Flux elucidation in Synechococcus 2973 reveals a fast and efficient routing of CO2 to biomass facilitated by the reincorporation of CO2 lost in photorespiration and peripheral metabolism.
Abstract
Synechococcus elongatus UTEX 2973 (Synechococcus 2973) has the shortest reported doubling time (2.1 h) among cyanobacteria, making it a promising platform for the solar-based production of biochemicals. In this meta-analysis, its intracellular flux distribution was recomputed using genome-scale isotopic nonstationary 13C-metabolic flux analysis given the labeling dynamics of 13 metabolites reported in an earlier study. To achieve this, a genome-scale mapping model, namely imSyu593, was constructed using the imSyn617 mapping model for Synechocystis sp. PCC 6803 (Synechocystis 6803) as the starting point encompassing 593 reactions. The flux elucidation revealed nearly complete conversion (greater than 96%) of the assimilated carbon into biomass in Synechococcus 2973. In contrast, Synechocystis 6803 achieves complete conversion of only 86% of the assimilated carbon. This high biomass yield was enabled by the reincorporation of the fixed carbons lost in anabolic and photorespiratory pathways in conjunction with flux rerouting through a nondecarboxylating reaction such as phosphoketolase. This reincorporation of lost CO2 sustains a higher flux through the photorespiratory C2 cycle that fully meets the glycine and serine demands for growth. In accordance with the high carbon efficiency drive, acetyl-coenzyme A was entirely produced using the carbon-efficient phosphoketolase pathway. Comparison of the Synechococcus 2973 flux map with that of Synechocystis 6803 revealed differences in the use of Calvin cycle and photorespiratory pathway reactions. The two species used different reactions for the synthesis of metabolites such as fructose-6-phosphate, glycine, sedoheptulose-7-phosphate, and Ser. These findings allude to a highly carbon-efficient metabolism alongside the fast carbon uptake rate in Synechococcus 2973, which explains its faster growth rate.
Photosynthesis produces a wide range of metabolites from only water, sunlight, and CO2. Cyanobacteria are oxygenic phototrophs that are more amenable to genetic transformations than microalgae and plants, making them ideal photosynthetic production platforms for industrially relevant chemicals and biofuels (Ducat et al., 2011; Quintana et al., 2011). While the industrial relevance of cyanobacteria has been restricted by their slow growth rates under photoautotrophic conditions, the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 (here referred to as Synechococcus 2973), which has a reported doubling time of 2.1 h under high-light and high-CO2 conditions, has emerged as a promising platform (Yu et al., 2015). The puzzling DNA sequence similarity with the well-studied and comparatively slower-growing cyanobacterium Synechococcus elongatus PCC 7942 (Synechococcus 7942) has made Synechococcus 2973 a model organism that could enable us to uncover the factors responsible for a faster photoautotrophic growth (Yu et al., 2015; Abernathy et al., 2017; Mueller et al., 2017). This has led to the rapid development of genetic tools, including a CRISPR/Cas9 system (Wendt et al., 2016). The faster growth rate and availability of genetic tools in Synechococcus 2973 make it a promising solar-based production platform for biochemicals. Photosynthesis-based Suc production already has been demonstrated in this organism (Song et al., 2016). Ongoing efforts to understand and engineer Synechococcus 2973 into a successful production host will benefit from an understanding of its metabolic flows supporting fast growth. This can be accomplished using 13C-metabolic flux analysis (13C-MFA; Long and Antoniewicz, 2014; Young, 2014; Niedenführ et al., 2015; Allen, 2016).
Isotope tracing using a one-carbon substrate results in uniform labeling of metabolites under an isotopic steady state (Shastri and Morgan, 2007). As a consequence of this, isotopic nonstationary 13C-metabolic flux analysis (INST-MFA) is needed to elucidate photoautotrophic flux distribution. This methodology has been used extensively to study the metabolism of cyanobacteria (Adebiyi et al., 2015), microalgae (Wu et al., 2015), and plants (Ma et al., 2014). In contrast to steady-state MFA, INST-MFA elucidates fluxes so as to recapitulate experimentally measured metabolite-labeling dynamics as well as steady-state labeling distributions (Nöh et al., 2007). The complexity of the underlying nonlinear least squares problem and the number of balance equations required to predict the labeling distribution of the metabolites has so far placed significant constraints on the scope of the metabolic reactions considered at one time. Generally, INST-MFA has been restricted to core models spanning central metabolism only (Nöh and Wiechert, 2011). Such simplifications introduce biases regarding pathway utilization (Gopalakrishnan and Maranas, 2015), which can result in inaccurate uncertainty calculations and limit meaningful inferences about energy metabolism in an organism. Furthermore, reaction lumping restricted the metabolite-labeling dynamics afforded by a core model, leading to significant disagreements between experimentally measured labeling dynamics and those predicted using simplified models (Nöh and Wiechert, 2011). In order to mitigate the effect of these factors on the accuracy of flux estimation, the scope of the INST-MFA technique has been expanded to genome-scale models with the implementation of an efficient algorithm for the simulation of metabolite-labeling dynamics (Gopalakrishnan et al., 2018). Meta-analysis of the labeling dynamics observed under photoautotrophic conditions in Synechocystis sp. PCC 6803 (Synechocystis 6803) using genome-scale INST-MFA showed that the genome-scale mapping model (GSMM), namely imSyn617, fitted the experimental data better than the core model (Gopalakrishnan et al., 2018). Interestingly, the flux distribution obtained using imSyn617 was qualitatively different from the one obtained using the core model, as indicated by the reliance on transaldolase (TAL) for the synthesis of Fructose-6-Phosphate (f6p), a bifurcated photorespiratory C2 cycle, and off-gassing of fixed CO2 by anabolic reactions outside core metabolism. These prediction departures from a core mapping model demonstrate the importance of using comprehensive GSMMs while performing INST-MFA to glean an accurate description of internal metabolic fluxes.
Genome-scale INST-MFA requires a high-quality genome-scale model (GSM; Gopalakrishnan and Maranas, 2015) spanning all the reactions encoded by the genome annotation without any presumption about their activity under a given condition. The GSM must contain a detailed biomass equation, and the synthesis pathways of macromolecular precursors such as amino acids, lipids, and nucleotides should be traced completely. The lumping of reactions should be kept to a minimum, and care should be taken to account for all the reactions in the amino acid and peripheral metabolism that produce central carbon intermediates such as acetate, fumarate, pyruvate, and CO2 as by-products. Once a GSM is available, the next step is to construct a GSMM describing the carbon mapping from the reactants to products for each reaction. Carbon-mapping information for reactions can be obtained from pathway databases such as MetaCyc (Caspi et al., 2018), existing GSMMs, and reaction mechanisms available from the literature. Atom-mapping algorithms such as Determination of Reaction Mechanisms (First et al., 2012), Minimum Weighted Edit-Distance (Latendresse et al., 2012), InfoChem-Map (Kraut et al., 2013), Canonical Labeling for Clique Approximation (Kumar and Maranas, 2014), and Reaction Decoder Tool (Rahman et al., 2016) also can be used to generate plausible mappings when this information is unavailable.
In this study, the photoautotrophic metabolism of Synechococcus 2973 under optimal growth conditions is elucidated using genome-scale INST-MFA. For this purpose, we constructed the GSMM imSyu593 using the GSM iSyu683 (Mueller et al., 2017). Flux variability analysis (FVA; Mahadevan and Schilling, 2003) was used to identify the reactions that are active under photoautotrophic conditions by constraining the model with experimentally measured bicarbonate uptake and growth rate. Flux elucidation using INST-MFA reveals a high Rubisco activity capable of reincorporating the fixed carbon lost in photorespiration and peripheral metabolism. The analysis also predicted that this organism relies entirely on the carbon-efficient phosphoketolase (PKetolase) pathway instead of the conventional pyruvate dehydrogenase (PDH) reaction for the synthesis of acetyl-CoA. The oxidative pentose phosphate pathway carried a negligible flux. These factors facilitate a nearly perfect routing of assimilated carbon toward biomass. Prediction departures from the core model (Abernathy et al., 2017) include the usage of TAL for sedoheptulose-7-phosphate (s7p) synthesis and PKetolase for acetyl-CoA synthesis. Comparison with the genome-scale flux distribution of Synechocystis 6803 (Gopalakrishnan et al., 2018) grown under its optimal growth conditions (Young et al., 2011) revealed interesting differences in pathway recruitment that were not apparent through the core model-based studies or their respective GSMs. Synechococcus 2973 has a higher overall carbon conversion (96%) compared with Synechocystis 6803 (86%). As a result, the biomass yield (per fixed carbon) of Synechococcus 2973 was 2% higher than that of Synechocystis 6803. This increase in the biomass yield accounted for 2.9% of the difference in the growth rate between these two species, with the remaining 97.1% due to differences in carbon uptake kinetics. The two species also differed in the choice of Calvin cycle reactions for the synthesis of f6p and s7p. In Synechococcus 2973, Ser and Gly were synthesized using the photorespiratory C2 cycle, whereas Synechocystis 6803 relied instead on the phosphoSer (PSER) pathway and C2 cycle. The new flux map obtained in this study provides valuable insight into a cyanobacterial strain of great industrial importance, directly supporting any rational redesigning of its metabolism to generate a superior production host.
RESULTS
The Genome-Scale Carbon-Mapping Model of Synechococcus 2973
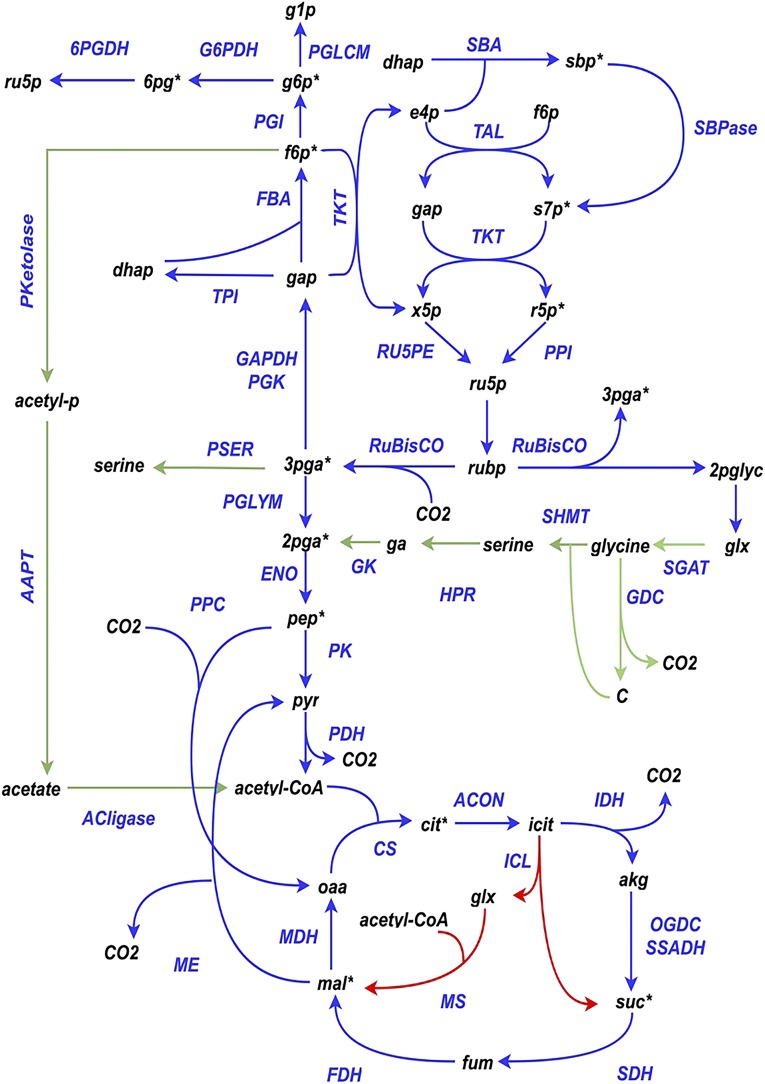
The elementary metabolite unit (EMU) network, obtained from imSyu593, capturing the carbon paths that can contribute to the labeling of the 13 fitted metabolites, spanned 232 metabolites and 352 reactions and consisted of 1,271 EMUs. Scale-up to imSyu593 introduces 41 novel carbon paths that were not included during flux elucidation using a core model (Abernathy et al., 2017). This includes acetyl-CoA production via the PKetolase pathway (Fig. 1), Ser production via the PSER pathway, and carbon recycling from peripheral metabolic pathways such as fatty acid biosynthesis, pantothenate and CoA biosynthesis, riboflavin metabolism, and porphyrin and chlorophyll biosynthesis. The glyoxylate shunt was excluded in imSyu593 based on the lack of genomic evidence (Fig. 1; Yu et al., 2015; Abernathy et al., 2017; Mueller et al., 2017). These differences in the carbon paths captured by the core model and imSyu593 cause significant differences in the predicted labeling dynamics. Compared with imSyn617, imSyu593 contains two novel carbon paths producing acetyl-CoA and erythrose-4-phosphate (e4p). The PKetolase pathway consisting of the enzymes PKetolase, ATP:acetate phosphotransferase, and acetate:CoA ligase allows acetyl-CoA production from the sugar phosphates (f6p and xylulose-5-phosphate) in the Calvin cycle while bypassing the lower glycolysis. The second novel carbon path was due to the presence of 5,6-dimethylbenzimidazole synthase (EC 1.13.11.79), which degraded FMN into dilaurate, dimethyl benzimidazole, and the central carbon intermediate e4p. This resulted in an alternative carbon path from ribose-5-phosphate (r5p) to e4p through riboflavin biosynthesis.
Figure 1.
Schematic of the central carbon metabolism of Synechococcus 2973. Reactions exclusive to the core model (Abernathy et al., 2017) and imSyu593 are highlighted in red and green, respectively. *, Metabolites whose labeling data were fitted in this study. 2pglyc, 2-Phosphoglycolate; 6PGDH, 6-phosphogluconate dehydrogenase; acetyl-p, acetyl phosphate; ACON, aconitase; akg, α-ketoglutarate; CS, citrate synthase; dhap, dihydroxyacetone phosphate; ENO, enolase; FBA, Fru bisphosphate aldolase; fbp, Fru bisphosphate; FDH, fumarate dehydrogenase; fum, fumarate; g1p, Glc-1-P; G6PDH, Glc-6-P dehydrogenase; gap, glyceraldehyde-3-phosphate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDC, Gly decarboxylase; GK, glycerate kinase; glx, glyoxalate; HPR, hydroxypyruvate reductase; ICL, isocitrate lyase; IDH, isocitrate dehydrogenase; MS, malate synthase; oaa, oxaloacetate; OGDC, 2-oxoglutarate decarboxylase; PGI, phosphoglucose mutase; PGK, phosphoglycerate kinase, PGLCM, phosphoglucomutase; PGLYM, phosphoglycerate mutase; PK, pyruvate kinase; PPC, phosphoenolpyruvate carboxylase; PPI, pentose phosphate isomerase; pyr, pyruvate; ru5p, ribulose-5-phosphate; RU5PE, ribulose-5-phosphate epimerase; rubp, ribulose-1,5-bisphosphate; SDH, succinate dehydrogenase; SGAT, Ser-glyoxylate aminotransferase; SHMT, Ser-hydroxymethyl transferase; SSDH, succinic semialdehyde dehydrogenase; TKT, transketolase; TPI, triosephosphate isomerase.
The Flux Distribution of Synechococcus 2973
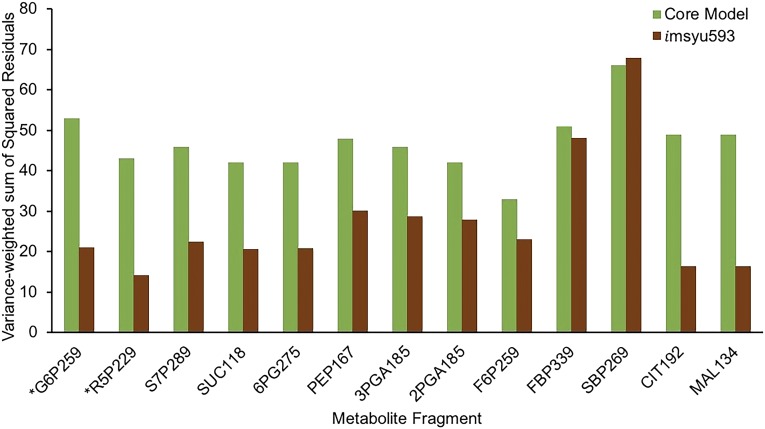
The estimation of intracellular fluxes using imSyu593 led to a better fit (sum of squared residuals [SSR] = 363, degrees of freedom = 402) of the labeling data when compared with that with the core model (SSR = 610, degrees of freedom = 535; Fig. 2). The F test was used to analyze the statistical significance of this improvement in the fit. The obtained F of 2.079 (P = 1.39 × 10−8) indicates with 95% confidence that the improvement is due to additional active carbon paths accounted for in imSyu593 compared with that in the core model and not due to the mere increase in the number of parameters. A large reduction in the SSR was observed for metabolites Glc-6-P, r5p, s7p, 6-phosphogluconate, 3-phosphoglycerate, 2-phosphoglycerate, phosphoenolpyruvate, citrate, succinate, and malate. The estimated 95% confidence intervals showed that 609 reactions were resolved with flux ranges narrower than 10 mmol g−1 dry weight h−1 for a bicarbonate uptake rate of 100 mmol g−1 dry weight h−1. Of this, only 334 reactions were resolved due to growth coupling, whereas the rest of the reactions were resolved due to MFA. Of the 78 unresolved reactions, 45 reactions are outside the purview of the EMU balances and are involved in pathways such as photosynthesis, oxidative phosphorylation, and the exchange and transport of oxygen, photons, protons, and water. The remaining 33 reactions, involved in pathways such as Ala and Asp metabolism, citrate cycle, Glu metabolism, glycolysis/gluconeogenesis, pentose phosphate pathway, purine metabolism, and starch and Suc metabolism, participate in unresolved futile cycles expending ATP. This is because the photosynthesis reactions responsible for generating ATP are not captured by the EMU balances and, therefore, are not constrained by the labeling data.
Figure 2.
Comparison of variance-weighted SSR obtained, in the case of Synechococcus 2973, using the core model and imSyu593 for the various metabolite fragments. Green bars correspond to the core model, and brown bars correspond to imsyu593. Lower SSR indicates a better recapitulation of the labeling dynamics for the corresponding fragment. *Fragments for which the difference in SSR is greater than 25.
Comparison of flux ranges generated using MFA and FVA (Supplemental Table S1) showed an average of 56% bound contraction (reduction in the MFA flux range relative to the FVA flux range). Reactions with the most impactful flux range reductions (greater than 99% bound contraction) were from central metabolism and included inactivated metabolic pathways such as the oxidative pentose phosphate pathway, lower tricarboxylic acid cycle, the PSER pathway, and glyoxylate degradation. This aids in the resolution of most reactions from central metabolism and decreases the uncertainty in biomass yield prediction using MFA by 74% relative to that using FVA. This 74% reduction in biomass flux range caused 334 growth-coupled reactions to reach a 74% bound contraction using MFA. The highly efficient carbon routing to biomass limited the possibility of the production of by-products such as sulfoquinovosyl-diacylglycerol, digalactosyl-diacylglycerol, and phosphatidylglycerol, contributing to the contraction of flux ranges in 43 peripheral metabolic reactions using MFA relative to FVA. Nevertheless, 78 reactions, including those involved in energy metabolism (photosynthetic light reactions and oxidative phosphorylation), remained poorly resolved by MFA as with FVA.
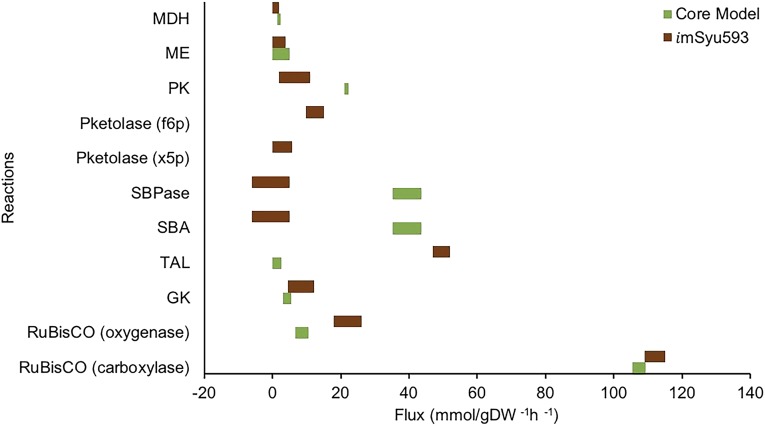
The flux distributions through the carbon fixation pathways obtained using imSyu593 and the core model were qualitatively different (Fig. 3). s7p was produced via the TAL reaction in imSyu593 as opposed to via the sedoheptulose-1,7-bisphosphate aldolase (SBA) and sedoheptulose-1,7-bisphosphatase (SBPase) reactions in the core model. To test the robustness of this inference, we removed TAL from imSyu593 and repeated the flux elucidation. Upon removing TAL from imSyu593, the minimum SSR increased to 368 and carbon flux was redirected through the SBA and SBPase reactions. The SSR increase was attributed to an increased disagreement between predicted and measured labeling dynamics of s7p and sedoheptulose-1,7-bisphosphate (sbp) fragments. The F test (F = 5.61, P = 0.018) indicated that the reduction in the SSR caused by including TAL was statistically significant and formed the basis for rejecting the model lacking TAL. The oxidative pentose phosphate pathway carried a negligible flux, which is consistent with flux predictions using the core model. The 95% confidence interval for this flux identified a lower bound of zero, indicating the dispensability of the oxidative pentose phosphate pathway under photoautotrophic conditions in Synechococcus 2973, which is consistent with the experimentally observed nonessentiality of the zwf gene in Synechococcus 2973 (Abernathy et al., 2017).
Figure 3.
Comparison of the flux ranges (95% confidence interval) for relevant reactions obtained using the core model (Abernathy et al., 2017) and imSyu593 under photoautotrophic conditions. Green bars correspond to the core model, and brown bars correspond to imsyu593. The left end of each bar indicates the lower bound, and the right end of each bar indicates the upper bound. The fluxes are normalized to a carbon uptake of 100 mmol g−1 dry weight (DW) h−1. See Figure 1 for abbreviations and details of individual reactions.
Flux elucidation using imSyu593 revealed that acetyl-CoA is synthesized predominantly from f6p via the PKetolase pathway instead of PDH as reported by the core model (Fig. 3). The PKetolase reaction splits f6p into e4p and acetyl phosphate, which is converted to acetyl-CoA via the ATP:acetate phosphotransferase and acetate:CoA ligase reactions (Fig. 1). No flux was seen through PDH, indicating that the PKetolase pathway is the sole source of acetyl-CoA in Synechococcus 2973. Since acetyl-CoA biosynthesis via the PKetolase pathway bypasses lower glycolysis, flux through lower glycolysis was reduced by 50% compared with that in the core model. PKetolase also catalyzes the conversion of xylulose-5-phosphate to glyceraldehyde-3-phosphate. However, this reaction was found to carry no flux. Removal of the f6p-utilizing PKetolase reaction resulted in a statistically acceptable fit with an increased SSR of 379. An F test (F = 18.15, P = 0.000025) indicated that the increase in the SSR was statistically significant enough to reject the model that lacks the PKetolase, demonstrating that the labeling data support that the PKetolase pathway was active under the experimental conditions. The pathway is usually not included in the core models of cyanobacterial metabolism; therefore, this is the first report that supports the operation of the native PKetolase pathway under photoautotrophic conditions. Flux through this pathway is corroborated by proteomic analysis reporting that the PKetolase enzyme level is comparable with that of phosphoenolpyruvate carboxylase, citrate synthase, and subunits of the PDH complex under photoautotrophic conditions in Synechococcus 2973 (Yu et al., 2015).
Rubisco oxygenase carried a 3-fold higher flux in imSyu593 compared with that in the core model. The ratio of Rubisco carboxylase to Rubisco oxygenase flux was found to be 83:17. High photorespiration was expected, since the nonstationary labeling experiment was performed under high-light conditions (Abernathy et al., 2017). Under similar conditions, previous studies have reported a 2.4-fold higher rate of photorespiratory oxygen uptake in Synechococcus 2973 compared with that in Synechococcus 7942 (Ungerer et al., 2018a). The high rate of photorespiration caused a large flux through the C2 cycle for phosphoglycolate salvage, which generates all of the Gly and Ser needed for biomass synthesis. The PSER pathway for Ser synthesis was found to be dispensable, as it was found to carry no flux.
In agreement with the core model predictions, malic enzyme (ME) was found to be dispensable and pyruvate was produced through pyruvate kinase. This departs from other cyanobacterial species, where the major route for pyruvate synthesis is via ME (Young et al., 2011; Jazmin et al., 2017). Consistent with the core model predictions, the tricarboxylic acid cycle was incomplete and exhibited a bifurcated topology, since there was no flux through 2-oxoglutarate decarboxylase and succinic semialdehyde dehydrogenase. This organism lacks the γ-aminobutyric acid shunt (Mueller et al., 2017). The flux and directionality of malate dehydrogenase (MDH) remain unresolved in this study. The genome annotation did not identify any gene that codes for MDH; therefore, this organism may not contain this enzyme. However, earlier labeling experiments with this species have shown the incorporation of carbons from malate into the amino acid Asp derived from oxaloacetate, suggesting the existence of either a previously uncharacterized MDH (Abernathy et al., 2017) or a pathway facilitating the conversion of malate to oxaloacetate.
The secretion rates of organic acids were negligible, and carbon loss in the form of CO2 evolution was insignificant. This implies that CO2 released in photorespiration and peripheral metabolism was reincorporated into the biomass through the Rubisco carboxylase reaction. Of the 100 mmol of bicarbonate taken up by Synechococcus 2793, 8 mmol was fixed by phosphoenolpyruvate carboxylase, indicating that at least 92 mmol should be fixed by Rubisco. However, Rubisco was found to fix 111 mmol of CO2, which exceeded the expected flux by 19 mmol. The excess CO2 fixed accounted for the 11 mmol of CO2 generated in the photorespiratory pathway and the 8 mmol of CO2 generated in growth-coupled anabolic pathways. The net carbon balance indicated that there is a nearly complete conversion (greater than 96%) of assimilated carbon into biomass, significantly higher than what was obtained for Synechocystis 6803 (86%). The biomass yield was predicted to be 2.48 g dry weight dmol−1 bicarbonate uptake. This is 2% higher than the predicted biomass yield in Synechocystis 6803 (Gopalakrishnan et al., 2018).
DISCUSSION
In this study, intracellular fluxes of the fast-growing cyanobacterium Synechococcus 2973 were recomputed using the recently developed genome-scale INST-MFA procedure (Gopalakrishnan et al., 2018), the dynamic labeling distribution of 13 intracellular metabolites (Abernathy et al., 2017), and the constructed GSMM imSyu593. Comparison with imSyn617 showed that imSyu593 is more exhaustive in terms of its coverage of cyanobacterial metabolism by accounting for carbon paths formed by pathways not included in imSyn617, such as the PKetolase pathway. The statistically significant improvement in the fit, with an F statistic of 2.079 (P = 1.39 × 10−8), indicates that the extra carbon paths included in imSyu593 are necessary to explain the labeling data.
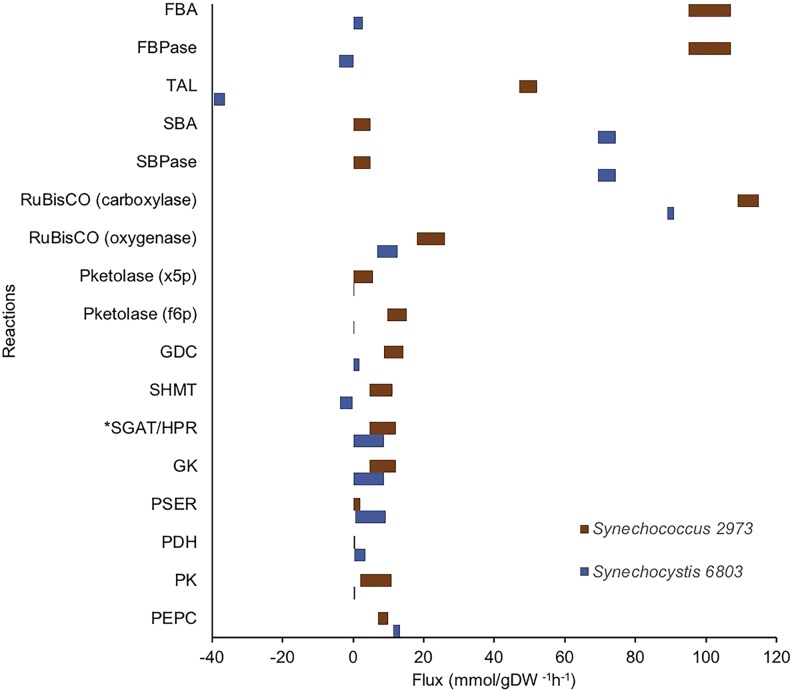
The complete reliance on the TAL reaction for the synthesis of s7p instead of the SBA/SBPase route is a striking deviation from the core model prediction. The use of TAL enzyme explains the observed faster labeling of s7p over that of sbp (Supplemental Fig. S1) in the labeling data (Abernathy et al., 2017). In the SBA/SBPase route, sbp is formed before s7p and sbp would label faster than s7p. Although a larger pool size for sbp would slow down its labeling, the resultant slow turnover of sbp would, in turn, slow down the labeling of all central carbon metabolites, since sbp is present in the Calvin-Benson-Bassham pathway (Nöh et al., 2007; McAtee et al., 2015). Thus, sbp is unlikely to have a larger pool size in this case, as we do not see this effect in the labeling data for downstream metabolites. In contrast, s7p is synthesized before sbp via TAL. However, under photoautotrophic conditions, the enzyme level of TAL is much smaller than that of SBA and SBPase (Yu et al., 2015). A faster labeling of s7p over sbp along with a lower TAL level can be explained with a metabolic channel resulting from the formation of a multienzyme complex consisting of SBA and SBPase. In such a multienzyme complex, there will be direct labeling of the s7p pool, bypassing sbp, since the product of the first enzyme, sbp, would be transferred directly to the second enzyme without labeling the sbp pool in the cytoplasm. A relatively higher flux through such a metabolic channel also might result in faster labeling of s7p compared with sbp with low levels of TAL. This explains both the labeling dynamics as well as the existing proteomics data (Yu et al., 2015); however, there is no direct experimental evidence for such a channel. In contrast, Synechocystis 6803 used the SBA/SBPase route to produce s7p and TAL to produce f6p from s7p and glyceraldehyde-3-phosphate (Fig. 4). This indicates that, even though the set of reactions in the Calvin cycle is conserved among cyanobacteria, the configuration in which they operate differs between species.
Figure 4.
Comparison of the normalized flux ranges (95% confidence interval) of selected reactions between Synechococcus 2973 and Synechocystis 6803 (Gopalakrishnan et al., 2018) under the photoautotrophic conditions. Brown bars correspond to Synechococcus 2973, and blue bars correspond to Synechocystis 6803. The left end of each bar indicates the lower bound, and the right end of each bar indicates the upper bound. The fluxes are normalized to a bicarbonate uptake of 100 mmol g−1 dry weight (DW) h−1. See Figure 1 for abbreviations and details on individual reactions.
PKetolase was reported to be active under mixotrophic conditions in cyanobacteria (Alagesan et al., 2013; Xiong et al., 2015). However, there are no reports on the activity of the native enzyme under the photoautotrophic conditions. Nevertheless, heterologous PKetolase enzymes were shown to increase the intracellular acetyl-CoA levels under photoautotrophic conditions in Synechocystis 6803 (Anfelt et al., 2015). Flux balance analysis predicted this reaction to be active under photoautotrophic conditions (Vu et al., 2012; Knoop et al., 2013). Interestingly, this pathway was not included in the mapping models of INST-MFA studies targeting the photoautotrophic metabolism of cyanobacteria. In this study, the f6p-utilizing variant of this pathway was found to be the major route for acetyl-CoA synthesis. The PKetolase route generates acetyl-CoA from two molecules of CO2, whereas the glycolysis route via PDH requires three CO2 molecules. This higher conservation of the fixed carbon reduces the number of CO2 molecules that need to be fixed by Rubisco to produce 1 mol of acetyl-CoA and confers both a kinetic and an energetic advantage for the production of acetyl-CoA-based products (Anfelt et al., 2015). Indeed, the overexpression of heterologous PKetolase enzyme has been shown to enhance the production of acetyl-CoA-based products such as n-butanol (Anfelt et al., 2015) in Synechocystis 6803 and acetone (Chwa et al., 2016) in Synechococcus 7942. The reliance of the wild-type Synechococcus 2973 on this pathway for the synthesis of acetyl-CoA makes it inherently suitable for the production of the above biochemicals.
The observed high flux through photorespiration causes oxidation of the fixed carbon while salvaging the 2-phosphoglycolate produced as the by-product of the Rubisco oxygenase reaction. However, this carbon loss is compensated by the high flux through the carboxylase reaction catalyzed by Rubisco, which reincorporates the oxidized carbon into biomass, resulting in an efficient carbon conversion into biomass. Indeed, the Rubisco carboxylase reaction carried a flux 20.6% higher than what is needed for a bicarbonate uptake of 100 mmol g−1 dry weight h−1. This allowed the organism to use the photorespiratory C2 cycle to produce the amino acids Gly and Ser without a significant decrease in the carbon conversion efficiency. In contrast, Synechocystis 6803 was not able to recycle the oxidized carbon, resulting in the liberation of 12% of the fixed carbon as CO2. As a result of this, Synechocystis 6803 used a bifurcated topology of the photorespiratory pathway that synthesized Gly and Ser without the loss of any fixed carbon. In Synechocystis 6803, Ser is synthesized using the PSER pathway, whereas Gly is produced from both the C2 cycle and the Ser-hydroxymethyl transferase reaction, consuming the Ser synthesized from the PSER pathway (Gopalakrishnan et al., 2018).
The nearly complete conversion (greater than 96%) of assimilated CO2 is the result of the reincorporation of CO2 liberated by photorespiration and other anabolic reactions, reduced flux through decarboxylating reactions such as the oxidative pentose phosphate pathway and PDH, and the usage of nondecarboxylating reactions such as PKetolase. A direct comparison with the biomass yield and growth rate of Synechocystis 6803 under its optimal growth conditions would enable us to quantify the relative contribution of the observed carbon-efficient metabolism to the growth rate difference between the two species. Such a comparative analysis between fast- and slow-growing species would shed light on the metabolic factors that might contribute to the faster growth rates. The carbon-efficient metabolism of Synechococcus 2973 resulted in a 2% higher biomass yield over that of Synechocystis 6803 (Gopalakrishnan et al., 2018). This difference in the biomass yield accounted for only 2.9% of the difference in the growth rate between Synechococcus 2973 (0.33 ± 0.05 h−1; Abernathy et al., 2017) and Synechocystis 6803 (0.09 h−1; Young et al., 2011). Thus, most of the difference in the growth rate between these two species is due to the higher CO2 uptake rate in Synecococcus 2973 (12.2 mmol g−1 dry weight h−1; Abernathy et al., 2017) compared with that of Synechocystis 6803 (3.7 mmol g−1 dry weight h−1; Young et al., 2011). This higher rate of CO2 fixation in Synecococcus 2973 requires a higher rate of ATP and NADPH generation. Studies guided by comparative genomics of Synecococcus 2973 and Synecococcus 7942 have shown that the single-nucleotide polymorphisms in ATP synthase and NAD+ kinase genes lead to high rates of ATP and NADPH generation in Synecococcus 2973 and that this contributes significantly to its faster growth rate (Ungerer et al., 2018b). This high rate of energy currency generation requires very high light intensities, and the optimal light intensity for Synecococcus 2973 (500 μE m−2 s−1) is higher than that for Synechocystis 6803 (300 μE m−2 s−1; Yu et al., 2015).
This meta-analysis using genome-scale INST-MFA showed that the fast-growing cyanobacterium Synechococcus 2973 possesses a more carbon-efficient metabolism when compared with that of Synechocystis 6803 and pointed out certain key features that contribute to this characteristic. Repeating the analysis on mutants lacking some of these features (e.g. PKetolase) will further improve our understanding about their role in increasing the carbon efficiency of photosynthetic metabolism. Also, it would be worthwhile to compare the genome-scale flux distribution of Synechococcus 2973 with that of the slow-growing close relative Synechococcus 7942 to elucidate the metabolic impact of the 55 single-nucleotide polymorphisms that exist between the genomes of these two strains (Yu et al., 2015). A comparison with the core model flux distribution of Synechococcus 7942 (Abernathy et al., 2017) revealed some interesting differences in central carbon metabolism. Synechococcus 7942 had zero flux through the photorespiratory pathway, whereas Synechococcus 2973 displayed a significant amount of photorespiration. There was a higher flux through the Calvin cycle in Synechococcus 2973 compared with Synechococcus 7942 in order to replenish the ribulose-1,5-bisphosphate lost in photorespiration. Synechococcus 2973 used TAL for the synthesis of s7p, whereas Synechococcus 7942 made use of the SBA/SBPase route. Since the PKetolase pathway was not included in the core model of Synechococcus 7942, there was a higher flux through glycolysis compared with that in Synechococcus 2973. Interestingly, both Synechococcus 2973 and Synechococcus 7942 used the pyruvate kinase enzyme instead of the ME for pyruvate production. Unfortunately, further comparison cannot be carried out, since the available Synechococcus 7942 fluxes are generated using the core model. Comparison of genome-scale flux distribution for these two species will reveal several other interesting differences. Since our analysis used the transition labeling data for a limited number of metabolites, the flux through certain reactions such as MDH could not be resolved. Usage of an expanded repertoire of metabolite-labeling data would resolve these uncertainties. The study used FVA to identify the active set of reactions in the GSM, and these reactions formed the metabolic network used for the genome-scale INST-MFA. However, a more precise way of identifying the active set of reactions would be to use the transcriptome and proteome data generated under similar conditions. The data also could act as additional constraints resulting in the enhanced resolution of the fluxes.
MATERIALS AND METHODS
Construction of imSyu593
The GSM used for flux elucidation was adapted from iSyu683 (Mueller et al., 2017) upon eliminating reactions incapable of carrying metabolic flux and reactions involved in loops by performing FVA (Mahadevan and Schilling, 2003) constrained by the measured growth rate and bicarbonate uptake rate (Supplemental Table S2; Abernathy et al., 2017). The genes involved in the PSER pathway were identified recently in Synechocystis sp. PCC 6803 (Klemke et al., 2015). BLAST analysis showed that the corresponding homologs are in Synechococcus elongatus UTEX 2973; therefore, this pathway was included in the model. Although iSyu683 does not include the MDH reaction, metabolite-labeling data suggest the presence of flux through this reaction (Abernathy et al., 2017), which led to the inclusion of MDH in the GSM and GSMM. The final GSM used for the analysis contained 700 reactions (Supplemental Table S3) and 667 metabolites (Supplemental Table S4). Apart from the metabolic reactions, reactions modeling the mixing of metabolically active and inactive pools were included for all the fitted metabolites. The GSMM generated for Synechococcus 2973, namely imSyu593, traces the carbon transitions through 593 reactions (Supplemental Table S5) in the GSM. The remaining reactions were outside the purview of the EMU balances and were not mapped, since they do not contribute to the labeling of the fitted metabolites. Carbon mapping for 519 shared reactions with Synechocystis 6803 were imported from the GSMM imSyn617 (Gopalakrishnan et al., 2018). Twenty-one reactions, mostly from fatty acid biosynthesis pathways, were mapped manually based on the reaction mechanism and the mapping information available in MetaCyc (Caspi et al., 2018). Mapping information for the remaining 50 reactions is unavailable in the literature and, therefore, was generated using the Canonical Labeling for Clique Approximation algorithm and verified manually (Kumar and Maranas, 2014).
Flux Estimation and Statistical Analysis of the Fit
The labeling data for flux elucidation were obtained from a previous study (Abernathy et al., 2017) where Synechococcus 2973 was grown under photoautotrophic conditions supporting optimal growth and 100% [13C]bicarbonate was used for the dynamic carbon-labeling experiment. The transition labeling data of the following 13 metabolites (Supplemental Table S6) were used for flux estimation: 2-phosphoglycerate, 6-phosphogluconate, r5p, Glc-6-P, phosphoenolpyruvate, s7p, succinate, malate, 3-phosphoglycerate, f6p, citrate, sbp, and Fru bisphosphate. The dynamic labeling data included the mass isotopomer distributions (MIDs) at time points 20, 30, 40, 60, 90, 150, 300, 600, 1,200, and 3,600 s. Scaled bicarbonate uptake rate (10 mmol g−1 dry weight h−1) and growth rate (0.27 h−1) also were fitted along with the labeling dynamics. Flux elucidation was performed as described earlier (Gopalakrishnan et al., 2018). This methodology uses the EMU-based decomposition of the carbon-mapping model to reduce the number of balance equations (ordinary differential equations [ODEs]) required to simulate the fragment MIDs (Antoniewicz et al., 2007). There were 1,271 EMUs after decomposing the imSyu593 to track the labeling of 13 metabolites that were fitted. This resulted in a total of 3,813 EMU mass fractions that need to be simulated to capture the labeling dynamics of the fitted metabolites. The ODEs capturing the labeling dynamics are then solved using Richardson’s extrapolation integrators after discretizing them in the time domain. A modified Levenberg-Marquardt algorithm was used to solve the nonlinear least squares problem that finds the genome-scale flux distribution that closely predicts the experimentally measured labeling dynamics. The numbers of free fluxes and pool sizes in the model were 202 and 217, respectively. Thus, for every step taken in the parameter space, a total 15.6 × 105 ODEs were solved to simulate 3,813 EMU fractions and calculate their sensitivity to 419 parameters. The statistical significance of the final fit was assessed by subjecting the SSR to a χ2 test (Antoniewicz et al., 2006). The F test was used to check the statistical significance of the improvement in the fit obtained using the GSMM over the core model. The 95% confidence interval for the estimated fluxes was calculated by determining the sensitivity of the minimum SSR as a function of individual fluxes (Antoniewicz et al., 2006). The estimated fluxes and confidence intervals are available in Supplemental Table S7.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of the relative fraction of M+0 isotopomer in sbp and s7p at various time points.
Supplemental Table S1. Comparison of flux ranges generated using MFA and FVA.
Supplemental Table S2. Experimental values of parameters such as growth rate and CO2 uptake.
Supplemental Table S3. The list of reactions part of the imSyu593.
Supplemental Table S4. The list of metabolites part of the imSyu593.
Supplemental Table S5. The atom-mapping information for imSyu593.
Supplemental Table S6. The MIDs of metabolites used for flux estimation.
Supplemental Table S7. Fluxes and confidence intervals estimated using imSyu593 in this study.
Footnotes
This work was supported by the National Science Foundation (NSF/MCB 1546840) and the U.S. Department of Energy (DE-SC0012722).
Articles can be viewed without a subscription.
References
- Abernathy MH, Yu J, Ma F, Liberton M, Ungerer J, Hollinshead WD, Gopalakrishnan S, He L, Maranas CD, Pakrasi HB, et al. (2017) Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol Biofuels 10: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebiyi AO, Jazmin LJ, Young JD (2015) 13C flux analysis of cyanobacterial metabolism. Photosynth Res 126: 19–32 [DOI] [PubMed] [Google Scholar]
- Alagesan S, Gaudana SB, Sinha A, Wangikar PP (2013) Metabolic flux analysis of Cyanothece sp. ATCC 51142 under mixotrophic conditions. Photosynth Res 118: 191–198 [DOI] [PubMed] [Google Scholar]
- Allen DK. (2016) Quantifying plant phenotypes with isotopic labeling & metabolic flux analysis. Curr Opin Biotechnol 37: 45–52 [DOI] [PubMed] [Google Scholar]
- Anfelt J, Kaczmarzyk D, Shabestary K, Renberg B, Rockberg J, Nielsen J, Uhlén M, Hudson EP (2015) Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb Cell Fact 14: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2006) Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8: 324–337 [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2007) Elementary metabolite units (EMU): A novel framework for modeling isotopic distributions. Metab Eng 9: 68–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, et al. (2018) The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res 46: D633–D639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwa JW, Kim WJ, Sim SJ, Um Y, Woo HM (2016) Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol J 14: 1768–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat DC, Way JC, Silver PA (2011) Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29: 95–103 [DOI] [PubMed] [Google Scholar]
- First EL, Gounaris CE, Floudas CA (2012) Stereochemically consistent reaction mapping and identification of multiple reaction mechanisms through integer linear optimization. J Chem Inf Model 52: 84–92 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Maranas CD (2015) 13C metabolic flux analysis at a genome-scale. Metab Eng 32: 12–22 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Pakrasi HB, Maranas CD (2018) Elucidation of photoautotrophic carbon flux topology in Synechocystis PCC 6803 using genome-scale carbon mapping models. Metab Eng 47: 190–199 [DOI] [PubMed] [Google Scholar]
- Jazmin LJ, Xu Y, Cheah YE, Adebiyi AO, Johnson CH, Young JD (2017) Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab Eng 42: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke F, Baier A, Knoop H, Kern R, Jablonsky J, Beyer G, Volkmer T, Steuer R, Lockau W, Hagemann M (2015) Identification of the light-independent phosphoserine pathway as an additional source of serine in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 161: 1050–1060 [DOI] [PubMed] [Google Scholar]
- Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R (2013) Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLOS Comput Biol 9: e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut H, Eiblmaier J, Grethe G, Löw P, Matuszczyk H, Saller H (2013) Algorithm for reaction classification. J Chem Inf Model 53: 2884–2895 [DOI] [PubMed] [Google Scholar]
- Kumar A, Maranas CD (2014) CLCA: Maximum common molecular substructure queries within the MetRxn database. J Chem Inf Model 54: 3417–3438 [DOI] [PubMed] [Google Scholar]
- Latendresse M, Malerich JP, Travers M, Karp PD (2012) Accurate atom-mapping computation for biochemical reactions. J Chem Inf Model 52: 2970–2982 [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR (2014) Metabolic flux analysis of Escherichia coli knockouts: Lessons from the Keio collection and future outlook. Curr Opin Biotechnol 28: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Jazmin LJ, Young JD, Allen DK (2014) Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc Natl Acad Sci USA 111: 16967–16972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5: 264–276 [DOI] [PubMed] [Google Scholar]
- McAtee AG, Jazmin LJ, Young JD (2015) Application of isotope labeling experiments and 13C flux analysis to enable rational pathway engineering. Curr Opin Biotechnol 36: 50–56 [DOI] [PubMed] [Google Scholar]
- Mueller TJ, Ungerer JL, Pakrasi HB, Maranas CD (2017) Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci Rep 7: 41569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenführ S, Wiechert W, Nöh K (2015) How to measure metabolic fluxes: A taxonomic guide for 13C fluxomics. Curr Opin Biotechnol 34: 82–90 [DOI] [PubMed] [Google Scholar]
- Nöh K, Wiechert W (2011) The benefits of being transient: Isotope-based metabolic flux analysis at the short time scale. Appl Microbiol Biotechnol 91: 1247–1265 [DOI] [PubMed] [Google Scholar]
- Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W (2007) Metabolic flux analysis at ultra short time scale: Isotopically non-stationary 13C labeling experiments. J Biotechnol 129: 249–267 [DOI] [PubMed] [Google Scholar]
- Quintana N, Van der Kooy F, Van de Rhee MD, Voshol GP, Verpoorte R (2011) Renewable energy from Cyanobacteria: Energy production optimization by metabolic pathway engineering. Appl Microbiol Biotechnol 91: 471–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SA, Torrance G, Baldacci L, Martínez Cuesta S, Fenninger F, Gopal N, Choudhary S, May JW, Holliday GL, Steinbeck C, et al. (2016) Reaction Decoder Tool (RDT): Extracting features from chemical reactions. Bioinformatics 32: 2065–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri AA, Morgan JA (2007) A transient isotopic labeling methodology for 13C metabolic flux analysis of photoautotrophic microorganisms. Phytochemistry 68: 2302–2312 [DOI] [PubMed] [Google Scholar]
- Song K, Tan X, Liang Y, Lu X (2016) The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl Microbiol Biotechnol 100: 7865–7875 [DOI] [PubMed] [Google Scholar]
- Ungerer J, Lin PC, Chen HY, Pakrasi HB (2018a) Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio 9: e02327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J, Wendt KE, Hendry JI, Maranas CD, Pakrasi HB (2018b) Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc Natl Acad Sci USA 115: E11761–E11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TT, Stolyar SM, Pinchuk GE, Hill EA, Kucek LA, Brown RN, Lipton MS, Osterman A, Fredrickson JK, Konopka AE, et al. (2012) Genome-scale modeling of light-driven reductant partitioning and carbon fluxes in diazotrophic unicellular cyanobacterium Cyanothece sp. ATCC 51142. PLOS Comput Biol 8: e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB (2016) CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Fact 15: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Xiong W, Dai J, Wu Q (2015) Genome-based metabolic mapping and 13C flux analysis reveal systematic properties of an oleaginous microalga Chlorella protothecoides. Plant Physiol 167: 586–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Lee TC, Rommelfanger S, Gjersing E, Cano M, Maness PC, Ghirardi M, Yu J (2015) Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Nat Plants 2: 15187. [DOI] [PubMed] [Google Scholar]
- Young JD. (2014) 13C metabolic flux analysis of recombinant expression hosts. Curr Opin Biotechnol 30: 238–245 [DOI] [PubMed] [Google Scholar]
- Young JD, Shastri AA, Stephanopoulos G, Morgan JA (2011) Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab Eng 13: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB (2015) Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep 5: 8132. [DOI] [PMC free article] [PubMed] [Google Scholar]