Abstract
Bamboo are woody grass species containing important economic and ecological values. Lei bamboo (Phyllostachys violascens) is a kind of shoot-producing bamboo species with the highest economic yield per unit area. However, identifying different varieties of Lei bamboo based on morphological characteristics is difficult. Microsatellites play an important role in plant identification and genetic diversity analysis and are superior to other molecular markers. In this study, we identified 18,356 expressed sequence tag-simple sequence repeat (EST-SSR) loci in Lei bamboo transcriptome data. A total of 11,264 primer pairs were successfully designed from unigenes of all EST-SSR loci, and 96 primer pairs were randomly selected and synthesized. A total of 54 primer pairs were used for classifying 16 Lei bamboo varieties and 10 different Phyllostachys species. The number of polymorphism alleles among the 54 primer pairs ranged from 3 to 12 for P. violascens varieties and 3 to 20 for Phyllostachys. The phylogenetic tree based on polymorphism alleles successfully distinguished 16 P. violascens varieties and 10 Phyllostachys species. Our study provides abundant EST-SSR resources that are useful for genetic diversity analysis and molecular verification of bamboo and suggests that SSR markers developed from Lei bamboo are more efficient and reliable than ISSR, SRAP or AFLP markers.
Keywords: Phyllostachys violascens, transcriptome, microsatellites, varieties, genetic diversity
Introduction
Bamboo is an economically important member of the woody grasses. It includes 88 genera and more than 1,400 species worldwide; 34 genera and 534 species are in China (Wu and Raven, 2006). Given the considerably fast growth, strong carbon fixation capability and edible shoots, bamboo has worldwide ecological and economic value.
The classification and nomenclature of plants are mainly based on morphological characteristics, such as roots, stems, leaves and flowers (Lichtenthaler, 1987). Flower morphology is the most important (PDCress and Staden, 1998). The identification and classification of bamboo is vital for germplasm collection and conservation (Soderstrom and Calderon, 1979). However, bamboo has a long juvenile phase, with the flowering interval of some species being up to 120 years (Janzen, 1976), which prevents the classification of bamboo when using only flower morphology. Moreover, morphological characteristics sometimes are not very reliable because they are affected by ecological factors, which leads to confused classification and reclassification of various accessions of bamboo (Das et al., 2007).
Microsatellites or simple sequence repeats (SSRs), characterized by high polymorphism, wide distribution in genomes, co-dominance and reproducibility, have been frequently used for genetic analysis (Goldstein and Schlötterer, 1999). For instance, 69 varieties of taro (Colocasia esculenta) were successfully separated by expressed sequence tag-SSR (EST-SSR) primers (You et al., 2015). Eleven varieties of Lycium were verified by EST-SSRs (Chen et al., 2017). SSR markers have been used to study genetic diversity, genetic distance and classification of bamboo species (Kaneko et al., 2008; Mukherjee et al., 2010; Lin et al., 2011; Jiang et al., 2013; Dong and Yang, 2014; Zhao et al., 2015; Abhishek et al., 2016).
Lei bamboo (Phyllostachys violascens) has higher economic value per unit area than other bamboo species with edible shoots (Liu et al., 2001). Lei bamboo has many varieties that differ in shoot time, shoot yield per unit area and pathogen resistance. Lin et al. (2011) classified different P. violascens varieties by using inter-simple sequence repeat (ISSR), sequence-related amplified polymorphism (SRAP), and amplified fragment length polymorphism (AFLP) markers. However, some varieties cannot be differentiated by just one molecular marker method (Lin et al., 2011). Therefore, the development of accurate and efficient molecular markers for classifying different varieties in Lei bamboo is important.
Previously, we sequenced the transcriptome of P. violascens (GenBank Accession No. SRX5137626) by using Illumina Truseq and assembled 132,678 unigenes. We identified EST-SSR loci as well as primer pairs based on these data. The major objective of this study was to develop efficient EST-SSR markers for classifying Lei bamboo varieties as well as Phyllostachys species. These markers may be applicable for bamboo taxonomic study, genetic diversity analysis and evolutionary research.
Materials and Methods
Plant Materials
Young leaves of 16 varieties of P. violascens (cv. Jianye, cv. Panggan, cv. Violascens, cv. Notata, cv. Zaoyuanzhu, cv. Flavistriatus, cv. Hongke, cv. Xiye, cv. Viridisulcata, cv. Linanensis, cv. Kuoyeqingtou, cv. Anhuihongke, cv. Flavicaginis, cv. Qingke, cv. Atrovaginis, cv. Anhuiensis) were sampled from the Bamboo Garden of Zhejiang Agriculture and Forestry University (Lin et al., 2011). Young leaf samples of nine Phyllostachys species (P. glabrata, P. verrucosa, P. bambusoides, P. aurea, P. edulis, P. virella, P. rivalis, P. parvifolia, and P. nidularia) were collected from the China Bamboo Expo Park in Huzhou, Zhejiang province, China.
Marker Loci Detection and SSR Primer Pair Design
MicroSAtellite (MISA)1 was used for SSR mining in the assembled contigs (Thiel et al., 2003). The minimum number of repeats used to select the SSRs was 10 for mononucleotide repeats, 6 for dinucleotide repeats, and 5 for tri-, tetra-, penta-, and hexanucleotide repeats. The Perl program Primer3.02 (Rozen and Skaletsky, 2000) was used to design Primer pairs with the design principles of length 18 to 27 bp and about 20 bp, melting temperature (Tm) 57°C to 63°C, and PCR product size 100 to 280 bp.
DNA Extraction and EST-SSR Marker Amplification
Total genomic DNA was extracted by using the cetyltrimethyl ammoniumbromide method (Porebski et al., 1997) and verified by electrophoresis on 1% agarose gel. PCR amplification was carried out in a 20-μl volume containing 50 ng template DNA, 2 μl of 10 × PCR buffer (Mg2+ free), 1.2 μl MgCl2 (20 mM), 1 μl sense primer (10 pmol), 1 μl anti-sense primer (10 pmol), 2 μl dNTP (10 mM), and 0.5 μl rTaq DNA polymerase (5 U, TAKARA). The PCR program consisted of an initial step of 95°C for 5 min, followed by 33 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 10 min (Rossetto, 2001). The PCR products were loaded on 1-mm-thick non-denaturing gels of 8% polyacrylamide (Acr/Bis = 29:1). The electrophoresis buffer contained 1 × TBE (100 mM Tris–HCl, 83 mM boric acid, 1 mM Na2EDTA, pH 8.0) (Han et al., 2010); the 20 bp DNA Ladder Dye Plus (Takara Biomedical Technology, Beijing) was used as a size standard.
PCR Product Sequencing
Silver nitrate stain gel was used for selecting suitable SSR primers (Panaud et al., 1996). The desired bands were accurately excised and subsequently purified by using the SanPrep Column DNA Gel Extraction Kit (Sangon, Shanghai) (Tang et al., 2010). The purified DNA bands were ligated with pMD-18-T vector (TaKaRa, Beijing) and sequenced by the Sanger method (Tang et al., 2010; Zhai et al., 2014). The PCR sequence similarity rate was more than 98% with transcriptome sequences applied.
Data Processing and Genetic Analysis
According to PAGE results, the absence or presence of bands was scored as zero or one in all SSR loci and two binary qualitative data matrices were generated (Rohlf, 2000; You et al., 2015). Dendrograms were constructed on the basis of these two binary qualitative matrices with the method unweight pair group method with arithmetic mean (UPGMA) selected for clustering and maximum number of tied trees set to 100. Before dendrogram construction, pairwise coefficients were calculated according to the Jaccard similarity coefficient method (Kosman and Leonard, 2005). The above-mentioned data processing, coupled with principal coordinates analysis (PCoA) involved using NTSYSpc v2.1 (Rohlf, 2000). The bootstrapping analysis repeat was 1,000 performed with the software FREETREE V.0.9.1.50 (Pavlícek et al., 1999). Bootstrap values more than 50 are listed on the dendrogram (Zhu et al., 2012; Zhai et al., 2013). In this study, we constructed three phylogenetic trees: 16 P. violascens varieties independently, 10 Phyllostachys species independently and combination with 16 P. violascens varieties and other 9 Phyllostachys species.
Results
Distribution and Frequency of SSR Markers
A total of 132,678 transcriptome sequences were examined by MISA software, and 18,356 EST-SSRs loci were identified in P. violascens. Among these SSRs, 2,239 (11.76%) unigenes contained more than one SSR locus, and 903 SSRs were presented in compound formation (Appendix 1 and Appendix 2). These SSRs were further divided into six different types based on unit size, that is, 9,099 mononucleotide repeats (mono-), 4,449 dinucleotide repeats (di-), 4,521 trinucleotide repeats (tri-), 235 tetranucleotide repeats (tetra-), 39 pentanucleotide repeats (penta-) and 13 hexanucleotide repeats (hexa-), accounting for 49.57, 24.24, 24.63, 1.28, 0.21, and 0.07% of total SSRs, respectively (Appendix 1).
The repeat motif length of SSR ranged from 10 to 60 bp. The most abundant was 10 bp (4,462, 24.31%), followed by 15 bp (3,362, 18.32%), 12 bp (2,166,13.0%), and 18 bp (1,878, 10.23%). The 60 bp length was the longest SSR loci (Appendix 1).
Within these SSRs, 95 motif types were detected (Appendix 1). The frequency distribution of the 20 most abundant SSR classical repeat motifs is in Table 1. Among those motif types, A/T was the most abundant (43.73%), followed by AG/CT (15.93%) and C/G (5.84%).
Table 1.
Frequency distribution of the 20 most frequent EST-SSR repeat motifs in Phyllostachys violascens.
| Repeats |
Number of repeats of the motif |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | motif | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 46 | Total | Frequency |
| 1 | A/T | – | – | – | – | – | 4314 | 1499 | 767 | 395 | 277 | 160 | 97 | 81 | 96 | 110 | 122 | 61 | 33 | 13 | 2 | 8027 | 43.73% | |
| 2 | AG/CT | – | 877 | 551 | 531 | 557 | 343 | 62 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | 1 | 2925 | 15.93% |
| 3 | CCG/CGG | 1177 | 376 | 91 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1647 | 8.97% |
| 4 | C/G | – | – | – | – | – | 148 | 135 | 99 | 84 | 77 | 71 | 61 | 72 | 77 | 101 | 71 | 52 | 15 | 7 | 2 | 1072 | 5.84% | |
| 5 | AC/GT | – | 360 | 188 | 140 | 102 | 105 | 33 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | 929 | 5.06% |
| 6 | AGG/CCT | 502 | 182 | 60 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 747 | 4.07% |
| 7 | AGC/CTG | 441 | 136 | 53 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 633 | 3.45% |
| 8 | AAG/CTT | 230 | 87 | 50 | 3 | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | 371 | 2.02% |
| 9 | AT/AT | – | 199 | 77 | 28 | 20 | 17 | 19 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 360 | 1.96% |
| 10 | ACC/GGT | 221 | 73 | 28 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 323 | 1.76% |
| 11 | AAC/GTT | 187 | 58 | 13 | 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 260 | 1.42% |
| 12 | CG/CG | – | 171 | 50 | 10 | 3 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 235 | 1.28% |
| 13 | ACG/CGT | 160 | 44 | 18 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 222 | 1.21% |
| 14 | ATC/ATG | 97 | 24 | 8 | 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 131 | 0.71% |
| 15 | AAT/ATT | 61 | 30 | 17 | 2 | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | 111 | 0.60% |
| 16 | ACT/AGT | 55 | 13 | 7 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 76 | 0.41% |
| 17 | AAAG/CTTT | 33 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 34 | 0.19% |
| 18 | ATCC/ATGG | 25 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 26 | 0.14% |
| 19 | AGGG/CCCT | 19 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 22 | 0.12% |
| 20 | AAGG/CCTT | 12 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 15 | 0.08% |
| Other | 155 | 27 | 1 | 1 | 2 | 2 | 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 190 | 1.04% | |
| Total | 3375 | 2665 | 1212 | 730 | 684 | 616 | 251 | 104 | 84 | 77 | 72 | 61 | 72 | 77 | 101 | 71 | 52 | 15 | 7 | 2 | 1 | 18356 | 100.00% | |
A total of 11,264 primer pairs were successfully designed by using Primer 3.0 and two Perl scripts of p3_in.pl and p3_out.pl3 from 15,600 unigenes. However, the software primer modeling failed for 6,189 unigene sequences (Appendix 2). We randomly selected 96 SSR primer pairs from all SSR primer pairs. Finally, 54 optimal primer pairs (Appendix 3) were used to study a series of filters: (1) unsatisfactory amplification, (2) unspecific amplification, and (3) sequence less than 98% similarity in comparison with the transcriptome. The optimized SSRs were used to analyze the genetic diversity of 16 P. violascens varieties and 10 Phyllostachys species.
Genetic Diversity of P. violascens
We used 54 primer pairs for the 16 P. violascens varieties, generating 399 alleles, which were treated as informative loci for dendrogram construction (Appendix 4). The number of SSR alleles was 3 to 12 for one primer pair, and each primer pair had 6.88 alleles, on average.
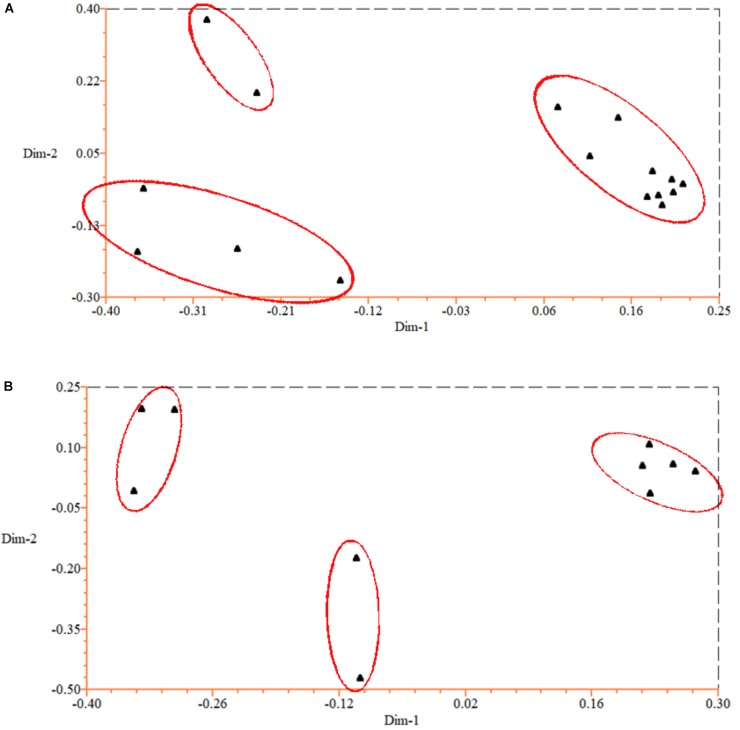
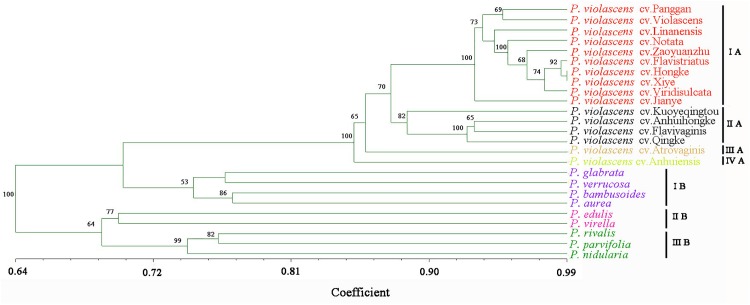
To test the genetic similarity between varieties, we used PCoA based on SSR data and found three groups among the 16 P. violascens varieties (Figure 1A). The phylogenetic tree was further constructed by using NTSYSpc (Figure 2A); the resulting Jaccard similarity coefficient ranged from 0.73 to 0.98. Four groups were uncovered in previous research according to genetic diversity of P. violascens varieties by combining ISSR, SRAP and AFLP marker analysis (Lin et al., 2011). For better comparison with previous study, the index 0.78 was selected and four main groups were detected in present study: groups I, II, III, and IV. Group I contained 10 varieties. Among the 10 varieties, P. violascens cv. Jianye and cv. Linanensis exhibited far genetic distances from other eight varieties; P. violascens cv. Hongke and cv. Xiye had very close genetic distance, with similarity index about 0.98. Among the four varieties in group II, the greater distant genetic distance was between P. violascens cv. Kuoyeqingtou and three other varieties. The groups III and IV contained only one variety each. These results were largely consistent with the PCoA results, with groups III and IV in the dendrogram clustered into one group on PCoA.
FIGURE 1.
Principal coordinates analysis (PCoA) of Phyllostachys violascens based on EST-SSR data for (A) 16 P. violascens varieties and (B) 10 Phyllostachys species.
FIGURE 2.
The dendrogram of bamboo species/varieties based on EST-SSR markers of P. violascens. Genetic relationship among (A) 16 varieties of P. violascens and (B) 10 species of Phyllostachys. The Roman numerals on the right side of the dendrogram represented the clustered groups. Bootstrap values over 50 were labeled on the branches of the dendrogram.
Genetic Diversity of Phyllostachys Bamboo
The above 54 primer pairs were used with the 10 Phyllostachys bamboo species to verify their cross-amplification potential in Phyllostachys. The SSR primer pairs of P. violascens were perfectly applied in other Phyllostachys species. The number of SSR alleles ranged from 3 to 20, with an average of 11.3 per primer pairs. A total of 656 loci were used to construct the dendrogram of Phyllostachys; no missing data were found in the study (Appendix 5). As shown in the phylogenetic tree (Figure 2B), the Jaccard similarity coefficient ranged from 0.67 to 0.84. In this study, the 10 Phyllostachys species were separated into three distinct groups: I, II, and III. Group I contained five species: P. violascens, P. glabrata, P. verrucosa, P. bambusoides, and P. aurea. The Jaccard similarity coefficient was up to 0.84 between P. violascens and P. glabrata. Group II included two bamboo species: P. edulis and P. virella. Group III contained three bamboo species: P. rivalis, P. parvifolia and P. nidularia. Similar grouping results were shown by PCoA (Figure 1B).
Discussion
Characterization of EST-SSRs in P. violascens
Simple sequence repeat markers have been widely used in fingerprint construction, genetic diversity analysis and molecular marker-assisted breeding because of their high polymorphism, repeatability and codominant inheritance (Ran et al., 2010; Feng et al., 2018; Shi et al., 2018; Wang et al., 2018). However, traditional methods for developing SSR markers are not very efficient (Tang et al., 2010). Next-generation sequencing technologies can generate massive data, which are good resources for developing SSR markers in many species including bamboo (Tang et al., 2010; Abhishek et al., 2016; Zhang et al., 2017). Nevertheless, SSRs in P. violascen have not been reported because of lack of transcriptome or genome sequences. In this study, we identified 18,356 EST-SSR loci from 15,600 unigenes, and a total of 11,264 primer pairs were designed based on the transcriptome of P. violascens. Overall, 54 primer pairs were successfully used for identifying 16 P. violascens varieties and 10 Phyllostachys species, so the transcriptome sequences were good resources for developing SSR markers.
The frequency of SSRs in P. violascens was 1/4.55 kB when including the mononucleotide repeats, which was close to wheat (1/5.46 kB) (Peng and Lapitan, 2005) but significantly higher than in Arabidopsis (1/13.83 kB) (Cardle et al., 2000). Various factors may affect SSR frequency, including search criteria, software, as well as species properties (Sorrells, 2005). Tectonic activities and climate fluctuations during species evolutionary history contributed to the production and accumulation of genetic variations. Multi-locus plastid phylogenic analyses of bamboo revealed that the Bambusoideae began to diversify at about 43.26 million years ago (Mya), followed by the rapid radiation of the Arundinarieae at about 12–14 Mya, which was supposed to be induced by the important strengthening of the East Asian monsoon in the late Miocene (Zhang et al., 2016). Therefore, the high frequency of SSRs detected in P. violascens may be due in part to the long and complex evolutionary process of bamboo.
The types of repeat motifs in this study were not uniformly distributed in the P. violascens transcriptome database (Table 1). Di- and tri-repeats were the most predominant when excluding mononucleotide repeats (Table 2). This result differed from Ma bamboo (Dendrocalamus latiflorus), in which tri-repeats are the most abundant (Abhishek et al., 2016). In addition, the proportion of tetra-, penta- and hexanucleotide repeats was significantly lower in P. violascens than D. latiflorus (Table 2). The differentiation between P. violascens and D. latiflorus was likely caused by distant genetic distance. Phyllostachys belong to the Arundinarieae, a kind of temperate woody bamboo, and Dendrocalamus is affiliated with tropical woody bamboo Bambuseae (Soreng et al., 2014). These two tribes diversified at the beginning of the bamboo evolutionary history and preserved distinct genetic elements (Zhang et al., 2012, 2016).
Table 2.
Comparison of frequency of microsatellites of Phyllostachys violascens, Dendrocalamus latiflorus, Arabidopsis thaliana, and Triticum aestivum.
| Plant | P. violascens | D. latiflorus | A. thaliana | T. aestivum |
|---|---|---|---|---|
| Total | 9,257 | 22,305 | 1,070 | 43,598 |
| Di- | 48.06% | 16.10% | 26.27% | 20.77% |
| Tri- | 48.84% | 47.70% | 73.04% | 74.26% |
| Tetra- | 2.54% | 26.10% | 0.72% | 3.36% |
| Penta- | 0.42% | 6.90% | 0 | 1.12% |
| Hexa- | 0.14% | 3.30% | 0 | 0.50% |
As shown in Table 1, the AG/CT motif was the most predominant di-repeat (17.11%), which was similar to pigeonpea (16.7%) (Dutta et al., 2011) but higher than wheat (8.7%) (Peng and Lapitan, 2005) and lower than taro (52.86%) (You et al., 2015). Among tri-repeats, CCG/CGG was the most abundant (5.91%), which was consistent with previous findings in taro (You et al., 2015), rice and maize (Cardle et al., 2000). The AGC/GCT type (2.94%) was the second most abundant tri-repeat, which was not common in taro (You et al., 2015), rice or maize (Cardle et al., 2000). Previous studies showed that the tri-repeat type of CCG/CGG was a rare motif in dicotyledonous plants but the most abundant repeat type among the tri-repeats in monocots (Varshney et al., 2002; You et al., 2015). Our results verified that many CCG/CGG repeats was a common feature of monocot plants and the high G/C content plays an important role in monocots (Morgante et al., 2002; Rota et al., 2005; Wang et al., 2010).
Genetic Diversity in 16 P. violascens Varieties
The genetic diversity of P. violascens germplasm has been unclear. The classification of 16 varieties of P. violascens previously involved using three distinct types of molecular markers - ISSR, SRAP and AFLP individually – but failed (Lin et al., 2011). When combining ISSR, SRAP and AFLP, these 16 varieties could be identified successfully (Lin et al., 2011). In the present study, the 16 varieties of P. violascens could also be distinguished by polymorphic EST-SSR markers, and significant genetic variations between different P. violascens varieties were uncovered. These results implied that SSR markers and the combination of ISSR, SRAP and AFLP markers were more sensitive than using the ISSR, SRAP and AFLP markers alone. Considering the convenience of experimental design, the SSR marker method is more applicable than ISSR, SRAP and AFLP combined.
In addition, both PCoA and phylogenetic analysis revealed similar grouping results for the 16 P. violascens varieties based on SSR markers (Figures 1A, 2A). The cluster results for SSR markers were similar to combining ISSR, SRAP and AFLP markers but differed little when using ISSR, SRAP, and AFLP markers alone (Lin et al., 2011). Hence, the SSR method and combining ISSR, SRAP and AFLP markers were more reliable than using ISSR, SRAP and AFLP markers alone. Meanwhile, the cluster result of SSR was more similar with AFLP than with ISSR and SRAP markers, which suggests that the AFLP method was more reliable than ISSR and SRAP methods in classifying P. violascens.
Transferability of EST-SSR Markers and Genetic Diversity Among 10 Phyllostachys Species
To test the versatility of EST-SSR markers derived from P. violascens, the 54 SSR primer pairs were further used to analyze the genetic diversity of 10 Phyllostachys species. Clear bands were generated and higher transfer rate (100%) was detected by using these SSR primer pairs as compared with Elymus sibiricus (22.40%) (Zhou et al., 2016), Chrysanthemum nankingense (20%) (Wang et al., 2013), Juglans mandshurica (30.8%) (Hu et al., 2016), and Neolitsea sericea (16.3%) (Chen et al., 2015), which implies the availability of SSR markers in the genus of Phyllostachys. Furthermore, a phylogenetic tree encompassing all 16 P. violascens varieties and 9 other Phyllostachys species used in this study was also built based on the EST-SSRs markers (Appendix 6). The results showed that all varieties of P. violascens were clustered into one main group and kept separate with the other Phyllostachys species. The internal branch structure of the common dendrogram was consistent with the separate phylogenetic analysis based on the 16 P. violascens varieties (Figure 2A) and 10 Phyllostachys species (Figure 2B), confirming the versatility and robustness of the EST-SSR markers in distinguishing different varieties of P. violascens and different Phyllostachys species. Therefore, these EST-SSR markers might be used for classifying different species in Phyllostachys and even different genera of bamboo.
Figure 2B shows that these 54 SSR primer pairs developed from P. violascens could successfully distinguish the 10 Phyllostachys species. Phyllostachys has been divided into Sect. Phyllostachys and Sect. Heterocladae according to the Flora of China (Wu and Raven, 2006). In this study, 10 species of Phyllostachys were divided into three groups by using SSR markers of P. violascens. Groups I and II belonged to Sect. Phyllostachys and group III belonged to Sect. Heterocladae. However, species from groups II and III were clustered together. One reasonable explanation was that Sect. Phyllostachys is not monophyletic. Alternatively, given the lack of informative characters, the existence of incomplete lineage sorting, and/or interspecies hybridization, it is not unexpected that bamboo species from different sections clustered together based on molecular markers and genome data (Zhang et al., 2016). As well, the classification of Sect. Phyllostachys and Sect. Heterocladae in the Flora of China mainly depended on morphological characteristics, such as whether the blade was horizontal, reflexed or erect and whether culm sheaths were covered with spots. Further studies of the classification of Sect. Phyllostachys and Sect. Heterocladae based on both morphological characteristics and molecular analysis are needed.
Author Contributions
KC and XL organized the research and wrote the experimental subjects. LZ and KZ participated in the research. LL and ZZ collected the sample and material. XL and WZ provided technical guidance and paper modification.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewers FB and FG, and handling Editor declared their shared affiliation.
Funding. The research was supported by funds from the Zhejiang University of Science and Technology, Major Program on Agricultural New Variety Breeding (2016C02056-8).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00050/full#supplementary-material
Frequency distribution of the EST-SSR repeat motifs in P. violascens.
The all SSR primer pairs of P. violascens.
The 54 primer pairs of P. violascens were applied in study.
The two-dimensional matrix of P. violascens varieties informative loci.
The two-dimensional matrix of Phyllostachys species informative loci.
The dendrogram of Phyllostachys species and P. violascens varieties based on EST-SSR markers of P. violascens.
References
- Abhishek B., Singh G., Raina A. S., Kaur J., Sharma R. K. (2016). Development of genic SSR marker resource from RNA-Seq data in Dendrocalamus latiflorus. J. Plant Biochem. Biotechnol. 25 179–190. 10.1007/s13562-015-0323-9 [DOI] [Google Scholar]
- Cardle L., Ramsay L., Milbourne D., Macaulay M., Marshall D., Waugh R. (2000). Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li R., Xia Y., Bai G., Guo P., Wang Z., et al. (2017). Development of EST-SSR markers in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilis Tsen et Lee) based on de novo transcriptomic assemblies. PLoS One 12:e0184736. 10.1371/journal.pone.0184736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Y., Cao Y. N., Yuan N., Nakamura K., Wang G. M., Qiu Y. X. (2015). Characterization of transcriptome and development of novel EST-SSR makers based on next-generation sequencing technology in Neolitsea sericea (Lauraceae) endemic to east asian land-bridge islands. Mol. Breed. 35:187 10.1007/s11032-015-0379-1 [DOI] [Google Scholar]
- Das M., Bhattacharya S., Basak J., Pal A. (2007). Phylogenetic relationships among the bamboo species as revealed by morphological characters and polymorphism analyses. Biologia Plantarum 51 667–672. 10.1007/s10535-007-0140-7 [DOI] [Google Scholar]
- Dong Y. R., Yang H. Q. (2014). Development of seventeen novel microsatellite markers of a wild and flowering bamboo Dendrocalamus membranaceus (Poaceae) and cross-amplification in Dendrocalamus genus. Conserv. Genet. Resour. 6 201–203. 10.1007/s12686-013-0055-x [DOI] [Google Scholar]
- Dutta S., Kumawat G., Singh B. P., Gupta D. K., Singh S., Dogra V., et al. (2011). Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 11:17. 10.1186/1471-2229-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Yi S. V., Zhang M., Zhou X. (2018). Development of novel EST-SSR markers for ploidy identification based on de novo transcriptome assembly for Misgurnus anguillicaudatus. PLoS One 13:e0195829. 10.1371/journal.pone.0195829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. B., Schlötterer C. (1999). Microsatellites: evolution and applications. Q. Rev. Biol. 83 633–634. [Google Scholar]
- Han Y. C., Teng C. Z., Hu Z. L., Song Y. C. (2010). An optimal method of DNA silver staining in polyacrylamide gels. Electrophoresis 28 1173–1175. [DOI] [PubMed] [Google Scholar]
- Hu Z., Zhang T., Gao X. X., Wang Y., Zhang Q., Zhou H. J., et al. (2016). De novo assembly and characterization of the leaf, bud, and fruit transcriptome from the vulnerable tree Juglans mandshurica for the development of 20 new microsatellite markers using illumina sequencing. Mol. Genet. Genomics 291 849–862. 10.1007/s00438-015-1147-y [DOI] [PubMed] [Google Scholar]
- Janzen D. H. (1976). Why bamboos wait so long to flower. Annu. Rev. Ecol. Syst. 7 347–391. 10.1146/annurev.es.07.110176.002023 [DOI] [Google Scholar]
- Jiang W. X., Zhang W. J., Ding Y. L. (2013). Development of polymorphic microsatellite markers for Phyllostachys edulis (Poaceae), an important bamboo species in china. Appl. Plant Sci. 1 391–393. 10.3732/apps.1200012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Franklin D. C., Yamasaki N., Isagi Y. (2008). Development of microsatellite markers for Bambusa arnhemica (Poaceae: Bambuseae), a bamboo endemic to northern australia. Conserv. Genet. 9 1311–1313. 10.1007/s10592-007-9467-z [DOI] [Google Scholar]
- Kosman E., Leonard K. (2005). Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol. Ecol. 14 415–424. 10.1111/j.1365-294X.2005.02416.x [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1987). [34] Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Lin X., Lou Y., Zhang Y., Yuan X., He J., Fang W. (2011). Identification of genetic diversity among cultivars of phyllostachys violascens using ISSR, SRAP and AFLP markers. Bot. Rev. 77 223–232. 10.1007/s12229-011-9078-8 [DOI] [Google Scholar]
- Liu L., Lin X., Ye L. (2001). Components of proteins in shoots of various cultivars of Phyllostachys praecox. J. Zhejiang For. Coll. 2001 55–57. [Google Scholar]
- Morgante M., Hanafey M., Powell W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30 194–200. 10.1038/ng822 [DOI] [PubMed] [Google Scholar]
- Mukherjee A. K., Ratha S., Dhar S., Debata A. K., Acharya P. K., Mandal S., et al. (2010). Genetic relationships among 22 taxa of bamboo revealed by ISSR and EST-based random primers. Biochem. Genet. 48 1015–1025. 10.1007/s10528-010-9390-8 [DOI] [PubMed] [Google Scholar]
- Panaud O., Chen X., Mccouch S. R. (1996). Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252 597–607. 10.1007/BF02172406 [DOI] [PubMed] [Google Scholar]
- Pavlícek A., Hrdá S., Flegr J. (1999). Free-Tree–freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. application in the RAPD analysis of genus Frenkelia. Folia Biologica 45 97–99. [PubMed] [Google Scholar]
- PDCress W. H., Staden J. V. (1998). Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 21 535–553. 10.1046/j.1365-3040.1998.00309.x 30669580 [DOI] [Google Scholar]
- Peng J. H., Lapitan N. L. (2005). Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct. Integr. Genomics 5 80–96. 10.1007/s10142-004-0128-8 [DOI] [PubMed] [Google Scholar]
- Porebski S., Bailey L. G., Baum B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15 8–15. 10.1007/BF02772108 [DOI] [Google Scholar]
- Ran L., Wu W. L., Yang Z., Li Y. H. (2010). SSR marker and its application to crop genetics and breeding. Genomics Appl. Biol. 29 137–143. [Google Scholar]
- Rohlf F. J. (2000). NTSYSPC: Numerical Taxonomy and Multivariate Analysis System. New York, NY: Exeter Software Setauket. [Google Scholar]
- Rossetto M. (2001). “Sourcing of SSR markers from related plant species,” in Plant Genotyping: The DNA Fingerprinting of Plants, ed. Henry R. J. (Wallingford, CT: CABI Publishing; ), 211–224. 10.1079/9780851995151.0211 [DOI] [Google Scholar]
- Rota M. L., Kantety R. V., Yu J. K., Sorrells M. E. (2005). Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics 6:23. 10.1186/1471-2164-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Shi X., Wu H., Li W., Guo W., Zheng Y., Yu S., et al. (2018). Development of polymorphic EST-SSR markers in Itea chinensis (Iteaceae) and cross-amplification in related species. Appl. Plant Sci. 6 e1013. 10.1002/aps3.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderstrom T. R., Calderon C. E. (1979). A commentary on the bamboos (Poaceae: Bambusoideae). Biotropica 11 161–172. 10.2307/2388036 [DOI] [Google Scholar]
- Soreng R. J., Davidse G., Peterson P. M., Zuloaga F. O., Judziewicz E. J., Filgueiras T. S. (2014). A worldwide phylogenetic classification of Poaceae (Gramineae). J. Syst. Evol. 53 117–137. 10.1111/jse.12150 [DOI] [Google Scholar]
- Sorrells M. E. (2005). Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23 48–55. 10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Tang D. Q., Lu J. J., Fang W., Zhang S., Zhou M. B. (2010). Development, characterization and utilization of genbank microsatellite markers in Phyllostachys pubescens and related species. Molecular Breed. 25 299–311. 10.1007/s11032-009-9333-4 [DOI] [Google Scholar]
- Thiel T., Michalek W., Varshney R. K., Graner A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106 411–422. 10.1007/s00122-002-1031-0 [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Thiel T., Stein N., Langridge P., Graner A. (2002). In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell. Mol. Biol. Lett. 7 537–546. [PubMed] [Google Scholar]
- Wang J., Ge Q., Li J., Gao H., Li J., Zhao F. (2018). Identification of novel EST-SSR markers by transcriptome sequencing in ridgetail white prawn Exopalaemon carinicauda. Genes Genomics 40 207–215. 10.1007/s13258-017-0571-2 [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Song X. M., Li Y., Hou X. L. (2013). In-silico detection of EST-SSR markers in three Brassica species and transferability in B. rapa. J. Hortic. Sci. Biotech. 88 135–140. 10.1080/14620316.2013.11512947 [DOI] [Google Scholar]
- Wang Z., Fang B., Chen J., Zhang X., Luo Z., Huang L., et al. (2010). De novo assembly and characterization of root transcriptome using illumina paired-end sequencing and development of cSSR markers in sweetpotato ( Ipomoea batatas). BMC Genomics 11:726. 10.1186/1471-2164-11-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. Y., Raven P. H. (2006). Flora of China. Vol 22 (Poaceae). Louis: Science press (Beijing) and mbg Press St. [Google Scholar]
- You Y. N., Liu D. C., Liu H. B., Zheng X. F., Diao Y., Huang X. F., et al. (2015). Development and characterisation of EST-SSR markers by transcriptome sequencing in taro (Colocasia esculenta (L.) Schoot). Molecular Breed. 35:134 10.1007/s11032-015-0307-4 [DOI] [Google Scholar]
- Zhai L., Liu L., Zhu X., Xu L., Jiang L., Gong Y. (2013). Development, characterization and application of novel expressed sequence tag-simple sequence repeat (EST-SSR) markers in radish (Raphanus sativus L.). J. Bio-technol. 12 921–935. [Google Scholar]
- Zhai L., Xu L., Wang Y., Cheng H., Chen Y., Gong Y., et al. (2014). Novel and useful genic-SSR markers from de novo transcriptome sequencing of radish (Raphanus sativus L.). Molecular Breed. 33 611–624. 10.1007/s11032-013-9978-x [DOI] [Google Scholar]
- Zhang X. Z., Zeng C. X., Ma P. F., Haevermans T., Zhang Y. X., Zhang L. N., et al. (2016). Multi-locus plastid phylogenetic biogeography supports the Asian hypothesis of the temperate woody bamboos (Poaceae: Bambusoideae). Mol. Phylogenet. Evol. 96 118–129. 10.1016/j.ympev.2015.11.025 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Wang Y. H., Shen S. K. (2017). De Novo Assembly of transcriptome and development of novel EST-SSR markers in rhododendron rex levl. through illumina Sequencing. Front. Plant Sci. 8:1664. 10.3389/fpls.2017.01664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. X., Zeng C. X., Li D. Z. (2012). Complex evolution in arundinarieae (Poaceae: Bambusoideae): incongruence between plastid and nuclear GBSSI gene phylogenies. Mol. Phylogenet. Evol. 63 777–797. 10.1016/j.ympev.2012.02.023 [DOI] [PubMed] [Google Scholar]
- Zhao H. S., Yang L., Peng Z. H., Sun H. Y., Yue X. H., Lou Y. F., et al. (2015). Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus phyllostachys. Sci. Rep. 5:8018. 10.1038/srep08018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Luo D., Ma L., Xie W., Wang Y., Wang Y., et al. (2016). Development and cross-species transferability of EST-SSR markers in Siberian wildrye (Elymus sibiricus L.) using Illumina sequencing. Sci. Rep. 6:20549. 10.1038/srep20549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. C., Raman H., Wu H., Lemerle D., Burrows G. E., Stanton R. (2012). Development of SSR markers for genetic analysis of silverleaf nightshade (Solanum elaeagnifolium) and related species. Plant. Mol. Biol. Rep. 31 248–254. 10.1007/s11105-012-0473-z [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency distribution of the EST-SSR repeat motifs in P. violascens.
The all SSR primer pairs of P. violascens.
The 54 primer pairs of P. violascens were applied in study.
The two-dimensional matrix of P. violascens varieties informative loci.
The two-dimensional matrix of Phyllostachys species informative loci.
The dendrogram of Phyllostachys species and P. violascens varieties based on EST-SSR markers of P. violascens.