Abstract
Congenital hypogonadotropic hypogonadism (CHH) is characterized by lack of normal pubertal development due to deficient gonadotropin-releasing hormone (GnRH) secretion or action, and is caused by genetic defects in several genes. Mutations in the CHD7 gene cause CHARGE syndrome (Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia and Ear abnormalities), but have also been found in patients with isolated CHH. The aim of this study was to identify CHD7 mutations in patients with CHH. Fifty Portuguese patients with CHH were screened for mutations in the CHD7 gene by DNA sequencing. Eight (16%) patients had CHD7 rare sequence variants that consisted of six missense (p.Gly388Glu, p.His903Pro, p.Thr1082Ile, p.Val1452Leu, p.Asp1854Gly, and p.Arg2065His) and two synonymous (p.Ser559Ser, and p.Ala2785Ala) mutations. Five of these mutations have never been reported before. Three CHD7 mutations occurred in patients that had mutations in additional CHH-genes. This study uncovered novel genetic variants that expand the known spectrum of mutations associated with CHH. The frequency of CHD7 mutations in this cohort was higher than that of other major CHH-genes and confirms the importance of including CHD7 in the genetic testing of CHH, even in the absence of additional CHARGE features.
Introduction
Congenital hypogonadotropic hypogonadism (CHH) is characterized by partial or complete lack of pubertal development, secondary to deficient gonadotropin-releasing hormone (GnRH) induced gonadotropin secretion1. The diagnosis is based on the existence of low levels of sex hormones associated with low or inappropriately normal levels of luteinizing hormone (LH) and follicle-stimulating hormone (FSH). CHH may occur associated with anosmia, a condition referred to as Kallmann syndrome (KS), or may occur without associated olfactory abnormalities, referred to as normosmic CHH (nCHH)1.
Monogenic or oligogenic defects are found in about 50% of patients with CHH, in genes that regulate the embryonic development or migration of GnRH neurons, or the synthesis, secretion or action of GnRH2. Mutations in the ANOS1 (KAL1) (MIM 300836), FGFR1 (KAL2) (MIM 136350) and GNRHR (MIM 138850) genes have been the most frequently reported, but there are now over 30 genes that have been associated with CHH. However, most of these genes are rarely involved in CHH and even the most commonly implicated genes usually account for less than 10% of cases2.
CHARGE syndrome is a rare disorder characterized by a variable combination of congenital anomalies that include Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia and Ear abnormalities3. Heterozygous loss-of-function mutations in the chromodomain helicase DNA-binding protein 7 (CHD7) gene (MIM 608892) are the major cause of CHARGE syndrome4. Hypogonadotropic hypogonadism and olfactory defects, which are the hallmarks of KS, are commonly observed in CHARGE syndrome5 and CHD7 mutations have been identified in patients with isolated CHH (i.e. without additional CHARGE features)6–11.
The aim of this study was to determine the frequency of CHD7 mutations in a cohort of patients with isolated CHH.
Material and Methods
Subjects
The study comprised 50 unrelated Portuguese patients with CHH (42 men and 8 women, mean age at diagnosis 21.4 years, range 14–45 years), 22 with KS and 28 with nCHH, recruited by Portuguese clinical endocrine centers. Inclusion criteria were patients with low or inappropriately normal serum FSH, LH and sex steroid levels, failure to enter spontaneous puberty by the age of 18 years or with medically induced puberty below this age, and absence of other pituitary hormone deficiencies. Olfactory function was assessed either by olfaction testing or self-reported by the patients, depending on the clinical center. None of the patients had diagnostic criteria for CHARGE syndrome12. In mutation-positive patients, additional family members were studied, when available. The control population consisted of >200 Portuguese unrelated healthy volunteers who were recruited among blood donors. The study was approved by the local research ethics committee (Faculty of Health Sciences, University of Beira Interior, Ref: CE-FCS-2012-012). Written informed consent was obtained from all subjects and all methods were performed in accordance with the relevant guidelines and regulations.
Genetic studies
Genomic deoxyribonucleic acid (DNA) was extracted from peripheral blood leucocytes using previously described methods13. Patients had already been screened for mutations in the ANOS1, FGFR1 and GNRHR genes, resulting in the discovery of three mutations in ANOS114, six mutations in FGFR115 and six mutations in GNRHR16. All 50 patients were subsequently screened for mutations in the CHD7 gene by polymerase chain reaction (PCR) amplification of the coding exons and exon-intron boundaries, and bi-directional sequencing using CEQ DTCS sequencing kit (Beckman Coulter, Fullerton, CA, USA) and an automated capillary DNA sequencer (GenomeLab TM GeXP, Genetic Analysis System, Beckman Coulter). Primer sequences for CHD7 were previously described by Song et al.17, except for the primer sequence of exons 2.4 and 3, which were designed using Primer 3 Plus18. Genomic sequence variants identified in patients were searched in the Exome Aggregation Consortium (ExAC) population variant database19, to assess their frequency in the general population. Variants found to be absent in the ExAC database or with frequencies <0.1% were further screened in a panel of at least 200 healthy Portuguese volunteers (400 alleles), using allele-specific PCR or sequence-specific restriction enzymes, to exclude the possibility that they represented common polymorphisms in the Portuguese population. Variants were considered to be pathogenic when they were simultaneously found to have an ExAC population frequency <0.1%, to be absent in the Portuguese control population, and to have a deleterious effect predicted by at least one of four bioinformatic programs (SIFT20, PolyPhen-221, Mutation Taster22 or Human Splicing Finder23). Sequence variant nomenclature followed standard guidelines24 and was based on the cDNA reference sequence for the CHD7 gene (GenBank accession NM_017780.3). Patients with pathogenic variants in CHD7 were further screened for mutations in additional genes related to the hypothalamic–pituitary–gonadal axis (PROKR2, PROK2, FGF8, GNRH1, KISS1R, TAC3 and TACR3), to identify possible cases of oligogenicity (primer sequences and PCR conditions are available upon request).
Results
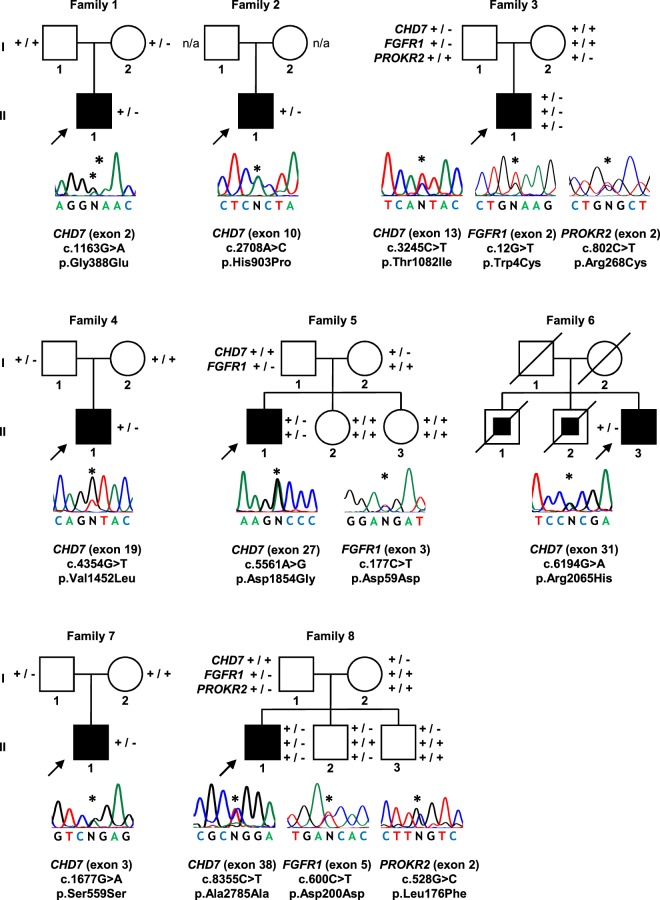
Sequence analysis of the entire coding region and exon–intron boundaries of CHD7 revealed 13 heterozygous variants that had frequencies <0.1% in the ExAC population database. Five of these variants were detected in normal Portuguese controls (p.Thr689Thr, 2 of 408 alleles; p.Lys729Glu, 1 of 436 alleles; c.2613 + 4 C > T, 1 of 454 alleles; p.Gly1479Gly, 1 of 398 alleles; p.Pro2072Pro, 1 of 450 alleles) and were excluded from further analysis. The remaining eight variants were found to be absent in the Portuguese control population and were predicted to cause changes in protein function, by at least one bioinformatic prediction program (Table 1). Thus, 16% (8 out of 50) of patients with CHH were considered to have pathogenic variants in CHD7. Mutations occurred in both KS (4/22, 18%) and nCHH (4/28, 14%). Mutations consisted of six missense (c.1163 G > A, p.Gly388Glu; c.2708 A > C, p.His903Pro; c.3245 C > T, p.Thr1082Ile; c.4354 G > T, p.Val1452Leu; c.5561 A > G, p.Asp1854Gly; c.6194 G > A, p.Arg2065His) and two synonymous (c.1677G > A, p.Ser559Ser; c.8355 C > T, p.Ala2785Ala) variants (Fig. 1). Conservation analysis revealed that, with the exception of p.Gly388Glu, all missense variants occurred at amino acids that were highly conserved across species (Supplemental Table 1).
Table 1.
Rare sequence variants predicted to be pathogenic by at least one computational program.
| CHD7 variant | Population allele frequency (ExAC/Portuguese)(a) | Computational programs that support a pathogenic effect(b) | Additional genetic variants |
|---|---|---|---|
| Missense | |||
| c.1163 G > A; p.Gly388Glu | 0.0008%/0% | SIFT, PP2, MT | |
| c.2708 A > C; p.His903Pro | 0%/0% | SIFT, PP2, MT | |
| c.3245 C > T; p.Thr1082Ile | 0%/0% | SIFT, PP2, MT | FGFR1 c.12 G > T and PROKR2 c.802 C > T(c) |
| c.4354 G > T; p.Val1452Leu | 0%/0% | MT | |
| c.5561 A > G; p.Asp1854Gly | 0%/0% | SIFT, PP2, MT | FGFR1 c.177 C > T |
| c.6194 G > A; p.Arg2065His | 0%/0% | SIFT, PP2, MT | |
| Synonymous | |||
| c.1677G > A; p.Ser559Ser | 0.0036%/0% | MT, HSF | |
| c.8355 C > T; p.Ala2785Ala | 0.0074%/0% | MT, HSF | FGFR1 c.600 C > T and PROKR2 c.528 G > C |
(a)ExAC Exome Aggregation Consortium frequency/Portuguese control population. (b)SIFT, Sorting Tolerant From Intolerant; PP2, PolyPhen-2; MT, Mutation Taster; HSF, Human Splicing Finder. (c)Patient previously reported by Gonçalves et al.15.
Figure 1.
CHD7 mutations identified in patients with CHH. Arrows represent index cases, filled symbols represent affected individuals, open symbols represent unaffected individuals, squares denote men, circles denote women, oblique lines through symbols represent deceased individuals. Filled squares within squares represent individuals reported to have heart defects associated with polydactyly. Patients from families 3, 5 and 8 had mutations in additional genes, thus representing cases of oligogenicity. Genotypes for additional family members, when available, are presented beside each individual (+, wild-type allele; −, mutated allele; n/a, not available for genetic studies). The position of each heterozygous mutation on the DNA sequence is indicated by an asterisk.
The p.Ser559Ser, p.Arg2065His and p.Ala2785Ala variants, have been previously identified in patients with CHARGE syndrome25. The remaining five variants have not yet been reported in patients with either CHH or CHARGE syndrome.
In six of the eight families, it was possible to determine the parental origin of the CHD7 mutation and in all six cases the mutation was transmitted by a parent who did not have CHARGE symptoms or a history of delayed puberty (Fig. 1). In three of these cases, patients presented mutations in additional CHH genes, namely one digenic mutation (CHD7/FGFR1) and two trigenic mutations (CHD7/FGFR1/PROKR2), thus representing cases of oligogenic inheritance (Fig. 1).
All CHD7 variants were submitted to the CHD7 mutation database at www.chd7.org.
The clinical characteristics of patients with identified CHD7 pathogenic variants are summarized in Table 2.
Table 2.
Clinical characteristics of patients with CHD7 rare sequence variants.
| Patient | Sex | Age of diagnosis (yrs) | Clinical presentation | Olfaction | Associated features | Basal hormone levels (laboratory normal reference range) | Brain MRI/CT | Sequence variants (heterozygous) |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 14 | Micropenis, cryptorchidism and delayed puberty | Anosmia | — | FSH 0.29 mIU/mL (1.1–13.6); LH < 0.07 mIU/mL (1.1–8.8); total T 0.34 ng/mL (2.8–11.1) | Pituitary hypoplasia | CHD7 c.1163 G > A, p.(Gly388Glu) |
| 2 | M | 14 | Micropenis and delayed puberty | Normal | — | n/a | Normal | CHD7 c.2708 A > C, p.(His903Pro) |
| 3 | M | 19 | Arrested puberty | Normal | Osteoporosis and osteopenia (lumbar spine Z-score −2.6; femoral neck Z-score −2.1). | FSH 2.6 mIU/mL (1.0–12.0); LH 2.0 mIU/mL (1.0–7.0); total T 0.32 ng/mL (2.60–10.00) | Normal | CHD7 c.3245 C > T, p.(Thr1082Ile) and FGFR1 c.12 G > T, p.(Trp4Cys) and PROKR2 c.802 C > T, p.(Arg268Cys) |
| 4 | M | 21 | Delayed puberty | Anosmia | Reversal of hypogonadism after testosterone replacement therapy (age 33 yrs). | FSH 1.7 mIU/mL (1.5–12.0); LH 0.9 mIU/mL (1.0–8.5); total T 0.7 ng/mL (2.6–16.0) | Normal | CHD7 c.4354 G > T, p.(Val1452Leu) |
| 5 | M | 17 | Micropenis, cryptorchidism and delayed puberty | Anosmia | Mild hearing deficit. | FSH < 0.3 mIU/mL (1.4–10.0); LH < 0.07 mIU/mL (1.5–9.3); total T < 0.2 ng/mL (2.4–8.3) | Normal | CHD7 c.5561 A > G, p.(Asp1854Gly) and FGFR1 c.177 C > T, p.(Asp59Asp) |
| 6 | M | 16 | Cryptorchidism and delayed puberty | Anosmia | Recurrent pyelonephritis. Renal transplant due to chronic renal failure (age 43 yrs). | FSH 3.4 mIU/mL (5.0–25.0); LH 1.9 mIU/mL (5.0–30.0); total T 0.87 ng/mL (2.50–7.50) | Normal | CHD7 c.6194 G > A, p.(Arg2065His) |
| 7 | M | 15 | Micropenis and delayed puberty | Normal | Psicomotor developmental delay. Strabismus. Surgery for deviated nasal septum and hypertrophy. Reversal of hypogonadism after testosterone replacement therapy (age 18 yrs). | FSH 0.71 mIU/mL (0.8–8.2); LH < 0.07 mIU/mL (0.7–7.2); total T 0.22 ng/mL (2.20–8.00) | Normal | CHD7 c.1677G > A, p.(Ser559Ser) |
| 8 | M | 17 | Delayed puberty | Normal | — | FSH 0.3 mIU/mL (0.8–8.2); LH < 0.1 mIU/mL (0.7–7.2); total T 0.05 ng/mL (2.20–8.00) | Pituitary hypoplasia | CHD7 c.8355 C > T, p.(Ala2785Ala) and FGFR1 c.600 C > T, p.(Asp200Asp) and PROKR2 c.528 G > C, p.(Leu176Phe) |
M, male; yrs, years; FSH, follicle stimulating hormone; LH, luteinizing hormone; T, testosterone; MRI, magnetic resonance imaging; CT, computerized tomography; n/a, not available. Hormone levels are only presented for gonadotrophins and testosterone, all other pituitary hormone measurements were normal.
Discussion
The overall prevalence of CHD7 mutations in this cohort of Portuguese patients with CHH was 16% (8 out of 50), with a similar distribution between the KS and nCHH forms. This is a high prevalence when compared to the contribution of other genes that have been historically considered as priorities in the genetic study of CHH, namely the ANOS1 (KAL1), FGFR1, and GNRHR genes26. Indeed, previous studies in the Portuguese population have shown that the ANOS1 (KAL1), FGFR1, and GNRHR genes are mutated in only 7.1%, 12.0%, and 12.5% of cases, respectively14–16. Studies in other populations have also shown an important contribution of CHD7 mutations in the aetiology of CHH, with frequencies ranging from 5.2% to 19.0%, of cases6–11. Thus, the CHD7 gene is becoming increasingly recognised as one of the most commonly mutated genes in CHH.
Although CHD7 mutations can cause both CHARGE syndrome and isolated CHH, it is likely that this variable phenotypic expression is related to the severity of the CHD7 mutations, as mutations in CHARGE syndrome are typically highly deleterious protein-truncating mutations, whereas CHD7 mutations in isolated CHH are typically missense8. Accordingly, in our CHH patients, all identified CHD7 mutations were either missense (n = 6) or synonymous (n = 2).
The eight CHD7 mutations identified in this study were found to be absent or very rare (<0.1%) in the ExAC population database19, absent in Portuguese controls, and predicted to be damaging by structure- and sequence-based bioinformatics programs20–23. Three of these mutations (one missense and two synonymous) have already been reported in patients with CHARGE syndrome25, and the remaining five missense mutations have not yet been reported in patients with either CHH or CHARGE syndrome. The missense mutations occurred at highly conserved amino acids. Furthermore, the p.His903Pro, p.Thr1082Ile and p.Val1452Leu mutations are located in chromodomain 2, the SNF2 domain and the helicase domain, respectively, which play important roles in the chromatin remodelling activity of the CHD7 protein6. The remaining missense mutations occurred outside these catalytic domains but were also predicted to affect protein function. The mechanisms by which the synonymous mutations exert their effect were not possible to determine. However, synonymous mutations - sometimes called ‘silent’ mutations - are widely acknowledged to be able to cause changes in protein expression, conformation and function, and have been implicated in several diseases27. It should be noted however that according to more stringent classification criteria recommended by the American College of Medical Genetics and Genomics (ACMG)28, only the p.Thr1082Ile variant is considered “likely pathogenic” and the remaining are considered “variants of uncertain significance”.
Three patients with CHD7 mutations presented additional defects in known CHH-genes. Thus, these represent cases of oligogenic inheritance, which is a frequent genetic finding in CHH29. Oligogenicity might explain some cases of incomplete penetrance, where carriers of a CHD7 mutation only express CHH in the presence of other mutated genes.
Our patients with CHD7 mutations did not undergo detailed radiological investigations to detect hypoplasia/agenesis of semicircular canals or of olfactory bulbs and tracts, which are currently major criteria for CHARGE syndrome30,31. Thus, we cannot exclude that patients have light forms of CHARGE syndrome. Other studies have shown that a subset of patients with apparently isolated CHH, in whom a CHD7 defect was demonstrated, were subsequently found to exhibit multiple CHARGE features and reclassified as having CHARGE syndrome32–34. Interestingly, three of our patients with CHD7 mutations had minor CHARGE features30,31, namely hearing deficit, renal anomalies and intellectual disability, respectively, but insufficient for a clinical diagnosis of CHARGE syndrome. It remains to be determined if these were coincidental findings or the result of the CHD7 mutations.
Our results should be viewed with caution as in vitro and in vivo functional tests are not readily available for CHD735, and therefore it was not possible to confirm the damaging effect of the observed CHD7 variants. In addition, a limited number of CHH-genes was analysed by Sanger sequencing and it remains to be determined if a more comprehensive genetic analysis (e.g. through whole exome sequencing) would uncover additional cases of oligogenicity and explain the incomplete penetrance observed in some families. Finally, CHH is clinically heterogeneous and sometimes overlaps with constitutional delay of growth and puberty (CDGP)36. Our study included patients with a wide range of ages and we cannot exclude the possibility that some of the younger patients, who underwent medically induced puberty, may represent cases of CDGP or mild forms of CHH that would have eventually developed spontaneous late puberty. Therefore, our results may not be directly comparable with those of other studies that used different inclusion criteria.
In conclusion, our study identified a high frequency of CHD7 mutations in patients with CHH and uncovered novel genetic variants that expand the known spectrum of mutations associated with CHH.
Supplementary information
Acknowledgements
This work was supported by the Portuguese Foundation for Science and Technology (PTDC/SAU-GMG/098419/2008) and by “Programa Operacional do Centro, Centro 2020” through the funding of the ICON project (Interdisciplinary Challenges On Neurodegeneration; CENTRO-01-0145-FEDER-000013)”. The authors are grateful to the following clinicians who contributed with patient samples and data: Ana Saavedra (Porto), Ana Varela (Porto), António Garrão (Lisboa), Carla Baptista (Coimbra), Carla Meireles (Guimarães), Carolina Moreno (Coimbra), Catarina Limbert (Lisboa) Cíntia Correia (Porto), Cláudia Nogueira (Porto), Duarte Pignatelli (Porto), Eduardo Vinha (Porto), Filipe Cunha (Porto), Francisco Carrilho (Coimbra), Luísa Barros (Coimbra), Luísa Cortez (Lisboa), Margarida Bastos (Coimbra), Maria João Oliveira (Porto), Mariana Martinho (Penafiel), Miguel Melo (Coimbra), Nuno Vicente (Coimbra), Patrícia Oliveira (Coimbra), Paula Freitas (Porto), Raquel Martins (Porto), Selma Souto (Porto), Susana Gama (Famalicão), and Teresa Martins (Coimbra).
Author Contributions
C.I.G., J.M.O. and M.C.L. conceived and designed the study. C.I.G. and F.M.P. performed the genetic studies of the patients. J.M.A., D.C., F.F., S.M., O.M. and B.D.P. collected samples and acquired clinical data of the patients with mutations. C.I.G. and M.C.L. drafted the article and all authors revised it critically for important intellectual content and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38178-y.
References
- 1.Boehm U, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism–pathogenesis, diagnosis and treatment. Nat Rev Endocrinol. 2015;11:547–564. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 2.Kim SH. Congenital Hypogonadotropic Hypogonadism and Kallmann Syndrome: Past, Present, and Future. Endocrinology and metabolism. 2015;30:456–466. doi: 10.3803/EnM.2015.30.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pagon RA, Graham JM, Jr., Zonana J, Yong SL. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. The Journal of pediatrics. 1981;99:223–227. doi: 10.1016/S0022-3476(81)80454-4. [DOI] [PubMed] [Google Scholar]
- 4.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian R, Crowley WF., Jr. Reproductive endocrine phenotypes relating to CHD7 mutations in humans. Am J Med Genet C Semin Med Genet. 2017;175:507–515. doi: 10.1002/ajmg.c.31585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HG, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramanian R, et al. Functionally compromised CHD7 alleles in patients with isolated GnRH deficiency. Proc Natl Acad Sci USA. 2014;111:17953–17958. doi: 10.1073/pnas.1417438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcos S, et al. The prevalence of CHD7 missense versus truncating mutations is higher in patients with Kallmann syndrome than in typical CHARGE patients. J Clin Endocrinol Metab. 2014;99:E2138–2143. doi: 10.1210/jc.2014-2110. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Gong C, Qin M, Liu Y, Tian Y. Clinical and genetic features of 64 young male paediatric patients with congenital hypogonadotropic hypogonadism. Clin Endocrinol (Oxf) 2017;87:757–766. doi: 10.1111/cen.13451. [DOI] [PubMed] [Google Scholar]
- 10.Ayers KL, et al. Variants in congenital hypogonadotrophic hypogonadism genes identified in an Indonesian cohort of 46,XY under-virilised boys. Hum Genomics. 2017;11:1. doi: 10.1186/s40246-017-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyama K, et al. Molecular genetic and clinical delineation of 22 patients with congenital hypogonadotropic hypogonadism. J Pediatr Endocrinol Metab. 2017;30:1111–1118. doi: 10.1515/jpem-2017-0035. [DOI] [PubMed] [Google Scholar]
- 12.Hefner MA, Fassi E. Genetic counseling in CHARGE syndrome: Diagnostic evaluation through follow up. Am J Med Genet C Semin Med Genet. 2017;175:407–416. doi: 10.1002/ajmg.c.31589. [DOI] [PubMed] [Google Scholar]
- 13.Lemos MC, Regateiro FJ. N-acetyltransferase genotypes in the Portuguese population. Pharmacogenetics. 1998;8:561–564. doi: 10.1097/00008571-199812000-00013. [DOI] [PubMed] [Google Scholar]
- 14.Goncalves CI, Fonseca F, Borges T, Cunha F, Lemos MC. Expanding the genetic spectrum of ANOS1 mutations in patients with congenital hypogonadotropic hypogonadism. Hum Reprod. 2017;32:704–711. doi: 10.1093/humrep/dew354. [DOI] [PubMed] [Google Scholar]
- 15.Goncalves C, et al. Novel FGFR1 mutations in Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism: evidence for the involvement of an alternatively spliced isoform. Fertil Steril. 2015;104:1261–1267 e1261. doi: 10.1016/j.fertnstert.2015.07.1142. [DOI] [PubMed] [Google Scholar]
- 16.Goncalves CI, et al. GNRHR biallelic and digenic mutations in patients with normosmic congenital hypogonadotropic hypogonadism. Endocr Connect. 2017;6:360–366. doi: 10.1530/EC-17-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song MH, et al. CHD7 mutational analysis and clinical considerations for auditory rehabilitation in deaf patients with CHARGE syndrome. PLoS One. 2011;6:e24511. doi: 10.1371/journal.pone.0024511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ExAC. Exome Aggregation Consortium. http://exac.broadinstitute.org (2018).
- 20.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 23.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 25.Bartels CF, Scacheri C, White L, Scacheri PC, Bale S. Mutations in the CHD7 gene: the experience of a commercial laboratory. Genet Test Mol Biomarkers. 2010;14:881–891. doi: 10.1089/gtmb.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semple RK, Topaloglu AK. The recent genetics of hypogonadotrophic hypogonadism - novel insights and new questions. Clin Endocrinol (Oxf) 2010;72:427–435. doi: 10.1111/j.1365-2265.2009.03687.x. [DOI] [PubMed] [Google Scholar]
- 27.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 28.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sykiotis GP, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci USA. 2010;107:15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hale CL, Niederriter AN, Green GE, Martin DM. Atypical phenotypes associated with pathogenic CHD7 variants and a proposal for broadening CHARGE syndrome clinical diagnostic criteria. American journal of medical genetics. Part A. 2016;170A:344–354. doi: 10.1002/ajmg.a.37435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legendre M, et al. Phenotype and genotype analysis of a French cohort of 119 patients with CHARGE syndrome. Am J Med Genet C Semin Med Genet. 2017;175:417–430. doi: 10.1002/ajmg.c.31591. [DOI] [PubMed] [Google Scholar]
- 32.Xu C, et al. Evaluating CHARGE syndrome in congenital hypogonadotropic hypogonadism patients harboring CHD7 variants. Genet Med. 2018;20:872–881. doi: 10.1038/gim.2017.197. [DOI] [PubMed] [Google Scholar]
- 33.Jongmans MC, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome–the clinical overlap with CHARGE syndrome. Clin Genet. 2009;75:65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergman JE, et al. The results of CHD7 analysis in clinically well-characterized patients with Kallmann syndrome. J Clin Endocrinol Metab. 2012;97:E858–862. doi: 10.1210/jc.2011-2652. [DOI] [PubMed] [Google Scholar]
- 35.van Ravenswaaij-Arts C, Martin DM. New insights and advances in CHARGE syndrome: Diagnosis, etiologies, treatments, and research discoveries. Am J Med Genet C Semin Med Genet. 2017;175:397–406. doi: 10.1002/ajmg.c.31592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassatella D, et al. Congenital hypogonadotropic hypogonadism and constitutional delay of growth and puberty have distinct genetic architectures. Eur J Endocrinol. 2018;178:377–388. doi: 10.1530/EJE-17-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.