Abstract
The mutational spectrum of deafness in Indochina Peninsula, including Vietnam, remains mostly undetermined. This significantly hampers the progress toward establishing an effective genetic screening method and early customized rehabilitation modalities for hearing loss. In this study, we evaluated the genetic profile of severe-to-profound hearing loss in a Vietnamese pediatric population using a hierarchical genetic analysis protocol that screened 11 known deafness-causing variants, followed by massively parallel sequencing targeting 129 deafness-associated genes. Eighty-seven children with isolated severe-to-profound non-syndromic hearing loss without family history were included. The overall molecular diagnostic yield was estimated to be 31.7%. The mutational spectrum for severe-to-profound non-syndromic hearing loss in our Vietnamese population was unique: The most prevalent variants resided in the MYO15A gene (7.2%), followed by GJB2 (6.9%), MYO7A (5.5%), SLC26A4 (4.6%), TMC1 (1.8%), ESPN (1.8%), POU3F4 (1.8%), MYH14 (1.8%), EYA1 (1.8%), and MR-RNR1 (1.1%). The unique spectrum of causative genes in the Vietnamese deaf population was similar to that in the southern Chinese deaf population. It is our hope that the mutation spectrum provided here could aid in establishing an efficient protocol for genetic analysis of severe-to-profound hearing loss and a customized screening kit for the Vietnamese population.
Introduction
The prevalence of infants born with moderate to severe bilateral hearing impairment is one in 1000, and the prevalence of infants with profound hearing loss, which causes severe speech impairment requiring cochlear implantation, is 4 in 10,0001. It has been shown that genetic causes account for about half of all known causes of these congenital hearing loss cases2,3. Hence, understanding the genetic etiology of hearing loss could aid practitioners to get a better understanding of the progression and risk of hearing loss in children, ultimately leading to the development of an optimal intervention strategy4–6. Most children with hearing loss were born to parents with normal hearing7, and genetic counseling for auditory rehabilitation and future family planning might be needed.
However, molecular genetic diagnosis of congenital hearing loss is not always feasible. Deafness is an extremely heterogeneous disorder; to date, more than 400 various types of syndromic hearing loss as well as more than 163 chromosomal loci and 115 genes of non-syndromic hearing loss (NSHL) have been identified (http://hereditaryhearingloss.org as of Apr 2018). It has been reported that not only are there numerous genetic variants responsible for hearing loss, but also the prevalence of the genetic variants is variable according to ethnicity and country of the study population. GJB2 variants are the most common cause of NSHL, and its detection rate differs depending on the ethnicity and country of origin; 20% in Caucasians of European descent8, 43% in Israelis9, 21~27% in Japan and Chinese10–12, 17% in Tunisians13, 17% in Iranian14, 14% in Australians15, and 10% in Koreans3,16. Furthermore, the GJB2 mutations were rarely found in African patients (0~1.0%)17–19. As such, the prevalence of GJB2 variants in hearing loss varied widely across different regions even within a single country. According to several genetic studies performed in Iran20, the prevalence of GJB2 varied from 0% to 38.3%, depending on the region. This diversity reflects the ethnic footprint of the Iranian population from neighboring countries. SLC26A4 (4.8~18%), MYO15A (9.6%), and MYO7A (~5%) were other prevalent causative genes in the Iranian population with NSHL20. In East Asian countries, mutations of SLC26A4 (10~13%) and mitochondrial DNA (1.8~2.1%) were frequently found following the mutation of GJB212,21. The mutations of GJB2, USH2A, MYO15A, and STRC genes were frequently found in the European population with autosomal recessive hearing loss22.
Knowing the mutational spectrum of deafness in a certain population offers a background data for the establishment of molecular genetic screening protocol for that population. The mutational spectrum of hearing loss in the population of Indochina Peninsula has not been studied yet; to the best of our knowledge, only information regarding the prevalence of GJB2 variants in Thailand is available23,24. Vietnam is the easternmost country on the Indochina Peninsula, sharing the region with Thailand, Laos, and Cambodia. In this study, we utilized the hierarchical genetic screening protocol that proved to be effective in East Asian populations to evaluate the genetic profile of severe-to-profound NSHL in the Vietnamese population, which in turn could provide the basis of health policy and medical processes for the treatment of hearing loss in the region.
Results
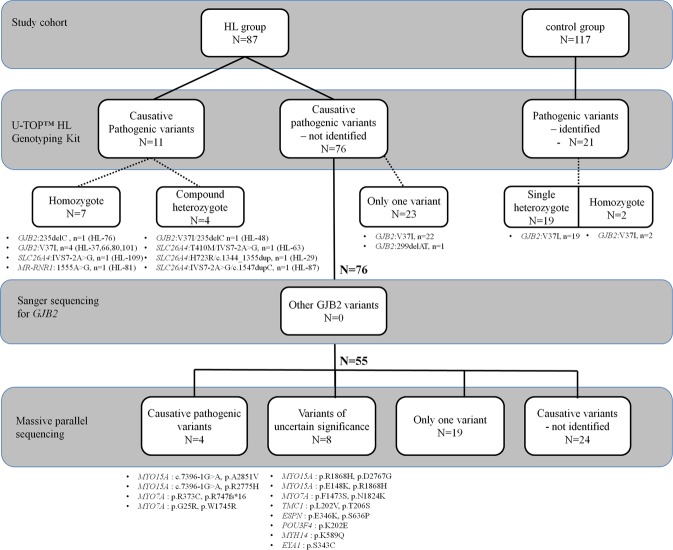
Three step protocol for genetic analysis
To evaluate the genetic profile of our Vietnamese population for severe-to-profound NSHL, we utilized a three-step genetic analysis protocol (Fig. 1). The first step was to screen for the 11 known deafness-causing variants using the U-TOPTM HL Genotyping Kit. The second step was to perform Sanger sequencing of all the coding regions of GJB2 in those who remained undiagnosed after the first step (N = 76). Finally, the third step involved massive parallel sequencing (MPS) in a subset of affected subjects (N = 55) that remained undiagnosed after the former two steps.
Figure 1.
Overview of three-step protocol for genetic analysis. Our entire cohort comprises eighty-seven patients with severe-to-profound non-syndromic hearing loss (HL group) and 117 normal participants (control group). In the first step, a total of eleven subjects are identified to have causative pathogenic mutations. The second step of Sanger sequencing for GJB2 does not additionally elucidate other GJB2 pathogenic variants than the ones already screened through the first step. Next, massive parallel sequencing is performed for 55 patients of the HL group. Four patients with causative (likely) pathogenic variants and eight patients with variants of uncertain significance are identified.
Among the 87 subjects segregating the severe-to-profound NSHL, complete molecular diagnosis was made on 11 subjects (N = 11/87, 12.6%) through the first screening step using the U_TOP HL genotyping kit (Fig. 1). These eleven subjects were divided into two groups. The first group (N = 9) included those with causative variants detected by the screening genotyping kit—c.235delC of GJB2 homozygotes (N = 1), p.V37I of GJB2 homozygotes (N = 4), p.V37I/c.235delC of GJB compound heterozygote (N = 1), c.IVS7-2A > G of SLC26A4 homozygotes (N = 1), p.T410M/c.IVS7-2A > G of SLC26A4 compound heterozygote (N = 1), and c.1555 A > G of MR-RNR1 (N = 1) (Fig. 1). The second group (N = 2) included those who underwent Sanger sequencing of SLC26A4 for further analysis—two compound heterozygotes of SLC26A4:p.H723R/c.1344_1355dup and c.IVS7-2A > G/c.1547dupC (Fig. 1). To address the pathogenic potential of p.V37I in this population, the Vietnamese control group with normal hearing was also screened using U-TOPTM HL Genotyping Kit, with a special focus on p.V37I. A total of 19 single heterozygotes of p.V37I of GJB2 and two homozygotes of p.V37I of GJB2 were identified from the control group with normal hearing (N = 117) (Figs 1 and S1). Resultantly, the frequency of p.V37I homozygote in the HL group (N = 4/87, 4.6%) was higher than that of the ethnicity-matched control group (N = 2/117, 1.7%), although the difference was not statistically significant (p = 0.154).
All the remaining 76 subjects underwent the second step – Sanger sequencing of GJB2 – to exclude other pathogenic variants, if any, elsewhere in GJB2 (Fig. 1). Single heterozygotes of p.V37I allele (N = 22) and single heterozygote of c.299delAT (N = 1) of GJB2, with no other pathogenic variants discovered elsewhere in GJB2 through GJB2 Sanger sequencing, were considered undiagnosed in the HL group (Fig. 1). None of these subjects carried the known non-coding region variants of GJB2 or the known large deletions of GJB6.
To further elucidate the causative variants in subjects who remained undiagnosed after the first two steps, the third step – MPS targeting the genes related to hearing loss – was performed (Fig. 1). Among the 55 subjects, four (7.3%) subjects turned out to have ‘causative pathogenic variants’ involving two genes (MYO15A, MYO7A). (Fig. 1 and Table 1). Meanwhile, eight subjects (N = 8/55, 14.5%) with severe-to-profound NSHL were classified as the group with variants of uncertain significance. Five subjects with variants in MYO15A, MYO7A, TMC1, and ESPN genes, which were inherited in autosomal recessive mode, were identified. In addition, one boy (HL-78) with a single novel variant of uncertain significance in POU3F4 with X-linked inheritance pattern and two subjects with a single variant in MYH14 and EYA1 genes with autosomal dominant inheritance pattern were identified (Fig. 1 and Table 1). In contrast, 19 subjects had only one variant with pathogenic potential or uncertain significance (34.5%) in the autosomal recessive deafness genes (see Supplementary Table S1) without reaching a final molecular genetic diagnosis, and the other 24 subjects (43.6%) did not carry any potentially pathogenic variant as documented by MPS (Fig. 1). In this study, the overall molecular diagnostic rate through our three-step protocol in our Vietnamese population with severe-to-profound NSHL was estimated to be 31.7% when the subjects with variants of uncertain significance were included and 19.0% when they were not.
Table 1.
Details of twelve deaf subjects having causative pathogenic variants or variants of uncertain significance after massive parallel sequencing targeting the genes related with hearing loss.
| Gene (GeneBank No.) | Family ID | Variant | Classification of variants | State | Depth (DP/AD) | Q call (Qual/MQ) | Prediction Algorithm | Conservation Score | MAF | Published reference (PMID) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation Taster | PolyPhen-2 | SIFT | PhyloP | GERP + + | ExAC, 1000 G | GnomAD* | ||||||||
| Causative (likely) pathogenic variants | ||||||||||||||
| MYO15A (NM_016239) | HL-92 | c.7396-1 G > A | LPa | Het | 61 | 60 | DC | NA | NA | 4.783 | 4.01 | A = 0.00002/1 (ExAC) | A = 0.0001 (2/14966) | This study |
| HL-92 | c.8552 C > T:p.Ala2851Val | Pa | Het | 80 | 60 | DC | PrD | D | 5.319 | 4.41 | T = 0.000008/1 (ExAC) | T = 0.000065, (1/15292) | This study | |
| HL-105 | c.7396-1 G > A | LPa | Het | 85 | 60 | DC | NA | NA | 4.783 | 4.01 | A = 0.00002/1 (ExAC) | A = 0.0001 (2/14966) | This study | |
| HL-105 | c.8324 G > A:p.Arg2775His | Pa | Het | 123 | 60 | DC | PrD | D | 5.89 | 5.1 | T = 0.000008/1 (ExAC) | T = 0.000 (0/245848) | 23767834, This study | |
| MYO7A (NM_000260) | HL-110 | c.1117 C > T:p.Arg373Cys | Pa | Het | 125 | 60 | DC | PrD | D | 4.201 | 5.11 | ND | ND | 22903915, This study |
| HL-110 | c.2239_2240delAG: p.Arg747fs*16 | Pa | Het | 65 | 60 | NA | NA | NA | 1.305_ 3.153 | 2.7 | ND | Del = 0.00002 (1/33480) | 22898263, This study | |
| HL-49 | c.73 G > A:p.Gly25Arg | Pa | Het | 392 | 60 | DC | PrD | D | 5.733 | 4.69 | A = 0.00002/2 (ExAC) | A = 0.000018 (2/110120) | 9002678, This study | |
| HL-49 | c.5233 T > G:p.Trp1745Arg | US | Het | 104 | 60 | DC | PrD | D | 1.028 | 5.13 | ND | ND | This study | |
| Variants of uncertain significance | ||||||||||||||
| MYO15A (NM_016239) | HL-108 | c.8300 A > G:p.Asp2767Gly | US | Het | 53 | 60 | DC | PrD | D | 4.864 | 5.1 | ND | ND | This study |
| HL-108 | c.5603 G > A:p.Arg1868His | US | Het | 62 | 60 | DC | PrD | T | 3.273 | 4.8 | A = 0.0001/15 (ExAC) A = 0.0004/2 (1000 G) | A = 0.0005 (11/18868) | This study | |
| HL-79 | c.442 G > A:p.Glu148Lys | US | Het | 195 | 60 | P | B | T | 1.087 | 5.25 | ND | ND | This study | |
| HL-79 | c.5603 G > A:p.Arg1868His | US | Het | 55 | 60 | DC | PrD | T | 1.048 | 4.8 | A = 0.0001/15 (ExAC) A = 0.0004/2 (1000 G) | A = 0.0005 (11/18868) | This study | |
| MYO7A (NM_000260) | HL-47 | c.4418 T > C:p.Phe1473Ser | US | Het | 257 | 60 | DC | PrD | D | 4.693 | 5.5 | ND | ND | This study |
| HL-47 | c.5472 C > G:p.Asn1824Lys | US | Het | 122 | 60 | DC | PrD | D | 1.98 | 3.49 | ND | ND | This study | |
| TMC1 (NM_138691) | HL-44 | c.604 C > G: p.Leu202Val | US | Het | 112 | 60 | DC | PrD | T | 1.871 | 5.78 | ND | ND | This study |
| HL-44 | c.616 A > T:p.Thr206Ser | US | Het | 110 | 60 | DC | PrD | T | 5.038 | 5.78 | ND | ND | This study | |
| ESPN (NM_031475) | HL-106 | c.1036 G > A:p.Glu346Lys | US | Het | 707 | 60 | DC | PrD | D | 4.523 | 3.77 | A = 0.00007/9 (ExAC) A = 0.0002/1 (1000 G) | A = 0.00009 (17/18870) | This study |
| HL-106 | c.1906T > C:p.Ser636Pro | US | Het | 23 | 24.8 | P | B | D | 1.289 | 5.1 | ND | ND | This study | |
| POU3F4 (NM_000307) | HL-78 | c.604 A > G:p.Lys202Glu | US | Hemi | 86 | 60 | D | PrD | NA | 4.676 | 5.31 | ND | ND | This study |
| MYH14 (NM_001145809) | HL-70 | c.1765A > C:p.Lys589Gln | US | Het | 270 | 60 | DC | PrD | D | 4.091 | 4.09 | C = 0.00002/2 (ExAC) | C = 0.000017, (2/111562) | This study |
| EYA1 (NM_000503) | HL-34 | c.1028 C > G: p.Ser343Cys | US | Het | (81/29) | (709/60) | DC | PrD | D | 5.842 | 5.86 | ND | ND | This study |
*Maximum minor allele frequency among all populations in gnomAD; DP, total depth; AD, alternative allele depth; Qual, SNP quality; MQ, mapping quality; LPa, likely pathogenic; Pa, pathogenic; US, uncertain significance; Het, Heterozygous; Hom, Homozygous; Hemi, Hemizygous; P, Polymorphism; DC, Disease causing; PrD, Probably damaging; PsD, Possibly damaging; D, Damaging; B, Benign; T, Tolerated; ND, not detected; NA, not applicable; PMID, PubMed ID (PMID is the unique identifier number used in PubMed.; PhyloP score from the Mutation Taster (http://www.mutationtaster.org/); in silico prediction Algorithm: Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml); SIFT (http://sift.jcvi.org/www/SIFT_chr_coords_submit.html); Conservation tools: GERP + + score in the UCSC Genome Browser (http://genome-asia.ucsc.edu/); ExAC, Exome Aggregation Consortium (http://exac.broadinstitute.org/); 1000 Genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/); GO-ESP, NHLBI GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS/); GnomAD, genome Aggregation Database (http://gnomad.broadinstitute.org/).
Mutation spectrum in Vietnamese population
The MYO15A variants were the most common cause of severe-to-profound NSHL in our Vietnamese population, and the prevalence of the MYO15A variants that were classified as (likely) pathogenic variants or uncertain significance was 7.2% (N = 4/55) (Table 2). Next, the autosomal recessive variants of GJB2 (6.9%, N = 6/87), MYO7A (5.5%, N = 3/55), and SLC26A4 (4.6%, N = 4/87) were identified as the next tier of causes in this population (Table 2). The frequency of variants in ESPN (1.8%, N = 1/55), TMC1 (1.8%, N = 1/55), POU3F4 (1.8%, N = 1/55), MYH14 (1.8%, N = 1/55), and MR-RNR1 (1.1%, N = 1/87) were less common in our Vietnamese population with severe or greater degree of HL.
Table 2.
Mutation spectrum of severe-to-profound non-syndromic hearing loss in Vietnamese pediatric population.
| Gene | Mode of inheritance | Causative pathogenic variants† | Variants of uncertain significance† | Prevalence (%) |
|---|---|---|---|---|
| MYO15A | AR | 2 (3.6%) | 2 (3.6%) | 7.2 |
| GJB2 | AR | 6 (6.9%)* | 6.9 | |
| MYO7A | AR | 2 (3.6%) | 1 (1.8%) | 5.5 |
| SLC26A4 | AR | 4 (4.6%) | 4.6 | |
| TMC1 | AR | 1 (1.8%) | 1.8 | |
| ESPN | AR | 1 (1.8%) | 1.8 | |
| POU3F4 | X-lined | 1 (1.8%) | 1.8 | |
| MYH14 | AD | 1 (1.8%) | 1.8 | |
| EYA1 | AD | 1 (1.8%) | 1.8 | |
| MR-RNR1 | mitochondrial | 1 (1.1%) | 1.1 |
†Number of diagnosis (prevalence, %); *, the homozygous p.V37I variants of GJB2 gene were included; AR, autosomal recessive; AD, autosomal dominant; No, number.
Discussion
In this study, we elucidated the mutation spectrum of severe-to-profound NSHL in the Vietnamese pediatric population for the first time. MYO15A, GJB2, MYO7A, and SLC26A4 variants were shown to be the leading causes in our cohort, and the estimated prevalence was 7.2%, 6.9%, 5.5%, and 4.6%, respectively. The variants of TMC1, ESPN, POU3F4, MYH14, EYA1, and MR-RNR1 genes followed those from the top tier deafness genes in the order of frequency as a molecular etiology of severe-to-profound NSHL in Vietnamese.
It may be worth noting that the mutation spectrum of this Vietnamese deaf population is unique and distinct. First, there was a significantly lower genetic load of GJB2 variants compared with other ethnic populations, including East Asians. The prevalence of the DFNB1 due to GJB2 variants in our Vietnamese pediatric population with severe-to-profound NSHL was calculated to be 6.9%, which could be further lowered to 2.3% if the p.V37I homozygotes were excluded. GJB2 variants are the leading genetic etiology of NSHL in many populations, even though the detection rate of GJB2 variants can significantly differ depending on the ethnicity and country. The prevalence of GJB2-associated hearing loss was quite variable, ranging from 0% in South Africa to 40~50% in Eastern Europe25. According to a nationwide genetic study about the GJB2 mutation spectrum in China, there was a significant difference in the detection rate of pathogenic GJB2 allele depending on the region within China, varying from 4% in Guangxi to 30.4% in Jiangsu, while the overall frequency of pathogenic GJB2 allele among moderate to severe hearing loss was 17.9%26. Interestingly, Guangxi, a city associated with a low prevalence of GJB2 variants, is in the southern part of China, bordering Vietnam. Vietnam is the easternmost country on the Indochina Peninsula, bordering China to the north, Laos to the northwest, Cambodia to the southwest, and Thailand across the Gulf of Thailand to the southwest. The population of Vietnam was approximately 94.6 million in 2016, and the ethnic group, “Kinh,” comprised 86% of the total population. Kinh has been known to originate from northern Vietnam. Recent phylogenetic studies revealed that the majority of Kinh originated from South China and a minority from Thailand and Indonesia27. Therefore, the low prevalence of detectable GJB2 variants could be viewed as the genetic similarity between the Kinh people and the people from southern parts of China. This suggests a close ethnic relationship between these groups. Low prevalence of detectable GJB2 variants does not necessarily indicate low prevalence of DFNB1 in these populations and might suggest the presence of unidentified variants, such as large genomic deletions residing somewhere in the DFNB1 locus, and thus affecting GJB2 expression and function. These variants would have evaded our current molecular diagnostic protocol. This hypothesis requires future confirmation. The genetic similarity with the southern parts of China can also be seen in the SLC26A4 mutation. Unlike the northern area of China, in which the prevalence of SLC26A4 mutation has been reported to range from 10 to 13%28–30, patients with severe-to-profound hearing from the Guangxi province showed limited prevalence of SLC26A4 variants (2.2%)31, similar to our Vietnamese cohort.
Second, the detection frequency of p.V37I in our Vietnamese population with severe-to-profound NSHL (18.4%) was significantly higher than that in East Asian populations (1~6.7%)26,32,33 and European populations (0.6%)25. This allele was also frequently detected in the population of Thailand, which is bordered by Vietnam (16.7%)34, questioning the causative role of this allele to severe to profound deafness in Vietnamese. Among the 23 subjects in the HL group with one p.V37I allele detected on the first screening step, four subjects had pathogenic or uncertain significance variants in other deafness genes unrelated to GJB2 (HL-63, SLC26A4:T410M/IVS7-2A > G; HL-49, MYO7A: p.G25R/ p.W1745R; HL-34, EYA1:p.S343C; HL-44, TMC1: p.L202V/p.T206S) (Table 1). The other 19 subjects showed either one (N = 10, HL-35, 46, 53, 57, 60, 72, 84, 86, 97, 99) or no (N = 9, HL-22, 27, 30, 33, 43, 67, 68, 69, 98) variant with pathogenic potential from other unrelated deafness genes (see Supplementary Table S1). Interpretation of causality between p.V37I homozygotes and severe-to-profound deafness warrants further caution because two homozygotes of p.V37I of GJB2 were also identified in the control group with normal hearing (Figs 1 and S1), and the prevalence of the homozygotes of p.V37I of GJB2 (1.7%) in the control group was not significantly different from that (4.6%) in the HL group (p = 0.154). However, the p.V37I variant has been previously reported to have a diverse clinical presentation, ranging from normal-to-profound hearing loss35,36, and the prevalence of p.V37I homozygote was, even if not statistically significant, higher in the HL group than in the control group (4.6% vs 1.7%) in our study. Given this, at least a subset of p.V37I homozygotes in the Vietnamese population can potentially cause severe-to-profound hearing loss. Both normal hearing from the p.V37I homozygotes in the control group and profound hearing loss from some p.V37I homozygotes in the HL group might be phenotypes of both extremes from the same genetic alteration, which is modulated by modifiers. To clarify the pathogenic role of p.V37I in severe-to-profound hearing loss, further study evaluating the influence of genetic modifiers on the phenotype of homozygous p.V37I variant or exploring the yet-to-be identified, pathogenic DFNB1 allele in trans with single heterozygous p.V37I, such as copy number variation (CNV)17,37–40, is warranted.
Third, MYO15A was shown to be the most frequent causative gene (7.2%) in our Vietnamese population, with a much higher prevalence than that in the European populations (3.1%)41. In fact, the MYO15A gene, which is a causative gene of DFNB3 (OMIM 600316) and has been known to be associated with moderate to severe hearing loss as well as profound hearing loss, is usually ranked as the third or fourth most common cause of DFNB deafness in many ethnicities42–45. The MYO7A variants also accounted for a significant proportion (5.5%) of severe-to-profound hearing loss in our Vietnamese population, which was higher than the estimates found in other studies42–44. MYO7A is located on 11p13.5, encoding the myosin 7 A protein. To date, there have been more than 280 different pathogenic or likely pathogenic mutations and 380 variants of uncertain significance reported from MYO7A (https://www.ncbi.nlm.nih.gov/clinvar as of Aug 2018), which have been associated with Usher syndrome 1B46, non-syndromic autosomal recessive hearing loss (DFNB2)47, and autosomal dominant hearing loss (DFNA11)48. Patients diagnosed with Usher syndrome type 1B having MYO7A variants typically show profound hearing loss at birth and progressive retinal degeneration49.
Another interesting finding is that the potentially pathogenic autosomal dominant variant of MYH14 was considered as a contributor to severe-to-profound hearing loss in our Vietnamese population. The MYH14 gene encodes myosin-14 protein and was reported to be an important causative gene for prelingual severe NSHL with autosomal dominant inheritance. This is unique considering the usual auditory phenotype of autosomal dominant deafness genes50. In our present study, we were unable to determine whether this MYH14 variant occurred de novo due to the lack of parental samples, even though auditory phenotypes of parents were allegedly normal. Therefore, we classified these as variants of uncertain significance. Nonetheless, the MYH14 variant was still considered the variant with pathogenic potential due to their extremely rare MAF in publicly available databases.
Currently, the diagnostic yield from the first two steps – U-TOPTM HL genotyping kit and Sanger sequencing of GJB2, respectively – does not seem high, only reaching 12.6% including the homozygous p.V37I variants of GJB2. The U-TOPTM HL genotyping kit, initially designed to screen East Asian subjects with hearing loss, does not appear to be suitable for the Vietnamese population with hearing loss, where MYO15A and MYO7A were the most frequent causative genes. Accordingly, an efficient screening kit customized for the mutation spectrum of this population should be designed. Moreover, we were able to detect the causative pathogenic variants or variants of uncertain significance using MPS in about 21.8% (N = 12/55) of our subjects with NSHL. In total, the diagnostic yield of our three-step molecular diagnostic protocol, including MPS, was estimated to be 31.7% in our Vietnamese population, which was inferior to other studies that used massively parallel sequencing3,18,22,41,51.
There were several limitations in this study, with respect to the confirmation of pathogenicity of detected variants and diagnostic rate of our molecular diagnostic protocol. We were unable to perform additional analyses, including a segregation study necessary to confirm the co-segregation of the variants with deafness among the family members and the trans-configuration of two compound heterozygous due to lack of parental samples in our cohort. Therefore, some genotypes obtained from the MPS analysis may not be conclusive, and the diagnostic yield of the three-step protocol employed in our study for molecular diagnosis of severe-to-profound hearing loss may be slightly overestimated. These issues could be addressed in a future study using segregation analysis or MPS with the inclusion of family members. Secondly, the variants of other deafness genes or CNVs, which were not included in the targeted deafness genes in this study or could not be detected with our diagnostic protocol, may have been missed. CNV has been identified as one of the frequent causes of ARNSHL. According to the literature, at least one CNV in a known deafness gene was found in 15.2% of patients with hearing loss, and causative CNVs were identified in 7.3% of these patients as homozygous, hemizygous in conjunction with a second pathogenic mutation, or biallelic CNVs37. Large genomic deletions, involving GJB2 or GJB6, have previously been reported as a causative mutation related to hearing loss38–40. Therefore, novel or unrevealed CNVs in the coding or noncoding region of GJB2 or GJB6, which would lead to null function, can explain severe-to-profound auditory phenotypes in our participants in a trans configuration with the single heterozygous p.V37I (N = 19/87, 21.8%). As aforementioned, the prevalence of GJB2 variants in our Vietnamese population was estimated to be only 2.3% when the p.V37I homozygotes were excluded. However, upon discovery of such occult genomic deletion, some of the single heterozygotes of p.V37I can later be classified as DFNB1, leading to an increase in the prevalence of DFNB1 in the Vietnamese population. Hence, in the next study, the presence of occult genomic deletion, if any, in the DFNB1 locus (either DFNB1A or DFNB1B) among these putative p.V37I carriers in a Vietnamese population will be explored by multiple ligation probe amplification (MLPA) covering the entire DFNB1 locus. Lastly, MPS test was not performed in 25 patients and their genetic diagnosis remained inconclusive, which may have been another limitation of this study.
In conclusion, our study is the first attempt to reveal the characteristics of mutation spectrum of severe-to-profound NSHL in the Vietnamese population, providing the basis for the establishment of a comprehensive molecular genetic diagnostic protocol. Furthermore, our data provide supportive evidence suggesting an ethnic relationship between the peoples of Vietnam and those in the southern parts of China.
Methods
Participants
Between September 2015 and May 2016, children who visited the Children Hospital 1 in Vietnam with delayed language development or decreased responses to sound stimuli were included in this study. A total of 87 children (48 males, 39 females) were selected and included. All participants underwent an audiological evaluation. The hearing thresholds at 0.5, 1, 2, and 4 kHz were evaluated with pure-tone audiometry, and the average threshold was calculated. In the cases where pure-tone audiometry could not be performed due to the subjects’ young age, the hearing level was determined using the auditory brainstem response threshold test or visual reinforcement audiometry. The inclusion criteria were severe-to-profound hearing loss with over 70 dB of hearing thresholds. Children who had syndromic hearing loss or familial history of hearing loss were excluded; to avoid any bias from acquired hearing loss, subjects who were older than 12 years were also excluded. The mean age was 4.6 ± 2.2 (0–11) years at the time of their initial visitation. Moreover, a total of 117 participants (40 males, 77 females) with normal hearing under 25 dB were included in the control group. The mean age of the control group was 33.5 ± 9.6 (20–66) years. The Institutional Review Boards (IRBs) of Seoul National University Bundang Hospital (SNUBH) in Korea (IRB-B-1007-105-402) and Research Center of Children Hospital 1 in Vietnam (CS/1/15/13) approved this study. All procedures were performed in accordance with relevant guidelines and regulations. We obtained written informed consent from all participants in accordance with the Declaration of Helsinki; in the case of children, written informed consent was obtained from their patient or guardian. After obtaining written informed consent, ten milliliters of whole blood for genetic analysis was obtained from all participants. All data generated or analyzed during this study are included in this published article and its supplementary information file.
U-TOP screening test
A total of 87 subjects in the HL group and 117 normal participants in the control group of our Vietnamese population were screened using the U-TOP™ HL Genotyping Kit (SeaSun Biomaterials, Daejeon, Korea), which is a real-time PCR-based MeltingArray genetic diagnostic kit (Fig. 1). This new screening tool was designed to detect variants known to be associated with mild-to-moderate hearing loss, as well as to screen variants associated with severe-to-profound hearing loss52. The molecular diagnostic platform, based on ethnicity-specific mutation spectrums of sensorineural hearing loss, contain 11 variants of 5 genes (GJB2: p.V37I, c.235delC, c.299delAT, p.R143W; SLC26A4: p.H723R, c.IVS7-2A > G, p.T410M, p.L676Q; MTRNR1: 1555 A > G; TMPRSS3: p.A306T; CDH23: p.P240L) showing high prevalence with varying degrees in the Korean population. The phenotype of the p.V37I variant of GJB2 was reported as mild to profound hearing loss53–55. Therefore, the homozygosity of p.V37I allele of GJB2 was considered as causative pathogenic variant in this study. The compound heterozygote of V37I and other pathogenic variant of GJB2, such as p.R143W, were proposed as possible causative variant of moderate to severe hearing loss55, and were included as causative pathogenic variant in this study.
When only one variant of either GJB2 or SLC26A4 was detected with the screening kit, Sanger sequencing for the entire coding region of the corresponding gene, beyond the coverage of the screening kit, was additionally performed to find the second variant. A rare novel variant of SLC26A4 in a trans configuration with a definitely pathogenic SLC26A4 variant was considered as ‘pathogenic’, as previously proposed56.
Sanger sequencing for GJB2
For 76 subjects whose causative genetic variants were not identified with a screening test, Sanger sequencing for the coding region of the GJB2 gene was performed (Fig. 1). Next, Sanger sequencing for previously reported, non-coding variants of GJB2 and evaluation of known genomic deletion of GJB6 was performed. In detail, Sanger sequencing for the four known pathogenic variant on the noncoding regions, c.-22-2A > C57, c.-23G > T58, c.−23 + 1 G > A59, and c.−259C > T60 was performed with two primer sets, as previously described61. Next, a multiplex breakpoint PCR assay was performed for two previously reported large genomic deletions (del[GJB6-D13S1830] and del[GJB6-D13S1854])40. To detect other structural variations on the 5-kb regions upstream of GJB2 within the DFNB1 locus, we also verified the raw data of MPS, if available, using the Integrative Genomic Viewer (http://www.broadinstitute.org/igv/home).
Massive parallel sequencing
A subset of subjects whose causal genetic variants were not identified with the U-TOPTM HL genotyping kit were evaluated with targeted exome sequencing (TES; Otogenetics, Norcross, GA, USA) and whole exome sequencing (WES; Otogenetics, Norcross, GA, USA). TES and WES were captured using the NimbleGen Sequence Catcher (Roche NimbleGen Inc., Madison, WI, USA) and SureSelect 50 Mb Hybridization and Capture kit, respectively. Bioinformatics analyses were performed, as previously described, against the targeted genes related to hearing loss (Fig. 1)62,63. In total, 32–69 million short reads (100-bp paired-end reads) were obtained via MPS. More than 85% of the target exon regions were covered by at least five sequence reads. The reads were mapped onto the UCSC hg19 reference genome. The non-synonymous single nucleotide polymorphisms (SNPs) were filtered with a depth ≥ 15. Minor allele frequency (MAF) of the variants was evaluated from publicly available databases, including ExAC (http://exac.broadinstitute.org/), 1000 Genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genome), GO-ESP (http://evs.gs.washington.edu/EVS/), and GnomAD (http://gnomad.broadinstitute.org/). Variants with MAF ≥ 0.5% were excluded, unless they were pathogenic according to the literature, ClinVar, or Deafness Variation Database (http://deafnessvariationdatabase.org/) (Accessed: May 26, 2018)64. We used a threshold of 0.5% for ARNSHL and 0.05% for ADNSHL65. We interpreted and classified the pathogenicity of our variants as one of the followings: Pathogenic, likely pathogenic, benign, likely benign, and uncertain significance according to the ACMG guidelines (Table 1 and Supplementary Table S1)66. Patient cohort was divided into four groups: (1) patients with causative (likely) pathogenic variants; (2) patients with variants of uncertain significance; (3) patients with only one variant of the gene with autosomal recessive inheritance pattern; (4) patients without detected causative variants (Fig. 1)22.
Supplementary information
Acknowledgements
We would like to thank the patients and health volunteers included in this study. We are grateful to all physicians and counselors for helping us care for the participants. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1D1A1B03034401) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1867, HI15C1632, and HI17C0952). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
J.H. wrote the main manuscript text. B.C. conceived the investigation, revised the manuscript for important intellectual content. P.N., D.O., J.H., A.K., M.K., H.P., L.T., N.D., J.K., J.L., S.O. and H.V. contributed to all aspects of the investigation, including methodological design, data collection and analysis, interpretation of the results. All authors reviewed and revised the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jae Joon Han and Pham Dinh Nguyen contributed equally.
Hoang Anh Vu and Byung Yoon Choi jointly supervised this work.
Contributor Information
Hoang Anh Vu, Email: hoangvuxinh@yahoo.com.
Byung Yoon Choi, Email: choiby2010@gmail.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38245-4.
References
- 1.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. New England Journal of Medicine. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Morton CC, Nance WE. Newborn hearing screening–a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 3.Park JH, et al. Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis. 2014;9:167. doi: 10.1186/s13023-014-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CC, Lee YC, Chen PJ, Hsu CJ. Predominance of genetic diagnosis and imaging results as predictors in determining the speech perception performance outcome after cochlear implantation in children. Archives of pediatrics & adolescent medicine. 2008;162:269–276. doi: 10.1001/archpediatrics.2007.59. [DOI] [PubMed] [Google Scholar]
- 5.Black J, Hickson L, Black B, Perry C. Prognostic indicators in paediatric cochlear implant surgery: a systematic literature review. Cochlear implants international. 2011;12:67–93. doi: 10.1179/146701010x486417. [DOI] [PubMed] [Google Scholar]
- 6.Brown KK, Rehm HL. Molecular diagnosis of hearing loss. Current protocols in human genetics, 2012;9.16:11–19.16. 16. doi: 10.1002/0471142905.hg0916s72. [DOI] [PubMed] [Google Scholar]
- 7.White KR. Early hearing detection and intervention programs: opportunities for genetic services. American journal of medical genetics. Part A. 2004;130A:29–36. doi: 10.1002/ajmg.a.30048. [DOI] [PubMed] [Google Scholar]
- 8.Denoyelle F, et al. Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet. 1997;6:2173–2177. doi: 10.1093/hmg/6.12.2173. [DOI] [PubMed] [Google Scholar]
- 9.Sobe T, et al. The prevalence and expression of inherited connexin 26 mutations associated with nonsyndromic hearing loss in the Israeli population. Hum Genet. 2000;106:50–57. doi: 10.1007/s004390051009. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, et al. GJB2, SLC26A4, and mitochondrial DNA12S rRNA hot-spots in 156 subjects with non-syndromic hearing loss in Tengzhou, China. Acta oto-laryngologica. 2016;136:800–805. doi: 10.3109/00016489.2016.1164893. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, et al. Mutation Spectrum of Common Deafness-Causing Genes in Patients with Non-Syndromic Deafness in the Xiamen Area, China. PloS one. 2015;10:e0135088. doi: 10.1371/journal.pone.0135088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usami S, Nishio SY, Nagano M, Abe S, Yamaguchi T. Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PLoS One. 2012;7:e31276. doi: 10.1371/journal.pone.0031276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masmoudi S, et al. Determination of the frequency of connexin26 mutations in inherited sensorineural deafness and carrier rates in the Tunisian population using DGGE. Journal of medical genetics. 2000;37:E39. doi: 10.1136/jmg.37.11.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghasemnejad T, Shekari Khaniani M, Zarei F, Farbodnia M, Mansoori Derakhshan S. An update of common autosomal recessive non-syndromic hearing loss genes in Iranian population. International journal of pediatric otorhinolaryngology. 2017;97:113–126. doi: 10.1016/j.ijporl.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Dahl HH, et al. Prevalence and nature of connexin 26 mutations in children with non-syndromic deafness. Med J Aust. 2001;175:191–194. doi: 10.5694/j.1326-5377.2001.tb143093.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, et al. Residual Hearing in DFNB1 Deafness and Its Clinical Implication in a Korean Population. PLoS One. 2015;10:e0125416. doi: 10.1371/journal.pone.0125416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabahuma RI, et al. Absence of GJB2 gene mutations, the GJB6 deletion (GJB6-D13S1830) and four common mitochondrial mutations in nonsyndromic genetic hearing loss in a South African population. International journal of pediatric otorhinolaryngology. 2011;75:611–617. doi: 10.1016/j.ijporl.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloan-Heggen CM, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasmelseed NM, et al. Low frequency of deafness‐associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Human mutation. 2004;23:206–207. doi: 10.1002/humu.9216. [DOI] [PubMed] [Google Scholar]
- 20.Beheshtian, M. et al. Heterogeneity of Hereditary Hearing Loss in Iran: a Comprehensive Review. Arch Iran Med19, 720–728, 0161910/aim.0010 (2016). [PMC free article] [PubMed]
- 21.Yuan Y, et al. Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. Journal of Translational Medicine. 2009;7:79. doi: 10.1186/1479-5876-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zazo Seco C, et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur J Hum Genet. 2017;25:308–314. doi: 10.1038/ejhg.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wattanasirichaigoon D, et al. High prevalence of V37I genetic variant in the connexin‐26 (GJB2) gene among non‐syndromic hearing‐impaired and control Thai individuals. Clinical genetics. 2004;66:452–460. doi: 10.1111/j.1399-0004.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- 24.Kudo T, et al. GJB2 (connexin 26) mutations and childhood deafness in Thailand. Otology & neurotology: official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2001;22:858–861. doi: 10.1097/00129492-200111000-00025. [DOI] [PubMed] [Google Scholar]
- 25.Chan DK, Chang KW. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope. 2014;124:E34–53. doi: 10.1002/lary.24332. [DOI] [PubMed] [Google Scholar]
- 26.Dai P, et al. GJB2 mutation spectrum in 2063 Chinese patients with nonsyndromic hearing impairment. Journal of translational medicine. 2009;7:26. doi: 10.1186/1479-5876-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pischedda S, et al. Phylogeographic and genome-wide investigations of Vietnam ethnic groups reveal signatures of complex historical demographic movements. Scientific reports. 2017;7:12630. doi: 10.1038/s41598-017-12813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, et al. Prevalence and range of GJB2 and SLC26A4 mutations in patients with autosomal recessive nonsyndromic hearing loss. Molecular medicine reports. 2014;10:379–386. doi: 10.3892/mmr.2014.2148. [DOI] [PubMed] [Google Scholar]
- 29.Duan SH, Zhu YM, Wang YL, Guo YF. Common molecular etiology of nonsyndromic hearing loss in 484 patients of 3 ethnicities in northwest China. Acta oto-laryngologica. 2015;135:586–591. doi: 10.3109/00016489.2015.1006334. [DOI] [PubMed] [Google Scholar]
- 30.Luo J, et al. Prevalence of Mutations in Deafness-Causing Genes in Cochlear Implanted Patients with Profound Nonsyndromic Sensorineural Hearing Loss in Shandong Province, China. Annals of human genetics. 2017;81:258–266. doi: 10.1111/ahg.12207. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, et al. Analysis common gene mutation spots of 127 non-syndromic deafness natients in Guangxi Drovince. Lin chuang er bi yan hou tou jing wai ke za zhi = Journal of clinical otorhinolaryngology, head, and neck surgery. 2015;29:1954–1958. [PubMed] [Google Scholar]
- 32.Han SH, et al. Carrier frequency of GJB2 (connexin-26) mutations causing inherited deafness in the Korean population. J Hum Genet. 2008;53:1022–1028. doi: 10.1007/s10038-008-0342-7. [DOI] [PubMed] [Google Scholar]
- 33.Tsukada K, Nishio S, Usami S. A large cohort study of GJB2 mutations in Japanese hearing loss patients. Clinical genetics. 2010;78:464–470. doi: 10.1111/j.1399-0004.2010.01407.x. [DOI] [PubMed] [Google Scholar]
- 34.Wattanasirichaigoon D, et al. High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals. Clinical genetics. 2004;66:452–460. doi: 10.1111/j.1399-0004.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- 35.Chai Y, et al. The homozygous p.V37I variant of GJB2 is associated with diverse hearing phenotypes. Clinical genetics. 2015;87:350–355. doi: 10.1111/cge.12387. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, Huang B, Wang G, Yuan Y, Dai P. The Relationship between the p.V37I Mutation in GJB2 and Hearing Phenotypes in Chinese Individuals. PLoS One. 2015;10:e0129662. doi: 10.1371/journal.pone.0129662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearer AE, et al. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6:37. doi: 10.1186/gm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldmann D, et al. A new large deletion in the DFNB1 locus causes nonsyndromic hearing loss. Eur J Med Genet. 2009;52:195–200. doi: 10.1016/j.ejmg.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Wilch E, et al. A novel DFNB1 deletion allele supports the existence of a distant cis-regulatory region that controls GJB2 and GJB6 expression. Clinical genetics. 2010;78:267–274. doi: 10.1111/j.1399-0004.2010.01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.del Castillo FJ, et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. Journal of medical genetics. 2005;42:588–594. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommen M, et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum Mutat. 2016;37:812–819. doi: 10.1002/humu.22999. [DOI] [PubMed] [Google Scholar]
- 42.Choi BY, et al. Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clinical genetics. 2009;75:237–243. doi: 10.1111/j.1399-0004.2008.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anwar S, et al. SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred’s syndrome in Pakistanis. Journal of human genetics. 2009;54:266–270. doi: 10.1038/jhg.2009.21. [DOI] [PubMed] [Google Scholar]
- 44.Kim SY, et al. Strong founder effect of p.P240L in CDH23 in Koreans and its significant contribution to severe-to-profound nonsyndromic hearing loss in a Korean pediatric population. Journal of translational medicine. 2015;13:263. doi: 10.1186/s12967-015-0624-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang MY, et al. Expansion of phenotypic spectrum of MYO15A pathogenic variants to include postlingual onset of progressive partial deafness. BMC medical genetics. 2018;19:29. doi: 10.1186/s12881-018-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weil D, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 47.Liu XZ, et al. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nature genetics. 1997;16:188–190. doi: 10.1038/ng0697-188. [DOI] [PubMed] [Google Scholar]
- 48.Liu XZ, et al. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nature genetics. 1997;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- 49.Lentz, J. & Keats, B. J. B. In GeneReviews((R)) (eds M. P. Adam et al.) (University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle, 1993).
- 50.Kim, B. J. et al. Discovery of MYH14 as an important and unique deafness gene causing prelingually severe autosomal dominant nonsyndromic hearing loss. The journal of gene medicine19, 10.1002/jgm.2950 (2017). [DOI] [PubMed]
- 51.Shearer AE, Smith RJ. Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care. Otolaryngol Head Neck Surg. 2015;153:175–182. doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han KH, et al. Establishment of a Flexible Real-Time Polymerase Chain Reaction-Based Platform for Detecting Prevalent Deafness Mutations Associated with Variable Degree of Sensorineural Hearing Loss in Koreans. PLoS One. 2016;11:e0161756. doi: 10.1371/journal.pone.0161756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huculak C, Bruyere H, Nelson TN, Kozak FK, Langlois S. V37I connexin 26 allele in patients with sensorineural hearing loss: evidence of its pathogenicity. American journal of medical genetics. Part A. 2006;140:2394–2400. doi: 10.1002/ajmg.a.31486. [DOI] [PubMed] [Google Scholar]
- 54.Pollak A, et al. M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. American journal of medical genetics. Part A. 2007;143a:2534–2543. doi: 10.1002/ajmg.a.31982. [DOI] [PubMed] [Google Scholar]
- 55.Kim SY, et al. Prevalence of p.V37I variant of GJB2 in mild or moderate hearing loss in a pediatric population and the interpretation of its pathogenicity. PLoS One. 2013;8:e61592. doi: 10.1371/journal.pone.0061592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi BY, et al. Hypo‐Functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: Genotype‐phenotype correlation or coincidental polymorphisms? Human mutation. 2009;30:599–608. doi: 10.1002/humu.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandia M, et al. A novel splice-site mutation in the GJB2 gene causing mild postlingual hearing impairment. PLoS One. 2013;8:e73566. doi: 10.1371/journal.pone.0073566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mani RS, et al. Functional consequences of novel connexin 26 mutations associated with hereditary hearing loss. Eur J Hum Genet. 2009;17:502–509. doi: 10.1038/ejhg.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirmaci A, Akcayoz-Duman D, Tekin M. The c.IVS1 + 1G > A mutation in the GJB2 gene is prevalent and large deletions involving the GJB6 gene are not present in the Turkish population. J Genet. 2006;85:213–216. doi: 10.1007/BF02935334. [DOI] [PubMed] [Google Scholar]
- 60.Matos TD, et al. A novel hearing-loss-related mutation occurring in the GJB2 basal promoter. Journal of medical genetics. 2007;44:721–725. doi: 10.1136/jmg.2007.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim SY, et al. Unraveling of Enigmatic Hearing-Impaired GJB2 Single Heterozygotes by Massive Parallel Sequencing: DFNB1 or Not? Medicine (Baltimore) 2016;95:e3029. doi: 10.1097/md.0000000000003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi BY, et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS One. 2013;8:e68692. doi: 10.1371/journal.pone.0068692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim NK, et al. Whole-exome sequencing reveals diverse modes of inheritance in sporadic mild to moderate sensorineural hearing loss in a pediatric population. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17:901–911. doi: 10.1038/gim.2014.213. [DOI] [PubMed] [Google Scholar]
- 64.Azaiez H, et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. American journal of human genetics. 2018;103:484–497. doi: 10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shearer AE, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. American journal of human genetics. 2014;95:445–453. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.