Abstract
MicroRNAs (miRNAs) play key roles in mammalian folliculogenesis (a complex process in which primordial follicles develop into mature oocytes) by inhibiting mRNA translation or by inducing its degradation, while the role of miRNA in folliculogenesis and regulation mechanism remain unclear. In this study, we explored the role of the p53/miR-27a/nuclear factor of activated T-cells 5 (NFAT5) signaling axis in mouse ovarian granulosa cell proliferation. Luciferase reporter assay, overexpression, site-directed mutagenesis, and chromatin immunoprecipitation (ChIP) assay results showed that the transcription factor p53 significantly decreased the expression level of miR-27a by binding to sites 4 (-646 to -637 bp) and 10 (-50 to -41 bp) of the miR-27a promoter. Moreover, miR-27a directly targeted the 3′-untranslated region of the target gene, NFAT5, to regulate its expression levels. p53 also upregulated the expression of NFAT5. Meanwhile, overexpression of NFAT5 strongly upregulated the mRNA and protein levels of the Wnt signaling genes, β-catenin and B-Cell CLL/Lymphoma 2 (Bcl-2). In addition, NFAT5 promoted mouse granulosa cell proliferation; this was confirmed by EdU/Hoechst immunostaining. Taken together, our findings define a novel pathway p53/miR-27a/NFAT5, and NFAT5 regulates mouse granulosa cell functions via activating Wnt signaling pathway.
Keywords: Mouse, Granulosa cell, p53, miR-27a, NFAT5, Wnt signaling pathway
Introduction
The generation of mammalian follicles is a continuous and complex process in which primordial follicles develop into pre-ovulatory follicles 1. Normally, only a limited number of follicles undergo this process and release oocytes, while the majority of follicles undergo atresia at different developmental stages 2. Granulosa cells (GCs) act as important mediators in ovarian follicle development and provide necessary growth factors and specific proteins to the oocytes, and the proliferation of GCs triggers the follicular growth and oocyte maturation 3,4. The complexity of folliculogenesis indicates that tightly regulated gene expression and interactions between many genes are necessary for successful oocyte development 5. However, the definitive regulatory mechanism underlying follicular development and GC proliferation remains unclear.
MiRNAs, a class of endogenous noncoding RNAs 19-23 nucleotides in length, regulate the expression level of target genes by complementary binding to the 3′-untranslated region (3′-UTR) of target genes thereby mediating translation inhibition or RNA degradation 6,7. MiRNAs play crucial roles in numerous cellular processes, including follicular development and atresia, cell proliferation, and apoptosis 8-11. The previous results of our laboratory showed that miR-27a was differentially expressed in pre-ovulatory ovarian follicles between Chinese Taihu and Large White sows 12. And we previously reported that miR-27a suppressed CHO cell proliferation by downregulating DKK2 expression 5. Thus, miR-27a has always been the focus of our research.
The Wnt signaling pathway is a main regulator of development in animals 13. Wnt signaling plays roles in fetal and ovarian development, gestation, and mammogenesis 14-15. Our previous studies have focused on Wnt signaling pathway, and investigated the regulation mechanism of Wnt signaling genes 5,16. p53 and nuclear factor of activated T-cells 5 (NFAT5) are two members of the Wnt signaling pathway. Moreover, the interaction among p53, miR-27a and NFAT5 was predicted by bioinformatics methods. Numerous studies have shown that p53 has a key role in tumor prevention 17. Moreover, the pregnancy rate and litter size of p53-/- female mice are significantly decreased. This indicates the important effect of p53 on mammalian reproduction 18. NFAT5, as a transcription factor, is a member of the Rel family and is involved in multiple biological processes, including embryonic development, cellular migration, and proliferation 19. Nevertheless, the function of NFAT5 in GCs is largely unknown.
Previously, we showed that miR-27a suppressed Chinese hamster ovary (CHO) cell proliferation by downregulating expression of the Wnt antagonist, Dickkopf-related protein 2 (DKK2) 5. The purpose of this study was to further analyze how the key genes of Wnt signaling pathway regulate miR-27a and how they are regulated by miR-27a. In the current study, we report that p53-mediated suppression of miR-27a increased the expression level of the target gene, NFAT5. We also demonstrated that NFAT5 induced sustained Wnt pathway activation through enhancing the expression of β-catenin and B-cell lymphoma/leukemia-2 (Bcl-2), thus facilitating mouse GC proliferation. Taken together, our results reveal a novel p53/miR-27a/NFAT5 pathway, and NFAT5 contributes to the proliferation of mouse GCs via activating Wnt signaling pathway.
Results
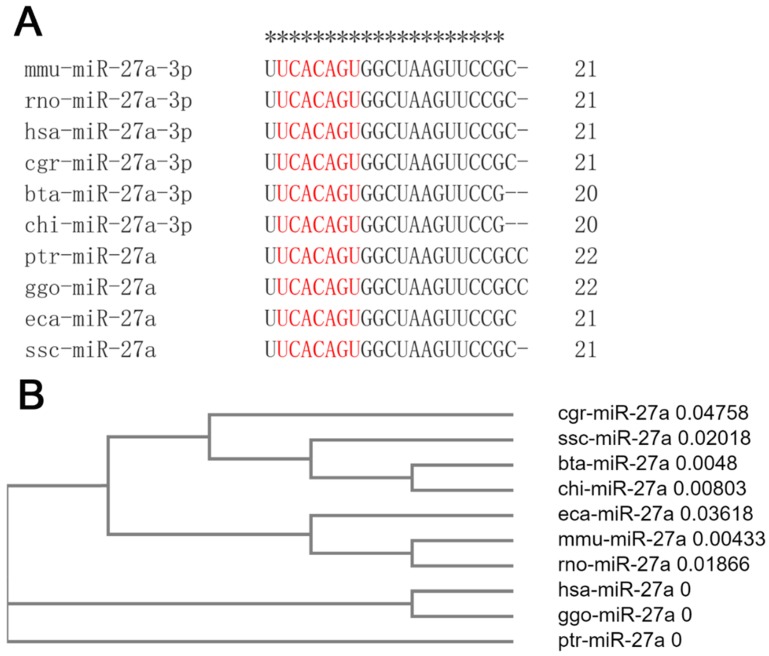
miR-27a is highly conserved in mammals
Mature sequences of miR-27a are highly conserved in mammals, including mice, rats, humans, hamsters, cattle, goats, chimpanzees, gorillas, horses, and pigs. The seed sequences of miR-27a were identical, although there were one or two base deletions at the end of the mammalian sequence (Figure 1A). Clustal Omega, a new multiple sequence alignment program, was used to build the phylogenetic tree of the pre-miRNA of miR-27a. The results revealed that, compared with the other species examined, the genetic relationships between mice and rats, cattle and goats, and humans and gorillas, were closer (Figure 1B).
Figure 1.
miR-27a was highly conserved in mammals. (A) The mature sequences of miR-27a were obtained from miRBase. The sequences of seed region were highlighted in red. mmu, mus musculus; rno, rattus norvegicus; hsa, homo sapiens; cgr, cricetulus griseus; bta, bos taurus; chi, capra hircus; ptr, pan troglodytes; ggo, gorilla gorilla; eca, equus caballus; ssc, sus scrofa. (B) The phylogenetic tree of miR-27a pre-miRNA. pre-miRNA sequences were obtained from NCBI.
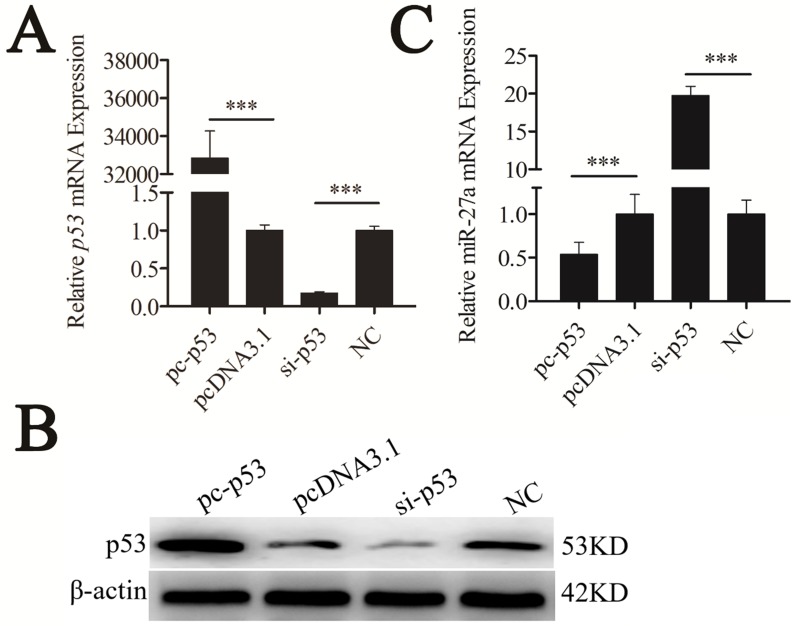
p53 represses expression of miR-27a
The most prominent property of the p53 protein is its action as a transcription factor because it possesses a functional transactivation domain 20. To determine whether p53 regulated the expression level of pre-miR-27a, mouse p53 cDNA (NM_001127233.1) was cloned into pcDNA3.1(+), named pc-p53. The pc-p53 plasmid and p53 siRNA (si-p53) were transfected into CHO-K1 cells. The quantitative real-time polymerase chain reaction (qRT-PCR) and western blot analyses revealed that p53 overexpression significantly promoted p53 mRNA and protein expression, respectively (P < 0.001). Transfection of si-p53 knocked down p53 mRNA and protein expression (P < 0.001) (Figure 2A, B), indicating that the recombinant plasmid in pc-p53 was expressed successfully.
Figure 2.
miR-27a was downregulated by p53 gene. CHO-K1 cells were transfected with pc-p53, pcDNA3.1 and si-p53, NC. (A) qRT-PCR was used to detect endogenous p53 mRNA 24 h after transfection. (B) Western blot analysis was used to detect p53 protein expression levels 48 h after transfection. (C) Twenty-four hours after transfection, the expression levels of miR-27a were determined by qRT-PCR. The data are represented as mean±S.D. of three independent experiments. *** P < 0.001.
The expression level of mature miR-27a was detected and significantly suppressed by p53 (Figure 2C). To confirm the repressive effect of p53 on miR-27a expression, we knocked down p53. This led to the increased expression of miR-27a (Figure 2C). These results demonstrated that miR-27a was downregulated by p53.
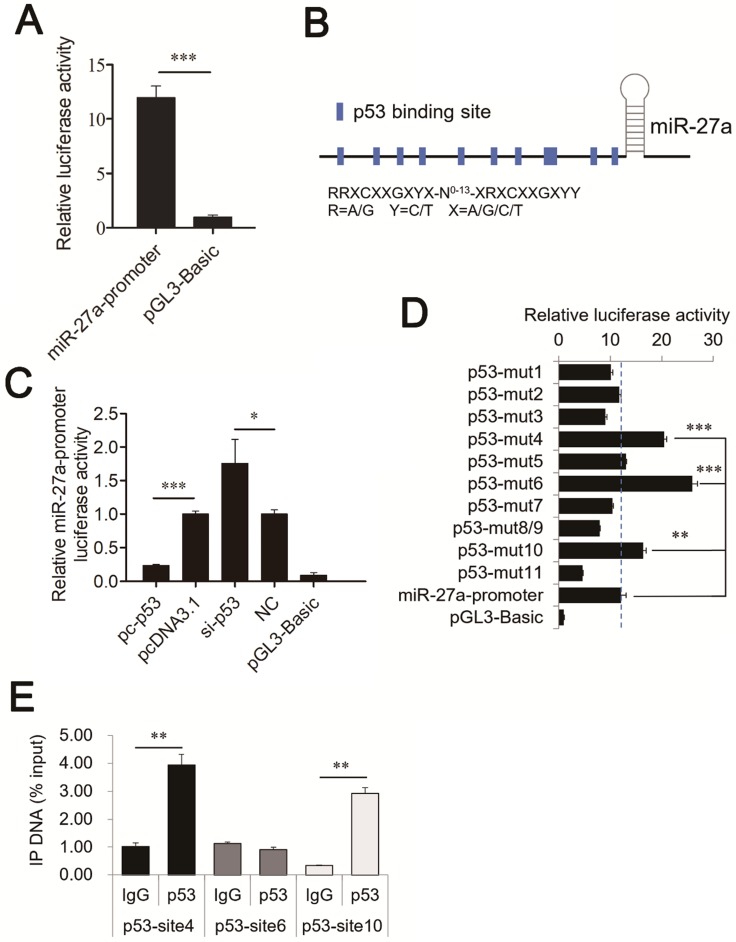
The miR-27a promoter contains p53 response elements
To identify the promoter of mouse miR-27a, a potential promoter (-1039 to -1 bp) was used to drive luciferase gene expression, and luciferase activity was determined. pGL3 plasmids containing the potential miR-27a promoter (miR-27a-promoter) were transiently transfected into CHO-K1 cells. The results revealed that a miR-27a-promoter vector was required for miR-27a transcriptional activity (Figure 3A). This demonstrates that the region from -1039 to -1 bp contains one or more cis-acting elements that could regulate miR-27a expression.
Figure 3.
Identification of the binding sites of p53 in the promoter regions of miR-27a. (A) Promoter activities of miR-27a promoter determined by luciferase assay. (B) Schematic diagram of the p53 binding sites in the miR-27a promoter. (C) Luciferase activity was analyzed 24h after CHO-K1 cells were co-transfected with miR-27a-promoter and pc-p53, pcDNA3.1, si-p53 or NC. (D) site-directed mutagenesis in the p53 binding sites of the miR-27a promoter were analyzed using luciferase assays. (E) ChIP assay of p53 binding to the miR-27a promoter in CHO-K1 cells in vivo. DNA isolated from immunoprecipitated material was amplified by qRT-PCR total chromatin was used as the input. Normal mouse IgG was used as a negative control. The data are represented as mean±S.D. of three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001.
To determine whether p53 is a verifiable transcription factor of miR-27a in vivo, binding site analysis with the TFBIND program revealed eleven putative p53 binding sites on the miR-27a promoter (Figure 3B). These p53 binding sites share highly conserved nucleotides with consensus sequences that can be depicted as RRXCXXGXYX-N0-13-XRXCXXGXYY, where R = A/G, Y = C/T, and X = A/G/C/T 21.
To identify p53 binding sites on the miR-27a promoter, we used luciferase assays to analyze the relationship between p53 and miR-27a. pc-p53 and pcDNA3.1, si-p53, and a nonsense control (NC) were co-transfected with the miR-27a-promoter vector into CHO-K1 cells. Overexpression of p53 significantly suppressed miR-27a promoter activity. In contrast, knockdown of p53 increased miR-27a promoter activity (Figure 3C).
The transcription factor p53 binds to the miR-27a promoter in vitro and in vivo
To confirm that p53 can bind to the promoter of mouse miR-27a, ten mutant pGL3 reporter constructs in which putative binding sites 1 (-1022 to -1013 bp), 2 (-853 to -844 bp), 3 (-801 to -787 bp), 4 (-646 to -637 bp), 5 (-553 to -544 bp), 6 (-451 to -442 bp), 7 (-403 to -394 bp), 8 (-250 to -241 bp) and 9 (-246 to -237 bp), 10 (-50 to -41 bp), and 11 (-17 to -8 bp) were created using site-directed mutagenesis. Sites 8 and 9 had overlapping sequences. Thus, they only yielded one mutant vector. Luciferase activity analysis of CHO-K1 cells revealed that mutating binding sites 4, 6, and 10 of the miR-27a promoter caused a substantial increase in promoter activity, while the mutating binding site 11 showed the opposite result (Figure 3D).
ChIP assays were performed in CHO-K1 cells to determine whether p53 could bind to binding sites 4, 6, and 10 of the miR-27a promoter in vivo. Chromatin was immunoprecipitated using a p53 antibody, and DNA fragments of the expected size were used as templates to perform qRT-PCR. As shown in Figure 3E, p53 interacted with the miR-27a promoter within binding sites 4 (-646 to -637 bp) and 10 (-50 to -41 bp). These results confirmed that the transcription factor p53 was capable of binding to these two binding sites in the mouse miR-27a promoter region, and repress miR-27a transcription.
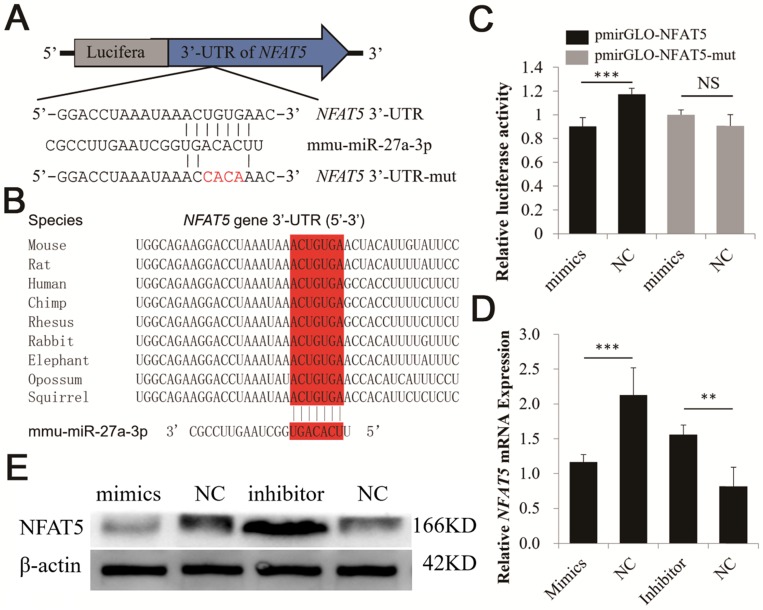
miR-27a directly targets NFAT5
TargetScan was used to explore potential targets for miR-27a. A putative binding site for miR-27a was predicted in the 3′-UTR of NFAT5. In addition, the miR-27a binding seed sequence (ACUGUGA) in the NFAT5 3′-UTR was very highly conserved in mammals, including mice, rats, humans, chimpanzees, rhesus monkeys, rabbits, elephants, opossums, and squirrels (Figure 4A, B).
Figure 4.
Identification of NFAT5 gene as a direct target of miR-27a in CHO-K1 cells. (A) Binding sites for miR-27a in the 3ˈ-UTR of NFAT5 gene predicted by TargetScan. Red characters indicate sequences that were mutated to abolish the interaction between miR-27a and the NFAT5 3ˈ-UTR. (B) The miR-27a binding site (Red) of different species in the NFAT5 3ˈ-UTR. (C) miR-27a mimics were transfected into CHO-K1 cells together with the pmirGLO-NFAT5 and pmirGLO-NFAT5-mut plasmid. Mimics NC was used as a negative control. (D) Endogenous NFAT5 mRNA levels were detected 24 h after CHO-K1 cells were transfected with mimics, mimics NC, inhibitor and inhibitor NC of miR-27a. (E) Western blot analysis was used to detect endogenous NFAT5 protein expression level 48 h after CHO-K1 cells were transfected with mimics, mimics NC, inhibitor and inhibitor NC of miR-27a. (F) Endogenous NFAT5 mRNA levels were detected 24 h after CHO-K1 cells were transfected with pc-p53, pcDNA3.1, si-p53 or NC. (G) Western blot analysis was used to detect endogenous NFAT5 protein expression level 48 h after CHO-K1 cells were transfected with pc-p53, pcDNA3.1, si-p53 or NC.The data are represented as mean ± S.D. of three independent experiments. ** P < 0.01, *** P < 0.001.
A dual-luciferase reporter system was used to analyze the interaction between miR-27a and NFAT5. We co-transfected miR-27a mimics and a dual-luciferase reporter recombinant plasmid (pmirGLO-NFAT5) containing the mouse 620 bp NFAT5 3′-UTR region into CHO-K1 cells. The results showed that the luciferase activity of pmirGLO-NFAT5 was significantly suppressed by miR-27a (Figure 4C). However, luciferase activity was unchanged when we co-transfected miR-27a mimics and pmirGLO-NFAT5-mut into CHO-K1 cells (Figure 4C). qRT-PCR and western blot analyses revealed that NFAT5 mRNA and protein expression levels were significantly suppressed after miR-27a mimics were transfected into CHO-K1 cells, whereas inhibition of miR-27a increased NFAT5 mRNA and protein expression levels (Figure 4D, E). Our data suggested that miR-27a required the 3′-UTR of NFAT5 to regulate its expression levels.
Relationship between NFAT5 and Wnt pathway
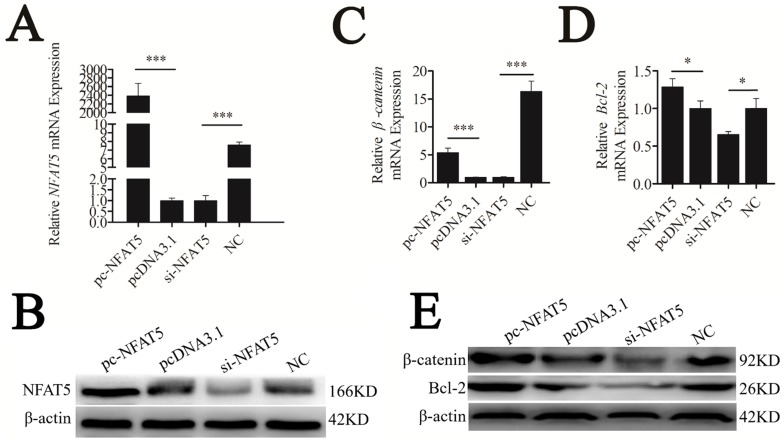
NFAT5 is a regulator of Wnt signaling 22. Thus, we hypothesized that it might play this role by regulating β-catenin and Bcl-2 expression. To test our hypothesis, mouse NFAT5 cDNA (NM_018823.2) was cloned into the pcDNA3.1 vector. The pc-NFAT5 plasmid was then transfected into CHO-K1 cells. NFAT5 overexpression significantly promoted the expression level of NFAT5 mRNA and protein, as determined using qRT-PCR and western blot analyses, respectively (P < 0.001). Furthermore, siRNA of NFAT5 (si-NFAT5) decreased NFAT5 mRNA and protein in CHO-K1 cells (Figures 5A, B). These results indicated that the recombinant plasmid and siRNA of pc-NFAT5 were successfully expressed.
Figure 5.
NFAT5 regulated the downstream genes of the Wnt signaling pathway. CHO-K1 cells were transfected with pc-NFAT5, pcDNA3.1 and si-NFAT5, NC. (A) qRT-PCR was used to detect endogenous NFAT5 mRNA 24 h after transfection. (B) Western blot analysis was used to detect NFAT5 protein expression levels 48 h after transfection. (C, D) Twenty-four hours after transfection, the expression levels of β-catenin and Bcl-2 were determined by qRT-PCR. (E) Western blot analysis was used to detect endogenous β-catenin and Bcl-2 protein expression level. The data are represented as mean±S.D. of three independent experiments. * P < 0.05, *** P < 0.001.
Along with overexpression of NFAT5, mRNA and protein levels of the Wnt signaling genes, β-catenin and Bcl-2, were strongly upregulated, whereas suppression of NFAT5 decreased these levels (Figure 5C-E). These results suggest that NFAT5 may enhance Wnt signaling by promoting the expression of β-catenin and Bcl-2.
NFAT5 promotes cells proliferation in mouse GCs
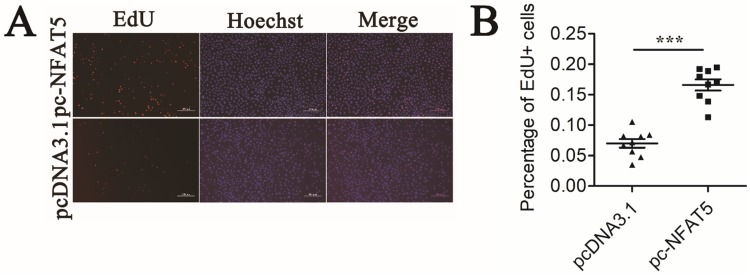
GCs proliferation is ultimately important in follicular growth and oocyte maturation 3. To study the effect of NFAT5 on the proliferation of GCs, we performed an EdU cell proliferation assay. Mouse GCs were transfected with the pc-NFAT5 or pcDNA3.1 plasmid. NFAT5 overexpression noticeably accelerated proliferation of GCs (Figure 6A). Quantitative analysis demonstrated that this change was statistically significant (P < 0.001) (Figure 6B). Taken together, these results suggest that the miR-27a target gene, NFAT5, may promote GC proliferation. EdU (red)/Hoechst (blue) immunostaining further confirmed this conclusion.
Figure 6.
Mouse GCs apoptosis induced by NFAT5 gene. (A) Mouse GCs were labeled with EdU (Red). Cell nuclei were also visualized by Hoechst staining (Blue). The images were representative of the results obtained. (B) The percentage of proliferating GCs was expressed as the ratio of EdU cells to total Hoechst cells. Data were presented as mean ± S.D. from nine independent experiments. *** P < 0.001.
Discussion
Folliculogenesis is a complex biological process. GCs provide necessary material to oocytes and play vital roles in ovarian follicle development. However, the definitive regulatory mechanism underlying follicular development and GC proliferation remains unclear.
MiRNAs, a type of small non-coding RNA, modulate the expression of protein-coding genes at the post-transcriptional level. The regulatory mechanism(s) of miRNAs in folliculogenesis has becomes an important area of research. Numerous previous studies have indicated that miRNAs have a vital role in folliculogenesis. miR-144 promotes mouse GC apoptosis and affects follicular atresia through the miR-144/cyclooxygenase-2/prostaglandin E2 pathway 12. miR-125a-3p is involved in mural granulosa cell migration prior to ovulation, and interfering with this signal in vivo hampers ovulation 23. In addition, miR-21 regulates apoptosis in mouse periovulatory granulosa cells 24.
Our laboratory previously used Solexa sequencing to show that miR-27a was differentially expressed in pre-ovulatory ovarian follicles between Chinese Taihu and Large White sows 12. By sequence analysis, we found that mature sequences of miR-27a were highly conserved in mammals (Figure 1A). In addition to diseases 25,26, miR-27a has a vital role in follicular development because it regulates numerous target genes. For example, miR-27a targets SMAD5 and regulates human GC apoptosis via the FasL-Fas pathway 27. miR-27a, which is differential expressed between human GCs from MI and MII oocytes, decreases the maturation rate of oocytes 28. Furthermore, we previously reported that miR-27a suppresses CHO cell proliferation by downregulating DKK2 expression 5. Thus, miR-27a has always been the focus of our research.
We have been focusing on the regulation of follicular development by the Wnt signaling pathway axis and miRNA. The Wnt transduction cascade is a crucial regulator in animal development 13. Fittingly, the transcription factor (p53) and target gene (NFAT5) of miR-27a are downstream of the Wnt signaling pathway.
Several previous studies have demonstrated that miRNA expression is regulated by many transcription factors 29,30. p53, the most extensively studied oncogene, codes for a sequence-specific DNA binding protein that regulates transcription 31,32. The p53 protein is composed of two N-terminal transactivation domains, a central DNA binding domain, a tetramerization domain, and a C-terminus regulatory domain 33. The DNA binding domain binds specifically to the cis-acting element of target genes and regulates gene transcriptional activity. To assess the transcription factors binding to the miR-27a promoter, eleven p53 binding sites were identified in the miR-27a promoter region using TFBIND software (Figure 3B). Importantly, the results of our study indicated that p53 bound directly to the miR-27a promoter and repressed its transcriptional activation. In contrast, another study found that the abundance of miR-27a was significantly increased after p53 activation in human cancer cells 34. Mutant, but not wild-type p53 directly bound to the miR-27a promoter and repressed miR-27a transcription in H1299 cells 35. The results of both previous studies are contrary to our observations that suggest the regulatory effect of p53 on miR-27a is entirely different in normal and cancerous cells. In addition, several other studies indicated that miR-27a could bind to the p53 3′-UTR and decrease the endogenous p53 expression level 36-38. Moreover, luciferase activity analysis revealed that mutating binding site 6 of the miR-27a promoter caused the highest promoter activity (Figure 3D). But curiously, the result of ChIP assay showed that p53 could not bind to binding site 6 of the miR-27a promoter in vivo. Although the transcription factor p53 cannot bind to the predicted p53 binding sites, the p53 binding sites may regulate the transcription activity of miR-27 by altering the binding of other adjacent transcription factors. Previous study could support the above viewpoint 39. Binding site 11 showed the opposite result which mutating binding site 11 caused a lower promoter activity compared to control group (Figure 3D). We thought the cause might be that the site mutations form a new unknown transcription factor binding site which inhibits the promoter activity of miR-27a.
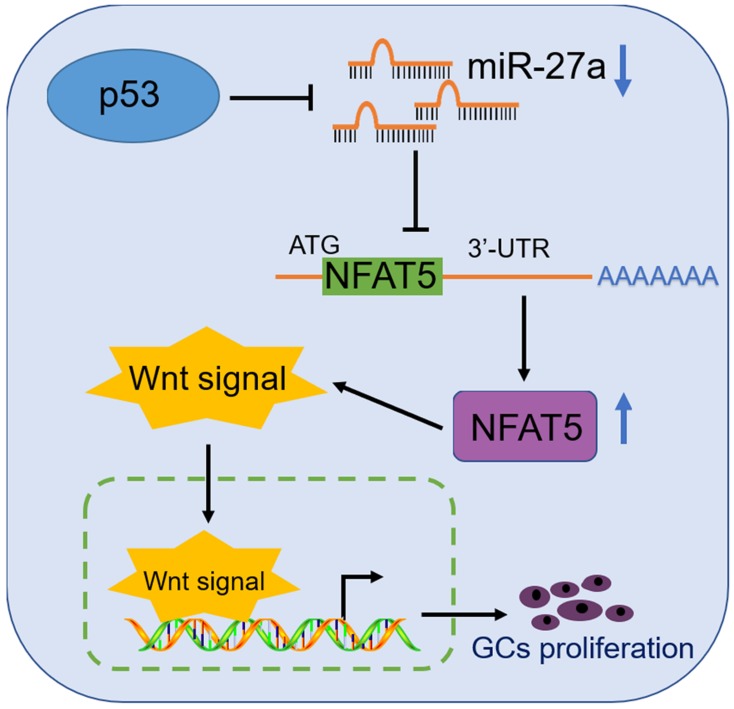
The expression of β-catenin and Bcl-2 as signals of the WNT signaling pathway was determined due to their relationship with folliculogenesis. β-Catenin regulates GC differentiation, and its over-activation leads to a Sertoli-to-granulosa-like cell transformation 40. Bcl-2 overexpression increases the number of primordial follicles 41, while its downregulation is involved in GC apoptosis which was induced by autophagosome accumulation 42. Thus, we chose β-catenin and Bcl-2 as target genes for folliculogenesis. Our results demonstrated that miR-27a downregulated the target gene, NFAT5 (Figure 4D, E), which promoted the expression of β-catenin and Bcl-2, as well as GC proliferation (Figures 5, 6). Our findings may provide a new p53/miR-27a/NFAT5 axis, and NFAT5 regulates mouse granulosa cell functions via activating Wnt signaling pathway (Figure 7).
Figure 7.
A schematic illustration of the proposed model depicting a p53/miR-27a/NFAT5 mediated regulatory pathway in mouse GCs.
Taken together, our results demonstrate that NFAT5 has a crucial role in GC proliferation and enhances the Wnt pathway via upregulating the expression of β-catenin and Bcl-2. In addition, NFAT5 is downregulated by the p53/miR-27a axis (Figure 7). These results suggest that a novel signaling pathway p53/miR-27a/NFAT5 is involved, and NFAT5 contributes to the proliferation of mouse GCs via activating Wnt signaling pathway. These conclusions have potential implications in improving follicular development through modifying the p53/miR-27a/NFAT5 pathway.
Materials and Methods
Cell culture
Mouse granulosa cells from pre-ovulatory ovarian follicles were obtained from murine ovaries after Kunming White female mice were treated for 48 h with 10 IU PMSG (SanSheng, China). The CHO-K1 cells were obtained from the China Centre for Type Culture Collection (Shanghai, China). Cells were cultured with Dulbecco's minimum essential medium/nutrient F-12 (DMEM/F-12) (Gibco, USA) supplied with 10% foetal bovine serum (Gibco) at 37 °C in a humidified atmosphere of 5% CO2.
Transfection and Dual-luciferase reporter assays
The Granulosa cells or CHO-K1 cells were plated and grown until they were 70-80% confluent at the time of transfection. miRNA or siRNA was transfected into the cells using Lipofectamine RNAiMAX Reagent (Invitrogen, USA). Plasmids were transfected into the cells using Lipofectamine 3000 (Invitrogen, USA). For dual-luciferase reporter assays, each wild-type or promoter region construct was transfected into the cells at 500 ng, together with 10 ng/well of pRL-TK (Promega, USA). After transfection for 24 h, the luciferase activities were measured with a PerkinElmer 2030 Multilabel Reader (PerkinElmer, USA).
Sequence analysis
Clustal Omega (Available online: https://www.ebi.ac.uk/Tools/msa/clustalo/) 43 was used to build the phylogenetic tree of pre-miRNA of miR-27a. Transcription factor binding sites in the promoter of mouse miR-27a were predicted using the TFBIND (http://tfbind.hgc.jp/) software. The potential target sites of miRNAs were predicted by TargetScan (http://www.targetscan.org/).
Plasmid construction
The promoter fragments of miR-27a was amplified using primers promoter-PF and promoter-PR. The purified PCR products were cloned into the pMD18-T vector (TaKaRa, China). Then the recombinant plasmids were double-digested with NheI and HindIII (Thermo) and ligated into the pGL3-Basic vector (Promega). Mouse p53 (NM_001127233.1) and NFAT5 (NM_018823.2) CDS were amplified and double-digested with EcoRI and XhoI (Thermo) and cloned into the pcDNA3.1(+) vector (Invitrogen, USA). The mouse NFAT5 3'-UTR were amplified, double-digested with PmeI and XhoI, and then cloned into the pmirGLO vector (Promega, USA). The binding site mutants were generated using a MutanBEST Kit (TaKaRa, China) and mutagenic primers. The primers are described in Table 1.
Table 1.
Primer sequences
| Name | Sequence (5'-3') |
|---|---|
| promoter-PF | CTAGCTAGCAACTCATCATGTAGCTTAAGCA |
| promoter-PR | CCCAAGCTTTTTGCTGTGGACCTTGCTCA |
| pc-p53-PF | CACTAGTCCAGTGTGGTGGAATTCATGACTGCCATGGAGGAGTCAC |
| pc-p53-PR | AACGGGCCCTCTAGACTCGAGTCAGTCTGAGTCAGGCCCCAC |
| pc-NFAT5-PF | ACTATAGGGAGACCCAAGCTGGCTAGCGCCACCATGCCCTCGGACTTCATCTCAT |
| pc-NFAT5-PR | GGTTTAAACGGGCCCTCTAGACTCGAGCTAAAAGGAGCCGGTTAAATTGTTC |
| p53-QPCR-PF | TCGTCCTTTGTCCCTTCTCAG |
| p53-QPCR-PR | CACCACCTCGGTCATGTACTCT |
| NFAT5-QPCR-PF | CTCCTCAGATCCAGTTGGTTCA |
| NFAT5-QPCR-PR | GCTGCATGTCTGGTTGGTTTAT |
| p53-mut-PF | TGGGGTGGGGTTATGTAGTCACTGGCTGGGGAGGAG |
| p53-mut-PR | CTCCTCCCCAGCCAGTGACTACATAACCCCACCCCA |
| mmu-U6-Loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATGGAACGCT |
| mmu-U6-PF | CTGGTAGGGTGCTCGCTTCGGCAG |
| mmu-U6-PR | CAACTGGTGTCGTGGAGTCGGC |
| mmu-miR-27a-Loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCGGAACT |
| mmu-miR-27a-PF | TCGGCAGGTTCACAGTGGCTAAG |
| mmu-miR-27a-PR | TCAACTGGTGTCGTGGAGTCGGC |
| ChIP-mut4-PF | CTCTCCATGGCCCCATTTG |
| ChIP-mut4-PR | CTGCTTCCCACTCTGCATCTATC |
| ChIP-mut6-PF | AGCATCCAGCAGCCTCGC |
| ChIP-mut6-PR | CACCCTAGCCCTCCTCTACTTC |
| ChIP-mut10-PF | TCACTGTGCCTCGGACGGCT |
| ChIP-mut10-PR | TTTGCTGTGGACCTTGCTCA |
Note: PF was upstream primer. PR was downstream primer. The part highlighted with red was enzyme site induced.
RNA interference
The synthesized siRNA of p53 (si-p53) and NFAT5 (si-NFAT5) and siRNA NC (RiboBio, China) was transfected into cells with Lipofectamine RNAiMAX (Invitrogen). After 48 and 72 h, cells were measured for Dual-luciferase reporter assays, qRT-PCR assays and western blot assays, respectively. The siRNA sequences are as follows: siRNA-p53 sense sequence: 5′-GAAUGAGGCCUUAGAGUUAdTdT-3′, antisense sequence: 3′-dTdTCUUACUCCGGAAUCUCAAU-5′; siRNA-NFAT5 sense sequence: 5′-GCUGGUGCUUUGAAUGUAAdTdT-3′; antisense sequenc: 3′-dTdTCGACCACGAAACUUACAUU-5′.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the EZ-ChIP™ Kit (Millipore, USA). Briefly, after crosslinking the chromatin with 1 % formaldehyde at 37 ◦C for 10 min and neutralizing with glycine for 5 min at room temperature, Cells were washed with cold PBS, scraped and collected on ice. Then, cells were harvested, lysed and sonicated. Nuclear lysates were sonicated 20 times for 10 s with 20 min intervals on ice water using a Vibra-Cell™ Processors VCX130 (Sonics, USA). An equal amount of chromatin was immuno-precipitated at 4 ◦C overnight with at least 1.5 μg of Anti-p53 (ab1101, Abcam, USA) and normal mouse IgG (Millipore, USA) antibodies. Immuno-precipitated products were collected after incubation with Protein A+G coated magnetic beads. The beads were washed, and the bound chromatin was eluted in ChIP elution buffer. Then the proteins were digested with Proteinase K for 4 h at 45 ◦C. Purified DNA from the samples and the input were analyzed for the presence of miR-27a promoter sequences containing putative p53 response elements using qRT-PCR. The primers used here are listed in Table 1.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA (including miRNA) was extracted from tissues or cells with TRIzol reagent (Invitrogen, USA). Cellular RNA was extracted 48 h after transfection. One μg of RNA was treated with 1 μl DNase I (Thermo Fisher, USA) to remove DNA contamination. Reverse transcription was performed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA). Random primers, oligo(dT)18 or miRNA specific stem-loop primers were added to initiate cDNA synthesis. qRT-PCR was performed on the Bio-Rad CFX96 system (Bio-Rad, USA) using iTaq Universal SYBR Green Supermix (Bio-Rad, USA). Primers used in the qRT-PCR were shown in Table 1. qRT-PCR conditions consisted of 1 cycle at 94 °C for 3 min, followed by 40 cycles at 94 °C for 20 sec, 60 °C for 20 sec, and 72 °C for 20 sec, with fluorescence acquisition at 74 °C. All PCRs were performed in triplicate and gene expression levels were quantified relatively to the expression of β-actin using Gene Expression Macro software (Bio-Rad) by employing 2-ΔΔCt value 44.
Western blotting analysis
Cell protein lysates were generated using RIPA Lysis Buffer (Beyotime, China). Cellular proteins were extracted 72 h after transfection. Western blotting was performed as reported previously 45. Five μg proteins were boiled in 5 × SDS buffer for 5 min, separated by SDS-PAGE, and transferred to PVDF membranes (Millipore, USA). Then, the membranes were blocked with skim milk and probed with Anti-p53 (A0263, abclonal, USA), Anti-NFAT5 (Bs-9473R, bioss,China), Anti-β-catenin (Ab32572, Abcam, USA), Anti-c-myc (Ab32072, Abcam, USA), Anti-Bcl-2 (12789-1-ap, Proteintech, China). β-actin (ab8226, Santa Cruz, USA) was used as a loading control. The results were visualized with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) and enhanced chemiluminescence. All blots were performed in triplicate and protein expression levels were quantified relatively to the expression of β-actin using the Image J 1.42q software (Wayne Rasband).
EdU proliferation assay
Proliferating granulosa cells were determined using the Cell-LightTM EdU DNA Cell Proliferation Kit (Ribobio, China). Briefly, at the indicated time point after cell transfection, 50 μM EdU was applied to the cultures and the cells were grown for 2h. Then, the cells were fixed with 4% formaldehyde in PBS for 30 min. After labeling, the granulosa cells were assayed according to the manufacturer's protocol. Analysis of granulosa cell proliferation (ratio of EdU+ to all granulosa cells) was performed using images of randomly selected fields obtained on a florescent microscope (Nikon Eclipse Ti-S, USA). Assays were done three times using triplicate wells.
Statistical analysis
Statistical analyses based on the two-tailed Student's t-test were performed using the Statistical Package for the Social Sciences software. Significance was determined at the 95% confidence interval. All data were expressed as the mean ± standard deviation (S.D.) from a representative experiment.
Acknowledgments
This work was supported financially by National Natural Science Foundation of China (No. 31501932) and Open Project of Key Laboratory of Animal Embryo Engineering and Molecular Breeding in Hubei Province (KLAEMB-2018-08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors were grateful to Editage Research Editing Service for manuscript editing.
References
- 1.Castrillon DH, Miao L, Kollipara R, Horner JW, Depinho RA. Suppression of Ovarian Follicle Activation in Mice by the Transcription Factor Foxo3a. Science. 2003;301(5630):215–8. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 2.Yu YS, Sui HS, Han Z Bin, Li W, Luo MJ, Tan JH. Apoptosis in Granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004;14(4):341–6. doi: 10.1038/sj.cr.7290234. [DOI] [PubMed] [Google Scholar]
- 3.Gebremedhn S, Salilew-Wondim D, Hoelker M, Rings F, Neuhoff C, Tholen E. et al. MicroRNA-183-96-182 Cluster Regulates Bovine Granulosa Cell Proliferation and Cell Cycle Transition by Coordinately Targeting FOXO1. Biol Reprod. 2016;94(6):127. doi: 10.1095/biolreprod.115.137539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril. 2015;103(2):303–16. doi: 10.1016/j.fertnstert.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Tao H, Wang L, Zhou J, Pang P, Cai S, Li J. et al. The transcription factor ccaat/enhancer binding protein β (C/EBPβ) and miR-27a regulate the expression of porcine Dickkopf2 (DKK2) Sci Rep. 2015;5:17972. doi: 10.1038/srep17972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 8.Wienholds E, Plasterk RHA. MicroRNA function in animal development. FEBS Letters. 2005;579(26):5911–22. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Sen A, Prizant H, Light A, Biswas A, Hayes E, Lee H-J. et al. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc Natl Acad Sci USA. 2014;111(8):3008–13. doi: 10.1073/pnas.1318978111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao R, Wu W, Zhou X, Liu K, Li B, Huang X. et al. Let-7g induces granulosa cell apoptosis by targeting MAP3K1 in the porcine ovary. Int J Biochem Cell Biol. 2015;68:148–57. doi: 10.1016/j.biocel.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Suh YS, Bhat S, Hong SH, Shin M, Bahk S, Cho KS. et al. Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat Commun. 2015;6:7693. doi: 10.1038/ncomms8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Lei B, Li H, Zhu L, Wang L, Tao H. et al. MicroRNA-144 is regulated by CP2 and decreases COX-2 expression and PGE2 production in mouse ovarian granulosa cells. Cell Death Dis. 2017;8(2):e2597. doi: 10.1038/cddis.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985–99. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H. Wnt/beta-Catenin Signaling in Development and Disease. Cell. 2006;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Parr BA, McMahon AP. Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature. 1998;395(6703):707–10. doi: 10.1038/27221. [DOI] [PubMed] [Google Scholar]
- 16.Tao H, Mei S, Zhang X, Peng X, Yang J, Zhu L. et al. Transcription factor C/EBPβ and 17β-estradiol promote transcription of the porcine p53 gene. Int J Biochem Cell Biol. 2014;47:76–82. doi: 10.1016/j.biocel.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 18.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450(7170):721. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 19.Aramburu J, Drews-Elger K, Estrada-Gelonch A, Minguillón J, Morancho B, Santiago V. et al. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem Pharmacol. 2006;72(11):1597–604. doi: 10.1016/j.bcp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–58. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates Period2 expression and the circadian clock. Nat Commun. 2013;4:1–11. doi: 10.1038/ncomms3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Zhou Y, Rychahou P, Liu C, Weiss HL, Evers BM. NFAT5 represses canonical Wnt signaling via inhibition of β-catenin acetylation and participates in regulating intestinal cell differentiation. Cell Death Dis. 2013;4(6):e671–9. doi: 10.1038/cddis.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman H, Chuderland D, Ninio-Many L, Hasky N, Kaplan-Kraicer R, Shalgi R. A novel regulatory pathway in granulosa cells, the LH/human chorionic gonadotropin-microRNA-125a-3p-Fyn pathway, is required for ovulation. FASEB J. 2015;29(8):3206–16. doi: 10.1096/fj.14-269449. [DOI] [PubMed] [Google Scholar]
- 24.Carletti MZ, Fiedler SD, Christenson LK. MicroRNA 21 Blocks Apoptosis in Mouse Periovulatory Granulosa Cells. Biol Reprod. 2010;83(2):286–95. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang W, Yu F, Yao H, Cui X, Jiao Y, Lin L. et al. MiR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene. 2014;33(20):2629–38. doi: 10.1038/onc.2013.214. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Liang H, Zhou G, Hu X, Liu Z, Jin F. et al. HIC1 and miR-23∼27∼24 clusters form a double-negative feedback loop in breast cancer. Cell Death Differ. 2017;24(3):421–432. doi: 10.1038/cdd.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie M, Yu S, Peng S, Fang Y, Wang H, Yang X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol Reprod. 2015;93(4):98. doi: 10.1095/biolreprod.115.130690. [DOI] [PubMed] [Google Scholar]
- 28.Kim YJ, Ku SY, Kim YY, Liu HC, Chi SW, Kim SH. et al. MicroRNAs transfected into granulosa cells may regulate oocyte meiotic competence during in vitro maturation of mouse follicles. Hum Reprod. 2013;28(11):3050–61. doi: 10.1093/humrep/det338. [DOI] [PubMed] [Google Scholar]
- 29.Di Gennaro A, Damiano V, Brisotto G, Armellin M, Perin T, Zucchetto A, A p53/miR-30a/ZEB2 axis controls triple negative breast cancer aggressiveness. Cell Death Differ; 2018. p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang M, Yao G, Yin M, Lü M, Tian H, Liu L. et al. Transcriptional cooperation between p53 and NF-κB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Mol Cell Endocrinol. 2013;370(1-2):119–29. doi: 10.1016/j.mce.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Laptenko O, Prives C. Transcriptional regulation by p53: One protein, many possibilities. Cell Death and Differentiation. 2006;13(6):951–61. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 32.Pinto M, Pickrell AM, Wang X, Bacman SR, Yu A, Hida A. et al. Transient mitochondrial DNA double strand breaks in mice cause accelerated aging phenotypes in a ROS-dependent but p53/p21-independent manner. Cell Death Differ. 2017;24(2):288–299. doi: 10.1038/cdd.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170(6):1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A. et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maqbool R, Lone SN, Ul Hussain M. Post-transcriptional regulation of the tumor suppressor p53 by a novel miR-27a, with implications during hypoxia and tumorigenesis. Biochem J. 2016;473(20):3597–610. doi: 10.1042/BCJ20160359. [DOI] [PubMed] [Google Scholar]
- 37.Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C. et al. Downregulation of miR-23a and miR-27a following Experimental Traumatic Brain Injury Induces Neuronal Cell Death through Activation of Proapoptotic Bcl-2 Proteins. J Neurosci. 2014;34(30):10055–71. doi: 10.1523/JNEUROSCI.1260-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towers CG, Guarnieri AL, Micalizzi DS, Harrell JC, Gillen AE, Kim J. et al. The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p. Nat Commun. 2015;6:10077. doi: 10.1038/ncomms10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11(7):1081–1098. doi: 10.1080/15548627.2015.1040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Zhang L, Hu Y, Chen M, Han F, Qin Y. et al. ß-catenin directs the transformation of testis Sertoli cells to ovarian granulosa-like cells by inducing Foxl2 expression. J Biol Chem. 2017;292(43):17577–17586. doi: 10.1074/jbc.M117.811349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaws J a, Hirshfield a N, Hewitt J a, Babus JK, Furth P a. Effect of bcl-2 on the primordial follicle endowment in the mouse ovary. Biol Reprod. 2001;64(4):1153–9. doi: 10.1095/biolreprod64.4.1153. [DOI] [PubMed] [Google Scholar]
- 42.Choi J, Jo M, Lee E, Choi D. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertil Steril. 2011;95(4):1482. doi: 10.1016/j.fertnstert.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Humana Press. 2014;1079:105–16. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Peng Y, Xiang H, Chen C, Zheng R, Chai J, Peng J. et al. MiR-224 impairs adipocyte early differentiation and regulates fatty acid metabolism. Int J Biochem Cell Biol. 2013;45(8):1585–93. doi: 10.1016/j.biocel.2013.04.029. [DOI] [PubMed] [Google Scholar]