Abstract
Aim:
Approximately 30% of acute myeloid leukemia (AML) patients carry FLT3 tyrosine kinase domain (TKD) mutations or internal tandem duplication (FLT3-ITD). Currently there is a paucity of compounds that are active against drug-resistant FLT3-ITD, which contains secondary mutations in the TKD, mainly at residues D835/F691.
Results:
HSD1169, a novel compound, is active against FLT3-ITD (D835 or F691). HSD1169 is also active against T-LAK cell-originated protein kinase (TOPK), a collaborating kinase that is highly expressed in AML cell lines. HSD1169 was active against MV4–11 and Molm-14 (FLT3-ITD cell lines) but not NOMO-1 or HL60 (FLT3-WT cell lines). HSD1169 was also active against sorafenib-resistant Molm13-res cell line (containing FLT3-ITD/D835Y).
Conclusion:
HSD1169 or an analog could become a therapeutic agent for AML containing drug-resistant FLT3-ITD.
Keywords: : acute myeloid leukemia, drug resistance, FLT3-ITD, secondary mutations, TOPK
The human genome encodes greater than 500 protein kinases, which constitute one of the largest gene families [1]. Protein kinases control key cellular processes, including cell cycle, transcription, migration, proliferation and apoptosis via phosphotransfer [1]. Many diseases, especially cancers, are driven by mutant or aberrantly expressed protein kinases [1,2] and such kinases have become drug targets [2,3]. Imatinib (Gleevec®) [4], an inhibitor of BCR-ABL and the first protein kinase inhibitor to be approved by the US FDA in 2001 for the treatment of chronic myeloid leukemia changed chronic myeloid leukemia from a lethal disease with a low 5-year survival rate to one with over 90% survival rate. Since the imatinib breakthrough, 29 other protein kinase inhibitor drugs [5] have been approved by the FDA [5]. In majority of the cases, the administration of tyrosine kinase inhibitors (TKIs) leads to remission but ultimately relapse occurs. During relapse, the cancer driving kinase usually attains a mutation (for aberrantly expressed kinase) or secondary mutation, which reduces the affinity of the kinase for the TKI. In addition, activation of compensatory signaling pathways may occur during relapse [6]. New generation TKIs, which are active against drug-resistant kinases are now being hotly pursued.
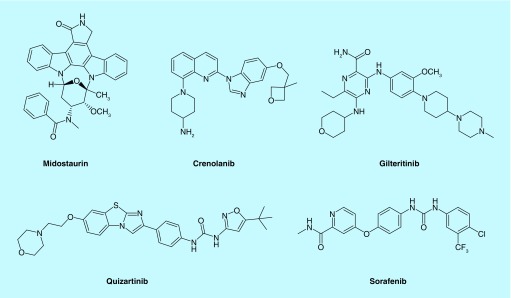
Our group has been interested in developing compounds that inhibit the proliferation of acute myeloid leukemia (AML) as AML patients have limited therapeutic options [7,8]. The 5-year survival rate for AML is one of the lowest (∼30% overall and less than 10% for patients over 60 years) among major cancers. In particular, about a third of AML patients harboring FLT3 internal tandem duplication (FLT3-ITD) or FLT3 containing point mutation in the tyrosine kinase domain (TKD) have poor prognosis. The FLT3 kinase has therefore emerged as an important anti-AML drug target [9,10]. Recently, midostaurin (Rydapt®), a potent inhibitor of FLT3-ITD was approved as an AML drug [11]. Other FLT3 inhibitors are also in different phases of clinical trials [12,13]. So far, several confounding issues regarding FLT3 inhibition as a viable strategy to cure AML remain to be resolved: midostaurin, the only approved FLT3 inhibitor so far is not a single agent and is used in combination with other cytotoxic drugs [11]; most FLT3 inhibitors, such as midostaurin, quizartinib, sorafenib and sunitinib, also inhibit c-Kit [14–17]. The concurrent inhibition of FLT3 and c-Kit leads to myelosuppression [18] but it has been challenging to develop potent FLT3 inhibitors that do not inhibit c-Kit also; most FLT3 inhibitors developed to date, such as midostaurin, ponatinib, sunitinib and sorafenib also potently inhibit VEGFRs [14,16]. VEGFRs are involved in several important processes, such as the formation and maintenance of the vasculature, platelet function, cell proliferation, migration and survival [19,20]. Kinase inhibitors that inhibit VEGFRs are known to cause cardiotoxicity as on-target toxicity [21] and are not ideal for elderly patients; most FLT3 inhibitors evaluated in the clinic so far do not achieve complete cure due to the emergence of secondary kinase mutations [22,23]. For example, several FLT3 inhibitors, such as quizartinib, ponatinib and sorafenib are weak inhibitors of FLT3 (D835C/H/Y) [23], which emerge only few months after FLT3 inhibitor treatment. Although there are a few FLT3 ligands that discriminate FLT3 from c-Kit (such as gilteritinib, which inhibits FLT3 with IC50 of 0.29 nM but only inhibits c-Kit with IC50 of 230 nM [24]) or that can inhibit certain forms of secondary mutations (e.g., crenolanib inhibits FLT3 and FLT3 D835Y with equipotent IC50 values of 1.3 and 6.9 nM, respectively [25]). So far, the discovery of an FLT3 inhibitor that solves all of the aforementioned issues has remained elusive.
In addition to FLT3 playing a central role in the pathogenesis of AML, other kinases (collaborating kinases) are also important for leukemogenesis. For example, T-LAK cell-originated protein kinase (TOPK) has been proposed as a target for AML, following the observation that it was highly expressed in most AML cell lines [26]. MV4–11 cell viability was decreased and apoptosis increased when TOPK was knocked down [26]. Furthermore, MV4–11 and MOLM13 cell lines were highly sensitive to the TOPK inhibitor OTS514 [26,27]. Based on the observations that singly targeting FLT3 or ‘collaborating kinases’ with inhibitors lead to pronounced growth inhibition in FLT3-driven AML cell lines, we rationalize that a multi FLT3/‘collaborating kinases’ inhibitor will not only have good potency against FLT3-ITD-driven AML but also such multitargeting inhibitor would have less propensity to encounter drug resistance, compared with an inhibitor that only targets FLT3 or the collaborating kinases. A challenge however is that the multitargeting kinase inhibitor should not inhibit ‘toxicity’ kinases, such as c-Kit and VEGFR, which could lead to cardiotoxicity or myelotoxicity.
Experimental procedures
General procedure for synthesis of compounds
A mixture of amine (1 mmol) and aldehyde (1 mmol) in 3 ml of absolute ethanol was refluxed for 2 h followed by addition of cyclic ketone or acyclic ketone (2.1 mmol) to the reaction mixture. A catalytic amount of concentrated hydrochloric acid was added and the reaction was continued to reflux for 6–12 h. After completion, sodium bicarbonate solution (3 ml) was added to the reaction mixture and extracted with ethyl acetate (50 ml × 2) and water (25 ml × 2). The organic layer was collected, dried (Na2SO4), concentrated under reduced pressure and purified by silica gel chromatography (dichloromethane:methanol 99:01≈80:20 or EtOAc:Hexane [50:50≈100:0]) to give the desired cyclized compound.
Cell lines & culture method
MV4–11, HL60 and K562 were routinely cultured in Iscove's Modified Dulbecco's Medium, Molm13-res and Molm14 were cultured in Roswell Park Memorial Institute (RPMI) 1640 and all media were supplemented with 10% fetal bovine serum. Cells were routinely cultured at 37°C with 5% CO2.
Antiproliferative activity
Molm-13-res cell line is a kind gift from Dr Sharyn Baker (The Ohio State University). MV4–11 and Molm-14 cell lines were kind gifts from Dr Mark Levis (Johns Hopkins University). HL60 and K562 were obtained from American Type Culture Collection (ATCC). Cells were seeded into 96-well microtiter plates at 2 × 104 cells/ml. Approximately 24 h after seeding, test compounds diluted into media were added to the cells to give a threefold serial dilution from 10 μM for IC50 determination. An equal volume of DMSO diluted in medium was used as vehicle control. After treatment, the plates were incubated at 37°C with 5% CO2 for 72 h and assayed for viability by adding CellTiter-Blue® Cell Viability Assay (Promega Corporation, WI, USA) for an additional 4 h. The plates were read with a BioTek Cytation 5 Cell Imaging Multi-Mode Reader (BioTek Instruments, VT, USA). Data were analyzed and graphed using a sigmoidal dose-response equation on GraphPad Prism 5 Software (Graphpad, CA, USA).
For NOMO-1 and MOLT-4, IC50 was determined using the services of Reaction Biology Corporation (PA, USA). Briefly, compounds were diluted in DMSO solution with 10-dose and threefold dilutions in a source plate starting at 20 mM. Using the Echo 550 (Labcyte Inc., CA, USA), 12.5 nl compounds were delivered into 384-well plates after which 25 μl of culture media containing 2000 of NOMO-1 or MOLT-4 cells was added to each of the wells in duplicates of the cell culture plate. The plates were incubated with the compounds at 37°C, 5% CO2 for 72 h followed by the addition of 25 μl of Cell Titer Glo 2.0 reagent to each well. The contents were mixed on an orbital shaker for 2 min and incubated at room temperature for 15 min to stabilize luminescent signal. Luminescence was recorded by Envision 2104 Multilabel Reader (PerkinElmer, CA, USA). The number of viable cells in culture was determined based on quantitation of the ATP present in each culture well. The IC50 curves were plotted and IC50 values were calculated using the GraphPad Prism 4 program based on a sigmoidal dose-response equation
In vitro kinase assays
The inhibition of FLT3-ITD kinase activity by HSD1169 and related compounds were performed using the ADP-Glo™ kinase assay system (Promega Corporation, WI, USA). Briefly, a 5 μl reaction containing compounds (100 nM), substrate (ATP and myelin basic protein (MBP) substrate at 10 μM and 0.1 mg/ml, respectively) and kinase (30 nM) was set up in duplicates in a 384-well white plate and incubated at room temperature for 3 h. As recommended by the manufacturer, 5 μl of the ADP-Glo reagent was added for 40 min followed by the addition of 10 μl of the kinase detection reagent for another 1 h at room temperature. Luminescence was measured using a BioTek Cytation 5 Cell Imaging Multi-Mode Reader.
The strength of binding of HSD1169 to FLT3 kinase mutants, ABL phosphorylated and nonphosphorylated was performed using the commercial KdELECT assay (DiscoverX Corporation, CA, USA) service.
Western blot analysis
MV4–11 cells were treated with HSD1169 at the indicated concentrations or with DMSO (0.1%). After the indicated time periods, cells were pelleted by centrifugation and lysed with M-PER™ Mammalian Protein Extraction Reagent (Life Technologies Corporation, CA, USA) supplemented with protease inhibitor cocktail (Roche) for total protein extraction. Cells were lysed for 10 min on ice with gentle intermittent shaking. The cell lysates were centrifuged at 6500 × g for 10 min at 4°C and the soluble proteins in the supernatant were saved. Protein concentrations of samples were determined using the bicinchoninic acid (BCA) assay. Total protein was separated on SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was then blocked with 5% Bovine seum albumin (BSA) in 1× TBST (Tris-buffered saline, 0.1% Tween 20 (20 mM Tris pH7.5, 150 mM NaCl and 0.1% Tween 20)) for 1 h at room temperature after which primary antibodies were incubated with the membrane following the manufacturer's recommendations. The following primary antibodies from Cell Signaling (MA, USA) were used: phospho-STAT5, STAT5, TOPK and β-actin.
RNA isolation & real-time PCR analysis
Aurum total RNA mini kit (Bio-Rad, CA, USA) was applied to extract RNA from MV4–11 cells treated with HSD1169 at the indicated concentrations for 24 h. SuperScript™ II Reverse Transcriptase and random primers were used for the reverse transcription of the extracted RNA to cDNA. Real-time PCR was performed by QuantiTect SYBR® Green PCR Kit and specific primers for TOPK and GADPH on a Bio-Rad CFX96® Real-Time System (Bio-Rad, CA, USA). The data were normalized to GAPDH Ct and analyzed using the 2(−ΔΔCT) method. Each condition was repeated in duplicate.
Results & discussion
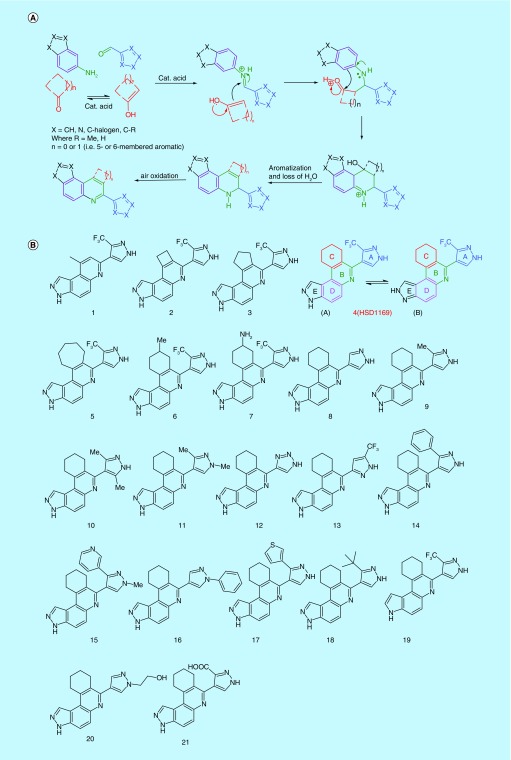
Novel chemical scaffold (8,9,10,11-tetrahydro-3H-pyrazolo[4,3-a]phenanthridine) preferentially inhibits FLT3-driven cell lines
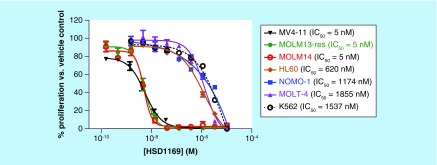
To discover compounds that are active against both FLT3-ITD and FLT3-ITD-harboring secondary mutations in the TKD, we screened our in-house synthesized compound library for new agents that inhibit the proliferation of FLT3- and FLT3 (ITD, D835Y)-driven AML cell lines (MV4–11 (FLT3-ITD), Molm-14 (FLT3-ITD), Molm-13-res (FLT3 (ITD, D835Y)) [28]. HSD1169 (see Figure 2B; synthesized via the Doebner reaction, see Figure 2A), which contains a novel 8,9,10,11-tetrahydro-3H-pyrazolo[4,3-a]phenanthridine scaffold was identified as a potent inhibitor of the three AML cell lines (IC50 5 nM). HSD1169 inhibited other non-FLT3-driven leukemia cell lines (K562, NOMO-1, HL60 and MOLT4) at significantly higher concentrations (620–1855 nM, Figure 3), strongly suggesting that HSD1169 is a FLT3 inhibitor. For the discovery of cell permeable inhibitors, phenotypic screening is superior to target-based screening, as an in vitro target inhibition screening could identify potent inhibitors that may not be cell permeable or not stable in complex cell environment. Additionally, phenotypic screening could unveil compounds that have novel modes of action that could not be predicted a priori [29]. However, a limitation of phenotypic screening is that it can be time consuming to identify the target of an active compound. This limitation is not severe for compounds that inhibit cancer cell proliferation because of the availability of a panel of cancer cell lines that are driven by various cancer drivers. For example, a compound that preferentially inhibits a FLT3-driven AML but not non-FLT3-driven AML will likely act via FLT3 signaling (at least partially).
Figure 2. . Novel kinase inhibitors synthesized.
(A) Mechanism for the one-flask synthesis of the compounds studied. (B) Structures of compounds evaluated in this study. Ring E of all of the compounds can exist as form (A) or form (B) as shown for HSD1169.
Figure 3. . Dose-dependent inhibition of leukemia cell lines’ proliferation by HSD1169.
Plot of the dose-dependent inhibition of indicated leukemia cell lines by HSD1169. Cultures were treated with a threefold dilution starting at 10 μM and incubated for 72 h. The percent proliferation was determined relative to the DMSO control for each compound and the data were fitted to a sigmoidal dose-response equation to obtain IC50 values. Error bars represent standard error of the mean of three replicates.
Figure 1. . FLT3 inhibitors in clinical trials or approved as acute myeloid leukemia drug.
Midostaurin was recently approved as an AML drug. The rest are in clinical trials.
AML: Acute myeloid leukemia.
Compound HSD1169 contains five rings (labeled as A, B, C, D and E in Figure 2). Structure–activity relationship studies indicated that the ring C in HSD1169 was important for both FLT3 kinase inhibition and proliferation inhibition (Table 1). Compound 1, which lacks ring C is a poor inhibitor of FLT3 and did not inhibit the growth of MV4–11 (see Table 1). Compounds 2, 3, 4 (HSD1169) and 5 contain 4-, 5-, 6- and 7-membered ring C and the order of activities against FLT3 enzymatic activity and AML cell lines proliferation is HSD1169∼5>3>>2. This indicates that six- or seven-membered ring C is optimum for FLT3 enzymatic and AML growth inhibitions. Substituted ring C compounds (6 and 7) were less active (see Table 1; entries vi and vii) in inhibiting AML cell growth than HSD1169. However, the methyl-substituted ring C (compound 6) was more active than the amino-substituted ring C (compound 7).
Table 1. . FLT3 kinase inhibition and antiproliferative activity of compounds.
| Entry | Compound | %FLT3 inhibition | IC50 (nM) | ||
|---|---|---|---|---|---|
| FLT3-ITD† | MV4–11 | Molm14 | Molm13-res | ||
| I | 1 | 22 | 797 | ND | ND |
| Ii | 2 | 21 | 1532 | ND | ND |
| Iii | 3 | 55 | 63 | 45.8 | 66.1 |
| Iv | 4 (HSD1169) | 80 | 5.4 | 4.9 | 5.1 |
| V | 5 | 87 | 9.1 | 16.6 | 13.7 |
| Vi | 6 | 77 | 13.7 | 26.8 | 20.7 |
| Vii | 7 | 71 | 132.5 | ND | ND |
| Viii | 8 | 53 | 38.4 | 61.5 | 40.2 |
| Ix | 9 | 54 | 61.5 | 64.1 | 86.7 |
| X | 10 | 54 | 159.4 | ND | ND |
| Xi | 11 | 21 | 744.2 | ND | ND |
| Xii | 12 | 56 | 154.5 | ND | ND |
| Xiii | 13 | 5 | >5000 | ND | ND |
| Xiv | 14 | 26 | 1449 | ND | ND |
| Xv | 15 | 6 | 1297 | ND | ND |
| Xvi | 16 | 9 | 1082 | ND | ND |
| Xvii | 17 | 19 | 772 | ND | ND |
| xviii | 18 | 20 | 484 | ND | ND |
| Xix | 19 | 8 | 898 | ND | ND |
| Xx | 20 | 27 | 1392 | ND | ND |
| Xxi | 21 | 6 | >5000 | ND | ND |
Compounds with IC50 greater than 100 nM against MV4–11 were not tested against Molm14 and Molm13-res.
†Percent inhibition was determined at 100 nM compound with 30 nM FLT3-ITD (obtained from SignalChem) in the presence of 10 μM ATP for 3 h.
ITD: Internal tandem duplication; ND: Not determined.
We next explored how the substitution pattern of the 1H-pyrazole group (ring A in HSD1169) affected both FLT3 enzymatic inhibition and AML growth inhibition. Compounds with hydrogen, methyl or trifluoromethyl groups at the 3-position of the pyrazole (compounds 8, 9 and HSD1169) were potent inhibitors of AML proliferation with the trifluoromethyl group being the best (compare entries viii, ix and iv in Table 1). Substituting the 3-position of the pyrazole ring with aromatic groups (pyridyl [compound 15] or thiophen-3-yl [compound 17]) led to inactive compounds. Other groups such as tert-butyl (compound 18) or carboxylic acid (compound 21) were also not tolerated at the 3-position of the pyrazole. The 1-position of the pyrazole did not tolerate substitution as compounds 11, 15, 16 and 20 with methyl, methyl, phenyl and 2-hydroxyethyl substitutions, respectively at that position were all inactive compounds (see entries xi, xv, xvi, and xx; Table 1). The 5-position of the pyrazole seems to tolerate substitution as compounds 9 (methyl group at position 3 and hydrogen at position 5) and 10 (methyl groups at both position 3 and 5 of the pyrazole) are both active compounds (compare entries ix and x; Table 1). Compound 13 (a 3,5-substituted pyrazole) and HSD1169 (a 3,4-substituted pyrazole) only differ from each other by the substitution pattern yet HSD1169 potently inhibits AML cell lines with IC50 of 5 nM whereas 13 is inactive even at 1000 nM. We next explored the replacement of the ring A pyrazole with other 5-membered heterocyles. Compound 12 (ring A is a triazole) was not as active as HSD1169. The nature of ring E was also critical for the inhibition of FLT3 and AML cell growth. Although HSD1169, an indazole derivative, was active, the analogous indole derivative (compound 19) did not inhibit FLT3 enzymatic activity or AML cell growth. Therefore, it appears that the different moieties of HSD1169 play important roles in the inhibition of FLT3 enzymatic activity and AML cell growth.
HSD1169 potently inhibits FLT3 TKD mutants & FLT3-ITD secondary TKD mutants
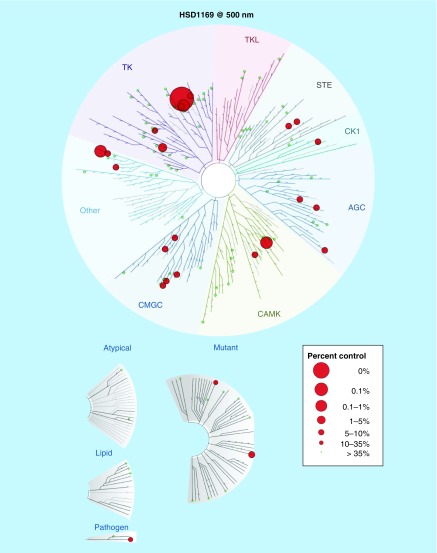
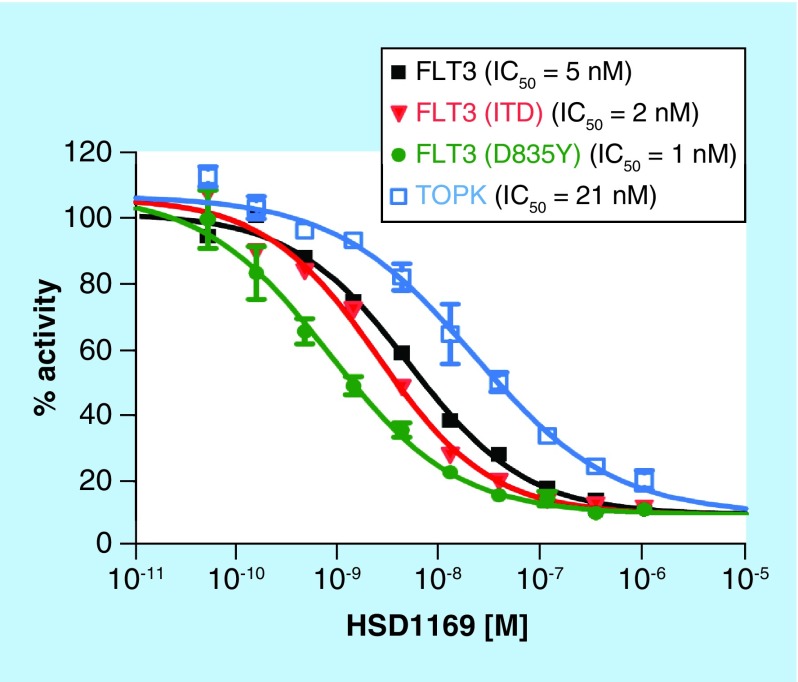
Kinase inhibition profiling using 97 disease-relevant kinases at DiscoverX indicated that HSD1169 is a selective kinase inhibitor and only potently inhibited a handful (JAK, MKNK2 and FLT3, including FLT3 kinase mutants), see Figure 4 & Table 2. AML patients that harbor FLT3-ITD or TKD mutation initially respond to current FLT3 inhibitors in clinical use or trial (midostaurin, sorafenib, quizartinib, crenolanib and gilteritinib) but ultimately relapse occurs due to FLT3 secondary mutation, which makes the FLT3 receptor less sensitive to the TKIs. The D835 and F691 mutations are two common mutations that occur at relapse. Quizartinib and sorafenib are not effective against both D835 and F691 secondary mutations whereas crenolanib and gilteritinib are effective against the D835Y secondary mutation. However all of the above FLT3 inhibitors are not effective against FLT3 (ITD, F691L) mutants (see Table 2). Thus there is a need for an FLT3 TKI that is effective against the F691L secondary mutants. Pleasingly, HSD1169 potently inhibits two common FLT3 secondary mutations FLT3 (ITD, D835V) and FLT3 (ITD, F691L) with Kd of 0.4 and 1.2 nM, respectively (see Table 2). We used Reaction Biology Corporation screening service to confirm that in addition to FLT3 binding, HSD1169 also potently inhibited FLT3 enzymatic activity. Consistent with the tight binding to FLT3 and mutants, HSD1169 inhibited the enzymatic activity of FLT3, FLT3-ITD and FLT3 D835Y (Figure 5). To the best of our knowledge, HSD1169 represents one of the best (if not the best) inhibitors of FLT3-ITD secondary mutation.
Figure 4. . HSD1169 is a selective kinase inhibitor.
HSD1169 has a strong affinity for FLT3, JAK2 and 3 and MKNK2 (with a percent inhibition greater than 90% at 500 nM of HSD1169) but not other over 90 tested kinases (DiscoverX ScanEdge kinases, which are distributed across the kinome).
Table 2. . HSD1169, crenolanib, sorafenib and midostaurin in vitro kinase binding.
| Kinase | HSD1169 ( Kd/nM)† | Crenolanib (Kd/nM)‡ | Sorafenib (Kd/nM)‡ | PKC412 (midostaurin) (Kd/nM)§ |
|---|---|---|---|---|
| FLT3 | 17 | 0.15 | 13 | 11 |
| FLT3-ITD | 0.78 | 0.26 | 95 | 11 |
| FLT3(D835H) | 4.4 | 0.16 | 11 | 6.8 |
| FLT3(D835Y) | 1.1 | 0.14 | 24 | 15 |
| FLT3(D835V) | 1.0 | 3.3 | 140 | – |
| FLT3(ITD, D835V) | 0.41 | 3.6 | 630 | – |
| FLT3(ITD, F691L) | 1.2 | 22 | 860 | – |
| ABL1-phosphorylated | 7200 | 430 | – | – |
| ABL1-nonphosphorylated | 5800 | 140 | 0.33 | – |
†This work: data obtained from KdELECT screening (DiscoverX).
‡Zimmerman et al. [28]. The authors also used KdELECT screening service at DiscoverX.
§Reference Kd provided by DiscoverX.
Figure 5. . HSD1169 potently inhibits FLT3 kinase and mutants as well as TOPK.
The effect of HSD1169 on the activity of 15 nM FLT3, 100 nM FLT3 (ITD), 0.5 nM FLT3 (D835Y) and 20 nM TOPK in the presence of 100 μM ATP (done at Reaction Biology Corporation). Enzyme activity in the presence of HSD1169 was normalized to the DMSO control and data fitted to a nonlinear regression using GraphPad Prism (GraphPad, CA, USA). Error bar represent standard error of the mean of duplicate reactions.
ITD: Internal tandem duplication; TOPK: T-LAK cell-originated protein kinase.
To have insights into the type of binding mode that HSD1169 engages with kinases, we determined the Kd for phosphorylated and nonphosphorylated ABL1. HSD1169 binds to nonphosphorylated ABL1 better (slightly) than phosphorylated ABL1 (Table 2), similar to crenolanib (a type I inhibitor) [28]. HSD1169 appears to be a type I kinase inhibitor.
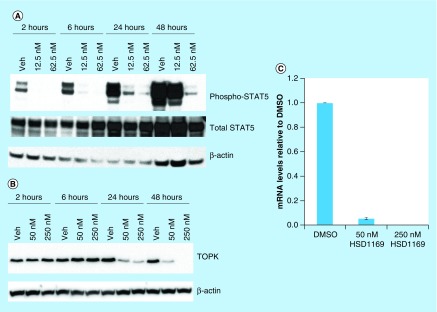
FLT3 signaling, which is amplified by collaborating kinases (such as TOPK) ultimately leads to the phosphorylation of downstream effectors, such as STAT5. We therefore investigated if the administration of HSD1169 to FLT3-driven cell line, such as MV4–11 would lead to the inhibition of STAT5. In agreement with the in vitro data that show potent inhibition of FLT3, western blot analysis showed that indeed HSD1169 inhibited the phosphorylation of STAT5, see Figure 6A.
Figure 6. . HSD1169 affects TOPK mRNA and protein levels in MV4–11 cells.
Western blot analysis of (A) phospho-STAT5 and total STAT5, and (B) TOPK after MV4–11 cells were treated with HSD1169 at the indicated times and concentrations. (C) MV4–11 cells were treated with HSD1169 and TOPK mRNA levels were assessed after 24 h.
TOPK: T-LAK cell-originated protein kinase.
TOPK is a particularly interesting target in AML and others had shown that inhibiting TOPK causes TOPK protein downregulation, probably via the downregulation of its mRNA [26]. For example, when MV4–11 cells were treated with siRNA of TOPK, a significant decrease in mRNA and protein expression was observed [26]. We therefore investigated if HSD1169 also caused the downregulation of TOPK protein and mRNA. Consistent with the work of Alachkar et al. [26] administration of HSD1169, which is also a TOPK inhibitor (Figure 5), to MV4–11 led to the downregulation of TOPK mRNA and reduction of TOPK protein in a dose-dependent manner (Figure 6B & C).
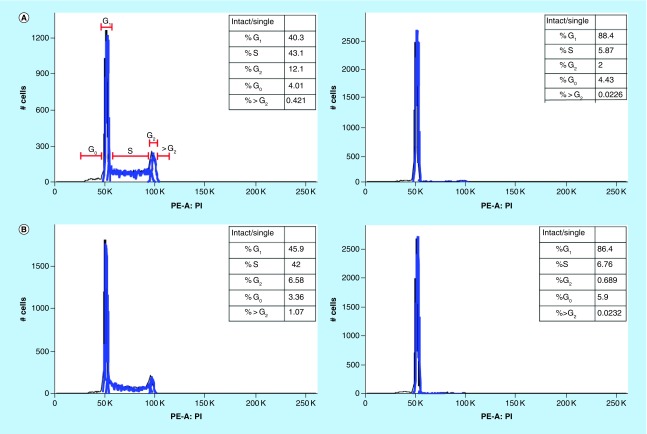
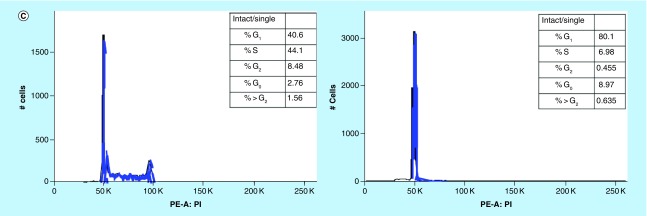
HSD1169 induces G1 cell cycle arrest in MV4–11 acute myeloid leukemia cells
To determine the effect of HSD1169 on cell cycle, we added different concentrations of HSD1169 to MV4–11 and the population of cells at the G0, G1, S and G2 phases at 24, 48 and 72 h. We observed a dose-dependent decrease in the population of cells in the S and G2 phases after 24 h and a corresponding dose-dependent increase in the percentage of cells in the G0/1 phase (Figure 7). Various FLT3 inhibitors cause cell cycle arrest at different phases. For example, AMG 925 (a dual FLT3/CDK4 inhibitor) [30] and AKN-028 (FLT3/CLK1/RPS6K inhibitor) [31] induce arrest in the G0/1 phase in Molm13 and MV 4–11 (both FLT3-ITD), respectively. Whereas PKC412 (midostaurin) did not affect cell cycle pattern in both MV4–11 and Molm13 [32]. It must be noted that in addition to FLT3, all of the reported FLT3 inhibitors also inhibit other kinases, so their effect on cell cycle will also depend on the other kinases that they inhibit.
Figure 7. . HSD1169 induces cell cycle arrest at the G0/1 phase in MV4–11 cells.
Cell cycle analysis of MV4–11 before (left panels) and after (right panels) treatment with HSD1169 (62.5 nM) for (A) 24 h, (B) 48 h and (C) 72 h. The various phases of the cell cycle are as indicated in (A).
Conclusion
In this report, we have revealed a new FLT3 inhibitor, HSD1169, which inhibits FLT3-ITD as well as the problematic D835 and F691 TKD mutants and secondary mutants FLT3 (ITD, D835V)/FLT3 (ITD, F691L). HSD1169 inhibits FLT3(ITD, F691L) with a Kd of approximately 1 nM whereas FLT3 inhibitors in clinical trials, such as crenolanib and sorafenib, inhibit this particular mutant FLT3 with Kd of 22 and 860 nM, respectively. In addition to FLT3 inhibition, HSD1169 also inhibits collaborating kinase, TOPK, which exacerbate AML. Yet HSD1169 does not potently bind to or inhibit over 90 other kinases that were tested. The in vitro inhibition of FLT3 correlated with the antiproliferative activity of HSD1169 against FLT3-driven AML cell lines at subnanomolar concentrations. We also showed that HSD1169 affected TOPK mRNA levels leading to an observed decrease in TOPK protein expression.
Future perspective
In the last few years, it has become clear that multitargeting kinase inhibitors perform better in the clinic than monotargeting inhibitors. However, a limitation of multitargeting is toxicity, if important kinases (such as VEGFR) that are relevant for essential organs such as heart and kidney are also inhibited. A challenge therefore is to develop a multitargeting but not ‘dirty’ kinase inhibitor (such as staurosporine, which is an overtly promiscuous kinase inhibitor and hence of limited clinical utility). We seem to have chanced upon such ideal ‘multitargeting’ yet clean kinase inhibitor, HSD1169. This inhibitor, HSD1169 or a close analog, represents a lead compound to develop AML patients for relapse. Ongoing work will focus on IND-enabling experiments of HSD1169 with an eye toward clinical translation.
Summary points.
FLT3-ITD kinases with secondary mutations, such as FLT3-ITD and D835/F691, are resistant to most developed FLT3 inhibitors.
Compounds that can inhibit targets other than FLT3-ITD could potentially improve acute myeloid leukemia treatment.
HSD1169, a very potent FLT3 inhibitor, is active against FLT3-ITD carrying TKD secondary mutations as well as TOPK.
HSD1169 potently inhibits the proliferation of drug-resistant acute myeloid leukemia cell lines like Molm13-res.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.future-science.com/doi/full/10.4155/fmc-2017-0298
Author's contributions
Conceptualization was done by HO Sintim. Investigation was done by N Dayal, C Opoku-Temeng, DE Hernandez, MA Sooreshjani and BA Carter-Cooper. C Opoku-Temeng helped in writing the original draft. Writing, review and editing was done by C Opoku-Temeng and HO Simtim. HO Sintim helped with funding acquisition. Supervision was done by HO Simtim and RG Lapidus.
Financial & competing interests disclosure
The authors thank Purdue University, Purdue Cancer Center, and Elks Foundation for funding. NMR and MS data were acquired by the NMR and MS facilities supported by NIH P30 CA023168. HO Sintim and RG Lapidus are cofounders of KinaRx, a start-up interested in translating AML therapeutics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Fleuren EDG, Zhang L, Wu J, Daly RJ. The kinome ‘at large’ in cancer. Nat. Rev. Cancer. 2016;16(2):83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 3.Giamas G, Stebbing J, Vorgias CE, Knippschild U. Protein kinases as targets for cancer treatment. Pharmacogenomics. 2007;8(8):1005–1016. doi: 10.2217/14622416.8.8.1005. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ. Imatinib as a paradigm of targeted therapies. Adv. Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 5.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 2015;36(7):422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Friedman R. Drug resistance in cancer: molecular evolution and compensatory proliferation. Oncotarget. 2016;7(11):11746–11755. doi: 10.18632/oncotarget.7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X, Zhou J, Wang C, et al. Identification of new FLT3 inhibitors that potently inhibit AML cell lines via an Azo Click-It/Staple-It approach. ACS Med. Chem. Lett. 2017;8(5):492–497. doi: 10.1021/acsmedchemlett.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larocque E, Naganna N, Ma X, et al. Aminoisoquinoline benzamides, FLT3 and Src-family kinase inhibitors, potently inhibit proliferation of acute myeloid leukemia cell lines. Future Med. Chem. 2017;9(11):1213–1225. doi: 10.4155/fmc-2017-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunwald MR, Levis MJ. FLT3 tyrosine kinase inhibition as a paradigm for targeted drug development in acute myeloid leukemia. Semin. Hematol. 2015;52(3):193–199. doi: 10.1053/j.seminhematol.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Beffinger M, Skwarska A. The role of FLT3 kinase as an AML therapy target. Curr. Pharm. Des. 2012;18(19):2758–2765. doi: 10.2174/138161212800626247. [DOI] [PubMed] [Google Scholar]
- 11.Levis M. Midostaurin approved for FLT3-mutated AML. Blood. 2017;129:3403–3406. doi: 10.1182/blood-2017-05-782292. [DOI] [PubMed] [Google Scholar]
- 12.Kiyoi H. FLT3 inhibitors: recent advances and problems for clinical application. Nagoya J. Med. Sci. 2015;77(1–2):7–17. [PMC free article] [PubMed] [Google Scholar]
- 13.Wander SA, Levis MJ, Fathi AT. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther. Adv. Hematol. 2014;5(3):65–77. doi: 10.1177/2040620714532123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 2003;9(1):327–337. [PubMed] [Google Scholar]
- 15.Cortes JE, Kantarjian H, Foran JM, et al. Phase I study of quizartinib administered daily to patients with relapsed or refractory acute myeloid leukemia irrespective of FMS-like tyrosine kinase 3-internal tandem duplication status. J. Clin. Oncol. 2013;31(29):3681–3687. doi: 10.1200/JCO.2013.48.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilhelm SM, Carter C, Tang L, et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 17.Gotlib J, Kluin-Nelemans HC, George TI, et al. Efficacy and safety of midostaurin in advanced systemic mastocytosis. N. Engl. J. Med. 2016;374(26):2530–2541. doi: 10.1056/NEJMoa1513098. [DOI] [PubMed] [Google Scholar]
- 18.Galanis A, Levis M. Inhibition of c-Kit by tyrosine kinase inhibitors. Haematologica. 2015;100(3):e77–e79. doi: 10.3324/haematol.2014.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes K, Roberts OL, Thomas AM, Cross MJ. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. 2007;19(10):2003–2012. doi: 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Cébe-Suarez S, Zehnder-Fjällman A, Ballmer-Hofer K. The role of VEGF receptors in angiogenesis; complex partnerships. Cell. Mol. Life Sci. 2006;63(5):601–615. doi: 10.1007/s00018-005-5426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen ZI, Ai DI. Cardiotoxicity associated with targeted cancer therapies. Mol. Clin. Oncol. 2016;4(5):675–681. doi: 10.3892/mco.2016.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fathi A, Levis M. FLT3 inhibitors: a story of the old and the new. Curr. Opin. Hematol. 2011;18(2):71–76. doi: 10.1097/MOH.0b013e3283439a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CC, Lin K, Stecula A, Sali A, Shah NP. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia. 2015;29(12):2390–2392. doi: 10.1038/leu.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori M, Kaneko N, Ueno Y, et al. Gilteritinib, a FLT3/AXL inhibitor, shows antileukemic activity in mouse models of FLT3 mutated acute myeloid leukemia. Invest. New Drugs. 2017;35(5):556–565. doi: 10.1007/s10637-017-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galanis A, Ma H, Rajkhowa T, et al. Crenolanib is a potent inhibitor of FLT3 with activity against resistance-conferring point mutants. Blood. 2014;123(1):94–100. doi: 10.1182/blood-2013-10-529313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alachkar H, Mutonga M, Malnassy G, et al. T-LAK cell-originated protein kinase presents a novel therapeutic target in FLT3-ITD mutated acute myeloid leukemia. Oncotarget. 2015;6(32):33410–33425. doi: 10.18632/oncotarget.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrated that TOPK collaborates with FLT3-ITD in acute myeloid leukemia (AML). Also showed that TOPK was a therapeutic target in AML.
- 27.Matsuo Y, Park JH, Miyamoto T, et al. TOPK inhibitor induces complete tumor regression in xenograft models of human cancer through inhibition of cytokinesis. Sci. Transl. Med. 2014;6(259):259ra145. doi: 10.1126/scitranslmed.3010277. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman EI, Turner DC, Buaboonnam J, et al. Crenolanib is active against models of drug-resistant FLT3-ITD-positive acute myeloid leukemia. Blood. 2013;122(22):3607–3615. doi: 10.1182/blood-2013-07-513044. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Crenolanib represents one of the promising AML drugs in clinical trial. It has shown activity against secondary FLT3-TKD mutations.
- 29.Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16(8):531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 30.Keegan K, Li C, Li Z, et al. Preclinical evaluation of AMG 925, a FLT3/CDK4 dual kinase inhibitor for treating acute myeloid leukemia. Mol. Cancer Ther. 2014;13(4):880–889. doi: 10.1158/1535-7163.MCT-13-0858. [DOI] [PubMed] [Google Scholar]
- 31.Eriksson A, Kalushkova A, Jarvius M, et al. AKN-028 induces cell cycle arrest, downregulation of Myc associated genes and dose dependent reduction of tyrosine kinase activity in acute myeloid leukemia. Biochem. Pharmacol. 2014;87(2):284–291. doi: 10.1016/j.bcp.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Odgerel T, Kikuchi J, Wada T, et al. The FLT3 inhibitor PKC412 exerts differential cell cycle effects on leukemic cells depending on the presence of FLT3 mutations. Oncogene. 2008;27(22):3102–3110. doi: 10.1038/sj.onc.1210980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.