Abstract
Glioma is one of the most common brain tumors, suggesting the importance of investigating the molecular mechanism of gliomas. We studied the roles of Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) in glioma. Expressions of RRM2 are higher in glioma tissues evidenced by TCGA data, western blot and immunohistochemistry. RRM2 is negatively correlated with glioma patient's survival. RNA-seq showed that genes involved in apoptosis, proliferation, cell adhesion and negative regulation of signaling were up-regulated upon RNAi-mediated knock-down of RRM2. Cell phenotypes specific for stably knocking down RRM2 were determined using stable transfection in vitro. In an in vivo model, knock-down of RRM2 inhibited tumor growth and caused suppression of AKT and ERK1/2 signalings. Interfering RRM2 also down-regulated the expression of cyclin A, cyclin B1, cyclin D1, Vimentin, and N-cadherin, and elevated E-cadherin expression. Moreover, overexpression of RRM2 failed to increase the expression of cyclin B1, cyclin D1, and N-cadherin when phosphorylation of AKT and ERK1/2 was suppressed by LY294002 or PD98059. These findings indicated that RRM2 is a positive regulator of glioma progression which contributes to the migration and proliferation of glioma cells through ERK1/2 and AKT signalings and might be a novel prognostic indicator for glioma patients.
Keywords: RRM2, Knock-down, G2/M phase arrest, ERK1/2, AKT
Introduction
Glioma is the most common type of primary brain tumor in China, the United States, and European countries 1, 2, accounting for about 80% of tumors starting in the brain per year 3. The treatment of gliomas includes surgical resection coupled with radiotherapy and chemotherapy 4. Unfortunately, gliomas often relapse in almost all patients, and the prognosis of glioblastoma (GBM, WHO grade IV glioma) patients remains dismal 5, 6. It is characterized as invasive, whose underlying molecular mechanisms need to be explored in detail 7, 8. Therefore, investigation of the mechanisms involved in tumor invasion, proliferation, and apoptosis may offer profound insights into the treatment of gliomas.
Deregulated cell cycle has been determined in many types of cancers, including GBM. Ribonucleotide reductase is an enzyme involved in the cell cycle. It is composed of two subunits, the regulatory subunit RRM1 and catalytic subunit RRM2 9, 10, which are essential for DNA replication and repair 11. An abnormally upregulated RRM2 expression has been demonstrated to promote fast cell division by increasing dNTP accumulation and is therefore involved in many kinds of cancers including colorectal cancer, non-small cell lung cancer (NSCLC), breast cancer, bladder cancer, ovarian cancer, pancreatic cancer, head and neck cancer12-16. RRM2 is also related to poor prognosis in cancers. For example, cervical cancer patients with positive RRM2 expression exhibited a higher recurrent rate and lower survival than those with negative RRM2 expression 17. RRM2 also can be considered as a prognostic marker for NSCLC and pancreatic cancer 18, 19. Besides, it has been reported that RRM2 dysregulation is associated with chemoresistance during cancer treatment. Downregulation of RRM2 could overturn the AKT-mediated tamoxifen resistance and improve therapeutic efficacy in breast cancer 20, and a previous study showed that patients with lower RRM2 expression could overcome the gemcitabine resistance in pancreatic cancer 21.
The primary objective of the presented study was to characterize the function of RRM2 on glioma cells. Here, the transcriptome of RRM2 shRNA-transfected U87 glioma cells was analyzed by RNA-Seq to explore the effects of RRM2 knock-down. The biological function and molecular mechanism of RRM2 on migration, proliferation and apoptosis were investigated both in vivo and in vitro. Finally, on the grounds that PI3K/AKT and MAPK signalings pathways play an important role in the development of cancers, including gliomas 22, the relationship between RRM2, PI3K/AKT and MAPK has been explored for further research. Therefore, this study will contribute to a deeper understanding of RRM2 and develop efficient strategies against RRM2 overexpressing gliomas.
Materials and Methods
Data acquisition from TCGA (the cancer genome atlas) and survival analysis
Gene expression quantification data (HTSeq - FPKM) in TCGA-GBM project was downloaded from the TCGA using the TCGAbiolinks R package 23. A total of 156 tumor samples and 5 normal paired normal sample were included in the RRM2 expression comparison. To evaluate the relationship between RRM2 expression and survival of GBM patients, Kaplan-Meier survival curve (overall) was analyzed using the SynTarget/BioProfiling database 24 for the TCGA_GBM dataset (121 patient samples).
Cell culture
Human glioma cell lines, U87 and LN-229, were purchased from ATCC (American Type Culture Collection) (Manassas, VA, USA). The cells were cultured in DMEM media supplemented with 0.5% Penicillin-Streptomycin Solution (Gibco, Waltham, MA, USA) and 10% fetal bovine serum (FBS) (Gibco, Waltham, MA, USA), maintained at 37 °C in humidified air with 5% CO2.
Histopathology and Immunohistochemistry
The adjacent normal and tumor samples of human patients were collected from Ba Yi Hospital, Nanjing, China. The study was performed with the approval of the Ethics Committee of Ba Yi Hospital. All tumor tissues were fixed in 10 % neutral formalin, trimmed appropriately in chemical hood after fixation (at least 24 hours). Then the samples were dehydrated and permeated with paraffin wax. Tissue samples were processed in melted paraffin in cassettes and frozen at -20°C until paraffin solidify completely. 4μm sections were prepared using microtome. The standard protocol used for the immunohistochemistry was provided in our previous study 25.The polyclonal anti-RRM2 and anti-Ki-67 (diluted at 1:100, Santa Cruz, CA) was incubated as the primary antibody overnight at 4°C. The slide without primary antibody incubation was used as negative control. After washing with phosphate-buffered saline (PBS), these tissues were incubated with the secondary antibodies, which were horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Envision kit, Dako, Denmark) following the manufacturer's instructions. At last, these sections were stained with hematoxylin. The TMA immunostaining images were captured using microscope from Carl Zeiss (Axio Observer A1, Jena, Germany) and analyzed by Image-Pro Plus 6.0 software.
Quantification of Ki67 and RRM2 expression
A total of 54 patients with brain tumors diagnosed at the department of Oncology-Pathology, Ba Yi Hospital, Nanjing, China. All histologic specimens were routinely stained for RRM2 and Ki-67 by immunohistochemistry. Tumors with histopathologic grading I and graded II were considered as low-grade tumors, whereas those graded III to IV were high-grade tumors. Permission to analyze the samples and correlate the results to patient data was obtained by the Ethics Committee of Ba Yi Hospital. Only distinct nuclear staining was accepted as a positive reaction for both markers. All cells with simultaneous nuclear staining and cytoplasmic RRM2 staining were regarded as positive for RRM2. The hot spot was arbitrarily defined as an area with 25 mm2 around the point with the highest concentration of cells immunoreactive for RRM2 or Ki-67 antigen in the tumor. In each area, the positive cells among 200 to 400 cells were counted in a X200 magnification manually. The RRM2and Ki-67 score were calculated as the percentage of positive cells. Data were analyzed using SPSS version 17.0 (SPSS, Chicago, Illinois). The correlation between RRM2 and Ki-67 was evaluated by Pearson correlation coefficient.
RRM2 knock-down in glioma cells
Plasmids of shRNAs targeting RRM2 and scrambled shRNA were purchased from GenePharma (Shanghai, China). The shRNA of RRM2 or scrambled was transfected into U87 or LN-229 cells using lipofectamine 2000 (Invitrogen, California, USA), followed by 40 days of selection with 600 μg/mL of puromycin. The RRM2 shRNAs (shRRM2#1 and shRRM2#2) target the sequences: GGU CCU ACA UUA GUG CCU UTT and GCU GUA CUA UCU GCG AAA UTT, respectively. RRM2 knock-down in these cells was confirmed by Western blot.
RNA library construction and sequencing
A total amount of 1μg qualified RNA per sample was used as input material for the library preparation. The sequencing libraries were generated using the VAHTS mRNA-seq v2 Library Prep Kit for Illumina® (Vazyme, NR601) following manufacturer's recommendations. Firstly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in Vazyme Frag/Prime Buffer. The cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers. Second strand cDNA synthesis was subsequently performed using buffer, dNTPs, DNA polymerase I and RNase H. Then, the cDNA fragments were end repaired with the addition of a single 'A' base at the 3'-end of each strand, ligated with the special sequencing adapters (Vazyme, N803) subsequently. The products were purified and size selected with VAHTSTM DNA Clean Beads (Vazyme, N411) in order to get appropriate size for sequencing. Following amplification, cDNA was purified and subjected to library construction for the paired-end 150 bp (PE150) sequencing on Illumina HiSeq X Ten platform.
Read alignment and differential expression analysis
Raw reads were trimmed to remove low-quality bases and adaptor sequences using Fastp (v0.18.0) 26 with default parameters. Ribosomal RNA were further removed by SortmeRNA (v2.1b) 27. After cleaning and quality checks, we obtained 45.7 and 56.2 million reads from control and RRM2 knock-down sample. Filtered reads were then aligned against human genome reference (hg38) by application of Hisat2 (v2.05) 28. Differentially expressed genes were deemed significant using GFOLD algorithm (v1.1.4) 29 which was more biologically meaningful for single replicate experiments. Genes with an absolute GFOLD ≠0 and fold change>1.3 were considered differentially expressed. Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO) over-representation test were performed using an R package clusterProfiler 30 (v3.9.2) to find enriched GO terms (p-value cutoff 0.05) in up- or down-regulated genes. GO terms were simplified using the simplify function of clusterProfiler. The data were plotted using the ggplot2 R package 31. All sequencing data were deposited in the sequence read archive (SRA) database and the BioProject ID is PRJNA505067.
Cell growth and colony formation assay
The growth of glioma cells was determined by the CCK-8 Kit (YEASEN, Shanghai, China) following the manufacturer's protocol. During the assay to examine colony formation, about 2,000 transfected cells per dish were plated in 6-well plate. After 14 days, the cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet for 30 minutes. Then gently wash it three times with PBS, the number of cell colonies was calculated.
Cell cycle analyzed by flow cytometry
60% of the digested Glioma cell lines which stably knocking out the gene of interest were added into the 6-well plate and incubating the cells for 12 hours overnight. Then Cells were collected by trypsinization after the PBS washing, fixed with 70% methanol for 5 hours at 4°C, and incubated with ribonuclease A for half an hour at 37°C. Their nuclei were then labeled in the dark with 100 ng/mL PI (propidium iodide). In the end, the cells were analyzed by a flow cytometer (Becton Dickinson, USA).
Cell apoptosis analyzed by flow cytometry
Detection of apoptotic cells was carried out using Annexin V-FITC/propidium iodide (PI)-labeled apoptosis detection kit (Beyotime, Shanghai, China). Briefly, glioma cells which stably transfected with shRRM2 or shNC were plated in 10 cm dishes and cultured in DMEM with 10% FBS. When the cells are over 90% in the dishes, the transfected cells were harvested and centrifuged for 5 min at 4°C, 250xg. Then, these cells were resuspended in cold binding buffer (10 mM HEPES buffer pH 7.4, 5 mM KCl, 150 mM NaCl, 1.8 mM CaCl2, and 1 mM MgCl2). These cells were firstly stained with FITC-labeled 25 ng/mL Annexin V in the dark for 30 min and then stained with 50 ng/mL propidium iodide for 5 min. These samples were run with a Becton Dickinson flow cytometer and the data were analyzed with Cell Quest software. In this study, the apoptotic cells were the population of Annexin V-positive and PI-negative cells. Annexin V and PI double positive cells were scored as necrotic or late apoptotic.
Wound-healing assay of glioma cells
The U87 or LN-229 cells which have been stably knocked down the RRM2 gene were planted in 6-well plates and the cells would grow to confluence after about 16 hours. Then the cells were cultured with serum-free DMEM for 12 h and then the monolayer was scratched by a 10-μL pipette tip. These cultured samples were photographed every 12 hours by a microscope from Carl Zeiss (Axio Observer A1, Jena, Germany). The number of the cells migrated to the wound was calculated.
Transwell migration assay
The glioma cells (U87 and LN-229) stably expressing shRRM2 or shNC were cultured in DMEM with 0.1% FBS for 12 hours. Then, 1 × 105 cells were placed on the upper layer of a cell culture insert with permeable membrane (8 μm pore size, Corning, Shanghai, China), and DMEM with 10% FBS was applied to the lower layer. After incubation at 37 °C for 14 hours, migrated cells in the lower layer were fixed with paraformaldehyde and stained with crystal violet to visualize nuclei, and non-migrated cells in the upper layer were removed. The number of migrating cells in six fields was counted under ×200 magnification. The experiments were repeated three times.
In vivo tumor growth
All animal procedures were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University (SYXK 2015-0015). All animals received human care and all animal experiments were conducted in accordance with the relevant guidelines and regulations. The animals were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). About 6 × 106 U87 cells (200 μL) stably transfected with shRRM2 or shNC were injected subcutaneously into the right flank of each Nude mouse (4 weeks old male nude mice, n=4). The mice were housed in cages with sawdust bedding in a specific pathogen-free room at a temperature of 25-26°C and a relative humidity of ~50%, light 12 h/day. Tumor volumes and body weight were measured by vernier calipers every five days. Tumor volume was assessed conforming to the formula: volume = (longest diameter x shortest diameter2)/232. Mice were sacrificed at the fortieth day after the beginning intratumoral injection of glioma cells. The tumors were harvested and stored at -80°C for further studies.
Western blot
The cells or tissues from in vitro or in vivo experiments were lysed in RIPA Buffer (1 mM EDTA pH 8.0, 50 mM Tris-HCl pH 8.0, 2% SDS, 5 mM DTT), and their protein concentration was determined by the BCA assay (Beyotime, Shanghai, China). The total protein (100 μg) was separated by 10%-15% SDS-PAGE gel and transferred to PVDF (polyvinylidene fluoride) membranes (Invitrogen, California, USA), and blocked with 5% skimmed milk powder in PBST (Phosphate Buffered Saline with Tween) or 5% w/v BSA, 1×TBS, 0.1% Tween at 4°C with gentle shaking about 12 hours. The membranes were immunoblotted with the primary antibodies for 4 hours at room temperature or overnight at 4°C and then incubated with the secondary antibodies for 2 hours. The corresponding semi-quantitative analysis was based on optical density with Image J software. In western blot analysis, cyclin B1, cyclin D1, BCL-2 (Boster Biological Technology), cyclin A, BAX, PARP, caspase-3, Vimentin (Proteintech), GAPDH (Bioworld), RRM2, E-cadherin, N-cadherin, phospho-AKT (T308), and phospho-ERK1/2 (T202/Y204) (Cell Signaling) were used for immunoblotting with secondary antibodies conjugated with horseradish peroxidase (Bioworld) and visualized with Bio-Rad ChemiDoc™ Imagers. All other chemical reagents were bought from Sigma (Shanghai, China).
Statistical analysis
All the results are shown as mean ± SD. Statistically significant differences between two groups were assessed by χ2 analysis and student's t-test using GraphPad Prism 7.0. P-value < 0.05 was considered to be statistically significant.
Results
1. RRM2 expression is enhanced in human glioma tissues
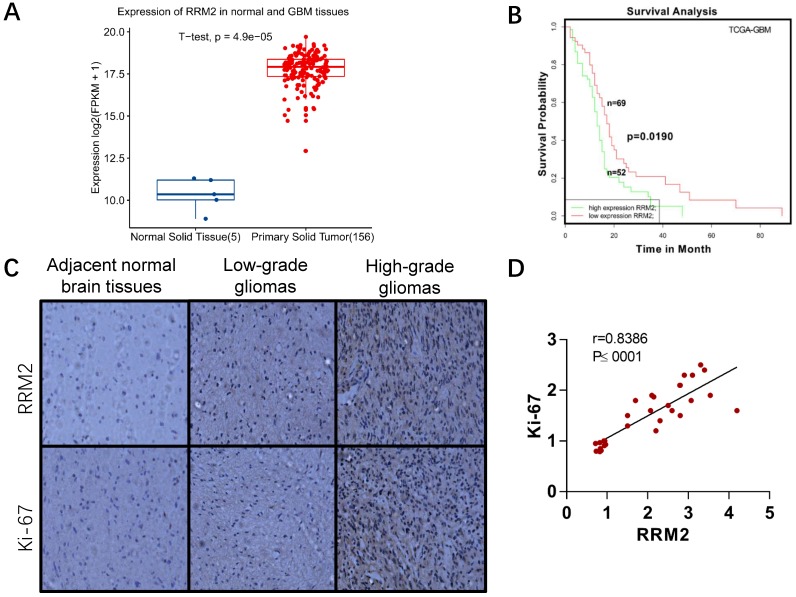
To evaluate the significance of RRM2 in glioma patients, we compared its expression between primary tumor and normal tissue group in TCGA-GBM project. RRM2 were statistically increased in tumor group as shown in Fig. 1A. It has been demonstrated that high RRM2 expression is an indicator of poor prognosis in many types of cancer 17-19, 33-35. Here, we analyzed the relationship between RRM2 expression and the survival of GBM patients. Survival data were retrieved from SynTarget/BioProfiling database (TCGA_GBM) 24. Patients with higher RRM2 expression rank than the average RRM2 expression rank across the TCGA_GBM dataset were classified as the high RRM2 group (52 cases), and the others were in the low RRM2 group (69 cases). The overall survival was compared between the two groups. The results showed that high RRM2 patients exhibited a significantly poorer survival outcome than the low RRM2 patients (Fig. 1B). Thus, RRM2 might be considered as a prognostic marker for GBM.
Fig 1.
Expression of RRM2 in human gliomas and normal brain tissues. A Expression of RRM2 in human normal brain tissues and GBM tissues from TCGA-GBM project. B Survival curve of the effect of the RRM2 gene on the survival time of GBM patients. C Immunohistochemistry (original magnification, ×200): Compared with adjacent normal human brain tissue and low-grade glioma, the expression of RRM2 and Ki-67 were significantly up-regulated in high-grade glioma. D The correlation between RRM2 and Ki-67 in human gliomas analyzed by the Pearson's correlation coefficient test.
We also detected the expression of RRM2 and Ki-67 protein in paraffin sections of glioma samples and adjacent normal samples of brain tissues. As shown in Fig. 1C-D, there was much more positive staining of RRM2 as well as Ki-67 staining in high-grade gliomas than in low-grade gliomas and adjacent normal brain tissues. It suggested that RRM2 expression was positively associated with Ki-67 expression and the grade of gliomas (Fig. 1D). Additionally, the correlation with high expression of RRM2 and high grade of gliomas was not affected by patients' age, gender, and tumor size (Table 1).
Table 1.
RRM2 expression and clinical characteristics of the 54 glioma patients.
| Clinicopathologic feature | n (%) | RRM2 | P-Value | ||
|---|---|---|---|---|---|
| Low | High | mean ± SEM | |||
| Age | P=0.266 | ||||
| ≤40 | 22 (40.74%) | 9 | 13 | 2.54±0.21 | |
| ≥40 | 32 (59.26%) | 14 | 18 | 2.60±0.18 | |
| Gender | P=0.2027 | ||||
| Female | 16 (29.62%) | 7 | 9 | 1.81±0.17 | |
| Male | 38 (70.37%) | 15 | 23 | 1.87±0.15 | |
| Surgery | P=0.3836 | ||||
| Biopsy | 8 (14.81%) | 3 | 5 | 2.31±0.24 | |
| Partial resection | 17 (31.48%) | 5 | 12 | 2.45±0.22 | |
| Gross total resection | 29 (53.71%) | 10 | 19 | 2.37±0.27 | |
| WHO Grade (%) | P<0.0001 | ||||
| ІІ | 16 (29.62%) | 6 | 10 | 1.71±0.23 | |
| ІІІ | 18 (33.33%) | 4 | 14 | 2.29±0.21 | |
| IV | 20 (37.05%) | 5 | 15 | 2.51±0.25 | |
| Low expression | 21 (39.00%) | 17 | 4 | 2.04±0.19 | P<0.0001 |
| High expression | 33 (61.00%) | 5 | 28 | 3.52±0.27 | |
| Tumor size | P=0.3388 | ||||
| <4cm | 23 (42.59%) | 11 | 12 | 2.45±0.22 | |
| ≥4cm | 31 (57.41%) | 13 | 18 | 2.39±0.23 | |
Statistical analyses were performed by the Pearson χ2 test. P < 0.05 was considered as statistically significant.
2. Effect of RRM2 knock-down on glioma using RNA-seq
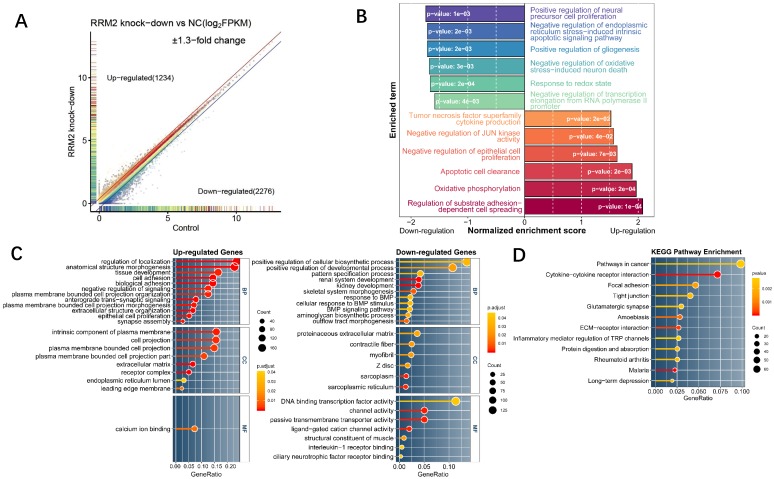
In order to characterize the function of RRM2 in glioma cell line, RNA-seq of U87 cell line on an RRM2 knock-down condition and a normal condition was designed to explore the global transcriptome changes. As a result, RNAi-mediated knock-down of RRM2 lead to 1234 up-regulated and 2276 down-regulated highly differentially expressed genes (Fig. 2A). To investigate the most significant biological function upon knock-down, GSEA and GO enrichment analyses were performed on the whole transcriptome and highly changed genes. GSEA for the whole transcriptome demonstrated that the genes whose expression was elevated upon RRM2 knock-down were significantly enriched for regulation of substrate adhesion-dependent cell spreading, apoptotic cell clearance, negative regulation of JUN kinase activity and negative regulation of epithelial cell proliferation (Fig. 2B). Meanwhile, as a complementary approach, GO enrichment was employed in order to improve our insight into these highly differentially expressed genes function. For up-regulated genes, cell adhesion, negative regulation of signaling, epithelial cell proliferation were the most enriched terms which have an extremely low adjusted p-value. However, positive regulation of cellular biosynthetic process has the maximum ratio of down-regulated genes and renal system development was the most significant (Fig. 2C). We also observed that these differentially expressed genes were mainly involved in the cancer pathway (Fig. 2D). Taken together, these results suggest that RRM2 may involve in tumorigenesis by affecting proliferation activity and apoptosis of glioma cell. Change in transcriptome indicated that the negative feedback regulation was activated upon RRM2 knock-down.
Fig 2.
Expression profile analysis of RRM2 knockdown in U87 glioma cells by RNA-Seq and bioinformatics. A Scatter plot for differentially expressed genes between RRM2 knock-down group and control. Grey points indicate the non-significant differentially expressed gene whose GFOLD value is zero. Colored points were significant differentially expressed genes (GFOLD≠0). With an additional strict cutoff of 1.3-fold change, 1234 and 2276 genes were determined as highly up- or down-regulated genes, respectively. B Differences in biological process scored by GSEA between the knock-down group and the control group. Shown are normalized enrichment scores (NES) indicating whether the GO gene sets is mobilized as a whole by RRM2 knock-down, either toward down-regulation (NES < 0) or toward up-regulation (NES > 0). C and D Gene ontology and KEGG pathway enrichment analysis which shows the distribution of terms exhibiting statistically significant differences (Fisher exact test, p-values were adjusted for multiple testing using the Benjamini-Hochberg procedure which were represented with the color scale). Only genes with at least 1.3-fold change were included to perform enrichment analysis.
3. Knock-down of RRM2 suppresses the migration of glioma cells
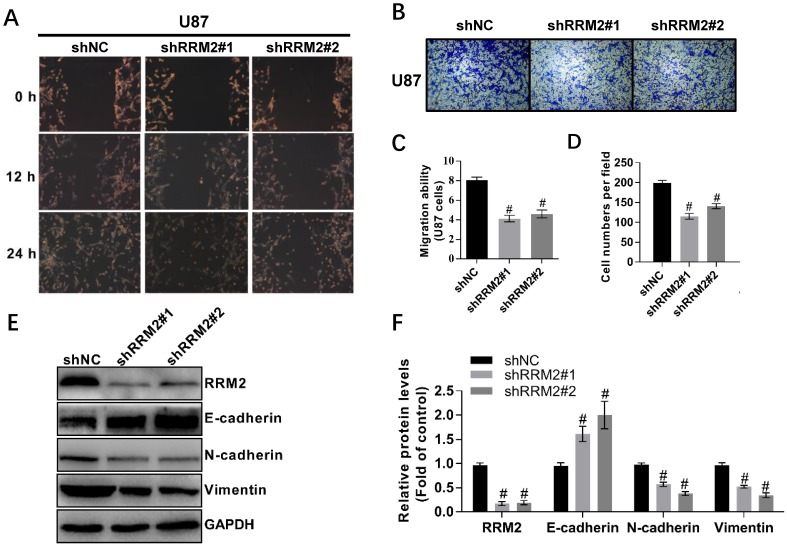
To investigate the effect of RRM2 on the migration of U87 and LN-229 cells, we used wound-healing and transwell assays. We took the representative photomicrographs of the assays every 12 hours after the monolayers scratched. Knock-down of RRM2 decreased the migration of U87 and LN-229 cells compared to the control. In addition, the transwell assay showed that the silence of RRM2 in U87 and LN-229 cells decreased the migrating cells (Fig. 3A-D, Supplementary Figure S1). To further explore the potential role of RRM2 in the migration of glioma cells, we investigated the involvement of RRM2 in regulating EMT biomarkers. Interfering with shRNAs upregulated E-cadherin levels and downregulated the levels of N-cadherin and vimentin (Fig. 3E-F). Therefore, RRM2 overexpression has the potential to facilitate the migration of glioma cells.
Fig 3.
Knock-down of RRM2 inhibits the migration of glioma cells. A and C Wound-healing assays with shNC (negative control), RRM2-shRNA#1 (shRRM2#1), and RRM2-shRNA#2 (shRRM2#2) transfected U87. Migration of the cells to the wound was observed at 0, 12, and 24 h with a microscope from Carl Zeiss (Axio Observer A1; ×200 magnification). Data shown are mean ± SD of three independent experiments, # P< 0.001, versus shNC (negative control). B and D Knock-down of RRM2 inhibited the migration ability of U87 cells by trans-well assays; Data shown are mean ± SD, # P< 0.001, versus shNC. E and F RRM2, E-cadherin, Vimentin, N-cadherin were detected by Western blot analysis and GAPDH was used as an internal control. data shown are mean ± SD of three independent experiments, # P< 0.001, versus shNC.
4. Knock-down of RRM2 discourages the proliferation of glioma cells
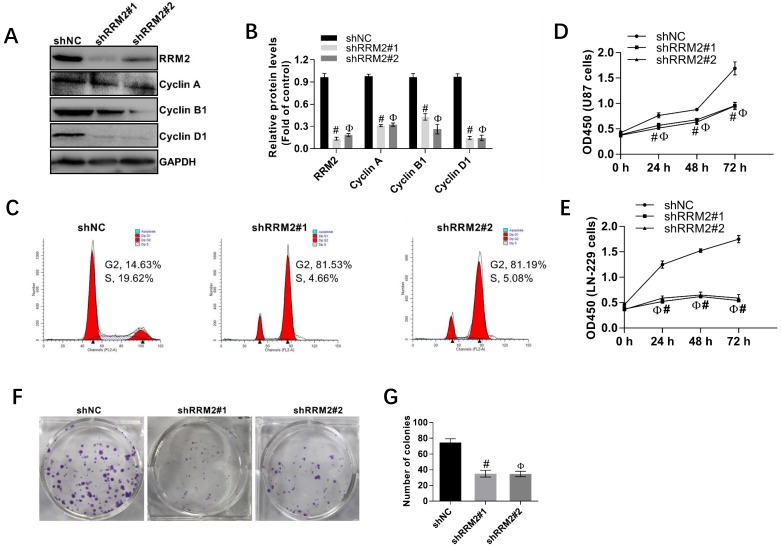
Human glioma cell lines, U87, and LN-229 cells were transfected with shNC, shRRM2#1, and shRRM2#2 and the knock-down efficiency of shRNAs was validated by Western blot. As depicted in Fig. 4A-B, when RRM2 expression was knocked down with shRNAs transfection in U87 cells, the expression of cyclin A, cyclin B1 and cyclin D1 were also downregulated. Furthermore, shRNAs increased the percent of U87 cells in G2 and M phase compared with the corresponding control (shNC) shown by the flow cytometry analysis (Fig. 4C). The CCK-8 assay also showed that knock-down of RRM2 remarkably attenuated the proliferation of U87 and LN-229 cells (Fig. 4D-E). Similarly, colony formation assay proved that low expression of RRM2 interfered with shRRM2 significantly repressed the proliferation of U87 cells (Fig. 4F-G). Taken together, these results suggest that downregulation of RRM2 significantly blocks the cell cycle developing from one stage to another, which leads to the inhibition of cell proliferation. Conversely, RRM2 overexpression can promote the proliferation of glioma cells.
Fig 4.
Interfering RRM2 inhibits the proliferation of cancer cells. A and B Western blot analysis showed that cyclin A, cyclin B1, and cyclin D1 were down-regulated in RRM2-transfected U87 cells. data expressed as mean ± SD of three independent experiments, # P<0.001 and Ф P<0.001, versus shNC. C RRM2 depletion significantly increased the proportion of U87 cells in G2 and M phase using flow cytometry. D and E Cell proliferation was detected by CCK-8 assay in U87 and LN229 cells; data expressed as mean ± SD of three independent experiments, # P< 0.001 and Ф P< 0.001, versus shNC. F and G Silencing of RRM2 inhibited the growth of U87 cells analyzed by colony-forming assay. The colonies were calculated. data expressed as mean ± SD of three independent experiments, # P< 0.001 and Ф P< 0.001, versus shNC.
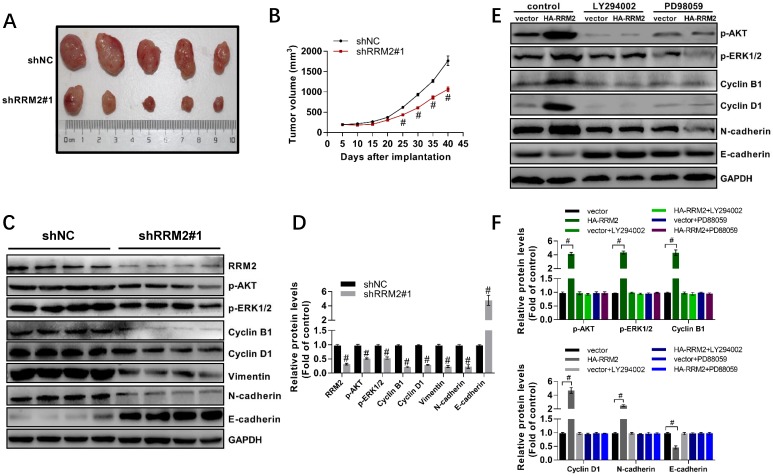
5. Knock-down of RRM2 inhibits proliferation of U87 cells in vivo and inactivates MAPK and AKT
In this study, we found that interfering RRM2 in nude mice could reduce the growth of U87 cells significantly and result in a smaller tumor size compared with the control (Fig. 5A,5B). Furthermore, much attention should be paid to that injection with glioma cells with stably-transfected shRRM2 down-regulated the levels of cyclin B1, cyclin D1, Vimentin, N-cadherin, phospho-ERK1/2 (T202/Y204) and phospho-AKT (T308), and increased E-cadherin levels in tumors of nude mice (Fig. 5C, 5D). Therefore, these results indicated that RRM2 knock-down can exert a strong antitumor effect in vivo, while RRM2 overexpression is shown to be capable of promoting tumor growth.
Fig 5.
RRM2 contributes to the proliferation of U87 cells and the activation of MAPK and AKT pathway in vivo. A RRM2-knock-down U87 cells were injected into nude mice and the tumors were harvested. B The volume of tumors was measured every 5 days (mean ± SD); # p< 0.001 versus shNC. C and D expression of indicated proteins were analyzed by western blot. data shown are mean ± SD, # p< 0.001 versus shNC. E and F U87 cells were transfected with plasmid used for overexpression of RRM2 protein. After 10 h transfection, the U87 cells were incubated with LY294002 (5 μM) or PD98059 (5 μM) for 12 h. Cells were then harvested for Western blot analysis. Data shown are mean ± SD, # p< 0.001 versus shNC.
To investigate the role of AKT and MAPK in RRM2 mediated migration, proliferation, and apoptosis of glioma cells, RRM2 was overexpressed in the U87 cells. 10 h after transfection, these cells were incubated with LY294002 or PD98059. The results showed that overexpression of RRM2 significantly elevated the phosphorylation of AKT and ERK1/2. Meanwhile, we found that overexpression of RRM2 increased the expression of cyclin B1, cyclin D1, and N-cadherin, and decreased the protein levels of E-cadherin. However, the effects induced by overexpression of RRM2 were blocked by LY294002 and PD98059 obviously (Fig. 5E-F). In another word, RRM2 failed to promote the migration and proliferation of U87 cells when the AKT and MAPK pathways were inhibited by LY294002 or PD98059.
Discussion
Gliomas were one of the most common and serious human cancers. According to the criteria of WHO, human gliomas are classified into grade I, II, III, and IV 36, 37. The diffusive infiltrative and invasive characteristics of cancer cells have been the main cause of the death of glioma patients 38. Therefore, it is extremely necessary to study the molecular mechanisms of gliomas progression and prognosis. This study could offer novel targets for developing therapeutic strategies for patients with glioma.
RRM2 is often overexpressed in many kinds of cancers, such as colorectal cancer, non-small cell lung cancer (NSCLC), breast cancer, bladder cancer, ovarian cancer, pancreatic cancer, head and neck cancer and lung cancer 12-16. RRM2 also can be considered as a prognostic marker for cancers including NSCLC, cervical cancer, and pancreatic cancer 17-19. Additionally, RRM2 is associated with chemo-resistance during cancer treatment. Previous studies have demonstrated that suppression of RRM2 is able to efficiently increase the survival of glioma patients and improve the chemo-resistance 20, 21. In the current study, we found that RRM2 was highly expressed in high-grade glioma tissue, which was shown by microarray analysis and IHC. The expression of RRM2 was significantly correlated with Ki-67 (Fig. 1D), a well-known indicator of cancer development and prognosis 39. Moreover, the data showed that low RRM2 expression exhibited better survival evidenced by microarray database. Thus, RRM2 may be a promising marker of glioma prognosis.
Nowadays, RNA-Seq was applied to comprehensively analyze the potential function of RRM2 in gliomas. RNA-Seq is a perfect method for transcriptome profiling that could offer more precise measurement of gene expressions 40. Here, we found that 1234 genes exhibited upregulated expression and 2276 genes exhibited downregulated levels in shRRM2-transfected U87 cells. The further GSEA analysis demonstrated that aberrant genes were distributed in regulation of substrate adhesion-dependent cell spreading, apoptotic cell clearance, negative regulation of JUN kinase activity and negative regulation of epithelial cell proliferation. GO terms of highly differentially expressed genes were also enriched in cell adhesion, negative regulation of signaling, epithelial cell proliferation, which were consistent with GSEA to a certain extent (Fig. 2A-2D). Together, the bioinformatics analysis indicated that a variety of biological events could be modulated by RRM2.
It has been reported that inhibition of AKT signaling always activates the intrinsic apoptotic pathway mediated by mitochondria in glioma cells 41, 42. Both AKT and ERK1/2 signalings play an essential role in invasion and apoptosis of glioma cells by modulating cyclin A, cyclin B1, cyclin D1, N-cadherin and E-cadherin, and cleaved caspase-3 22. Here, the results suggested that interfering RRM2 obviously inhibited the growth of U87 cells in nude mice, and knock-down of RRM2 decreased the tumor volume (Fig. 5A-5B). In in vivo study, we found a downregulation in the phosphorylation of ERK1/2 and AKT, as well as the expression of cyclin B1, cyclin D and N-Cadherin. Contrarily, the levels of E-cadherin, a repressor of cell invasion, were strikingly elevated (Fig. 5C-5D). Next, overexpression of RRM2 was able to enhance the phosphorylation of AKT and ERK1/2, and increase the protein levels of their downstream targets including cyclin B1, cyclin D1, and N-cadherin. However, overexpression of RRM2 failed to upregulate the expression of cyclin B1, cyclin D1, and N-cadherin when the phosphorylation of AKT and ERK1/2 was inhibited by LY294002 or PD98059 (Fig. 5E-5F). Consequently, RRM2 regulated the invasion and apoptosis of glioma cells depending on AKT and ERK1/2 pathways, indicating that the role of RRM2 in progression and prognosis of glioma could be mediated by AKT and ERK1/2 signalings.
In in vitro study, we also investigated the role of RRM2 in migration, proliferation, and apoptosis of glioma cells. We found that knock-down of RRM2 suppressed the growth of glioma cells, and RRM2 may perform as a regulator of cell cycle during G2-M phase (Fig. 4C-4G). To explore the regulating mechanisms of RRM2 in cell cycle of U87 cells, we determined the expression of cyclin A, cyclin B1, and cyclin D1 (Fig. 4A-4B), which are essential regulators of cell cycle 22, and we observed that their expression was decreased after knock-down of RRM2. Meanwhile, knock-down of RRM2 significantly suppressing the migration of U87 cells evidenced by trans-well assay and wound-healing assay (Fig. 3A-3D). Knock-down of RRM2 also significantly elevated the expression of E-cadherin (Fig. 3E-3F), a suppressor of glioma development and invasion 43. Additionally, more apoptotic cells were detected by flow cytometry in the shRRM2-transfected cells than the negative control, accompanied by upregulation of cleaved caspase-3 and downregulation of BCL-2 (Fig. 3E-3F), indicating that an important role of RRM2 in balancing apoptosis and proliferation.
In conclusion, RRM2 plays an important role in many molecular pathological processes evidenced by RNA-Seq. RRM2 enhances growth, migration and inhibits apoptosis of glioma cells in vivo and in vitro through upregulating phosphorylation of ERK1/2 and AKT. Suppression of RRM2 may be considered as a potential target for the treatment of RRM2-overexpressing gliomas.
Supplementary Material
Supplementary figures.
Supplementary table.
Acknowledgments
Authors' contributions
Y.B.Y. and J.M.J. conceived and designed the experiments. S.H.Z., Z.H., Y.B.Y., S.J.W., Z.Y.N., X.T.Y., Y.Z.N., X.J.C. and Y.Z.H. carried out the experiments. Y.B.Y., Z.H. and X.Z.P. analyzed the data. H.M. contributed the reagents and materials. S.H.Z., Z.H., Y.B.Y., J.M.J., W.C.Y., and Z.Y.S drafted the manuscript. All authors reviewed the final manuscript.
Funding
This project was sponsored by Nanjing Medical University (2013NJMU008, 13JC007).
Declarations
Ethics approval and consent to participate: all the experimental procedures were approved by the animal care committee of Nanjing Medical University, Nanjing, Jiangsu, 211166, China. Consent for publication: This manuscript has been approved by all authors for publication.
References
- 1.Porter KR, McCarthy BJ, Freels S, Kim Y, Davis FG. Prevalence estimates for primary brain tumors in the United States by age, gender, behavior, and histology. Neuro-Oncology. 2010;12:520–7. doi: 10.1093/neuonc/nop066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang T, Tang GF, Lin Y, Peng XX, Zhang X, Zhai XW. et al. Prevalence estimates for primary brain tumors in China: a multi-center cross-sectional study. Chinese Med J-Peking. 2011;124:2578–83. [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C. et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro-Oncology. 2016;18:v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 5.Bartek J, Mistrik M, Bartkova J. Thresholds of replication stress signaling in cancer development and treatment. Nat Struct Mol Biol. 2012;19:5–7. doi: 10.1038/nsmb.2220. [DOI] [PubMed] [Google Scholar]
- 6.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From Molecular Pathology to Targeted Treatment. Annu Rev Pathol-Mech. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 7.Meyer MA. Malignant gliomas in adults. New Engl J Med. 2008;359:1850. doi: 10.1056/NEJMc086380. - [DOI] [PubMed] [Google Scholar]
- 8.Vehlow A, Cordes N. Invasion as target for therapy of glioblastoma multiforme. Bba-Rev Cancer. 2013;1836:236–44. doi: 10.1016/j.bbcan.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Torrents E, Aloy P, Gibert I, Rodriguez-Trelles F. Ribonucleotide reductases: Divergent evolution of an ancient enzyme. J Mol Evol. 2002;55:138–52. doi: 10.1007/s00239-002-2311-7. [DOI] [PubMed] [Google Scholar]
- 10.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–53. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 11.Nordlund N, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 12.Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885–92. doi: 10.1111/j.1365-2559.2010.03725.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu XY, Zhou BS, Xue LJ, Yen F, Chu PG, Un F. et al. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer. 2007;6:374–81. doi: 10.3816/CCC.2007.n.007. [DOI] [PubMed] [Google Scholar]
- 14.Rahman MA, Amin RMR, Wang DS, Koenig L, Nannapaneni S, Chen ZJ. et al. RRM2 Regulates Bcl-2 in Head and Neck and Lung Cancers: A Potential Target for Cancer Therapy. Clin Cancer Res. 2013;19:3416–28. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida Y, Tsunoda T, Doi K, Tanaka Y, Fujimoto T, Machida T. et al. KRAS-mediated Up-regulation of RRM2 Expression Is Essential for the Proliferation of Colorectal Cancer Cell Lines. Anticancer Res. 2011;31:2535–9. [PubMed] [Google Scholar]
- 16.Xia GG, Wang HC, Song ZL, Meng QC, Huang XY, Huang XY. Gambogic acid sensitizes gemcitabine efficacy in pancreatic cancer by reducing the expression of ribonucleotide reductase subunit-M2 (RRM2) J Exp Clin Cancer Res; 2017. p. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su YF, Wu TF, Ko JL, Tsai HT, Tee YT, Chien MH, The Expression of Ribonucleotide Reductase M2 in the Carcinogenesis of Uterine Cervix and Its Relationship with Clinicopathological Characteristics and Prognosis of Cancer Patients. PLoS One; 2014. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mah V, Alavi M, Marquez-Garban DC, Maresh EL, Kim SR, Horvath S, Ribonucleotide Reductase Subunit M2 Predicts Survival in Subgroups of Patients with Non-Small Cell Lung Carcinoma: Effects of Gender and Smoking Status. PLoS One; 2015. p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Meng L, Wang XW, Ma GY, Chen JH. Expression of RRM1 and RRM2 as a novel prognostic marker in advanced non-small cell lung cancer receiving chemotherapy. Tumor Biol. 2014;35:1899–906. doi: 10.1007/s13277-013-1255-4. [DOI] [PubMed] [Google Scholar]
- 20.Wu P, Dugoua JJ, Eyawo O, Mills EJ. Traditional Chinese medicines in the treatment of hepatocellular cancers: a systematic review and meta-analysis. J Exp Clin Cancer Res; 2009. p. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ZJ, Liu XF, Ho RLKY, Lam CWK, Chow MSS. Precision or Personalized Medicine for Cancer Chemotherapy: Is there a Role for Herbal Medicine. Molecules; 2016. p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan YY, Zhang YL, Chen LJ, Liu Y, Feng YL, Yan JY. The Critical Role of Rab31 in Cell Proliferation and Apoptosis in Cancer Progression. Mol Neurobiol. 2016;53:4431–7. doi: 10.1007/s12035-015-9378-9. [DOI] [PubMed] [Google Scholar]
- 23.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amelio I, Tsvetkov PO, Knight RA, Lisitsa A, Melino G, Antonov AV. SynTarget: an online tool to test the synergetic effect of genes on survival outcome in cancer. Cell Death Differ. 2016;23:912. doi: 10.1038/cdd.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu AB, Wu B, Guo JS, Luo WR, Wu D, Yang HL, Elevated expression of CDK4 in lung cancer. J Transl Med; 2011. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics. 2012;28:2782–8. doi: 10.1093/bioinformatics/bts515. [DOI] [PubMed] [Google Scholar]
- 30.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiebig HH, Berger DP, Winterhalter BR, Plowman J. In vitro and in vivo evaluation of US-NCI compounds in human tumor xenografts. Cancer treatment reviews. 1990;17:109–17. doi: 10.1016/0305-7372(90)90034-d. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Liu XY, Warden CD, Huang YS, Loera S, Xue LJ, Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer; 2014. p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Zhang HW, Du YH, Gu XM. Prognostic significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with advanced non-small cell lung cancer receiving chemotherapy. Tumor Biol. 2014;35:12679–88. doi: 10.1007/s13277-014-2592-7. [DOI] [PubMed] [Google Scholar]
- 35.Tang H, Xiao GH, Behrens C, Schiller J, Allen J, Chow CW. et al. A 12-Gene Set Predicts Survival Benefits from Adjuvant Chemotherapy in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2013;19:1577–86. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollon T, Hervey-Jumper SL, Sagher O, Orringer DA. Advances in the Surgical Management of Low-Grade Glioma. Semin Radiat Oncol. 2015;25:181–8. doi: 10.1016/j.semradonc.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzo D, Ruggiero A, Martini M, Rizzo V, Maurizi P, Riccardi R. Molecular Biology in Pediatric High-Grade Glioma: Impact on Prognosis and Treatment. Biomed Res Int; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang T, Zhao B, Li XC, Wan JH. ARPP-19 promotes proliferation and metastasis of human glioma. Neuroreport. 2016;27:960–6. doi: 10.1097/WNR.0000000000000638. [DOI] [PubMed] [Google Scholar]
- 39.Su C, Liu C, Zhao L, Jiang J, Zhang J, Li S. et al. Amide Proton Transfer Imaging Allows Detection of Glioma Grades and Tumor Proliferation: Comparison with Ki-67 Expression and Proton MR Spectroscopy Imaging. American Journal of Neuroradiology. 2017;38:1702–9. doi: 10.3174/ajnr.A5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mure H, Matsuzaki K, Kitazato KT, Mizobuchi Y, Kuwayama K, Kageji T. et al. Akt2 and Akt3 play a pivotal role in malignant gliomas. Neuro Oncol. 2010;12:221–32. doi: 10.1093/neuonc/nop026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chautard E, Ouedraogo ZG, Biau J, Verrelle P. Role of Akt in human malignant glioma: from oncogenesis to tumor aggressiveness. J Neurooncol. 2014;117:205–15. doi: 10.1007/s11060-014-1382-9. [DOI] [PubMed] [Google Scholar]
- 43.Zheng KB, Wang GY, Li CH, Shan XS, Liu HP. Knockdown of ILK inhibits glioma development via upregulation of E-cadherin and downregulation of cyclin D1. Oncol Rep. 2015;34:272–8. doi: 10.3892/or.2015.3983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.
Supplementary table.