Summary
Slit-Robo signaling has been characterized as a repulsive signal for precise axon pathfinding and cell migration during embryonic development. Here, we describe a role for Sox2 in the regulation of Robo1 in Schwann cells and for Slit3-Robo1 signaling in controlling axon guidance within the newly formed nerve bridge following peripheral nerve transection injury. In particular, we show that macrophages form the outermost layer of the nerve bridge and secrete high levels of Slit3, while migratory Schwann cells and fibroblasts inside the nerve bridge express the Robo1 receptor. In line with this pattern of Slit3 and Robo1 expression, we observed multiple axon regeneration and cell migration defects in the nerve bridge of Sox2-, Slit3-, and Robo1-mutant mice. Our findings have revealed important functions for macrophages in the peripheral nervous system, utilizing Slit3-Robo1 signaling to control correct peripheral nerve bridge formation and precise axon targeting to the distal nerve stump following injury.
Keywords: Sox2, Slit3, Robo1, peripheral nerve, injury, regeneration, macrophages, Schwann cell, fibroblast, migration
Graphical Abstract
Highlights
-
•
Sox2 regulates Robo1 receptor expression in Schwann cells
-
•
Macrophages form the outermost layer of the nerve bridge and secrete Slit3
-
•
Ectopic Schwann cell migration/axon regrowth in bridge of Slit3+/−:Robo1+/− mice
Dun et al. show that after peripheral nerve injury, macrophages form the outermost layer of the nerve bridge and secrete Slit3. Migrating Schwann cells inside the nerve bridge express the Robo1 receptor. The Slit3-Robo1 repulsive signaling between macrophages and Schwann cells plays a critical role in adult peripheral nerve regeneration.
Introduction
Nerve transection injury following trauma often generates a nerve gap between the proximal and distal nerve stumps, which prevents correct re-targeting of regenerating axons into the distal nerve. Consequently, effective repair following transection remains a significant challenge in order to achieve precise axon re-targeting into the distal nerve stump and regain full nerve function following such an injury (Moore et al., 2009). It is well known that during development, precise axon targeting is controlled by several families of axon guidance molecules, such as Netrins, Slits, Ephrins, and Semaphorins (Bashaw and Klein, 2010). The synergistic effect of these molecules has an incredible ability to control precise axon targeting over long distances during development. Recent studies have found that Netrins, Slits, Ephrins, and Semaphorins are differentially and topographically expressed in the injured peripheral nervous system (PNS) (Giger et al., 2010, Dun and Parkinson, 2017), but their functions in axon targeting during peripheral nerve regeneration are largely unknown.
Our previous study has shown that EphrinB-EphB2-dependent signaling between nerve fibroblasts and Schwann cells leads to an increase of the Sox2 protein in Schwann cells following peripheral injury and that axon regeneration in the nerve bridge of EphB2 knockout mice was disorganized (Parrinello et al., 2010). Here, we study the effects of the loss of Sox2 in Schwann cells during peripheral nerve regeneration and show a dramatic regeneration defect of axonal pathfinding and Schwann cell migration in the sciatic nerve bridge of Sox2 knockout mice. We further relate this regeneration defect to Sox2 regulation of Robo1 expression in Schwann cells. Robo1 is a crucial axon guidance receptor for the secreted Slit glycoproteins (Blockus and Chédotal, 2016). So far, three Slit ligands (Slit1–3) and four Robo receptors (Robo1–4) have been identified (Blockus and Chédotal, 2016). In mammals, Slit1–3 bind to Robo1 and Robo2 with high affinity, but Robo3 and Robo4 are not high-affinity receptors (Blockus and Chédotal, 2016, Koch et al., 2011, Zelina et al., 2014). Binding of Slit1–3 to Robo1–2 typically initiates a repulsive signal to control axon pathfinding and cell migration during embryonic development (Blockus and Chédotal, 2016). We recently showed that Slit1–3 and Robo1–2 are highly expressed in the adult mouse peripheral nervous system (Carr et al., 2017). A wider investigation of Slit1–3 and Robo1–2 expression in the mouse sciatic nerve bridge following injury shows that macrophages form the outermost layer of the nerve bridge and secrete high levels of Slit3, while migrating Schwann cells and fibroblasts inside the nerve bridge express the Robo1 receptor. Using Sox2, Slit1–3, and Robo1–2 gene mutant mice, we observe axon regeneration and cell migration defects in Schwann cell-specific Sox2 knockout and Slit3+/−, Slit3−/−, and Robo1+/− mice. Our findings revealed that macrophage-derived Slit3 is required for interaction with Robo1 on Schwann cells and fibroblasts in the nerve bridge to control the trajectories of their migration. The loss of Slit3-Robo1-dependent signaling in the nerve bridge leads to ectopic Schwann cell and fibroblast migration and consequent incorrect axon targeting during regeneration following peripheral nerve transection injury.
Results
Axon Pathfinding and Schwann Cell Migration Defects in Sox2 Knockout Mice Following Nerve Transection
The Sox2 transcription factor is expressed by Schwann cells, is downregulated as they differentiate within the peripheral nervous system, but is then upregulated following nerve injury (Le et al., 2005, Parrinello et al., 2010, Roberts et al., 2017). By immunostaining, we first confirmed that Sox2 protein is detectable 48 h after injury (Figures S1A–S1C), and by day 7, 73% ± 6% Schwann cells in the distal nerve stump express Sox2 (Figures S1D–S1F and S1L). To understand Sox2 function in Schwann cells, we analyzed events of Wallerian degeneration and axon pathfinding following injury in mice with a Schwann cell-specific loss of Sox2 (Sox2 KO) (Figures S1J–S1K) by using the P0-CRE line and a conditional Sox2 allele (Feltri et al., 1999, Taranova et al., 2006, Favaro et al., 2009). DNA recombination in Sox2 KO mice was confirmed by PCR analysis (Figure S1K). The lack of Sox2 protein expression in Schwann cells of Sox2 KO mice was confirmed by immunolabeling (Figures S1G–S1I) and western blot (Figure S1M) in injured sciatic nerves; the immunolabeling and western blot data indicated an approximate 100% efficiency of DNA recombination. Myelination in postnatal (Figures S1Q–S1V) and adult (Figures S1W–S1Z) Sox2 KO nerves was normal with no changes in the G ratio or numbers of myelinated fibers. Following transection injury, distal Sox2 KO nerves showed unchanged downregulation of myelin proteins (Figures S1M–S1P) and normal macrophage recruitment (Figures S2A–S2C) but slightly reduced levels of Schwann cell proliferation (Figures S2D–S2F). Following crush injury, Sox2 KO nerves showed a small decrease in the speed of axon regeneration and functional recovery (Figures S2J–S2K), but remyelination of the nerve was unchanged (Figures S2L–S2R). We further tested the role of Sox2 in myelin protein downregulation following injury by the use of a Schwann cell-specific Sox2-overexpressing mouse, which shows increased Sox2 expression following injury (Roberts et al., 2017), and showed that there are no changes in the rate of loss of myelin proteins in the distal nerve of Sox2-overexpressing mice (Figures S2S–S2X). Thus, our data revealed that Sox2 is not required for myelin protein downregulation in Schwann cells after peripheral nerve injury that is in contrast to its negative regulatory role during developmental myelination and remyelination following injury in vivo (Roberts et al., 2017).
Next, we examined the effects of Sox2 loss upon axon pathfinding in the nerve bridge following transection injury. At both 10 and 14 days following transection, we saw large numbers of axons leaving the nerve bridge (Figures 1B and 1D) and a completely abnormal nerve bridge formation at three months post-injury (Figure 1F). Comparing both the number of axon bundles at the mid-point of the nerve bridge and axon density in the distal nerve stump at 14 days following injury showed that regenerating axons correctly crossing the nerve bridge and entering the distal nerve are both significantly reduced in Sox2 KO mice (Figures 1G–1L). Migrating Schwann cells inside the nerve bridge are essential for guiding regenerating axons back to the distal nerve stump (Cattin et al., 2015, Parrinello et al., 2010, Rosenberg et al., 2014). To see if the axon regeneration defects in Sox2 KO mice are caused by ectopic Schwann cell migration, we GFP-labeled Schwann cells by crossing Sox2 KO animals with proteolipid protein (PLP)-GFP mice (Mallon et al., 2002). Abnormal Schwann cell (GFP+) migration in the nerve bridge of Sox2 KO animals could be observed at 6 days following transection with regenerating axons following the ectopic migrating Schwann cells (Figures 2A–2C). In contrast to the normal Schwann cell cord formation in control nerves, which connect the proximal and distal nerve stumps (Figure 2A), ectopic-migrating Schwann cells in Sox2 KO nerves did not form correct Schwann cell cords connecting the proximal and the distal nerve stumps (Figures 2B and 2C). Ectopic-migrating Schwann cells and misdirected regenerating axons in Sox2 KO nerves could be easily observed leaving the nerve bridge at 14 days after injury, with Schwann cells in most cases apparently proceeding in front of axons (Figures 2E and 2F).
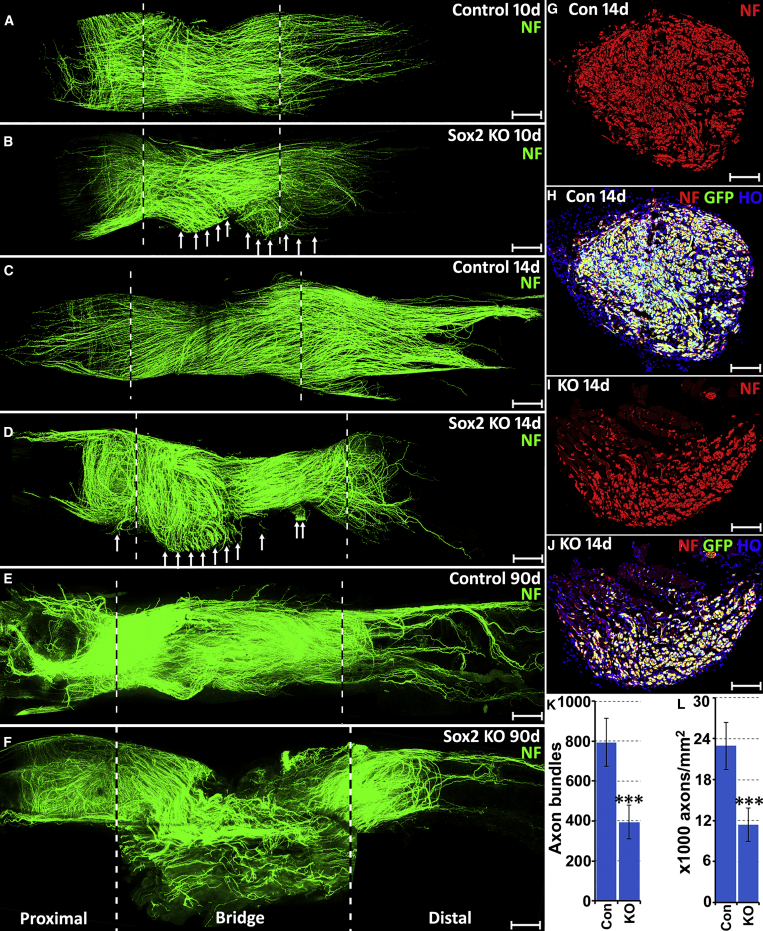
Figure 1.
Axon Guidance Defects in the Nerve Bridge of Sox2 KO Mice
(A–F) Whole sciatic nerves stained with neurofilament (NF, green) antibody to show the pattern of regenerating axons in the nerve bridge of control and Sox2 KO mice at 10 (A and B), 14 (C and D), and 90 (E and F) days following transection injury. The nerve bridge is indicated between two dashed lines. Regenerating axons leaving the nerve bridge in Sox2 KO mice at 10 and 14 days are indicated by white arrows in (B) and (D). An unrepaired nerve bridge is still presented in Sox2 KO mice even at 90 days (F).
(G–J) Neurofilament (NF) antibody staining shows axon bundles (red) in the middle of the nerve bridge in control (G and H) and Sox2 KO mice at 14 days (I and J); Schwann cells are labeled with GFP in both control (H) and Sox2 KO (J) mice. Scale bar in (A–F) represents 300 μm and in (G–J) represents 6 μm.
(K and L) Quantification of numbers of axon bundles in the middle of the nerve bridge (K) and axon density (L) in the distal nerve stump of control and Sox2 KO mice. n = 3; ∗∗∗ indicate p < 0.001 compared with controls.
Several z series were captured on a Zeiss LSM510 confocal microscope in (A)–(F), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
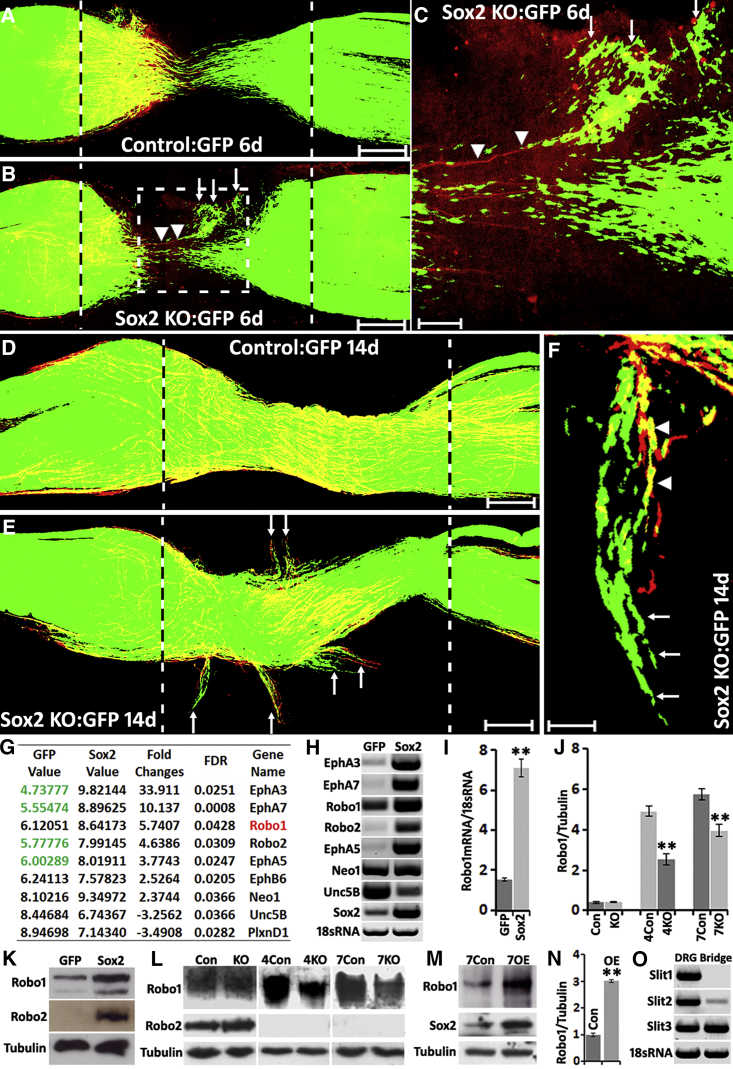
Figure 2.
Ectopic Schwann Cell Migration in the Nerve Bridge of Sox2 KO Mice and Sox2 Regulating Robo1 Expression in SCs
(A) Schwann cell (GFP+) migration from both proximal and distal nerve stumps in control mice 6 days after sciatic nerve transection injury.
(B) Ectopic Schwann cell migration (white arrows) in the nerve bridge of Sox2 KO mice 6 days after transection injury.
(C) Higher magnification image from (B, dotted-line square) showing regenerating axons (labeled with neurofilament, red, indicated by arrowheads) following the ectopic migrating Schwann cells (white arrows) and leaving the nerve bridge.
(D) Schwann cells stayed in the nerve bridge in control mice at 14 days following sciatic nerve transection injury.
(E) Ectopic migrating Schwann cells (white arrows) leaving the nerve bridge in Sox2 KO mice at 14 days after injury.
(F) Ectopic migrating Schwann cells (white arrows) localizing in front of regenerating axons (indicated by arrowheads) of Sox2 KO mice.
Scale bar in (A, B, D and E) represents 200 μm, in (C) represents 60 μm, and in (F) represents 30 μm.
(G and H) Microarray data (G) and RT-PCR (H) from control and Sox2-overexpressing Schwann cells. Values in (G) represent the fluorescence intensity of dye-labeled cDNA fragments that have hybridized to the probes on the microarray chip.
(I) qRT-PCR validation of Robo1 mRNA upregulation in Sox2-overexpressing Schwann cells, n = 3.
(J) Quantification of Robo1 protein levels in the distal nerve stump of control (Con) and Sox2 KO mice at 4 and 7 days after injury, n = 3.
(K) Western blots showing Robo1 and Robo2 protein upregulation in Sox2-overexpressing Schwann cells.
(L) Western blots comparing Robo1 and Robo2 protein levels in the distal nerve stump of control and Sox2 KO mice at 4 and 7 days after injury.
(M) Western blot comparing Robo1 protein levels in the distal nerve stump of control (Con) and Sox2-overexpressing (OE) mice at 7 days following injury.
(N) Quantification of Robo1 protein levels from (M), n = 3.
(O) RT-PCR showing that Slit3 is highly expressed in the nerve bridge at 7 days after injury; dorsal root ganglion (DRG) samples have been used as positive controls.
∗∗ in (I), (J), and (N) indicate p < 0.01 compared with controls.
Several z series were captured on a Zeiss LSM510 confocal microscope in (A), (B), (D), and (E), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
Sox2 Regulates Robo1 Receptor Expression in Schwann Cells
This strong phenotype in the nerve bridge of Sox2 KO mice led us to examine potential Sox2 targets that are required for the regulation of Schwann cell migration and axonal guidance. Using GFP control and Sox2/GFP-expressing adenoviruses (Le et al., 2005), we conducted an in vitro microarray analysis of control and Sox2-overexpressing Schwann cells and identified a total of 544 genes significantly regulated by Sox2 (p < 0.05, Table S1). Gene map annotator and pathway profiler analysis from Gene Ontology revealed 9 classic axon guidance receptors in the axon guidance signaling pathway (Figure 2G) to be regulated by Sox2, all of which are known to have important functions in regulating cell migration, including glial cells (Brose and Tessier-Lavigne, 2000, Bagri and Tessier-Lavigne, 2002, Tsai and Miller, 2002, Wang et al., 2013). The Sox2 regulation of these axon guidance receptors was further validated by RT-PCR (Figure 2H). Although overexpressing Sox2 in Schwann cells significantly upregulates EphA3, EphA5, EphA7, and Robo2, they are barely detectable in control Schwann cells, which express physiological levels of Sox2 (Figure 2H). In contrast, Robo1, EphB6, Neo1, Unc5B, and PlxnD1 are all expressed in control Schwann cells (Figures 2G and 2H). Robo1, EphB6, and Neo1 are further upregulated by overexpressing Sox2, while Unc5B and PlxnD1 are downregulated (Figures 2G and 2H), of which Robo1 has the highest fold change (Figures 2G–2I). The Sox2 regulation of Robo1 and Robo2 in control and Sox2-overexpressing Schwann cells was validated by qRT-PCR (Figure 2I) and western blot (Figure 2K) analysis. As Slit-Robo signaling has recently been shown to regulate Schwann cell migration (Wang et al., 2013), we chose to study the Slit-Robo signaling system further in our experiments.
Previously, Wang et al. (2013) showed Robo1 expression in rat Schwann cells, and we also showed by immunohistochemistry that Robo1 is expressed in Schwann cells of the adult mouse sciatic nerve (Carr et al., 2017). To confirm that the Robo1 protein is expressed in dedifferentiated Schwann cells following injury, we immuno-stained Robo1 on longitudinal sciatic nerve sections from adult PLP-GFP mice at 7 days after transection injury. This not only confirmed that migrating Schwann cells in the nerve bridge express the Robo1 receptor (Figures 3A–3D) but also confirmed that Schwann cells in the distal nerve stump express Robo1 (Figures 3H and 3I). We also confirmed Robo1 expression on cultured rat Schwann cells by Robo1 staining (Figure 3J) and western blot (Figure 2K). Inside the nerve bridge, Robo1 is also highly expressed in regenerating axons (Figures S3A–S3C) and vimentin-positive fibroblasts (Figures 3E–3G) but is weakly expressed in endothelial cells (Figures S3D–S3G).
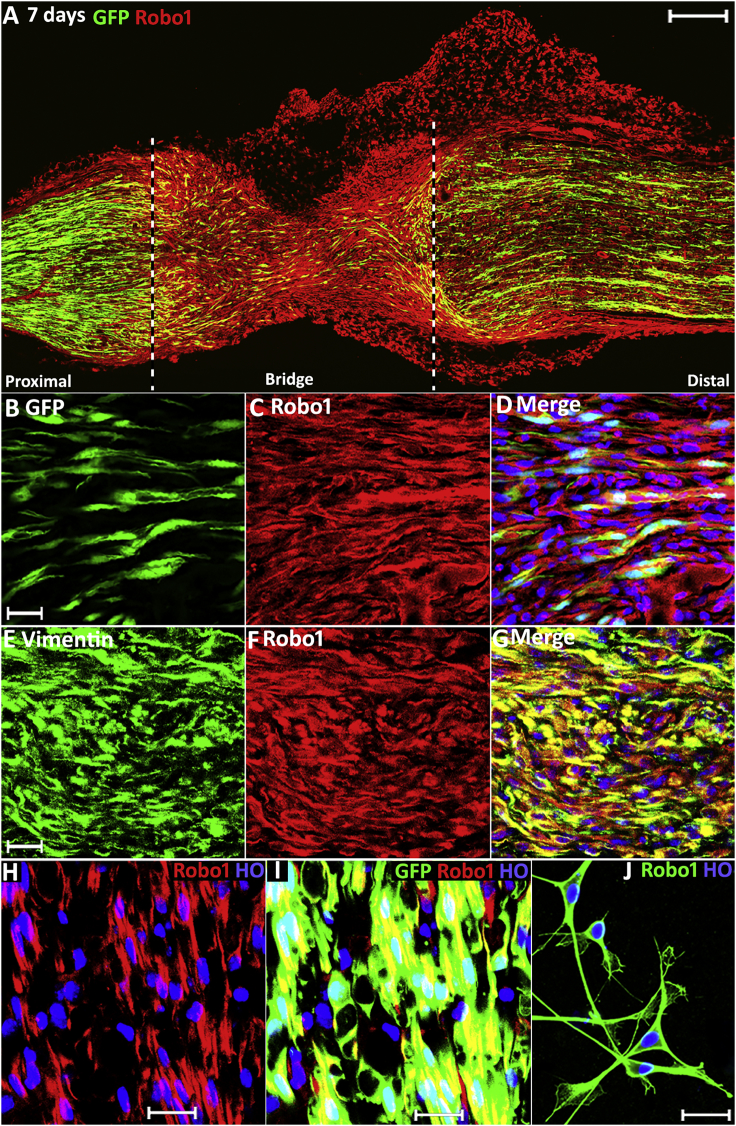
Figure 3.
Robo1 Expression in the Nerve Bridge
(A) Robo1 staining on a longitudinal nerve bridge section at 7-days post-injury from a PLP-GFP mouse showing that Robo1 is highly expressed in migrating cells of the nerve bridge.
(B) Higher magnification image showing migrating GFP+ Schwann cells inside the nerve bridge.
(C) Higher magnification image showing Robo1 positive cells inside the nerve bridge.
(D) Merged image showing Schwann cells inside the nerve bridge express Robo1.
(E) Vimentin staining showing migrating fibroblasts in the nerve bridge.
(F) Robo1 staining showing Robo1 positive cells inside the nerve bridge.
(G) Merged image showing migrating fibroblasts in the nerve bridge express Robo1.
(H) Robo1 staining in the distal nerve stump of PLP-GFP mice.
(I) Merged image showing Schwann cells in the distal nerve stump express high levels of Robo1.
(J) Robo1 staining in cultured rat Schwann cells.
Scale bar in (A) represents 200 μm, in (B)–(G) represents 20 μm, and in (H)–(J) represents 10 μm.
Several z series were captured on a Zeiss LSM510 confocal microscope in (A), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
After confirming that Sox2 regulates Robo1 expression in Schwann cells, we next used western blots to compare the levels of Robo1 protein in the distal nerve stump of control and Sox2 KO mice at 4- and 7-days post-injury. Western blot results revealed that Robo1 is upregulated in the distal nerve stump of both control and Sox2 KO mice following sciatic nerve transection injury (Figure 2L). Quantification of the Robo1 protein levels from control and Sox2 KO mice showed that the expression of Robo1 is about 50% decreased in the distal nerve stump of Sox2 KO mice at 4-days post-injury compared to control mice (Figure 2J). To further confirm that Sox2 regulates Robo1 in Schwann cells, we performed the same experiment in Sox2-overexpressing mice, which show increased Sox2 expression following injury (Figure 2M) (Roberts et al., 2017). Supporting our results in Sox2 KO mice, the Robo1 protein level is significantly higher (3-fold) in Sox2-overexpressing mice than control mice (Figures 2M and 2N). We also studied Robo2 expression in cultured Schwann cells, in control and Sox2 KO mice by western blot. Consistent with our published findings that Robo2 is an axonal protein in sciatic nerve (Carr et al., 2017), western blot results showed that Robo2 is not expressed in cultured Schwann cells (Figure 2K) and the distal nerve stump of both control and Sox2 KO mice following axonal breakdown (Figure 2L).
Macrophages in the Outermost Layer of the Nerve Bridge Express Slit3
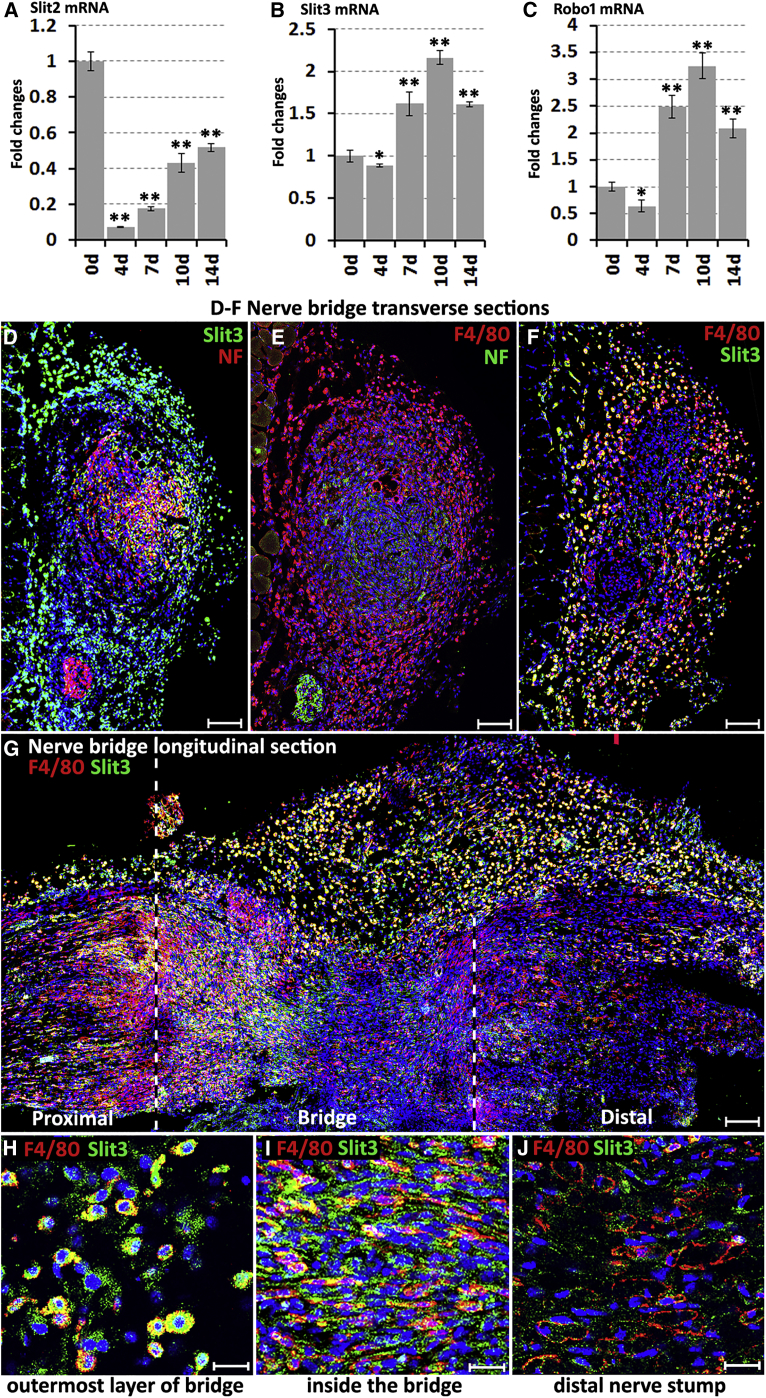
Given the results with Sox2 regulating Robo1 expression in Schwann cells and ectopic Schwann cell migration in the nerve bridge of Sox2 KO mice, we next studied the expression of Robo1 ligands Slit1–3 in the nerve bridge. Recently, we showed that Slit2 and Slit3 mRNAs are expressed in the intact mouse sciatic nerve but Slit1 mRNA is undetectable (Carr et al., 2017), and others reported that cultured Schwann cells express Slit2 and Slit3 but not Slit1 (Wang et al., 2013). Lastly, our microarray and RT-PCR data also showed Slit2 and Slit3 expression in Schwann cells, although they are not significantly regulated by Sox2 (data not shown). Examining Slit1–3 mRNA by RT-PCR in the mouse sciatic nerve bridge 7 days after injury revealed that Slit1 mRNA is undetectable and Slit2 expression is very weak, whereas Slit3 is highly expressed (Figure 2O). We further studied the time course of Slit2, Slit3, and Robo1 mRNA levels in the nerve bridge by qRT-PCR, showing that Slit2 mRNA levels are low in the nerve bridge compared to intact nerve (0 days) from 4 to 14 days (Figure 4A), the period that regenerating axons cross the nerve bridge after mouse nerve transection injury (Dun and Parkinson, 2015), while both Slit3 and Robo1 mRNAs are upregulated in the nerve bridge from 7 days following injury (Figures 4B and 4C).
Figure 4.
Macrophages in the Outermost Layer of the Nerve Bridge Express Slit3
(A–C) Time course of Slit2 (A), Slit3 (B), and Robo1 (C) mRNA fold changes in the nerve bridge following injury, n = 3. ∗p < 0.05, ∗∗p < 0.01 compared with the intact nerve.
(D) Slit3 and neurofilament (NF) double staining on nerve bridge transverse section showing that Slit3-positive cells form the outermost layer of the nerve bridge.
(E) F4/80 and NF double staining showing that the outermost layer of the nerve bridge is formed by a high density of macrophages.
(F and G) Slit3 and F4/80 double staining on a transverse (F) and longitudinal (G) nerve bridge sections confirming that macrophages in the outermost layer of the nerve bridge express Slit3.
(H–J) Higher magnification images from (G) showing that macrophages in the outermost layer of the nerve bridge express high levels of Slit3 (H), macrophages in the middle of the nerve bridge also express Slit3 (I), but macrophages in the distal nerve stump express low levels of Slit3 (J).
Blue signal in (D)–(J) represents cell nuclei staining (Hoechst). Scale bar in (D)–(G) represents 100 μm and in (H)–(J) represents 10 μm.
Several images were captured on a Zeiss LSM510 confocal microscope in (D) and (E) and combined into one image using Adobe Photoshop software (Adobe Systems). Several z series were captured on Zeiss LSM510 confocal microscope in (G), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
The nerve bridge consists of newly formed tissue connecting the proximal and distal nerve stumps and comprises macrophages, fibroblasts, endothelial cells, migrating Schwann cells, and regenerating axons (Cattin et al., 2015). Using cell-type-specific markers to identify Slit3-positive cells in the nerve bridge by immunohistochemistry, we identified macrophages (F4/80+) in the outermost layer of the nerve bridge expressing high levels of Slit3 (Figures 4D–4H). Slit3 is also expressed in macrophages inside the nerve bridge (Figures 4G and 4I). In contrast, macrophages in the distal nerve stump do not express high levels of Slit3 (Figure 4J), indicating that macrophages in the nerve bridge may have a different phenotype from those in the distal nerve stump. Inside the nerve bridge, Slit3 is also expressed in migrating Schwann cells (Figures S3H–S3J) and regenerating axons (Figures S3K–S3M) but is not expressed in endothelial cells (Figures S3N–S3P).
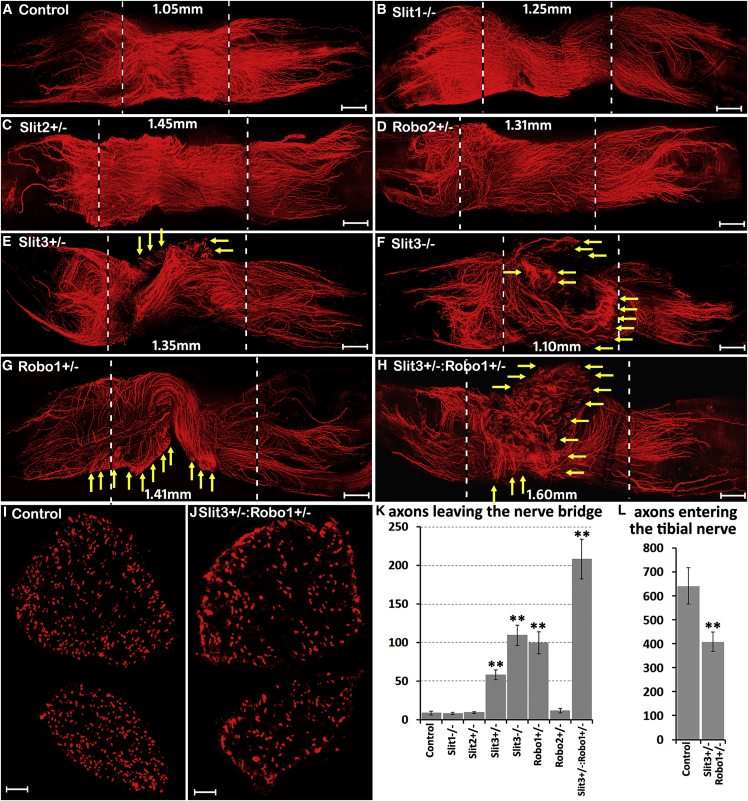
Axon Pathfinding Defects in Slit3 and Robo1 Gene Mutant Mice
Expression of Slit3 by macrophages in the outermost layer of the nerve bridge suggests a model whereby Slit3 could regulate the pathfinding of Robo1- and Robo2-expressing axons and the migration trajectories of Robo1-expressing Schwann cells and fibroblasts. To further test the roles of Slit-Robo signaling in the nerve bridge during regeneration, we transected the sciatic nerves of Slit1–3 (Plump et al., 2002, Yuan et al., 2003) and Robo1–2 gene mutant mice (Andrews et al., 2008, Grieshammer et al., 2004) and examined the axon regeneration pattern in the nerve bridge by whole-mount staining (Dun and Parkinson, 2015). Upon sciatic nerve transection, the nerve trunk retracts and generates a gap; the length of this gap varies from 1.0 mm to 2.5 mm in our experiments (Figures 5 and 6). Although the gap length is variable, which affects the total number of axons crossing the nerve bridge, examination of the axon regeneration pattern in the nerve bridge 14 days following sciatic nerve transection injury showed significant axon regeneration defects in the nerve bridge in Slit3+/−, Slit3−/−, and Robo1+/− mice but none were observed in Slit1−/−, Slit2+/−, and Robo2+/− mice (Figures 5A–5G). We next generated Slit3+/−:Robo1+/− mice and found significantly more misguided axons in Slit3+/−:Robo1+/− mice compared to Slit3+/− and Robo1+/− mice (Figures 5H and 5K). A comparison of the total number of regenerated axons in the distal tibial nerve following injury also revealed a significant reduction in Slit3+/−:Robo1+/− mice compared to controls (Figures 5I, 5J, and 5L). Thus, the regeneration defect in the nerve bridge observed in Slit3+/− and Slit3−/− mice but not in Slit1−/− and Slit2+/− mice is consistent with our finding that Slit3 is highly expressed by macrophages in the outermost layer of the nerve bridge. The regeneration defect in the nerve bridge observed in Robo1+/− mice but not in Robo2+/− mice suggests that, rather than having a direct effect on regenerating axons, Slit3 is required to interact with Robo1 on migrating Schwann cells and fibroblasts inside the nerve bridge to regulate correct nerve bridge formation.
Figure 5.
Axon Guidance Defect in Slit3 and Robo1 Gene Mutant Mice
Whole sciatic nerve stained with neurofilament antibody (red) to show the pattern of regenerating axons in the nerve bridge 14 days after transection injury.
(A) In control animals, regenerating axons stay in the middle of the nerve bridge and grow straight toward the distal nerve stump.
(B–D) Regenerating axons in Slit1−/− (B), Slit2+/− (C), and Robo2+/− mice (D) show a similar regeneration pattern as the control mice.
(E–H) Regenerating axons leave the nerve bridge (indicated by yellow arrows) in Slit3+/− (E), Slit3−/− (F), Robo1+/− (G), and Slit3+/−:Robo1+/− (H) mice.
(I and J) Neurofilament antibody staining on tibial nerve transverse sections from control (I) and Slit3+/−: Robo1+/− (J) mice 14 days after injury.
(K) Total number of axons leaving the nerve bridge in control and Slit and Robo gene mutant mice, n = 3.
(L) Quantification of regenerated axon numbers in the tibial nerve of control and Slit3+/−:Robo1+/− mice, n = 3.
The nerve bridge is indicated between two dashed lines and the bridge length has been labeled in (A)–(H). ∗∗p < 0.01 compared with controls. Scale bar in (A)–(H) represents 250 μm and in (I)–(J) represents 50 μm.
Several z series were captured on a Zeiss LSM510 confocal microscope in (A)–(H), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
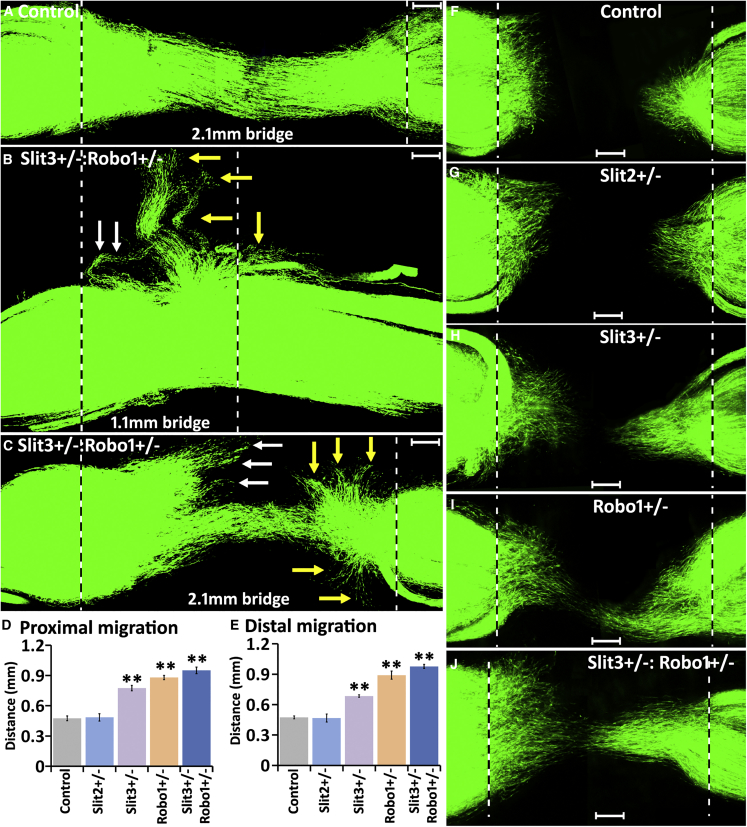
Figure 6.
Slit3-Robo1 Signaling Controls the Trajectory and the Distance of Schwann Cell Migration Inside the Nerve Bridge
(A–C) Fourteen days after transection injury, in control mice, GFP+ Schwann cell cords connect the proximal and distal nerve stumps. In Slit3+/−:Robo1+/− mice (B and C), a large population of Schwann cells leave the nerve bridge from both the proximal nerve end (indicated by white arrows) and the distal nerve end (indicated by yellow arrows). Bridge length in (A) and (C) is 2.1 mm and in (B) is 1.1 mm.
(D and E) The distance of leading Schwann cells inside the nerve bridge from the proximal nerve end (D) and from the distal nerve end (E) 5 days after nerve transection, n = 3. ∗∗p < 0.01 compared with controls.
(F–J) Schwann cell migration in control (F), Slit2+/− (G), Slit3+/− (H), Robo1+/− (Ι), and Slit3+/−:Robo1+/− (J) mice 5 days after transection injury.
Scale bar in (A)–(C) and (F)–(J) represents 200 μm.
Several z series were captured on a Zeiss LSM510 confocal microscope in (A) –(C) and (F) –(J), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
To confirm that the regeneration defects observed in Slit3−/− and Robo1+/− mice are not linked to any developmental defect, we examined the sciatic nerve structure and expression of myelin basic protein (Mbp) and myelin protein zero (Mpz) in 2-month-old Slit2+/−, Slit3−/−, and Robo1+/− mice. The results showed that sciatic nerve development is apparently normal in Slit2+/−, Slit3−/−, and Robo1+/− mice (Figures S4 and S5A–S5E). Western blotting showed that the Slit3 protein is undetectable in the sciatic nerve from Slit3−/− mice and Robo1 protein levels in Robo1+/− mice and Slit3 protein levels in Slit3+/− mice are approximately 50% of controls (Figures S5H–S5K). As Slit3 is highly expressed in macrophages on the outermost layer of the nerve bridge and a previous report showed that Slit3 promotes monocyte migration (Geutskens et al., 2010), we examined macrophage recruitment in the nerve bridge of Slit3−/− mice at 7 days after nerve transection injury. Counts of macrophage numbers in the outermost layer of the nerve bridge indicated that there was no significant difference in macrophage recruitment between control and Slit3−/− mice (Figures S5L–S5R). Thus, axon regeneration defects seen in Slit3−/−, Slit3+/−, and Robo1+/− mice are not due to either a developmental defect of the sciatic nerve or reduced macrophage recruitment after injury but rather by the disturbance of Slit3-Robo1 repulsive signaling in the nerve bridge due to decreased levels of Slit3 or Robo1 in these mice.
Although both transection and crush injury will induce a similar distal degeneration of the nerve, crush injury is a better model to measure the speed of axon growth and functional recovery because the axon path at the injury site is preserved (Nguyen et al., 2002). Therefore, we further studied peripheral nerve regeneration on Slit1–3 and Robo1–2 gene mutant mice after crush injury. Staining the whole nerve with neurofilament antibody and measuring the axon regrowth revealed that the distance of axon regeneration is similar among control, Slit1−/−, Slit2+/−, Slit3−/−, Robo1+/−, Robo2+/−, and Robo1+/−:Robo2+/− mice (Figure S6C). We further examined the functional recovery in Robo1+/− mice by measuring the static sciatic nerve functional index (SSI) after crush injury. We observed no significant difference in the functional recovery of control and Robo1+/− mice (Figure S6D). By western blot we found that Slit1, Slit2 (Figure S6A), and Robo2 (Figure 2L) proteins are not expressed in the distal nerve stump. By qRT-PCR and immunostaining analysis, we showed that Slit3 mRNA is downregulated in the distal nerve stump (Figure S6B) and macrophages inside the distal nerve stump do not express Slit3 (Figure 4J). Thus, our crush injury data together with the Slit and Robo expression pattern in the distal nerve stump indicate that Slit-Robo signaling does not play an important role in the distal nerve stump during regeneration following nerve crush. Instead, Slit3-Robo1 signaling regulates correct nerve bridge formation and precise axon targeting after peripheral nerve transection injury.
Slit3-Robo1 Signaling Controls Trajectory and Speed of Schwann Cells Migration in the Nerve Bridge
Above, we showed that regenerating axons leave the nerve bridge in Slit3 and Robo1 gene mutant mice (Figure 5); furthermore, ectopic Schwann cell migration occurred in Sox2 KO mice following transection injury (Figure 2) and Sox2 regulates the Robo1 receptor expression in Schwann cells (Figure 2). These findings indicate that the Slit3-Robo1 signaling between macrophages and migrating Schwann cells may control the trajectory of Schwann cell migration in the nerve bridge. Because there are more regenerating axons leaving the nerve bridge in Slit3+/−:Robo1+/− mice (Figure 5H, K), we crossed the PLP-GFP mice with Slit3+/−:Robo1+/− mice to label Schwann cells with GFP in both control and Slit3+/−:Robo1+/− mice and examine the trajectories of Schwann cell migration in the nerve bridge 14 days following injury. In control nerves, migrating Schwann cells stayed inside the nerve bridge and formed Schwann cell cords connecting the proximal and distal nerve ends (Figure 6A). However, in Slit3+/−:Robo1+/− mice, a large population of Schwann cells was observed leaving the nerve bridge from both the proximal and the distal nerve stumps, no matter the length of the nerve bridge (Figures 6B and 6C). Previously, Wang et al. (2013) demonstrated that recombinant Slit2 protein could repel Schwann cells in vitro. We also generated Schwann cell GFP-labeled control and Slit2+/− mice, but we did not observe any aberrant Schwann cell migration in Slit2+/− mice (Figures S6E and SF), further confirming that Slit2 is not required for normal Schwann cell migration inside the nerve bridge.
Previous reports have shown that cultured mouse bone marrow macrophages as well as peritoneal macrophages express Slit3 (Tanno et al., 2007, Kim et al., 2018). Therefore, we performed Schwann cell and bone marrow-derived macrophage co-cultures to further confirm a direct Slit3-Robo1 repulsive signaling between Schwann cells and macrophages. We first confirmed that cultured bone marrow macrophages from Slit3+/+ mice express Slit3 but not bone marrow macrophages from Slit3−/− mice (Figure S7A). In Schwann cell and Slit3+/+ macrophage co-culture, we observed the formation of Schwann cell clusters 24 h after the initial mixed cell seeding (Figures S7C–S7H). In Schwann cell and Slit3+/+ macrophage co-culture, Schwann cells often grew on top of each other and lost their typical bi-polar processes (Figure S7G). In contrast, in Schwann cell and Slit3−/− macrophage co-cultures, the formation of Schwann cell clusters was not observed and the bi-polar processes of Schwann cells were still clearly visible (Figures S7B and S7I–L). Thus, our co-culture experiments further confirmed a direct Slit3-dependent repulsive signaling between Schwann cells and macrophages.
Consistent with the well-established effects of Slit-Robo repulsive signaling for cell migration, our data indicated that macrophage-derived Slit3 acts as a repellent to keep Robo1-expressing Schwann cells inside the nerve bridge. Thus Slit3 could also interact with Robo1 on Schwann cells to regulate the speed of Schwann cell migration within the nerve bridge. Therefore, we examined Schwann cell migration in the nerve bridge of Schwann cell labeled (GFP+) control, Slit2+/−, Slit3+/−, Robo1+/−, and Slit3+/−:Robo1+/− mice (Figures 6D–6J). From measurements of the distance of leading Schwann cells from the proximal and the distal nerve ends 5 days after injury, we found that Schwann cells migrated significantly faster in Slit3+/−, Robo1+/−, and Slit3+/−:Robo1+/− mice than in control or Slit2+/− mice (Figures 6D–6J), indicating that Slit3 acts an repellent on Robo1-expressing Schwann cells and slows down Schwann cell migration inside the nerve bridge. This process may be important to allow the substrate formation within the bridge and prevent aberrant and too rapid Schwann cell migration.
Schwann cells undergo dedifferentiation and reprogramming before they can migrate from both proximal and distal nerve stumps, with migration starting at 4d post-nerve transection in the mouse (Dun and Parkinson, 2015, Cattin et al., 2015, Parrinello et al., 2010). To confirm the faster migration observed in Slit3 and Robo1 gene mutant mice is not caused by a more rapid dedifferentiation of Schwann cells following injury, we used western blotting to compare the downregulation of myelin proteins Mbp and Mpz in the distal nerve stump of control, Slit2+/−, Slit3−/−, and Robo1+/− mice 4 days after sciatic nerve transection. We also compared the upregulation of cJun and the activation of the ERK1/2 mitogen-activated protein (MAP) kinase between control and Robo1+/− mice. The results showed that Schwann cell myelin loss and cJun and phospho-ERK1/2 activation in Slit2+/−, Slit3−/−, and Robo1+/− mice were not significantly different than control animals (Figures S5A–S5G), thus eliminating this possibility of faster Schwann cell dedifferentiation in these nerves driving earlier cell migration.
Aberrant Fibroblast Migration in Slit3 and Robo1 Gene Mutant Mice
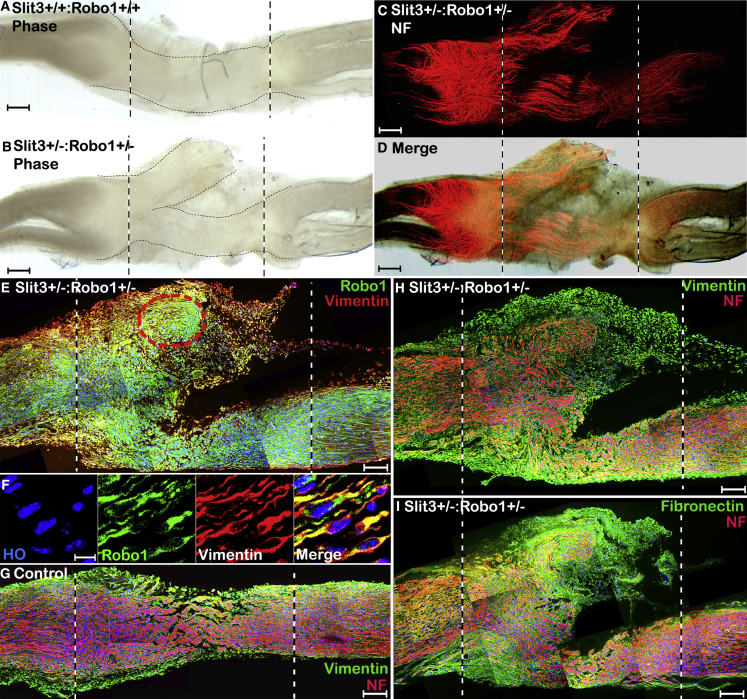
Fibroblasts are one of the major cell types forming the nerve bridge, and they migrate into the nerve bridge much earlier than endothelial cells and Schwann cells (Parrinello et al., 2010, Williams et al., 1983). Our immunostaining showed that Robo1 is also highly expressed in migrating fibroblasts of the nerve bridge (Figures 3E–3G), indicating that macrophage-derived Slit3 may also act as a repellent on Robo1-expressing fibroblasts to control nerve bridge formation. To reveal this Slit3-Robo1 function between macrophages and fibroblasts, we labeled for Robo1, vimentin, fibronectin, and neurofilament on longitudinal nerve bridge sections from control and Slit3+/−:Robo1+/− mice. This staining result revealed that Robo1-positive fibroblasts formed extra nerve bridge tissue in Slit3+/−:Robo1+/− mice (Figure 7) and that the misdirected axons have extended into this extra nerve bridge tissue (Figure 7H and 7I). Thus, it appears that macrophage-derived Slit3 is also essential for controlling the trajectories of fibroblast migration and correct nerve bridge formation.
Figure 7.
Ectopic Fibroblast Migration in Slit3+/−:Robo1+/− Mice
(A and B) Phase contrast images showing extra nerve bridge tissue formed in Slit3+/−:Robo1+/− mice (B) but not in control mice (A).
(C) NF staining showing misguided axons in the nerve bridge of Slit3+/−:Robo1+/− mice.
(D) Merged image of (B) and (C) shows that misguided axons in Slit3+/−:Robo1+/− extended into extra nerve bridge tissue.
(E) Double staining of Robo1 with fibroblast marker vimentin on longitudinal nerve bridge sections from Slit3+/−:Robo1+/− mice 14 days after transection injury showing Robo1-positive fibroblasts are the major cells forming the extra nerve bridge tissue in Slit3+/−:Robo1+/− mice.
(F) Higher magnification image from (E) (red circled area) showing fibroblasts in the extra nerve bridge tissue express high levels of Robo1.
(G) Double staining of NF with fibroblast marker vimentin in the nerve bridge of control mice.
(H and I) Double staining of NF with fibroblast marker vimentin (H) or fibronectin (I) showing that the extra nerve bridge tissue was formed largely by fibroblasts and the misdirected axons have extended into the extra nerve bridge tissue.
Scale bar in (A)–(D) represents 100 μm, in (E) and (G)–(I) represents 200 μm, and in (F) represents 10 μm.
Several z series were captured on a Zeiss LSM510 confocal microscope in (C), (E), and (G) –(I), covering the entire field of interest. The individual series were then flattened into a single image for each location and combined into one image using Adobe Photoshop software (Adobe Systems).
Normal Blood Vessel Regeneration in Slit3 and Robo1 Gene Mutant Mice
Endothelial cell migration and blood vessel regeneration are also early key events for correct nerve bridge formation (Cattin et al., 2015). Recently, we showed that blood vessels of the intact sciatic nerve express Robo1 (Carr et al., 2017). After injury, Robo1 is still highly expressed in vessels of the proximal and the distal nerve stumps (Figures S3D–S3G and S6G–S6J) but was downregulated in endothelial cells of newly formed blood vessels in the nerve bridge (Figures S6H and S6J). We next whole-mount stained nerves from control, Slit3+/−, Robo1+/−, and Slit3+/−:Robo1+/− mice with CD31 antibody after sciatic nerve transection injury to examine endothelial cell migration and vessel formation in the nerve bridge. We did not observe any endothelial cells leaving the nerve bridge in Slit3+/−, Robo1+/−, and Slit3+/−:Robo1+/− mice (Figures S6K–S6N). Thus, Schwann cell migration and axon regeneration defects observed in Slit3+/−, Robo1+/−, and Slit3+/−:Robo1+/− mice appear not to be caused by aberrant blood vessel regeneration. Blood vessel regeneration in the nerve bridge has been shown to be regulated by vascular endothelial growth factor (VEGF) secreted from macrophages (Cattin et al., 2015). The majority of blood vessels regenerate just beneath the outermost layer of Slit3-positive macrophages (Williams et al., 1983). Our findings may suggest that migrating endothelial cells inside the nerve bridge downregulate Robo1 expression to desensitize the Slit3 repulsive function in order to utilize macrophage-derived VEGF for blood vessel regeneration. Thus, the differential and topographical regulation of Robo1 in different cell types of the nerve bridge may be key to successful peripheral nerve regeneration.
Discussion
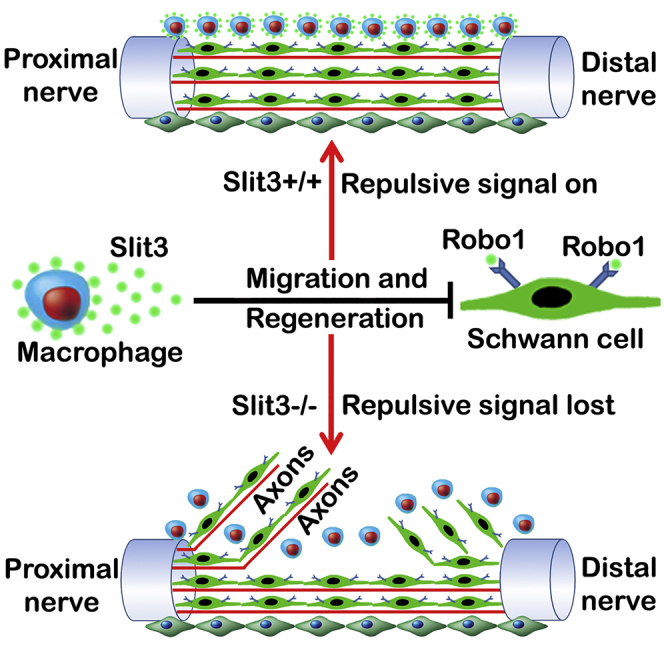
Schwann cells are a regenerative cell type, and their plasticity facilitates the repair of the peripheral nervous system after damage (Jessen and Mirsky, 2016). Our previous work showed that EphrinB-EphB2 signaling between fibroblasts and Schwann cells regulates Sox2 expression in Schwann cells and that Sox2 regulates cell sorting in the nerve bridge (Parrinello et al., 2010). In this study, we show that Sox2 also acts to increase Robo1 expression in Schwann cells and the Slit3-Robo1 signaling between macrophages and Schwann cells control the trajectories of Schwann cell migration in the peripheral nerve bridge. The directionality of regenerating axons within the nerve bridge is known to be determined by migratory Schwann cells from both nerve stumps (Rosenberg et al., 2014, Cattin et al., 2015). Regenerating axons extend into the nerve bridge in random directions when they lack Schwann cell guidance (Rosenberg et al., 2014). Our whole-mount sciatic staining of Sox2 KO nerve showed that Schwann cells still localize in front of misguided axons; therefore, the abnormal axon guidance seen in the nerve bridge of Sox2 KO animals is not due to axons no longer being associated with Schwann cells. In accordance with Sox2 regulation of Robo1 expression in Schwann cells and Slit3 expression by the macrophages surrounding the nerve bridge, we further showed that Schwann cells leave the nerve bridge in Slit3 and Robo1 gene mutant mice following injury. Thus, a disruption of Slit3-Robo1 signaling within the bridge, in either Sox2, Slit3, or Robo1 mutant mice, results in ectopic Schwann cell migration and the axon regeneration defect observed following nerve transection injury. This work has identified a role for Sox2 in Schwann cells, through regulation of the Robo1 receptor and a mechanism involving Slit3 and Robo1 in macrophage-mediated guidance of Schwann cell migration and axonal pathfinding within the nerve bridge.
Our data show that macrophage-derived Slit3 functions as a repellent to keep migrating Schwann cells inside the nerve bridge. The fast Schwann cell migration in Slit3 and Robo1 gene mutant mice (Figures 6D and 6E) also indicates that Slit3 acts as a repellent on Robo1-expressing Schwann cells to regulate Schwann cell migration. Aberrant Schwann cell migration could be easily observed at later time points in Slit3+/−:Robo1+/− (Figures 6B and 6C) mice but Schwann cells appear to be perfectly aligned in the nerve bridge with little indication of misdirection when they just start to migrate (Figures 6F and 6G). In the early stages of nerve bridge formation, the substrate formed for Schwann cell migration appears much narrower than either the proximal or distal nerve stump tissue (Figures 2D, 2E, 6A, and 6C). Early migrating Schwann cells may follow the narrow path and cross the nerve gap even in Slit3+/−:Robo1+/− mice. However, in the later stages of Schwann cell migration when there are many more Schwann cells migrating out from both the proximal and the distal nerve stumps with a widespread area of migration, not all migrating Schwann cells are able to follow the narrow substrate. Schwann cells unable to follow the narrow substrate will leave the nerve bridge if Slit3-Robo1 signaling is disrupted. Our observation in the Sox2, Slit3, and Robo1 gene mutant mice indicates that macrophage-derived Slit3 is a key molecule to keep this population of migrating Schwann cells in the nerve bridge when they are unable to follow an initial narrow migration path.
Previous studies have reported that fibroblasts are one of the major cell types forming the nerve bridge following injury and they migrate into the nerve bridge much earlier than endothelial cells and Schwann cells (Parrinello et al., 2010, Williams et al., 1983). Our staining also showed that fibroblasts are one of the major cell types in the nerve bridge expressing the Robo1 receptor (Figures 3D–3G). The phase contrast image of the Slit3+/−:Robo1+/− injured nerve indicated that extra nerve bridge tissue was formed in Slit3+/−:Robo1+/− mice, which was not aligned to the distal nerve stump (Figures 7A–7D). Taking advantage of the fact that Robo1-positive cells could still be stained by the Robo1 antibody in Slit3+/−:Robo1+/− mice, we were able to show that Robo1-positive fibroblasts are also a major cell type forming the extra nerve bridge in addition to Robo1-positive Schwann cells. Thus, macrophage-derived Slit3 also interacts with Robo1-expressing fibroblasts inside the nerve bridge to control the trajectory of their migration. Fibroblasts inside the nerve bridge express ephrinB2, another axon guidance molecule, and Schwann cells express its receptor EphB2. EphrinB2-EphB2 signaling between fibroblasts and Schwann cells results in Schwann cell sorting and forms Schwann cell cords in the nerve bridge to guide regrowing axons across the nerve gap (Parrinello et al., 2010). Therefore, in addition to the direct repulsive effect of macrophage-derived Slit3 on migrating Schwann cells in the nerve bridge, we cannot rule out that the aberrant fibroblast migration could also provide a substrate and direct Schwann cells leaving the nerve bridge.
Macrophages are an important cell type for peripheral nerve regeneration (Chen et al., 2015). The best characterized roles for macrophages in peripheral nerve regeneration are in the processes of Wallerian degeneration and executing clearance of both axonal and myelin debris (Martini et al., 2008). Infiltrating macrophages also secrete pro-inflammatory and anti-inflammatory factors that have both direct and indirect effects on neuronal survival and axon outgrowth (Zigmond, 2012). In the case of transection injury, macrophages are one of the earliest cell types arriving into the nerve gap and actively take part in the formation of the nerve bridge (Cattin et al., 2015, Rosenberg et al., 2012). They can be found in the nerve gap within 24 h of nerve transection and secrete VEGF to promote the formation of new blood vessels within the bridge (Cattin et al., 2015). In this study, we demonstrated that macrophages form the outermost layer of the nerve bridge and express the axon guidance molecule Slit3 to control both nerve bridge formation and axon regeneration after peripheral nerve transection injury. In support of our findings, previous studies have also shown that macrophage expression of Slit3 both at the spinal cord and cerebellum lesions (Wehrle et al., 2005). By immunostaining with both F4/80 (pan-macrophage) and CD206 (M2 macrophage) antibodies, we found that macrophages have a different morphology and cell size in the proximal nerve stump, the nerve bridge, and the distal nerve stump (Figures 4H–4J and S5O-S5R). Interestingly, macrophages in the outermost layer of the nerve bridge express high levels of Slit3 (Figures 4H and 4I) but macrophages in the distal nerve stump express little Slit3 (Figure 4J), indicating that macrophages may have different phenotypes in different environments within the injured peripheral nerve.
Successful peripheral nerve regeneration requires the coordination of multiple cell types in the nerve bridge. Cell coordination between macrophages and Schwann cells (Martini et al., 2008), macrophages and endothelial cells (Cattin et al., 2015), Schwann cells and fibroblasts (Parrinello et al., 2010), and Schwann cells and perineurial cells (Lewis and Kucenas, 2014) have all been reported. Here, we propose that macrophages utilize Slit3-Robo1 signaling to control the trajectories of Schwann cell and fibroblast migration within the nerve bridge. Our findings have revealed an important macrophage function in regulating both nerve bridge formation and controlling the target specificity of peripheral axon regeneration. We believe this macrophage function could potentially be manipulated in the future to promote successful peripheral nerve repair.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sox2 for western blotting | Novus Biological | Cat# NB110-37235; RRID:AB_792070 |
| Sox2 for immunostaining | Millipore | Cat# AB5603; RRID:AB_2286686 |
| myelin basic protein | Sigma | Cat# SAB2500665; RRID:AB_10605117 |
| myelin protein zero | Santa Cruz | Cat# sc-13912; RRID:AB_648794 |
| β2A Tubulin | Santa Cruz | Cat# sc-134229; RRID:AB_2210517 |
| cJun | Becton Dickinson | Cat# 610327; RRID:AB_397717 |
| pERK1/2 | Cell Signaling | Cat# 9102; RRID:AB_330744 |
| Iba1 | Wako | Cat# 019-19741; RRID:AB_839504 |
| Ki67 | Abcam | Cat# ab15580; RRID:AB_443209 |
| F4/80 | Abcam | Cat# ab6640; RRID:AB_1140040 |
| Krox20 | Covance | Cat# PRB-236P-100; RRID:AB_291594 |
| Slit1 | Sigma | RRID: SAB1307048 |
| Slit2 | Chemicon | Cat# AB5701; RRID:AB_2286130 |
| Slit3 | Sigma | Cat# SAB2104337; RRID:AB_10668990 |
| Slit3 | R&D | Cat# AF3629; RRID:AB_2189998 |
| Robo1 | Abcam | Cat# ab7279; RRID:AB_449561 |
| Robo2 | Santa Cruz | Cat# sc-25673, RRID:AB_2269780 |
| Neurofilament heavy chain | Abcam | Cat# ab4680, RRID:AB_304560 |
| CD31 | BD Biosciences | Cat# 550274; RRID:AB_393571 |
| S100 | DAKO | Cat# Z0311; RRID:AB_10013383 |
| CD68 | Abcam | Cat# ab53444; RRID:AB_869007 |
| CD206 | R&D | Cat# AF2535; RRID:AB_2063012 |
| Vimentin | Abcam | Cat# ab24525; RRID:AB_778824 |
| Fibronectin | Abcam | Cat# ab2413; RRID:AB_2262874 |
| GAPDH | EMD Millipore | Cat# MAB374; RRID:AB_2107445 |
| Bacterial and Virus Strains | ||
| GFP adenoviruses | Le et al., 2005 | N/A |
| Sox2/GFP adenoviruses | Le et al., 2005 | N/A |
| Critical Commercial Assays | ||
| Affymetrix array analysis | Patterson Institute for Cancer Research (Manchester, UK) | N/A |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE123915 |
| Experimental Models: Organisms/Strains | ||
| Sox2flox/flox mice | Taranova et al., 2006, Favaro et al., 2009 | N/A |
| Rosa26R-Sox2IRESGFP mice | Lu et al., 2010 | N/A |
| Slit1−/− mice | Plump et al., 2002 | N/A |
| Slit2+/− mice | Plump et al., 2002 | N/A |
| Slit3+/− mice | Yuan et al., 2003 | N/A |
| Robo1+/− mice | Andrews et al., 2008 | N/A |
| Robo2+/− mice | Grieshammer et al., 2004 | N/A |
| mP0-TOTACRE mice | Feltri et al., 1999 | N/A |
| PLP-GFP | Mallon et al., 2002 | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xin-peng Dun (xin-peng.dun@plymouth.ac.uk).
Experimental Model and Subject Details
Animals
All work involving animals was carried out according to Home Office regulation under the UK Animals Scientific Procedures Act 1986. Ethical approval for all experiments was granted by Plymouth University Animal Welfare and Ethical Review Board. Wistar rat and C57BL/6 mouse breeding pairs were purchased from Charles River UK limited. Sox2 conditional knockout mice (Sox2fl/fl) have been described previously (Taranova et al., 2006, Favaro et al., 2009). The P0-CRE (mP0-TOTACRE) mice have been described previously (Feltri et al., 1999). Mice with Schwann cell-specific loss of Sox2 were generated by breeding of Sox2fl/fl with the P0-CRE line. The generation of Sox2 overexpressing (Sox2 OE) mice was as described (Roberts et al., 2017, Lu et al., 2010). Slit1 and Slit2 (Plump et al., 2002), Slit3 (Yuan et al., 2003), Robo1 (Andrews et al., 2008) and Robo2 (Grieshammer et al., 2004) gene knockout mice were maintained as heterozygous mice for breeding to generate experimental mice. Homozygous PLP-GFP mice (Mallon et al., 2002) were crossed with Sox2 KO, Slit2, Slit3, Robo1 heterozygous mice and Slit3/Robo1 double heterozygous mice to generate Schwann cell labeled (GFP) strains.
Primary cell lines
Primary rat Schwann cells were cultured from postnatal day 3 Wistar rat pups, sex of rat pups was not identified at this age. Primary mouse macrophages were cultured from bone marrow of 6 weeks old male Slit3+/+ and male Slit3−/− mice.
GFP or Sox2/GFP adenovirus
GFP and Sox2/GFP adenoviruses were obtained from Professor Jeffrey Milbrandt (Le et al., 2005).
Method Details
Cell culture
Schwann cells were prepared from sciatic nerve and brachial plexus of postnatal day 3 Wistar rat pups as previously described (Roberts et al., 2017). Schwann cells were cultured in low glucose (1g/ml) DMEM containing 3% FBS, 10ng/ml NRG-1 (R&D, Cat No. 396-HB-050) and 2uM forskolin (Sigma, Cat No. 344270). Bone marrow macrophages were cultured from bone marrow of six weeks old Slit3+/+ and Slit3−/− mice as previously described (Trouplin et al., 2013). Coverslips (13mm) were coated with 0.1mg/ml Poly-L-lysine hydrobromide (Sigma, P6282) in 24 well plates for Schwann cells and bone marrow macrophages co-culture, Schwann cells and bone marrow macrophages were mixed with 5:3 ratio and a total 16 × 104 cells were seeded per coverslip in low glucose DMEM containing 3% FBS. In Schwann cell and Slit3+/+ macrophage co-cultures, Schwann cells grow on top of each other and the counting of the total number of Schwann cells in the clusters is not accurate. Instead, the area of Schwann cells on the co-culture coverslips is therefore quantified in ImageJ.
Peripheral nerve surgery
Two month old male and female mice were anaesthetised with isoflurane, the right sciatic nerve was exposed and for the nerve cut procedure transected at approximately 0.5 cm distal to the sciatic notch and no re-anastamosis of the severed nerve was performed. This allowed analysis of axon pathfinding and cell migration in the nerve bridge that forms between the retracted proximal and distal nerve stumps. For nerve crush experiments, the sciatic nerve was crushed once for 30 s using a pair of delicate forceps (Fine Science Instruments, 0.4mm tip angled, 11063-07), and again for 30 s at the same site but orthogonal to the initial crush. Overlying muscle was sutured and the skin was closed with an Autoclip applier. All animals undergoing surgery were given appropriate post-operative analgesia (0.025% bupivacaine solution, topically applied above the muscle suture before applying the surgical clip) and monitored daily. At the indicated time points post-surgery for each experiment described, animals were euthanased humanely by CO2 in accordance with UK Home Office regulations.
Adenoviral infection and Affymetrix array analysis
Schwann cells were infected with control GFP or Sox2/GFP adenovirus (Le et al., 2005, Parrinello et al., 2010) for 24 hours in defined medium (DM) and then changed to DM without virus. After 48 hours of adenovirus infection, total RNA was purified with miRNeasy mini kit (QIAGEN, 217004) and on-column DNase digestion was also performed. Triplicate mRNA samples purified from GFP or Sox2/GFP adenovirus-infected Schwann cells were analyzed on an Affymetrix 1.0 ST Rat Exon array by the Patterson Institute for Cancer Research (Manchester, UK). Affymetrix data was analyzed using the Affymetrix Power Tools software. Regulated transcripts were identified using a false discovery rate controlled p value of < 0.05. The putative functional role of Sox2 targets was then analyzed by Gene Ontology analysis with the Gene Map Annotator and Pathway Profiler (GenMAPP) software package. The 9 classic axon guidance receptors were identified from axon guidance signaling pathways.
mRNA purification, cDNA synthesis, RT-PCR and qRT-PCR
Total mRNA was extracted with miRNeasy Mini Kit (QIAGEN, 217004) and first stand cDNA was synthesized with M-MLV reverse transcriptase (Promega, M368) and random hexamer primers (Promega, C1181). RT-PCR was performed in the G-Storm GS4M, qRT-PCR was performed in the PCR LightCycler480 Real-Time PCR Instrument (Roche Applied Science) using SYBR Green I Master with the following primers: Sox2 forward: 5′-AGACGCTCATGAAGAAGGATAAG-3′, reverse: 5′-GAGCTGGTCATGGAGTTGTACTG-3′, Slit1 forward: 5′-CTGCTCCCCGGATATGAACC-3′, reverse: 5′-TAGCATGCA CTCACACCTGG-3′, Slit2 forward: 5′-AACTTGTACTGCGACTGCCA-3′, reverse: 5′-TCCTCATCACTGCAGACAAACT-3′, Slit3 forward: 5′-AGTTGTCTGCCTTCCGACAG-3′, reverse: 5′-TTTCCATGGAGGGTCAGCAC-3′, Robo1 forward: 5′-GCTGGCGACATGGGATCATA-3′, reverse: 5′-AATGTGGCGGCTCTTGAACT-3′, Robo2 forward: 5′-CGAGCTCCTCCACAGTTTGT-3′, reverse: 5′-GTAGGTTCTGGCTGCCTTCT-3′, EpHA3 forward: 5′- GCTGGCAGAAAGACAGGAAC-3′, reverse: 5′-CGGGAACTGGACCATTCTTA-3′. EpHA5 forward: 5′-GGTACCTGCCAAGCTCCTTC-3′, reverse: 5′-ATTCCATTGGGGCGATCTGG-3′. EpHA7 forward: 5′-ATCTGAAACCGGGAACAGTG-3′ reverse: 5′-GTAGGTTTTGGTGCCTGGAA-3′. Unc5B forward: 5′-ACTGCACTTTCACCCTGGAG-3′, reverse: 5′-GACTAGCTTTGGTGGCGAAG-3′. Neo1 forward: 5′-TGAACCAGTTGTGGGAAACA-3′, reverse 5′-CCACCACGATAGTGATGACG −3′, 18sRNA forward: 5′-GAGAAACGGCTACCACATCC-3′, reverse: 5′-GGACACTCAGCTAAGAGCATCG-3′. GAPDH forward: 5′-AAGGTCATCCCAGAGCTGAA-3′, reverse: 5′-CTGCTTCACCACCTTCTTGA-3′. Cross point (Cp) values were calculated by using the software of the LightCycler480 Real-Time PCR Instrument. Relative mRNA level were calculated by the −2ΔΔCT method (Livak and Schmittgen, 2001) using GAPDH as a reference gene for normalization. All reactions were carried out in triplicate for statistical analysis.
Immunohistochemistry and immunocytochemistry
Sciatic nerves were dissected out and fixed overnight in 4% paraformaldehyde (PFA) at 4°C. Samples were then washed in phosphate buffered saline (PBS, 3 × 10 minutes) and dehydrated in 30% sucrose (in PBS) overnight at 4°C. Subsequently, samples were embedded in OCT medium and sectioned on a cryostat at a thickness of 12μm. Cultured bone marrow macrophages, rat Schwann cells and bone marrow macrophages co-cultures on coverslips were fixed 15 minutes in 4% paraformaldehyde (PFA) at 4°C and then washed in PBS (3 × 10 minutes). Sections or cells were permeabilised with 0.25% Triton X-100 plus 1% bovine serum albumin (BSA) in PBS for 30 minutes and then blocked with blocking buffer (3% BSA plus 0.05% Triton X-100 in PBS) for 45 minutes at room temperature. Sections were incubated with primary antibodies (diluted in blocking buffer) overnight at 4°C. Next day, sections were washed with PBS (3 × 10 minutes) and then incubated with species specific secondary antibodies plus Hoechst dye (diluted in blocking buffer) for 1 hour at room temperature. Finally, sections were washed with PBS (3 × 10 minutes) and mounted with Citiflour (Agar Scientific, R1320) for imaging with a Leica SP8 confocal microscope.
Western blot
Nerve samples were directly sonicated into 1X SDS loading buffer. Proteins were separated on 7.5% or 12% SDS polyacrylamide running gels and then transferred onto a polyvinylidene fluoride (PVDF, 0.45μm) transfer membrane using the wet transfer method. Membranes were blocked with 5% fat free milk in TBST (Tris buffered saline plus 0.1% Tween-20) for one hour at room temperature. Primary antibodies were diluted (1:500) in 5% milk (in TBST) and the membranes were incubated in primary antibodies overnight at 4°C. The next day, the membranes were washed in TBST (3x10 minutes) and then incubated with HRP conjugated secondary antibody (1:5000 in 5% milk TBST) for one hour at room temperature. After three 10 minute TBST washes (10 minutes each), Pierce ECL western blotting substrate was added onto the membrane and incubated for five minutes to develop the chemiluminescent signal. Amersham Hyperfilm ECL films were used to capture the intensity of chemiluminescent signal. Exposed films were then developed in a Compact X4 automatic processor. The intensity of protein bands was quantified using the free ImageJ software available from https://imagej.nih.gov/ij/.
Whole nerve neurofilament antibody and CD31 antibody staining
At the described time points following surgery, nerves were removed and fixed in 4% paraformaldehyde for 5 hours at 4°C. Following fixation, nerves were then washed in PTX (1% Triton X-100 (Sigma, T9284) in PBS three times for 10 minutes each time. To ensure better antibody penetration for nerve crush samples, the epineurium was removed in these preparations after washing in PTX. Nerves were subsequently incubated with blocking solution (10% fetal bovine serum (FBS) in PTX) overnight at 4°C. The following day, nerves were transferred into primary antibodies in PTX containing 10% FBS and incubated for 72h at 4°C with gentle rocking. Primary antibodies used for the experiments are neurofilament heavy chain (1:1000, Abcam, ab4680) and CD31 (1:100, BD Biosciences, 550274). After the incubation, nerves were washed with PTX three times for 15 minutes each wash, followed by washing in PTX for 6 hours at room temperature, with a change of PTX every 1 hour. Secondary antibodies (1:500, Invitrogen) were diluted in PTX containing 10% FBS, and incubated with the nerves for 48h at 4°C with rocking. Next, nerves were washed in PTX three times for 15 minutes each, followed by washing in PTX for 6 hours at room temperature, changing the PTX each hour, and then washed overnight without changing PTX at 4°C. Nerves were cleared sequentially with 25%, 50%, 75% glycerol (Sigma, G6279) in PBS between 12–24h for each glycerol concentration. Following clearing, nerves were mounted in CitiFluor (Agar Scientific, R1320) for confocal imaging. The length of regenerating axons at day 7 after crush injury was measured by the neurofilament staining from crush site to the front of regenerating axons under an epifluorescent microscope.
Mouse static sciatic index (SSI)
The mouse static sciatic index (SSI) measurement (Baptista et al., 2007) was used to assess functional recovery in control, Sox2 KO and Robo1+/− mice after sciatic nerve crush injury. The SSI measurement was made by an individual blinded to the genotype of the animals. 4 animals were used for each genotype. Briefly, mice were placed in a clear acrylic display box (15cm x15cm x 15cm) and filmed with a digital camera from underneath. These videos were examined on computer and selected frames were used for measurements of the print length and toe spread. The print length factor (PLF) and toe spread factor (TSF) was calculated as described previously (Baptista et al., 2007). SSI was calculated with the formula SSI = 101.3 × TSF−54.03 × PLF−9.5.
Transmission electron microscopy and low vacuum scanning microscopy
Sciatic nerves were fixed in 2.5% glutaraldehyde in 0.1M cacodylate buffer (pH7.2) for 24 hours. The nerves were rinsed in cacodylate buffer and post-fixed in 1% osmium tetroxide in the same cacodylate buffer for 24 hours. Nerves were rinsed in cacodylate buffer, dehydrated in an ascending ethanol series and embedded in agar low viscosity resin blocks. For low vacuum scanning microscopy, resin block surfaces were polished and analyzed using a JEOL 6610 LV-SEM machine. For transmission electron microscopy, ultra-thin sections were cut with a Leica EM UC7 ultramicrotome and stained with uranyl acetate and lead citrate. Sections were examined and photographed using a JEOL 1200EX or 1400 transmission electron microscope. 200 myelinated fibers per animal were measured for myelin thickness and axon perimeter using ImageJ software and 3 mice of each genotype were used for the measurement. G-ratio was calculated as axonal diameter/fiber diameter.
Quantification and Statistical Analysis
The statistical significance was analyzed using the Student’s t test by comparing the test groups with the control groups. All data are represented in the figures as mean values ± SEM. P values are indicated with single asterisk (∗ < 0.05), double asterisks (∗∗ < 0.01) and triple asterisks (∗∗∗ < 0.001) on graphs. Where graphs are not labeled with an asterisk, any differences between the test groups and the control groups were non-significant. The n number for each experiment has been stated in figure legends, in most cases n is defined as the number of mice from the same genotype and same experiment. Sample size was not predetermined by statistical methods and randomization was not applied. Because of the small sample sizes (n < 5 for most statistical comparisons), assumptions of how well normality and equal variances fit the data could not be reliably assessed. No samples or data were excluded from the analysis. Samples for western blotting and qRT-PCR were prepared by grouping nerves from three mice together for each time point to create a pooled sample. We have used pooled biological replicates for the repetition of these experiments.
Data and Software Availability
All data used in this manuscript are available upon request. The accession numbers for the raw microarray data reported in this paper is GEO: GSE123915.
Acknowledgments
This work was supported by The National Natural Science Foundation of China (81371353) to X.D. and UK Wellcome Trust (088228/Z/09/Z) to D.B.P. and A.C.L. We would like to thank Dr. William Andrews and Professor Marc Tessier-Lavigne for providing the Slit1–2 and Robo1–2 mice, Professor David M. Ornitz and Professor Alain Chédotal for providing the Slit3 mice, Dr. Larysa H. Pevny and Dr. Silvia K. Nicolis for providing the Sox2fl/fl mice, Professor Laura Feltri and Larry Wrabetz for providing the P0-CRE mice, Professor Mark W. Onaitis for providing the Sox2-overexpressing mice, and Professor Wendy B. Macklin for providing the PLP-GFP mice. We thank Jack Kelly for help submitting the microarray data.
Author Contributions
X.D., A.C.L., and D.B.P. designed research and interpreted data; X.D., L.C., P.K.W., R.W.B., L.K.D., T.M., and S.L.R. performed experiments; and X.D. and D.B.P. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: February 5, 2019
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.081.
Supplemental Information
References
- Andrews W., Barber M., Hernadez-Miranda L.R., Xian J., Rakic S., Sundaresan V., Rabbitts T.H., Pannell R., Rabbitts P., Thompson H. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 2008;313:648–658. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Bagri A., Tessier-Lavigne M. Neuropilins as Semaphorin receptors: in vivo functions in neuronal cell migration and axon guidance. Adv. Exp. Med. Biol. 2002;515:13–31. [PubMed] [Google Scholar]
- Baptista A.F., Gomes J.R., Oliveira J.T., Santos S.M., Vannier-Santos M.A., Martinez A.M. A new approach to assess function after sciatic nerve lesion in the mouse—adaptation of the sciatic static index. J. Neurosci. Methods. 2007;161:259–264. doi: 10.1016/j.jneumeth.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J., Klein R. Signaling from axon guidance receptors. Cold Spring Harb. Perspect. Biol. 2010;2:a001941. doi: 10.1101/cshperspect.a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blockus H., Chédotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- Brose K., Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Carr L., Parkinson D.B., Dun X.P. Expression patterns of Slit and Robo family members in adult mouse spinal cord and peripheral nervous system. PLoS One. 2017;12:e0172736. doi: 10.1371/journal.pone.0172736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A.L., Burden J.J., Van Emmenis L., Mackenzie F.E., Hoving J.J., Garcia Calavia N., Guo Y., McLaughlin M., Rosenberg L.H., Quereda V. Macrophage-induced blood vessels guide Schwann cell-mediated regeneration of peripheral nerves. Cell. 2015;162:1127–1139. doi: 10.1016/j.cell.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Piao X., Bonaldo P. Role of macrophages in Wallerian degeneration and axonal regeneration after peripheral nerve injury. Acta Neuropathol. 2015;130:605–618. doi: 10.1007/s00401-015-1482-4. [DOI] [PubMed] [Google Scholar]
- Dun X.P., Parkinson D.B. Visualizing peripheral nerve regeneration by whole mount staining. PLoS One. 2015;10:e0119168. doi: 10.1371/journal.pone.0119168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun X.P., Parkinson D.B. Role of Netrin-1 signaling in nerve regeneration. Int. J. Mol. Sci. 2017;18:E491. doi: 10.3390/ijms18030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R., Valotta M., Ferri A.L., Latorre E., Mariani J., Giachino C., Lancini C., Tosetti V., Ottolenghi S., Taylor V., Nicolis S.K. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- Feltri M.L., D’Antonio M., Previtali S., Fasolini M., Messing A., Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N Y Acad. Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- Geutskens S.B., Hordijk P.L., van Hennik P.B. The chemorepellent Slit3 promotes monocyte migration. J. Immunol. 2010;185:7691–7698. doi: 10.4049/jimmunol.0903898. [DOI] [PubMed] [Google Scholar]
- Giger R.J., Hollis E.R., 2nd, Tuszynski M.H. Guidance molecules in axon regeneration. Cold Spring Harb. Perspect. Biol. 2010;2:a001867. doi: 10.1101/cshperspect.a001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U., Le Ma, Plump A.S., Wang F., Tessier-Lavigne M., Martin G.R. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev. Cell. 2004;6:709–717. doi: 10.1016/s1534-5807(04)00108-x. [DOI] [PubMed] [Google Scholar]
- Jessen K.R., Mirsky R. The repair Schwann cell and its function in regenerating nerves. J. Physiol. 2016;594:3521–3531. doi: 10.1113/JP270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.J., Lee Y.S., Lee S.Y., Baek W.Y., Choi Y.J., Moon S.A., Lee S.H., Kim J.E., Chang E.J., Kim E.Y. Osteoclast-secreted SLIT3 coordinates bone resorption and formation. J. Clin. Invest. 2018;128:1429–1441. doi: 10.1172/JCI91086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A.W., Mathivet T., Larrivée B., Tong R.K., Kowalski J., Pibouin-Fragner L., Bouvrée K., Stawicki S., Nicholes K., Rathore N. Robo4 maintains vessel integrity and inhibits angiogenesis by interacting with UNC5B. Dev. Cell. 2011;20:33–46. doi: 10.1016/j.devcel.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Le N., Nagarajan R., Wang J.Y., Araki T., Schmidt R.E., Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. USA. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.M., Kucenas S. Perineurial glia are essential for motor axon regrowth following nerve injury. J. Neurosci. 2014;34:12762–12777. doi: 10.1523/JNEUROSCI.1906-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu Y., Futtner C., Rock J.R., Xu X., Whitworth W., Hogan B.L., Onaitis M.W. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon B.S., Shick H.E., Kidd G.J., Macklin W.B. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J. Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini R., Fischer S., López-Vales R., David S. Interactions between Schwann cells and macrophages in injury and inherited demyelinating disease. Glia. 2008;56:1566–1577. doi: 10.1002/glia.20766. [DOI] [PubMed] [Google Scholar]
- Moore A.M., Kasukurthi R., Magill C.K., Farhadi H.F., Borschel G.H., Mackinnon S.E. Limitations of conduits in peripheral nerve repairs. Hand (N. Y.) 2009;4:180–186. doi: 10.1007/s11552-008-9158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q.T., Sanes J.R., Lichtman J.W. Pre-existing pathways promote precise projection patterns. Nat. Neurosci. 2002;5:861–867. doi: 10.1038/nn905. [DOI] [PubMed] [Google Scholar]
- Parrinello S., Napoli I., Ribeiro S., Wingfield Digby P., Fedorova M., Parkinson D.B., Doddrell R.D., Nakayama M., Adams R.H., Lloyd A.C. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143:145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump A.S., Erskine L., Sabatier C., Brose K., Epstein C.J., Goodman C.S., Mason C.A., Tessier-Lavigne M. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Roberts S.L., Dun X.P., Doddrell R.D.S., Mindos T., Drake L.K., Onaitis M.W., Florio F., Quattrini A., Lloyd A.C., D’Antonio M., Parkinson D.B. Sox2 expression in Schwann cells inhibits myelination in vivo and induces influx of macrophages to the nerve. Development. 2017;144:3114–3125. doi: 10.1242/dev.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A.F., Wolman M.A., Franzini-Armstrong C., Granato M. In vivo nerve-macrophage interactions following peripheral nerve injury. J. Neurosci. 2012;32:3898–3909. doi: 10.1523/JNEUROSCI.5225-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A.F., Isaacman-Beck J., Franzini-Armstrong C., Granato M. Schwann cells and deleted in colorectal carcinoma direct regenerating motor axons towards their original path. J. Neurosci. 2014;34:14668–14681. doi: 10.1523/JNEUROSCI.2007-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno T., Fujiwara A., Sakaguchi K., Tanaka K., Takenaka S., Tsuyama S. Slit3 regulates cell motility through Rac/Cdc42 activation in lipopolysaccharide-stimulated macrophages. FEBS Lett. 2007;581:1022–1026. doi: 10.1016/j.febslet.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Taranova O.V., Magness S.T., Fagan B.M., Wu Y., Surzenko N., Hutton S.R., Pevny L.H. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouplin V., Boucherit N., Gorvel L., Conti F., Mottola G., Ghigo E. Bone marrow-derived macrophage production. J. Vis. Exp. 2013;22:e50966. doi: 10.3791/50966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.H., Miller R.H. Glial cell migration directed by axon guidance cues. Trends Neurosci. 2002;25:173–175. doi: 10.1016/s0166-2236(00)02096-8. discussion 175–176. [DOI] [PubMed] [Google Scholar]
- Wang Y., Teng H.L., Huang Z.H. Repulsive migration of Schwann cells induced by Slit-2 through Ca2+-dependent RhoA-myosin signaling. Glia. 2013;61:710–723. doi: 10.1002/glia.22464. [DOI] [PubMed] [Google Scholar]
- Wehrle R., Camand E., Chedotal A., Sotelo C., Dusart I. Expression of netrin-1, slit-1 and slit-3 but not of slit-2 after cerebellar and spinal cord lesions. Eur. J. Neurosci. 2005;22:2134–2144. doi: 10.1111/j.1460-9568.2005.04419.x. [DOI] [PubMed] [Google Scholar]
- Williams L.R., Longo F.M., Powell H.C., Lundborg G., Varon S. Spatial-temporal progress of peripheral nerve regeneration within a silicone chamber: parameters for a bioassay. J. Comp. Neurol. 1983;218:460–470. doi: 10.1002/cne.902180409. [DOI] [PubMed] [Google Scholar]
- Yuan W., Rao Y., Babiuk R.P., Greer J.J., Wu J.Y., Ornitz D.M. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc. Natl. Acad. Sci. USA. 2003;100:5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelina P., Blockus H., Zagar Y., Péres A., Friocourt F., Wu Z., Rama N., Fouquet C., Hohenester E., Tessier-Lavigne M. Signaling switch of the axon guidance receptor Robo3 during vertebrate evolution. Neuron. 2014;84:1258–1272. doi: 10.1016/j.neuron.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Zigmond R.E. Cytokines that promote nerve regeneration. Exp. Neurol. 2012;238:101–106. doi: 10.1016/j.expneurol.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this manuscript are available upon request. The accession numbers for the raw microarray data reported in this paper is GEO: GSE123915.