Abstract
Identifying fastidious pathogens in patients with prosthetic joint infection (PJI) by culture is challenging. Metagenomic next-generation sequencing (mNGS) is a novel culture-independent approach that is associated with a higher likelihood for identifying pathogens. We present a case where mNGS was implemented to identify Parvimonas micra, a rarely reported and difficult-to-culture PJI pathogen.
Keywords: Prosthetic joint infection, Parvimonas micra, Metagenomic next-generation sequencing
Introduction
Result of conventional culture is one of the most important criteria in establishing a microbiologic diagnosis of prosthetic joint infection (PJI) 1. The sensitivity of cultures to detect pathogens remains limited due to biofilm formation and prior use of antimicrobials. Negative microbiology results might lead to inappropriate treatment.
Metagenomic next-generation sequencing (mNGS) is a novel technology that uses high-throughput sequencing and bioinformatics analysis to detect all microbial species in a given sample if present, and had been successfully applied in identifying pathogens from synovial fluid and sonicate fluid samples in patients with PJIs 2, 3, 4. Importantly, mNGS is culture-independent. Herein, we present a case of culture-negative chronic PJI, in which the causative pathogen, Parvimonas micra, was detected by mNGS. Ultimately, the infection was controlled by revision surgery and therapy with penicillin.
Case presentation
A 65-year-old female underwent a left total hip arthroplasty (THA) due to developmental dysplasia of the left hip seven years prior to her current presentation. After surgery, she suffered from mild and intermittent left hip pain that was exacerbated by movement. She developed a gradually enlarging mass in her left thigh three years earlier. She was admitted to a local hospital for further examination. C-reactive protein (CRP) was 2.89 mg/L (normal range 0-8 mg/L) and erythrocyte sedimentation rate (ESR) was 23 mm/h (normal range 0-25 mm/h). X-rays and computerized tomography scans showed a defect in the anterior cortex of the left femur (Figure 1). MRI showed effusion around the femur and a heterogeneous mass connected to the femur defect via an underlying tract (Figure 2). Analysis of synovial fluid revealed a white blood cell count of 42,478 × 106/L with 92.3%neutrophils. Seven-day aerobic and anaerobic cultures in blood culture bottles revealed no growth. No antibiotic was administrated.
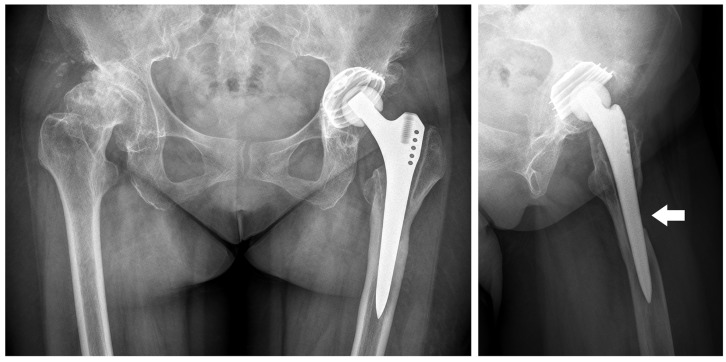
Figure 1.
X-ray films show no sign of migration, but a periprosthetic defect at the anterior cortex of left femur from lateral view (white arrow).
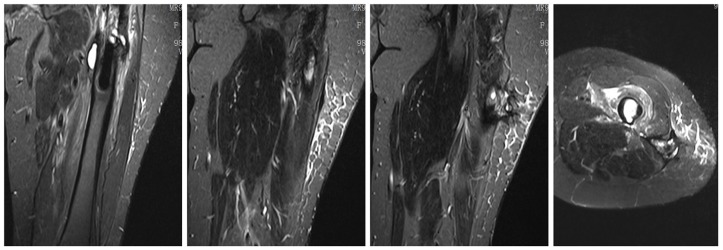
Figure 2.
Coronal and horizontal plane of magnetic resonance T2-weighted images show effusion around the femur connected to subcutaneous tissue through an underlying tract.
She was transferred to the author's hospital for further diagnosis and treatment one month later. A physical examination revealed deep tenderness and percussion pain over her left inguinal area and greater trochanter. A firm mass of 5cm ×5cm size is palpated at left thigh. The flexion movement was limited < 70°. Four-milliliter cloudy synovial fluid was collected by arthrocentesis of left hip. Analysis of 500-μL synovial fluid showed that the white blood cell counts were 17,200×106/L with 91.2% neutrophils. Two-milliliter of synovial fluid was cultured aerobically in a BACTEC Peds Plus bottle (Becton Dickinson, USA) for 7 days, and the culture results were negative. We employed 16S rRNA gene PCR as previously described to detect the pathogenic bacteria from synovial fluid 5, yielding negative result.
Meanwhile, we analyzed the residual 500-μL synovial fluid by mNGS, including experiment procedures as follows: extraction of total DNA, conduction of DNA libraries, sequencing on the BGISEQ-500 platform (BGI-Tianjin, Tianjin, China). The raw data from sequencing were analyzed by a bioinformatics pipeline developed by BGI. The bioinformatic analysis included filtering out low-quality reads, removing human host sequences, and aligning to the in-house built Microbial Genome Database. Detailed information of sequencing and bioinformatic analysis is listed in the supplementary materials.
The analysis of mNGS yielded a total of 20,004,285 single-end reads. The number of microbial reads was 370, the number of bacterial reads was 228 (87.7% of microbial reads). Parivimonas had the highest relative abundance of 16.0% and 13 stringently mapped reads in genus-level, while P. micra had the highest coverage rate of 0.18% within corresponding genus and 13 stringently mapped reads in species-level. P. micra was hypothesized as the potential pathogenic organism. However, the abundance and number of reads mapped to Parivimonas was not much higher than other mapped organisms (e.g., Brucella), which diminished our certainty of distinguishing real pathogens from contamination.
The patient presented with an infected arthroplasty based on imaging and laboratory results. Therefore, we decided to proceed with a two-stage revision surgery. During revision surgery, we observed a 2cm×3cm bone defect at the anterolateral region of the femur connecting with granulation tissues in the vastus lateralis muscle. A frozen section of periprosthetic tissue revealed an average neutrophil count of >20 per high power field from 6 periprosthetic samples. We performed explantation and debridement accordingly. We implanted an articulating antibiotic loaded bone cement spacer, containing 4g vancomycin per 40g cement (Figure 3).
Figure 3.
A set of antibiotic-loaded articulate spacers that were implanted.
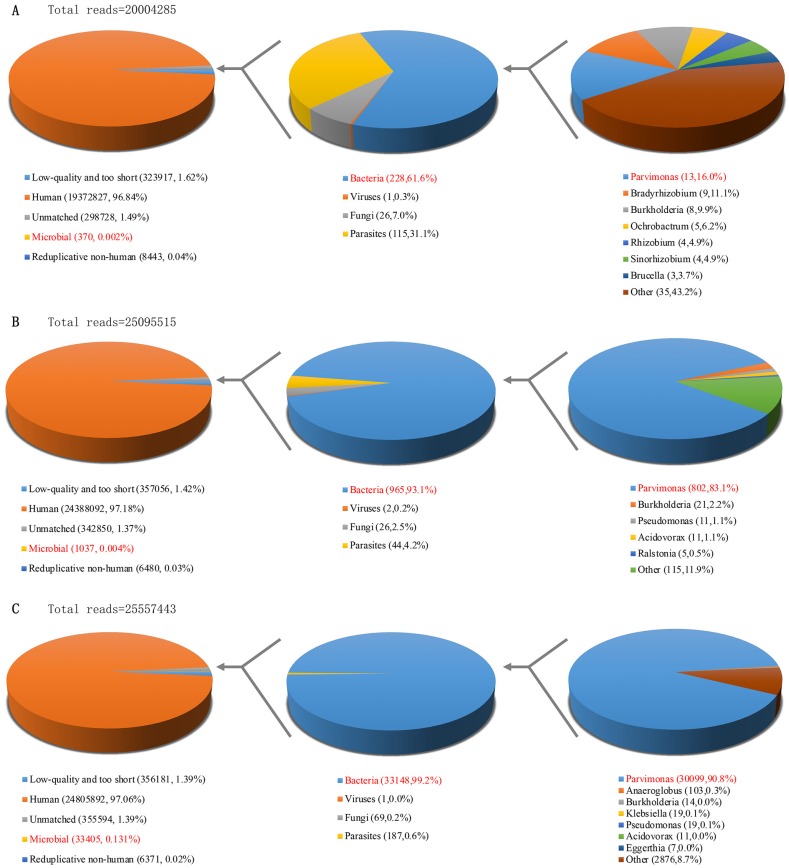
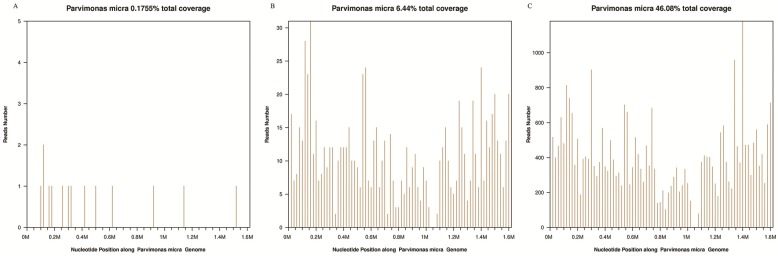
We later performed sonication of explanted prostheses using a previously published workflow 5. Aerobic and anaerobic cultures of the synovial fluid, sonicate fluid, and periprosthetic tissue via blood culture bottle and sheep blood agar plated at 37℃ for 14 days did not reveal any organism(s) growth. However, mNGS yielded positive results from intraoperative synovial fluid and sonicate fluid, with P. micra as the predominant microorganism. The number of unique reads of P. micra was 802 (83.1% of microbial reads) and coverage rate of P. micra was 6.44% in synovial fluid. The number of unique reads of P. micra was 30099 (90.8% of microbial reads) and coverage rate of P. micra was 46.08% in sonicate fluid (Figure 4). Therefore, we regarded P. micra as the definite causative pathogen.
Figure 4.
Taxonomic classification of reads by bioinformatic analysis of sequenced data from preoperative synovial fluid (A), intraoperative synovial fluid (B) and sonicate fluid (C).
We performed a verification experiment using 16S rRNA gene PCR. Sonicate fluid yielded a positive result, but intraoperative synovial fluid and tissues remained negative. Alignment of Sanger sequencing result with the GenBank database revealed a 97.9% similarity of P. micra, from which the causative organism was validated.
Following consultation with an infectious disease specialist, we prescribed a therapeutic regimen consisting of intravenous piperacillin / tazobactam for two weeks, followed by oral amoxicillin / clavulanate potassium for 8 weeks. At a 12-month follow-up, the patient had improved function in the left hip without coxalgia. Her CPR and ESR were within normal limits. Due to these satisfactory results, she decided not to pursue a second stage revision surgery.
Discussion
P. micra (previously classified as Peptostreptococcus micros) is a gram-positive obligate anaerobic coccus. P. micra is known as a symbiotic bacteria that exists commonly in human oral cavity and the gastrointestinal tract. It can cause gingival or periodontal infections, but extra-oral infections are rare (e.g. epidural abscess and spondylitis). To our knowledge, there are limited P. micra-caused PJI cases that have been reported, two of which were related to dental procedures 6, 7. The culture for P. micra was frequently negative due to the difficulty of culturing this anaerobic bacterium and prior use of antibiotic. Employing highly-sensitive molecular techniques is critical for detecting P. micra.
mNGS is an innovative technology that can sequence most of the genomes present in a clinical specimen with the purpose of identifying potential pathogens. Compared with PCR-based diagnostic technology, mNGS has many advantages, such as capability of simultaneously identifying bacteria, fungi in a single assay and obtaining quantified abundance of microorganisms for distinguishing causative pathogens. mNGS is particularly well-suited in identifying fastidious and rare pathogens like Mycoplasma salivarium 8. Recently, P. micra has been successfully detected by mNGS of sonicate fluid 4 or synovial fluid 2 from a patient with PJI, where culture-based detection did not yield an organism. The case we presented provides an example of how mNGS might improve the yield for detecting uncommon pathogens in patients with PJI.
From our original mNGS results, a variety of microorganisms were identified that were regarded as contaminant or background microorganisms originating from skin or deep tissues (Cutibacterium acnes, coagulate-negative staphylococci), reagents (Streptococci and Acinetobacter), the environment (Ralstonia and Burkholderia), or other sources. It is of utmost importance to eliminate sources of contamination by employing negative controls in the same batch, using DNA-free reagents, and even spiking purified nucleic acid into specimen 9. mNGS has the advantage of quantitative analysis that can distinguish pathogens from contaminant when compared to PCR techniques 10. However, insufficient sequencing data of the pathogenic organism may make it difficult to distinguish pathogen-related from contaminant-related reads. Validation by repeated molecular tests is necessary.
Furthermore, we applied mNGS on a variety of specimen types: synovial fluid and sonicate fluid. Remarkably, the analysis from sonicate fluid yielded greater than 30-fold higher reads of P. micra than synovial fluid, without increasing background organisms. It is plausible that sonication can enrich microorganism abundance in samples by dislodging biofilms from the surface of implants 11. Our intriguing results suggest that sonicate fluid is potentially advantageous for distinguishing real pathogen from contaminant organisms compared to synovial fluid; however, further controlled trials are needed.
Although we did not implement sequence alignment of reads against an antibiotic resistance gene database due to insufficient coverage of the reference genome (Figure 5), the majority of patients infected with P. micra were successfully treated with penicillin, metronidazole or clindamycin 12. We adopted the suggested regimen by using a penicillin-based regimen to effectively control the infection, avoiding the use of empiric administration of broad-spectrum antimicrobial regimens typically used in cases of culture-negative PJI. Thus, we were able to provide the patient with a targeted and cost-effective treatment plan that was ultimately successful.
Figure 5.
The aligned unique reads yielded from preoperative synovial fluid (A), intraoperative synovial fluid (B) and sonicate fluid (C) covered 0.18%, 6.44% and 46.08% of the Pavimonas micra reference genome, respectively. Y-axis was reads number. x-axis was genome length. The bar was reads number per 1kb in the genome.
Conclusions
In summary, of the presented case report of a patient with PJI due to P. micra may indicate that mNGS may have a potential for identifying uncommon pathogens in culture-negative PJIs.
Supplementary Material
Supplementary methods and figure.
Acknowledgments
This paper is supported by Natural Science Foundation of Fujian Province (grant number 2018I0006 and 2018Y4003).
Author Contributions
Writing and Figure Organization, H.Z. and Z.C., 16s Ribosome RNA Gene PCR Performing, W.Q., Surgery performing, L.W. and Z.W., Microbial Culture, Y.B., English Editing and Manuscript Reviewing, F.X. and Z.W., Metagenomic Next-Generation Sequencing, X.L., L.Y. and N.X. All authors reviewed the final version of the manuscript.
Consent
Written informed consent was obtained from the patient for publication of this case report. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Institutional Ethics Committee of authors' affiliated institutions.
References
- 1.Parvizi J, Gehrke T. International consensus on periprosthetic joint infection: let cumulative wisdom be a guide. J Bone Joint Surg Am. 2014;96:441. doi: 10.2106/JBJS.N.00023. [DOI] [PubMed] [Google Scholar]
- 2.Ivy MI, Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Hanssen AD, Abdel MP, Direct Detection and Identification of Prosthetic Joint Infection Pathogens in Synovial Fluid by Metagenomic Shotgun Sequencing. J Clin Microbiol; 2018. p. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD. et al. Identification of Prosthetic Joint Infection Pathogens Using a Shotgun Metagenomics Approach. Clin Infect Dis. 2018;67:1333–8. doi: 10.1093/cid/ciy303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Street TL, Sanderson ND, Atkins BL, Brent AJ, Cole K, Foster D. et al. Molecular Diagnosis of Orthopedic-Device-Related Infection Directly from Sonication Fluid by Metagenomic Sequencing. J Clin Microbiol. 2017;55:2334–47. doi: 10.1128/JCM.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Z, Wu Q, Fang X, Li W, Zhang C, Zeng H. et al. Comparison of culture and broad-range polymerase chain reaction methods for diagnosing periprosthetic joint infection: analysis of joint fluid, periprosthetic tissue, and sonicated fluid. Int Orthop. 2018;42:2035–40. doi: 10.1007/s00264-018-3827-9. [DOI] [PubMed] [Google Scholar]
- 6.Bartz H, Cb N, Bollmann C, Kuhl M, Zimmermann S, Heeg K. et al. Micromonas (Peptostreptococcus) micros: unusual case of prosthetic joint infection associated with dental procedures. International Journal of Medical Microbiology. 2005;294:465–70. doi: 10.1016/j.ijmm.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Stoll T, Stucki G, Brühlmann P, Vogt M, Gschwend N, Michel BA. Infection of a total knee joint prosthesis by peptostreptococcus micros and propionibacterium acnes in an elderly RA patient: Implant salvage with longterm antibiotics and needle aspiration/irrigation. Clinical Rheumatology. 1996;15:399–402. doi: 10.1007/BF02230366. [DOI] [PubMed] [Google Scholar]
- 8.Thoendel M, Jeraldo P, Greenwoodquaintance KE, Chia N, Abdel MP, Steckelberg JM, A Possible Novel Prosthetic Joint Infection Pathogen, Mycoplasma salivarium, Identified by Metagenomic Shotgun Sequencing. Clinical Infectious Diseases; 2017. p. 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice C. et al. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch Pathol Lab Med. 2017;141:776–86. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 10.Hartley JC, Harris KA. Molecular techniques for diagnosing prosthetic joint infections. J Antimicrob Chemother. 2014;69(Suppl 1):i21–4. doi: 10.1093/jac/dku249. [DOI] [PubMed] [Google Scholar]
- 11.Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR. et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med. 2007;357:654–63. doi: 10.1056/NEJMoa061588. [DOI] [PubMed] [Google Scholar]
- 12.George IA, Pande A, Parsaei S. Delayed infection with Parvimonas micra following spinal instrumentation. Anaerobe. 2015;35:102–4. doi: 10.1016/j.anaerobe.2015.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods and figure.