Lu C, Pan MH.. 2015. BmREEPa is a novel gene that facilitates BmNPV entry into silkworm cells. PLoS ONE 10, e0144575 ( 10.1371/journal.pone.0144575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westenberg M, Uijtdewilligen P, Vlak JM. 2007. Baculovirus envelope fusion proteins F and GP64 exploit distinct receptors to gain entry into cultured insect cells. J. Gen. Virol. 88, 3302–3306. ( 10.1099/vir.0.83240-0) [DOI] [PubMed] [Google Scholar]
- 37.Sabin LR, Hanna SL, Cherry S. 2010. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 22, 4–9. ( 10.1016/j.coi.2010.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong M, Zuo H, Zhu F, Hu Z, Chen L, Yang Y, Lv P, Yao Q, Chen K. 2018. The interaction between baculoviruses and their insect hosts. Dev. Comp. Immunol. 83, 114–123. ( 10.1016/j.dci.2018.01.019) [DOI] [PubMed] [Google Scholar]
- 39.Yuan C, Xing L, Wang M, Wang X, Yin M, Wang Q, Hu Z, Zou Z. 2017. Inhibition of melanization by serpin-5 and serpin-9 promotes baculovirus infection in cotton bollworm Helicoverpa armigera. PLoS Pathog. 13, e1006645 ( 10.1371/journal.ppat.1006645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washburn JO, Kirkpatrick BA, Volkman LE. 1996. Insect protection against viruses. Nature 383, 767 ( 10.1038/383767a0) [DOI] [Google Scholar]
- 41.Ardisson-Araujo DM, Rohrmann GF, Ribeiro BM, Clem RJ. 2015. Functional characterization of hesp018, a baculovirus-encoded serpin gene. J. Gen. Virol. 96, 1150–1160. ( 10.1099/vir.0.000041) [DOI] [PubMed] [Google Scholar]
- 42.Cao Q, Zhu SY, Wu Y, Liu Y, Zhu J, Wang W. 2012. The effect of a small conotoxin-like ctx gene from Autographa californica nuclear polyhedrosis virus (AcMNPV) on insect hemolymph melanization. Pol. J. Microbiol. 61, 183–189. [PubMed] [Google Scholar]
- 43.Mitchell JK, Friesen PD. 2012. Baculoviruses modulate a proapoptotic DNA damage response to promote virus multiplication. J. Virol. 86, 13 542–13 553. ( 10.1128/JVI.02246-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandergaast R, Schultz KL, Cerio RJ, Friesen PD. 2011. Active depletion of host cell inhibitor-of-apoptosis proteins triggers apoptosis upon baculovirus DNA replication. J. Virol. 85, 8348–8358. ( 10.1128/JVI.00667-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clem RJ, Fechheimer M, Miller LK. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254, 1388–1390. ( 10.1126/science.1962198) [DOI] [PubMed] [Google Scholar]
- 46.Yamada H, Kitaguchi K, Hamajima R, Kobayashi M, Ikeda M. 2013. Novel apoptosis suppressor Apsup from the baculovirus Lymantria dispar multiple nucleopolyhedrovirus precludes apoptosis by preventing proteolytic processing of initiator caspase Dronc. J. Virol. 87, 12925–12934. ( 10.1128/JVI.02065-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crook NE, Clem RJ, Miller LK. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67, 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Byers NM, Vandergaast RL, Friesen PD. 2016. Baculovirus inhibitor-of-apoptosis Op-IAP3 blocks apoptosis by interaction with and stabilization of a host insect cellular IAP. J. Virol. 90, 533–544. ( 10.1128/JVI.02320-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda M, Yamada H, Ito H, Kobayashi M. 2011. Baculovirus IAP1 induces caspase-dependent apoptosis in insect cells. J. Gen. Virol. 92, 2654–2663. ( 10.1099/vir.0.033332-0) [DOI] [PubMed] [Google Scholar]
- 50.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86, 13 729–13 734. ( 10.1128/JVI.02041-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehrabadi M, Hussain M, Matindoost L, Asgari S. 2015. The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J. Virol. 89, 8182–8192. ( 10.1128/JVI.00802-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Q, Qiu L, Li G. In press. Baculovirus-encoded MicroRNAs: a brief overview and future prospects. Curr. Microbiol. ( 10.1007/s00284-018-1443-y) [DOI] [PubMed] [Google Scholar]
- 53.O'Reilly DR, Miller LK. 1989. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245, 1110–1112. ( 10.1126/science.2505387) [DOI] [PubMed] [Google Scholar]
- 54.O'Reilly DR, Miller LK. 1991. Improvement of a baculovirus pesticide by deletion of the EGT gene. Nat. Biotech. 9, 1086–1089. ( 10.1038/nbt1191-1086) [DOI] [Google Scholar]
- 55.Kamita SG, Nagasaka K, Chua JW, Shimada T, Mita K, Kobayashi M, Maeda S, Hammock BD. 2005. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl Acad. Sci. USA 102, 2584–2589. ( 10.1073/pnas.0409457102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Houte S, Ros VI, Mastenbroek TG, Vendrig NJ, Hoover K, Spitzen J, van Oers MM.. 2012. Protein tyrosine phosphatase-induced hyperactivity is a conserved strategy of a subset of baculoviruses to manipulate lepidopteran host behavior. PLoS ONE 7, e46933 ( 10.1371/journal.pone.0046933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katsuma S, Koyano Y, Kang W, Kokusho R, Kamita SG, Shimada T. 2012. The baculovirus uses a captured host phosphatase to induce enhanced locomotory activity in host caterpillars. PLoS Pathog. 8, e1002644 ( 10.1371/journal.ppat.1002644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoover K, Grove M, Gardner M, Hughes DP, McNeil J, Slavicek J. 2011. A gene for an extended phenotype. Science 333, 1401 ( 10.1126/science.1209199) [DOI] [PubMed] [Google Scholar]
- 59.Han Y, van Houte S, Drees GF, van Oers MM, Ros VI.. 2015. Parasitic manipulation of host behaviour: baculovirus SeMNPV EGT facilitates tree-top disease in Spodoptera exigua larvae by extending the time to death. Insects 6, 716–731. ( 10.3390/insects6030716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ros VI, van Houte S, Hemerik L, van Oers MM.. 2015. Baculovirus-induced tree-top disease: how extended is the role of egt as a gene for the extended phenotype? Mol. Ecol. 24, 249–258. ( 10.1111/mec.13019) [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, et al. 2018. Host miRNAs are involved in hormonal regulation of HaSNPV-triggered climbing behaviour in Helicoverpa armigera. Mol. Ecol. 27, 459–475. ( 10.1111/mec.14457) [DOI] [PubMed] [Google Scholar]
- 62.Ohkawa T, Majima K, Maeda S. 1994. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J. Virol. 68, 6619–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, Kuzio JA, Possee RD. 1997. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238, 243–253. ( 10.1006/viro.1997.8816) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.